Abstrct
Objectives
To investigate the association between proton magnetic resonance spectroscopy (1H-MRS) metabolic features and the grade of gliomas, and to establish a machine-learning model to predict the glioma grade.
Methods
This study included 112 glioma patients who were divided into the training (n = 74) and validation (n = 38) sets based on the time of hospitalization. Twenty-six metabolic features were extracted from the preoperative 1H-MRS image. The Student's t-test was conducted to screen for differentially expressed features between low- and high-grade gliomas (WHO grades II and III/IV, respectively). Next, the minimum Redundancy Maximum Relevance (mRMR) algorithm was performed to further select features for a support vector machine (SVM) classifier building. Performance of the predictive model was evaluated both in the training and validation sets using ROC curve analysis.
Results
Among the extracted 1H-MRS metabolic features, thirteen features were differentially expressed. Four features were further selected as grade-predictive imaging signatures using the mRMR algorithm. The predictive performance of the machine-learning model measured by the AUC was 0.825 and 0.820 in the training and validation sets, respectively. This was better than the predictive performances of individual metabolic features, the best of which was 0.812.
Conclusions
1H-MRS metabolic features could help in predicting the grade of gliomas. The machine-learning model achieved a better prediction performance in grading gliomas than individual features, indicating that it could complement the traditionally used metabolic features.
Keywords: Proton magnetic resonance spectroscopy, Machine learning, Glioma grading, Support vector machine
Highlights
-
•
1H-MRS metabolic features could help in predicting the grades of gliomas.
-
•
The machine-learning model performed better than individual metabolic features.
-
•
The use of machine-learning model can complement the metabolic features.
1. Introduction
Diffuse gliomas represent 80% of malignant brain tumors (Ceccarelli et al., 2016). Adult diffuse gliomas are classified into grades II to IV according to histological criteria (Louis et al., 2016). Patients with low-grade gliomas (grade II) typically survive for more than five years after diagnosis, whereas those with high-grade gliomas (grades III-IV) survive for approximately 1–3 years after diagnosis (Jiang et al., 2016). Hence, predicting histological grades of diffuse gliomas with high accuracy could highlight tumor invasive growth patterns, which might allow for a precise assessment of tumor biological behavior and aid in clinical therapeutic decision making for the precise management of glioma patients (Su et al., 2018).
Proton magnetic resonance spectroscopy (1H-MRS) is a noninvasive tool for investigating the spatial distribution of the metabolic changes within the brain (Yamasaki et al., 2011). It can provide information on the neuronal integrity and on the levels of neurotransmitters including N-acetylaspartate (NAA), a neuronal marker, choline (Cho), which is involved in the synthesis and degradation of cell membranes, creatine (Cr), which reflects the energetic activity in the cells, lipid (Lip), a product of membrane phospholipids and necrotic debris, and lactate (Lac), which correlates with anaerobic glycolysis. In many studies, 1H-MRS has been shown to determine the tumor type and grade (Dou et al., 2015). 1H-MRS adds relevant diagnostic value exceeding that obtained from structural MR imaging alone. High-grade gliomas, particularly glioblastomas (grade IV), display unique metabolic features when compared with normal brain tissues and low-grade gliomas (Clark et al., 2016).
However, the accuracy of 1H-MRS in distinguishing between low- and high-grade gliomas needs further improvement. Since biomedical metabolic information is acquired through the quantification of the 1H-MRS data, proper and optimized quantification techniques will influence the objectivity and robustness of its results. Single metabolic features and peak height ratios of the intra-tumor metabolites have already been computed using 1H-MRS (Bulik et al., 2013). However, the optimum method for predicting the grade of the glioma and whether a machine-learning model of 1H-MRS metabolic features has a superior diagnostic performance to individual features are unclear.
Thus, we designed this retrospective study to investigate which 1H-MRS parameter is most effective in differentiating between low- and high-grade gliomas, and to determine whether using a machine-learning model of 1H-MRS parameters is more effective in differentiating low-grade gliomas from high-grade gliomas than using individual parameters.
2. Materials and methods
2.1. Patients
Ethical approval for this retrospective study was provided by the medical ethics committee of Beijing Tiantan Hospital, Capital Medical University, and the need for informed consent was waived. A total of 112 patients who were treated at our hospital between March 2016 and December 2017 were included in this study. The inclusion criteria were as follows: (1) patients with pathological diagnoses of primary gliomas of grade II-IV based on the WHO classification; (2) patients underwent both preoperative T2-weighted MRI examination and preoperative 1H-MRS examination; (3) patients with no history of preoperative therapy; and (4) patients whose clinical characteristics were available.
The patients were divided into training and validation sets based on the time of hospitalization. Seventy-four patients treated between March 2016 and February 2017 (43 WHO II glioma, 9 WHO III glioma, and 22 WHO IV glioma) were included into the training set to build the imaging signature and to development the model. Thirty-eight patients treated between March 2017 and December 2017 (19 WHO II glioma, 11 WHO III glioma, and 8 WHO IV glioma) were assigned into an independent validation set for model validation.
2.2. Histopathological analysis
Tissue samples obtained by subtotal/total resection were histopathologically examined by an experienced neuropathologist. The glioma was graded based on the WHO classification, and was dichotomized into two groups: the low- and high-grade glioma groups. Grade II gliomas were regarded as low grade, whereas grade III and IV gliomas were regarded as high grade. The histopathology is the ground truth for comparison to 1H-MRS.
2.3. Magnetic resonance imaging
All patients underwent MR imaging using a 3.0 T MR system (Siemens Magnetom Prisma, Germany) with a 20-channel head/neck coil. The MR protocols were as follows: (1) The parameters used for the turbo spin echo (TSE) sequence based T2-weighted imaging were TR/TE: 5000 ms/105 ms, slices: 33, slice thickness: 3 mm with 0.9 mm gap, field of view: 220 mm × 220 mm, and matrix size: 448 × 358.4. (2) The parameters used for the MR spectra obtained using the multi-voxel chemical shift imaging 1H-MRS method were TR/TE: 1700 ms/135 ms, field of view: 160 mm × 160 mm, volume of interest (VOI): 80 mm × 80 mm, and number of excitations (NEX): 3. The VOI of 1H-MRS was placed on the axial T2-weighted image including as much of the lesion as possible and the contralateral normal-appearing brain parenchyma while avoiding the subcutaneous fat, bones and sinuses.
2.4. Imaging processing
A neuroradiologist (Shaowu Li with twenty years of experiences in MR imaging interpretation) delineated the tumor area on T2-weighted images using the software SPIN (Signal Processing in NMR, Detroit, Michigan, USA). Then the voxels were carefully positioned to include the largest tumor region.
The spectroscopy data of each voxel was processed using the imaging software, Syngo.via from Siemens Healthineers. The preprocessing procedure included spatial filtering, 3D Fourier transformation for spatial localization, baseline subtraction after fitting with polynomial functions, and phase correction. Subsequently, the resonance intensities of the individual spectra were determined by calculating the integral of areas under the peaks of the chemical dislocation graphs.
The intra-tumor metabolites, Cho, NAA, Cr, Lac, and Lip, were sampled from total area of the tumor and normalized based on the corresponding metabolites in the contralateral normal-appearing brain parenchyma (normalized metabolic parameter = original metabolic parameter/contralateral metabolic parameter). The mean concentrations were then analyzed by obtaining the intensities of the spectra and the ratios between the spectra. Twenty-six metabolic features were included: Cho, Cr1, NAA, Lac, lipid1 (Lip1), lipid2 (Lip2), (Lip1 + Lip2), (Lac + Lip1 + Lip2), Cho/Cr1 ratio, NAA/Cr1 ratio, Lac/Cr1 ratio, Lip1/Cr1 ratio, Lip2/Cr1 ratio, (Lip1 + Lip2)/Cr1 ratio, (Lac + Lip1 + Lip2)/Cr1 ratio, Cho/NAA ratio, Lip1/NAA ratio, Lip2/NAA ratio, Lac/NAA ratio, (Lip1 + Lip2)/NAA ratio, (Lac + Lip1 + Lip2)/NAA ratio, Lip1/Cho ratio, Lip2/Cho ratio, Lac/Cho ratio, (Lip1 + Lip2)/Cho ratio, and (Lac + Lip1 + Lip2)/Cho ratio.
2.5. Feature selection and individual feature analysis
For feature selection, we first screened the features that were significantly (P < 0.05) differentially expressed between the low- and high-grade gliomas using the Student's t-test. A ROC curve analysis was performed on each differentially expressed feature to evaluate its predictive performance in grading gliomas. Next, the minimum Redundancy Maximum Relevance (mRMR) feature selection algorithm was used to further identify an imaging signature for grading the gliomas in the training set (De Jay et al., 2013). The mRMR algorithm attempts to optimize the maximum relevance and minimum redundancy simultaneously. The Student's t-test was conducted on the features selected using the mRMR algorithm to observe whether they were differentially expressed between the low- and high-grade glioma groups in the validation set. All the P values derived from multiple testing were adjusted using the Bonferroni correction.
2.6. Machine-learning model establishment
A support vector machine (SVM) fitted with the final selected features was used in the training set. The SVM classifier, a specific type of supervised machine-learning method, is designed to classify the data points by maximizing the margins between the classes in a high-dimensional space (Cortes and Vapnik, 1995). The SVM classifier outputted a decision value for each patient that could be used to predict the grade of the glioma. The ROC curves were plotted and the AUC was calculated to illustrate the diagnostic performance of the predictive model. An optimal cutoff value was identified when sensitivity plus specificity was maximal. The established predictive model was subsequently evaluated in an independent validation set.
2.7. Statistical analysis
The mRMR algorithm, SVM classifier, and ROC curve analysis were implemented using the R software, version 3.3.2 (R Core Team (2016). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/). The code of the mRMR algorithm and SVM classifier was provided in the supplementary material. The clinical characteristics of the training and validation sets were compared using the Student's t-test and Chi-square test. P < 0.05 was considered statistically significant.
3. Results
3.1. Radiological and clinical characteristics
As shown in Table 1, the 1H-MRS metabolites examined were assigned as creatine (Cr1) at 3.02 to 3.12 ppm, choline (Cho) at 3.17 to 3.27 ppm, N-acetylaspartate (NAA) at 1.98 to 2.08 ppm, and lactate (Lac) at 1.10 to 1.50 ppm. Lipid peak included Lip1 defined at 0.70 to 1.00 ppm and Lip2 defined at 1.25 to 1.60 ppm, respectively.
Table 1.
Metabolites and peak integration range.
| Metabolites | Metabolites abbreviation | Integration range (ppm) |
|---|---|---|
| Creatine | Cr1 | 3.02–3.12 |
| Choline | Cho | 3.17–3.27 |
| N-acetylaspartate | NAA | 1.98–2.08 |
| Lipid1 | Lip1 | 0.70–1.00 |
| Lipid2 | Lip2 | 1.25–1.60 |
| Lactate | Lac | 1.10–1.50 |
Abbreviation: ppm: parts per million.
In this study, 62 and 50 patients diagnosed by pathological grading with low- and high-grade gliomas, respectively were enrolled and divided into the training (n = 74) and validation (n = 38) sets. Table 2 shows the patient characteristics. There were no significant differences between the training and validation sets in age (P = 0.6506), sex (P = 0.3225) or high/low grade (P = 0.4138), which indicated the justifiability of using these two sets.
Table 2.
Patient characteristics.
| Number | Mean Age (years) | Sex (male/female) | Grade (high/low) | |
|---|---|---|---|---|
| Training set | 74 | 42.76 | 48/26 | 31/43 |
| Validation set | 38 | 43.84 | 21/17 | 19/19 |
| P value | 0.6506a | 0.3225b | 0.4138b |
Student's t-test.
Chi-square test.
3.2. Feature selection and individual feature analysis
First, the performance of each 1H-MRS metabolic feature in predicting the glioma grade was assessed using the training set. As shown in Table 3, the expression of the following features were found to be significantly different (P < 0.05) between the low- and high-grade glioma groups: NAA, Lip2, (Lip1 + Lip2), (Lac + Lip1 + Lip2), Cho/Cr1 ratio, Lip2/Cr1 ratio, (Lip1 + Lip2)/Cr1 ratio, (Lac + Lip1 + Lip2)/Cr1 ratio, Cho/NAA ratio, Lip1/NAA ratio, Lip2/NAA ratio, (Lip1 + Lip2)/NAA ratio, and (Lac + Lip1 + Lip2)/NAA. The corresponding AUCs of the differentially expressed features were calculated (Table 3). The Lip2/NAA ratio and (Lip1 + Lip2)/NAA ratio yielded AUC values of 0.812 and 0.810, respectively, which were the most predictive of the glioma grade among the features assessed. Their accuracies in predicting the glioma grade were 71.62% and 72.97%, respectively.
Table 3.
Statistics of intra-tumor 1H-MRS metabolic parameters in the training set.
| Metabolic parameters (normalized) | Low-grade gliomas (n = 43) | High-grade gliomas (n = 31) | P value | Adjust P | AUC |
|---|---|---|---|---|---|
| Cho | 1.581 ± 0.076 | 1.953 ± 0.269 | 0.1328 | 0.2302 | |
| Cr1 | 1.017 ± 0.034 | 0.983 ± 0.084 | 0.6778 | 0.8366 | |
| NAA | 0.539 ± 0.023 | 0.430 ± 0.023 | 0.0018⁎⁎ | 0.0094⁎⁎ | 0.7254 |
| Lac | 3.411 ± 0.590 | 3.719 ± 0.512 | 0.7079 | 0.8366 | |
| Lip1 | 3.323 ± 0.831 | 3.528 ± 0.899 | 0.8687 | 0.9411 | |
| Lip2 | 2.144 ± 0.228 | 3.940 ± 0.495 | 0.0006⁎⁎⁎ | 0.0078⁎⁎ | 0.7592 |
| Lip1 + Lip2 | 2.190 ± 0.285 | 3.460 ± 0.346 | 0.0058⁎⁎ | 0.0168⁎ | 0.7382 |
| Lac + Lip1 + Lip2 | 2.234 ± 0.215 | 3.396 ± 0.344 | 0.0036⁎⁎ | 0.0117⁎ | 0.6962 |
| Cho/Cr1 | 1.673 ± 0.087 | 2.501 ± 0.292 | 0.0028⁎⁎ | 0.0104⁎ | 0.6572 |
| NAA/Cr1 | 0.521 ± 0.020 | 0.522 ± 0.036 | 0.992 | 0.992 | |
| Lac/Cr1 | 4.188 ± 0.796 | 11.80 ± 5.181 | 0.094 | 0.1748 | |
| Lip1/Cr1 | 3.564 ± 0.811 | 4.763 ± 0.979 | 0.3466 | 0.5006 | |
| Lip2/Cr1 | 2.659 ± 0.430 | 9.002 ± 2.895 | 0.0135⁎ | 0.0319⁎ | 0.7449 |
| (Lip1 + Lip2)/Cr1 | 2.633 ± 0.412 | 7.557 ± 2.209 | 0.0130⁎ | 0.0319⁎ | 0.7397 |
| (Lac + Lip1 + Lip2)/Cr1 | 2.727 ± 0.369 | 8.227 ± 2.743 | 0.0228⁎ | 0.0494⁎ | 0.6992 |
| Cho/NAA | 4.832 ± 0.723 | 12.19 ± 2.591 | 0.0026⁎⁎ | 0.0104⁎ | 0.7089 |
| Lip1/NAA | 8.516 ± 2.025 | 13.89 ± 3.479 | 0.1608 | 0.2613 | |
| Lip2/NAA | 6.287 ± 0.906 | 27.42 ± 7.127 | 0.0009⁎⁎⁎ | 0.0078⁎⁎ | 0.8124 |
| Lac/NAA | 12.19 ± 2.843 | 25.37 ± 5.858 | 0.0309⁎ | 0.0618 | |
| (Lip1 + Lip2)/NAA | 6.199 ± 0.913 | 23.37 ± 6.098 | 0.0017⁎⁎ | 0.00936⁎⁎ | 0.8102 |
| (Lac + Lip1 + Lip2)/NAA | 6.796 ± 0.986 | 22.89 ± 5.114 | 0.0006⁎⁎⁎ | 0.0078⁎⁎ | 0.7824 |
| Lip1/Cho | 2.985 ± 0.830 | 1.788 ± 0.322 | 0.2429 | 0.3715 | |
| Lip2/Cho | 2.167 ± 0.578 | 2.609 ± 0.369 | 0.5571 | 0.7623 | |
| Lac/Cho | 2.313 ± 0.395 | 2.641 ± 0.567 | 0.6252 | 0.8128 | |
| (Lip1 + Lip2)/Cho | 2.202 ± 0.584 | 2.283 ± 0.290 | 0.9117 | 0.9482 | |
| (Lac + Lip1 + Lip2)/Cho | 2.126 ± 0.488 | 2.272 ± 0.321 | 0.8192 | 0.9261 |
Note: Data were expressed as the mean ± standard error of mean (SEM).
The bold words and number mean that they are statistically significant.
P < 0.05
P < 0.01, and
P < 0.001 by Student's t-test.
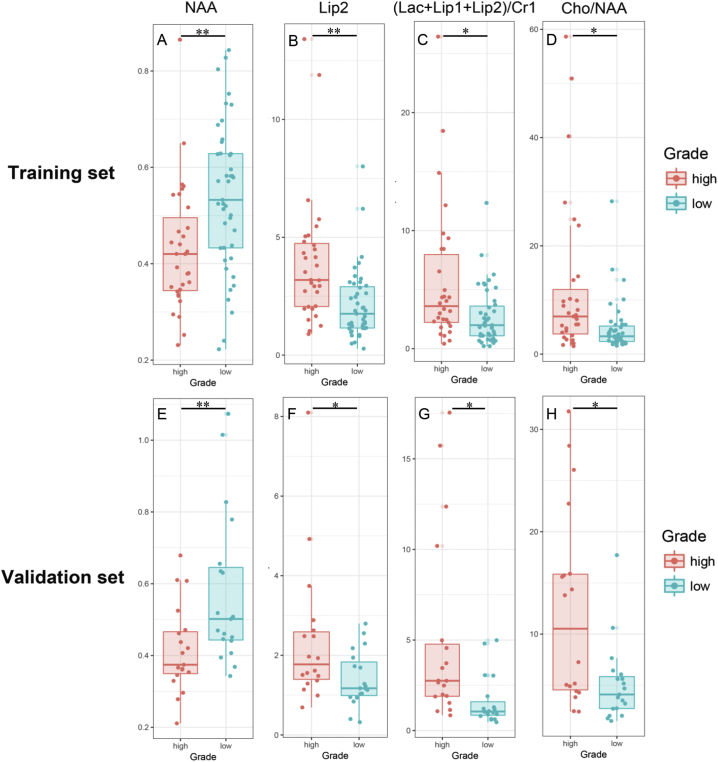
Four 1H-MRS metabolic features were further selected using the mRMR algorithm for the establishment of a machine-learning model. The selected four features were NAA, Lip2, (Lac + Lip1 + Lip2)/Cr1 ratio, and Cho/NAA ratio. Their distribution is shown in Fig. 1. In the training set, the NAA was found to be significantly higher in the low-grade glioma group than in the high-grade glioma group (P = 0.0094). The Lip2, (Lac + Lip1 + Lip2)/Cr1 ratio, and Cho/NAA ratio were significantly higher in the high-grade glioma group than in the low-grade glioma group (P = 0.0078, P = 0.0494, and P = 0.0104, respectively). Similarly, in the validation set, the NAA was significantly higher in the low-grade glioma group than in the high-grade glioma group (P = 0.0072), and the Lip2, (Lac + Lip1 + Lip2)/Cr1 ratio, and Cho/NAA ratio were significantly higher in the high-grade glioma group than in the low-grade glioma group (P = 0.0324, P = 0.0381, and P = 0.0394, respectively). Thus, the differential expression of these four features was significantly different between the low- and high-grade gliomas in both sets.
Fig. 1.
The values of NAA, Lip2, (Lac + Lip1 + Lip2)/Cr1 ratio, and Cho/NAA ratio within the tumor regions in the training (A-D) and validation sets (E-H). The Student's t-test was used to obtain the statistical difference between the high- (red) and low-grade gliomas (green). The data are presented as means ± standard deviation of the parameters, NAA (A and E), Lip2 (B and F), (Lac + Lip1 + Lip2)/Cr1 ratio (C and G), and Cho/NAA ratio (D and H).
* indicates P < 0.05; ** indicates P < 0.01.
3.3. Machine-learning model establishment
3.3.1. Training set
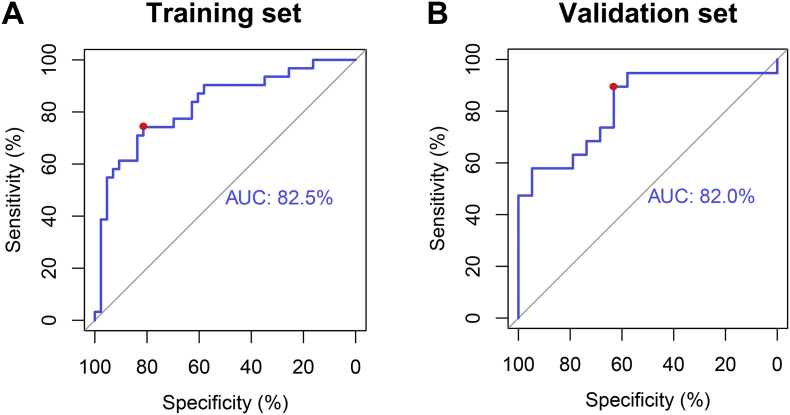
Using the training set, a machine-learning model was developed based on the four selected metabolic features and the SVM classifier. For each patient, the SVM classifier outputted a decision value to predict the glioma grade, and a ROC curve was delineated. The AUC was 0.825 in the ROC curve analysis. The optimal cutoff point (0.4) exhibited a sensitivity, specificity, and accuracy of 74.2%, 81.4%, and 78.38%, respectively (Fig. 2A). These results demonstrated that the use of this model was superior to the use of the Lip2/NAA ratio (AUC = 0.812) and (Lip1 + Lip2)/NAA ratio (AUC = 0.810) for distinguishing between low- and high-grade gliomas.
Fig. 2.
The ROC curves for the glioma-grading predictive model in the training and validation set. A: In the training set, the AUC is 82.5%. At the optimal cutoff value (0.4), the sensitivity, specificity, and accuracy are 74.2%, 81.4%, and 78.38%, respectively (red dot). B: In the validation set, the AUC of the predictive model is 82.0%. At the best cutoff point (0.8), the sensitivity, specificity, and accuracy were 89.5%, 63.2%, and 76.31%, respectively (red dot).
3.3.2. Validation set
The machine-learning model was then applied to the validation set. The glioma grades could be predicted effectively. The AUC was 0.820 in the ROC curve analysis. At the best cutoff point (0.8), sensitivity, specificity, and accuracy were 89.5%, 63.2%, and 76.31%, respectively (Fig. 2B).
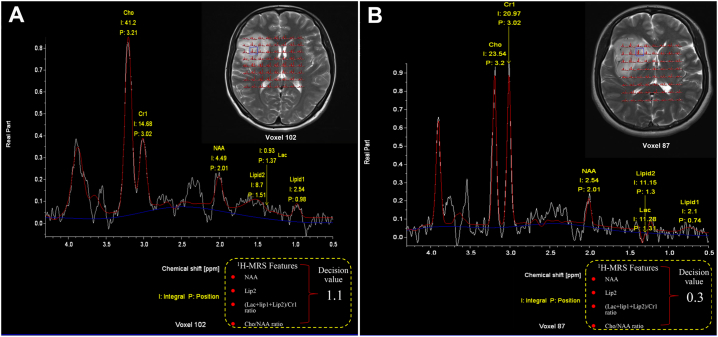
The prediction processes of two representative cases are shown in Fig. 3. The first case was a 44-year-old female patient with a WHO grade II glioma. She was accurately classified into the low-grade group using the SVM decision value, which was high (1.1). The second case was a 41-year-old female patient with a WHO grade IV glioma. She was accurately classified into the high-grade group using the SVM decision value, which was low (0.3).
Fig. 3.
Axial T2 images with 1H-MRS voxels of two representative patients. After classification, a decision value is outputted from the predictive model. (A) A 44-year-old woman with a WHO grade II right frontal-insular astrocytoma is accurately classified into low-grade glioma with a high decision value (1.1). (B) A 41-year-old woman with a right frontal glioblastoma is accurately classified into high-grade glioma with a low decision value (0.3). The metabolite spectra were obtained from the blue box in the tumor region.(For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
We investigated the performance of a series of 1H-MRS metabolic features in differentiating between low- and high-grade gliomas. We further identified a grade-predictive signature using the mRMR algorithm. Subsequently, a machine-learning model was created using the selected features and the SVM classifier. The model achieved an AUC of 0.825 and 0.820 in the training and validation sets, respectively. This model performed better than any individual metabolic feature, indicating that it can be used as a non-invasive approach for better classifying the glioma grades and that it could complement the traditionally used 1H-MRS metabolic parameters.
Accurate preoperative grading of gliomas is important to estimate the prognosis and plan the treatment: the treatment strategies applied to high-grade gliomas need to be more aggressive than those applied to low-grade gliomas (Fouke et al., 2015). Conventional MR imaging with gadolinium-based contrast agents has an established role in characterizing cerebral tumors and is considered the reference standard for preoperative diagnostic evaluation in some instances. Nevertheless, contrast enhancement is not specific for malignancy and primarily reflects the message of the contrast material across a disrupted blood-brain barrier (Vogelbaum et al., 2012). Several studies have demonstrated that the absence of enhancement does not necessarily imply that the glioma is low-grade (White et al., 2005).
Previous studies have revealed that the perfusion images perform well in grading gliomas. A pioneer study found that histogram analysis of normalized cerebral blood volume could grade gliomas with high sensitivity (90%), better than the established hot-spot method (55%–76%) (Emblem et al., 2008a). Another study revealed that combining the tumor blood volume histogram signatures derived from perfusion images and SVM could achieve a true positive rate of 0.76 and a true negative rate of 0.82 in grading gliomas (Emblem et al., 2008b). Additionally, using a SVM model with a radial basis function kernel exhibited better performance (true positive rate 0.83; true negative rate 0.91) (Zollner et al., 2010), which was better than our model. However, certain patients could be allergic to the contrast agent used in the perfusion imaging, which limited the application of this approach (Behzadi et al., 2018). Without the limitation of contrast agent, 1H-MRS could also be used in grading gliomas (Kalpathy-Cramer et al., 2014) and predicting the survival of gliomas (Andronesi et al., 2017), similar to the perfusion imaging (Emblem et al., 2014; Emblem et al., 2015).
1H-MRS displays the oncometabolic state of the tumor where conventional MRI cannot make such distinctions (Dowling et al., 2001). The levels of N-acetylaspartate and creatine are reduced, whereas those of choline and choline-containing compounds are increased in tumors when compared with their respective levels in the normal brain tissue (McKnight et al., 2007). A low NAA level indicates the loss of the neuronal cell viability, whereas a high Cho level infers an increased cellular turnover (Bulik et al., 2013). The metabolites lactate at 1.33 ppm and mobile lipids, with its most prominent peaks at 0.9 and 1.3 ppm, were also observed. Lactate accumulation in brain tumors, caused by increased glycolysis, is associated with ischemic changes in the poorly perfused tumor parenchyma, or increased necrotic tissue (Fulham et al., 1992). While several studies suggest that high-grade gliomas can be distinguished from low-grade tumors using higher Lac peaks (Yamasaki et al., 2011), others show no significant correlations (Shimizu et al., 1996; Sibtain et al., 2007). Lipids exist as macromolecules in the cell membranes and myelin sheaths in the normal brain tissue. When the brain tissue is damaged or disrupted, as occurs during the growth of a brain tumor, the lipid macromolecules are transformed into mobile lipids. Lipids correlates with the presence of necrosis when the cells die and their membranes break down (Remy et al., 1997). Among all the gliomas, Lip peaks are more frequently found in glioblastomas (Kuesel et al., 1994).
The results are usually expressed as ratios between the cerebral metabolites rather than as absolute concentrations. Single metabolite ratios, such as Cho/Cr ratio and Cho/NAA ratio, have been used to reveal significant differences between low- and high-grade gliomas in most studies (Rao et al., 2013; Wang et al., 2016). There are also studies in which high-grade gliomas were detected by calculating the ratio of lipids and lactate (LL)/Cr (Kim et al., 2006; Yoon et al., 2014). However, it is not known whether combined ratios could improve the diagnostic accuracy. The highest accuracy in differentiating between low- and high-grade gliomas in our study was 72.97% for the (Lip1 + Lip2)/NAA ratio, yet it increased to 78.38%, along with an increased in the AUC from 0.8102 to 0.825, when the predictive model was used. The combination of NAA, Lip2, (Lac + Lip1 + Lip2)/Cr1 ratio, and Cho/NAA ratio used in our predictive model was more useful than any single 1H-MRS metabolic feature in differentiating the glioma grade preoperatively.
Furthermore, although the role of 1H-MRS quantitative measurements in the diagnosis and monitoring of glial tumors have been studied, there were no independent validation sets in most of the previous studies. In our study, an SVM-based machine-learning model was created using the training set and was validated using the independent validation set. An SVM can automatically learn the distinguishing patterns using the existing data and create a corresponding model (Zacharaki et al., 2009). Compared with the diagnostic performance of this, the predictive model we developed might be a superior index for distinguishing between low- and high-grade gliomas.
There were some limitations to this study. First, the data available was limited. Further studies are needed to confirm the efficacy of this predictive model. Secondly, as an isolated modality, 1H-MRS could be combined with other advanced imaging techniques such as perfusion and diffusion MR information to improve the diagnostic accuracy.
In conclusion, we found that some 1H-MRS metabolic features could help in predicting the grade of gliomas. Using the mRMR algorithm and the SVM classifier, we established a machine-learning model that achieved a better prediction performance in identifying the tumor grade than those achieved using individual features. This indicates that it could complement the traditionally used 1H-MRS metabolic features in predicting glioma grades.
Conflict of interest
The authors of this manuscript declare no relationships with any companies whose products or services may be related to the subject matter of the article.
Acknowledgement
This work was supported by the National High Technology Research and Development Program of China (863 Program, No. 2015AA020504), National Key Research and Development Program of China (2018YFC0115604), National Natural Science Foundation of China (No. 81501186), Beijing Municipal Science & Technology Commission (No. Z171100000117002) and Innovation Fund Project of Beijing Neurosurgical Institute (Institute Youth-2014017).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.nicl.2019.101835.
Contributor Information
Tao Jiang, Email: taojiang1964@163.com.
Shaowu Li, Email: lys5@sina.com.
Appendix A. Supplementary data
Supplementary material
References
- Andronesi O.C., Esmaeili M., Borra R.J.H., Emblem K., Gerstner E.R., Pinho M.C., Plotkin S.R., Chi A.S., Eichler A.F., Dietrich J., Ivy S.P., Wen P.Y., Duda D.G., Jain R., Rosen B.R., Sorensen G.A., Batchelor T.T. Early changes in glioblastoma metabolism measured by MR spectroscopic imaging during combination of anti-angiogenic cediranib and chemoradiation therapy are associated with survival. npj Precis. Oncol. 2017;1 doi: 10.1038/s41698-017-0020-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behzadi A.H., Zhao Y., Farooq Z., Prince M.R. Immediate allergic reactions to gadolinium-based contrast agents: a systematic review and meta-analysis. Radiology. 2018;286:471–482. doi: 10.1148/radiol.2017162740. [DOI] [PubMed] [Google Scholar]
- Bulik M., Jancalek R., Vanicek J., Skoch A., Mechl M. Potential of MR spectroscopy for assessment of glioma grading. Clin. Neurol. Neurosurg. 2013;115:146–153. doi: 10.1016/j.clineuro.2012.11.002. [DOI] [PubMed] [Google Scholar]
- Ceccarelli M., Barthel F.P., Malta T.M., Sabedot T.S., Salama S.R., Murray B.A., Morozova O., Newton Y., Radenbaugh A., Pagnotta S.M., Anjum S., Wang J., Manyam G., Zoppoli P., Ling S., Rao A.A., Grifford M., Cherniack A.D., Zhang H., Poisson L., Carlotti C.G., Jr., Tirapelli D.P., Rao A., Mikkelsen T., Lau C.C., Yung W.K., Rabadan R., Huse J., Brat D.J., Lehman N.L., Barnholtz-Sloan J.S., Zheng S., Hess K., Rao G., Meyerson M., Beroukhim R., Cooper L., Akbani R., Wrensch M., Haussler D., Aldape K.D., Laird P.W., Gutmann D.H., Network T.R., Noushmehr H., Iavarone A., Verhaak R.G. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse Glioma. Cell. 2016;164:550–563. doi: 10.1016/j.cell.2015.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark P.M., Mai W.X., Cloughesy T.F., Nathanson D.A. Emerging approaches for targeting metabolic vulnerabilities in malignant Glioma. Curr Neurol. Neurosci. Rep. 2016;16:17. doi: 10.1007/s11910-015-0613-6. [DOI] [PubMed] [Google Scholar]
- Cortes C., Vapnik V. Support-vector networks. Mach. Learn. 1995;20:273–297. [Google Scholar]
- De Jay N., Papillon-Cavanagh S., Olsen C., El-Hachem N., Bontempi G., Haibe-Kains B. mRMRe: an R package for parallelized mRMR ensemble feature selection. Bioinformatics. 2013;29:2365–2368. doi: 10.1093/bioinformatics/btt383. [DOI] [PubMed] [Google Scholar]
- Dou W., Zhang M., Zhang X., Li Y., Chen H., Li S., Lu M., Dai J., Constans J.M. Convex-envelope based automated quantitative approach to multi-voxel 1H-MRS applied to brain tumor analysis. PLoS One. 2015;10 doi: 10.1371/journal.pone.0137850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling C., Bollen A.W., Noworolski S.M., McDermott M.W., Barbaro N.M., Day M.R., Henry R.G., Chang S.M., Dillon W.P., Nelson S.J., Vigneron D.B. Preoperative proton MR spectroscopic imaging of brain tumors: correlation with histopathologic analysis of resection specimens. AJNR Am. J. Neuroradiol. 2001;22:604–612. [PMC free article] [PubMed] [Google Scholar]
- Emblem K.E., Nedregaard B., Nome T., Due-Tonnessen P., Hald J.K., Scheie D., Borota O.C., Cvancarova M., Bjornerud A. Glioma grading by using histogram analysis of blood volume heterogeneity from MR-derived cerebral blood volume maps. Radiology. 2008;247:808–817. doi: 10.1148/radiol.2473070571. [DOI] [PubMed] [Google Scholar]
- Emblem K.E., Zoellner F.G., Tennoe B., Nedregaard B., Nome T., Due-Tonnessen P., Hald J.K., Scheie D., Bjornerud A. Predictive modeling in glioma grading from MR perfusion images using support vector machines. Magn. Reson. Med. 2008;60:945–952. doi: 10.1002/mrm.21736. [DOI] [PubMed] [Google Scholar]
- Emblem K.E., Due-Tonnessen P., Hald J.K., Bjornerud A., Pinho M.C., Scheie D., Schad L.R., Meling T.R., Zoellner F.G. Machine learning in preoperative glioma MRI: survival associations by perfusion-based support vector machine outperforms traditional MRI. J. Magn. Reson. Imaging. 2014;40:47–54. doi: 10.1002/jmri.24390. [DOI] [PubMed] [Google Scholar]
- Emblem K.E., Pinho M.C., Zollner F.G., Due-Tonnessen P., Hald J.K., Schad L.R., Meling T.R., Rapalino O., Bjornerud A. A generic support vector machine model for preoperative glioma survival associations. Radiology. 2015;275:228–234. doi: 10.1148/radiol.14140770. [DOI] [PubMed] [Google Scholar]
- Fouke S.J., Benzinger T., Gibson D., Ryken T.C., Kalkanis S.N., Olson J.J. The role of imaging in the management of adults with diffuse low grade glioma: a systematic review and evidence-based clinical practice guideline. J. Neuro-Oncol. 2015;125:457–479. doi: 10.1007/s11060-015-1908-9. [DOI] [PubMed] [Google Scholar]
- Fulham M.J., Bizzi A., Dietz M.J., Shih H.H., Raman R., Sobering G.S., Frank J.A., Dwyer A.J., Alger J.R., Di Chiro G. Mapping of brain tumor metabolites with proton MR spectroscopic imaging: clinical relevance. Radiology. 1992;185:675–686. doi: 10.1148/radiology.185.3.1438744. [DOI] [PubMed] [Google Scholar]
- Jiang T., Mao Y., Ma W., Mao Q., You Y., Yang X., Jiang C., Kang C., Li X., Chen L., Qiu X., Wang W., Li W., Yao Y., Li S., Li S., Wu A., Sai K., Bai H., Li G., Chen B., Yao K., Wei X., Liu X., Zhang Z., Dai Y., Lv S., Wang L., Lin Z., Dong J., Xu G., Ma X., Cai J., Zhang W., Wang H., Chen L., Zhang C., Yang P., Yan W., Liu Z., Hu H., Chen J., Liu Y., Yang Y., Wang Z., Wang Z., Wang Y., You G., Han L., Bao Z., Liu Y., Wang Y., Fan X., Liu S., Liu X., Wang Y., Wang Q., Chinese Glioma Cooperative G. CGCG clinical practice guidelines for the management of adult diffuse gliomas. Cancer Lett. 2016;375:263–273. doi: 10.1016/j.canlet.2016.01.024. [DOI] [PubMed] [Google Scholar]
- Kalpathy-Cramer J., Gerstner E.R., Emblem K.E., Andronesi O., Rosen B. Advanced magnetic resonance imaging of the physical processes in human glioblastoma. Cancer Res. 2014;74:4622–4637. doi: 10.1158/0008-5472.CAN-14-0383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.H., Chang K.H., Na D.G., Song I.C., Kwon B.J., Han M.H., Kim K. 3T 1H-MR spectroscopy in grading of cerebral gliomas: comparison of short and intermediate echo time sequences. AJNR Am. J. Neuroradiol. 2006;27:1412–1418. [PMC free article] [PubMed] [Google Scholar]
- Kuesel A.C., Sutherland G.R., Halliday W., Smith I.C. 1H MRS of high grade astrocytomas: mobile lipid accumulation in necrotic tissue. NMR Biomed. 1994;7:149–155. doi: 10.1002/nbm.1940070308. [DOI] [PubMed] [Google Scholar]
- Louis D.N., Perry A., Reifenberger G., von Deimling A., Figarella-Branger D., Cavenee W.K., Ohgaki H., Wiestler O.D., Kleihues P., Ellison D.W. The 2016 World Health Organization classification of Tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- McKnight T.R., Lamborn K.R., Love T.D., Berger M.S., Chang S., Dillon W.P., Bollen A., Nelson S.J. Correlation of magnetic resonance spectroscopic and growth characteristics within grades II and III gliomas. J. Neurosurg. 2007;106:660–666. doi: 10.3171/jns.2007.106.4.660. [DOI] [PubMed] [Google Scholar]
- Rao P.J., Jyoti R., Mews P.J., Desmond P., Khurana V.G. Preoperative magnetic resonance spectroscopy improves diagnostic accuracy in a series of neurosurgical dilemmas. Br. J. Neurosurg. 2013;27:646–653. doi: 10.3109/02688697.2013.771724. [DOI] [PubMed] [Google Scholar]
- Remy C., Fouilhe N., Barba I., Sam-Lai E., Lahrech H., Cucurella M.G., Izquierdo M., Moreno A., Ziegler A., Massarelli R., Decorps M., Arus C. Evidence that mobile lipids detected in rat brain glioma by 1H nuclear magnetic resonance correspond to lipid droplets. Cancer Res. 1997;57:407–414. [PubMed] [Google Scholar]
- Shimizu H., Kumabe T., Tominaga T., Kayama T., Hara K., Ono Y., Sato K., Arai N., Fujiwara S., Yoshimoto T. Noninvasive evaluation of malignancy of brain tumors with proton MR spectroscopy. AJNR Am. J. Neuroradiol. 1996;17:737–747. [PMC free article] [PubMed] [Google Scholar]
- Sibtain N.A., Howe F.A., Saunders D.E. The clinical value of proton magnetic resonance spectroscopy in adult brain tumours. Clin. Radiol. 2007;62:109–119. doi: 10.1016/j.crad.2006.09.012. [DOI] [PubMed] [Google Scholar]
- Su C., Jiang J., Zhang S., Shi J., Xu K., Shen N., Zhang J., Li L., Zhao L., Zhang J., Qin Y., Liu Y., Zhu W. Radiomics based on multicontrast MRI can precisely differentiate among glioma subtypes and predict tumour-proliferative behaviour. Eur. Radiol. 2018;29:1986–1996. doi: 10.1007/s00330-018-5704-8. [DOI] [PubMed] [Google Scholar]
- Vogelbaum M.A., Jost S., Aghi M.K., Heimberger A.B., Sampson J.H., Wen P.Y., Macdonald D.R., Van den Bent M.J., Chang S.M. Application of novel response/progression measures for surgically delivered therapies for gliomas: response assessment in Neuro-oncology (RANO) working group. Neurosurgery. 2012;70:234–243. doi: 10.1227/NEU.0b013e318223f5a7. (discussion 243-234) [DOI] [PubMed] [Google Scholar]
- Wang Q., Zhang H., Zhang J., Wu C., Zhu W., Li F., Chen X., Xu B. The diagnostic performance of magnetic resonance spectroscopy in differentiating high-from low-grade gliomas: a systematic review and meta-analysis. Eur. Radiol. 2016;26:2670–2684. doi: 10.1007/s00330-015-4046-z. [DOI] [PubMed] [Google Scholar]
- White M.L., Zhang Y., Kirby P., Ryken T.C. Can tumor contrast enhancement be used as a criterion for differentiating tumor grades of oligodendrogliomas? AJNR Am. J. Neuroradiol. 2005;26:784–790. [PMC free article] [PubMed] [Google Scholar]
- Yamasaki F., Kurisu K., Kajiwara Y., Watanabe Y., Takayasu T., Akiyama Y., Saito T., Hanaya R., Sugiyama K. Magnetic resonance spectroscopic detection of lactate is predictive of a poor prognosis in patients with diffuse intrinsic pontine glioma. Neuro-Oncol. 2011;13:791–801. doi: 10.1093/neuonc/nor038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J.H., Kim J.H., Kang W.J., Sohn C.H., Choi S.H., Yun T.J., Eun Y., Song Y.S., Chang K.H. Grading of cerebral glioma with multiparametric MR imaging and 18F-FDG-PET: concordance and accuracy. Eur. Radiol. 2014;24:380–389. doi: 10.1007/s00330-013-3019-3. [DOI] [PubMed] [Google Scholar]
- Zacharaki E.I., Wang S., Chawla S., Soo Yoo D., Wolf R., Melhem E.R., Davatzikos C. Classification of brain tumor type and grade using MRI texture and shape in a machine learning scheme. Magn. Reson. Med. 2009;62:1609–1618. doi: 10.1002/mrm.22147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zollner F.G., Emblem K.E., Schad L.R. Support vector machines in DSC-based glioma imaging: suggestions for optimal characterization. Magn. Reson. Med. 2010;64:1230–1236. doi: 10.1002/mrm.22495. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material