Abstract
Photopharmacology relies on molecules that change their biological activity upon irradiation. Many of these are derived from known drugs by replacing their core with an isosteric azobenzene photoswitch (azologization). The question is how many of the known bioactive ligands could be addressed in such a way. Here, we systematically assess the space of molecules amenable to azologization from databases of bioactive molecules (DrugBank, PDB, CHEMBL) and the Cambridge Structural Database. Shape similarity scoring functions (3DAPfp) and analyses of dihedral angles are employed to quantify the structural homology between a bioactive molecule and the cis or trans isomer of its corresponding azolog (“azoster”) and assess which isomer is likely to be active. Our analysis suggests that a very large number of bioactive ligands (>40 000) is amenable to azologization and that many new biological targets could be addressed with photopharmacology. N-Aryl benzamides, 1,2-diarylethanes, and benzyl phenyl ethers are particularly suited for this approach, while benzylanilines and sulfonamides appear to be less well-matched. On the basis of our analysis, the majority of azosters are expected to be active in their trans form. The broad applicability of our approach is demonstrated with photoswitches that target a nuclear hormone receptor (RAR) and a lipid processing enzyme (LTA4 hydrolase).
Short abstract
Azobenzene isosters (azosters) are common motifs in drugs and bioactive compounds. Substitution with azobenzenes could lead to thousands of candidates for photopharmacology.
Introduction
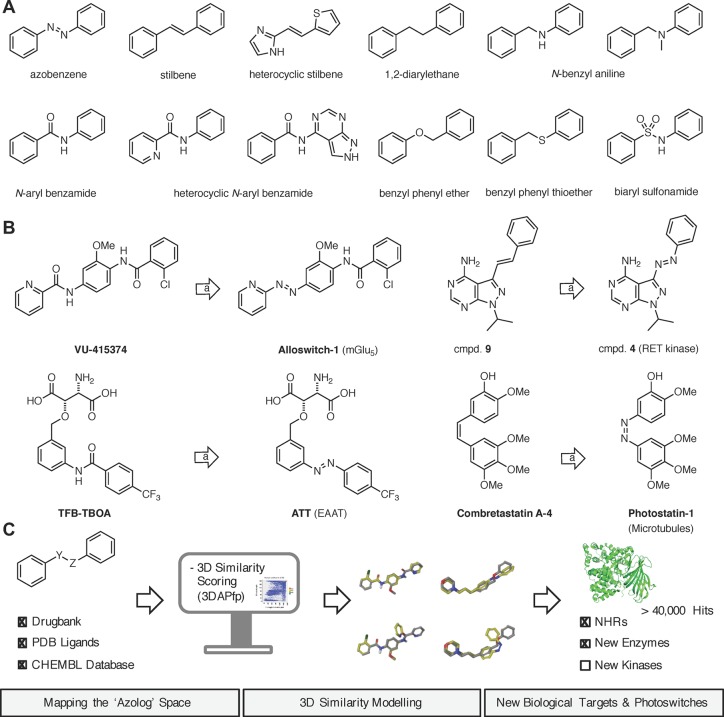
Many biological targets can be modulated by small molecules, which can be modified with a photoswitch to obtain optical control over their function. This approach, termed photopharmacology, has been successfully applied to ion channels, receptors, enzymes, transporters, and elements of the cytoskeleton.1−3 Many drugs and bioactive molecules possess motifs that resemble azobenzenes; i.e., they feature two arenes separated by a two-atom linker (Figure 1A). Substitution of this linker with a diazene unit (−N=N−) allows for the incorporation of a photoswitch with minimal structural perturbation of the pharmacophore. We call this approach “azologization” and the corresponding isosteric molecules “azosters”.4,5 Ideally, only one isomer of the azolog exhibits the bioactivity of the parent drug, while the other is inactive. A number of stilbenes,6−12N-aryl benzamides,13,14 benzyl phenyl ethers,15,16 and benzyl anilines17 have been successfully azologed. Other types, such as 1,2-diaryl ethanes, biaryl sulfonamides, N-alkyl benzylanilines, could also be amenable to this approach (Figure 1B).
Figure 1.
Azologization of bioactive molecules. (A) The structure of azobenzene in its trans-form and azosteric motifs found in bioactive molecules. (B) Selected examples of azosters drawn from the literature. (C) Systematic mapping of the photopharmacology space.
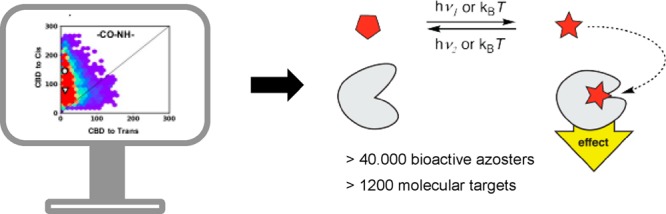
The success of azologization strategies to date raises the question how many bioactive molecules could be put under optical control in such way. To this end, we assessed the suitability of known bioactive ligands for azologization by virtual screening. Candidates from the PDB, ChEMBL, and DrugBank databases were evaluated based on computed structural homology (3DAPfp) and analysis of the dihedral angles of a central four-atom linker (Figure 1C).18 This analysis leads to the identification of over 40 000 bioactive molecules that should be amenable to azologization, many of which open new target classes for photopharmacology.
Results and Discussion
Identification of Bioactive Azologs
To confine our search to molecules with known bioactivity, we turned to the databases PDB ligand,19 DrugBank,20 and CHEMBL.21 PDB ligand contains experimentally determined 3D coordinates of molecules in their target-bound conformations, whereas DrugBank and CHEMBL combine 2D data (SMILES strings) with comprehensive drug target information. We also analyzed the Cambridge Structural Database (CSD), the largest source of small molecule single crystal X-ray structures.22 To identify possible azologable compounds in these databases, we considered all molecules with up to 50 non-hydrogen atoms and selected those containing a pair of aromatic or heteroaromatic ring systems separated by a two-atom linker (e.g., −CH=CH–, −CH2–CH2–, −CH2–NH–, −CONH–, −CH2–O–, −CH2–S–, −SO2–NH–, and others). In total, we identified more than 180 000 bioactive molecules that meet these criteria (Table 1), the majority of which were N-aryl benzamides (>60 000), benzyl phenyl ethers (>23 000), biaryl sulfonamides (>19 000), and benzyl anilines (>17 000). We also found more than 7000 stilbenes. The 3D coordinates of these molecules were either extracted from the databases (PDB and CSD) or calculated with the 3D structure generator CORINA (Drugbank and ChEMBL).
Table 1. Overview of Databases.
| no.
of cmpds as per top five linker types |
||||||||
|---|---|---|---|---|---|---|---|---|
| database | no. of cmpds | no. of azologable cmpdsa | –CO–NH– | –NH–CH2– | –SO2–NH– | –O–CH2– | –CH=CH– | no. of linker types |
| PDB ligand | 1,272,030 | 2,027 | 603 | 356 | 152 | 236 | 72 | 44 |
| CSD | 764,008 | 9,081 | 1,056 | 331 | 654 | 515 | 1,303 | 277 |
| DrugBank | 7,133 | 340 | 91 | 54 | 44 | 40 | 16 | 33 |
| ChEMBL | 1,678,393 | 167,688 | 59,522 | 16,263 | 18,354 | 22,724 | 6,856 | 192 |
In each database, only unique molecule entries (after counterion removal) containing acyclic linkers were considered. See Experimental Section for details.
Analysis of PDB Ligands and CSD Structures
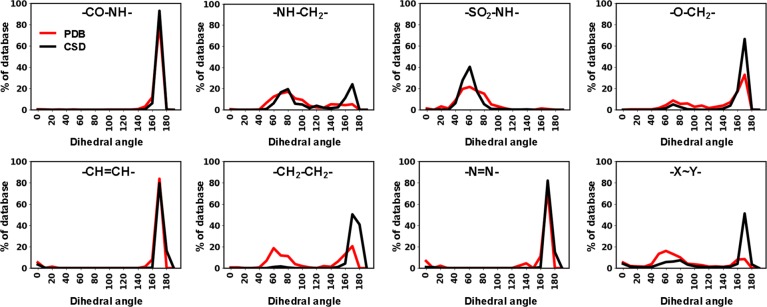
We first turned to the azologable PDB ligands because their target-bound conformations are known and can be directly compared to that of the respective cis or trans azologs. We also decided to include the experimental structures from the CSD into this analysis which are usually obtained in much higher resolution. To assess which compounds and linker-types are best suited for azologization, we compared the dihedral angles (ω) defined by the C–X–Y–C linkers (Figure 2). Molecules with ω-values close to 180° correspond to trans-azologs, while ω-values close to 0° match the angles in cis-azologs.
Figure 2.
Dihedral angle analysis of PDB and CSD compounds with azologable linkers.
The majority of PDB/CSD structures had dihedral angles close to 180° corresponding to the respective trans-azologs. For biaryl sulfonamides, benzyl anilines, and benzyl phenyl ethers we observed that a considerable number of representatives had dihedral angle values around 60°. Since this gauche confirmation neither fits the geometry of trans- nor cis-azologs, these compounds are likely less amenable to azologization.
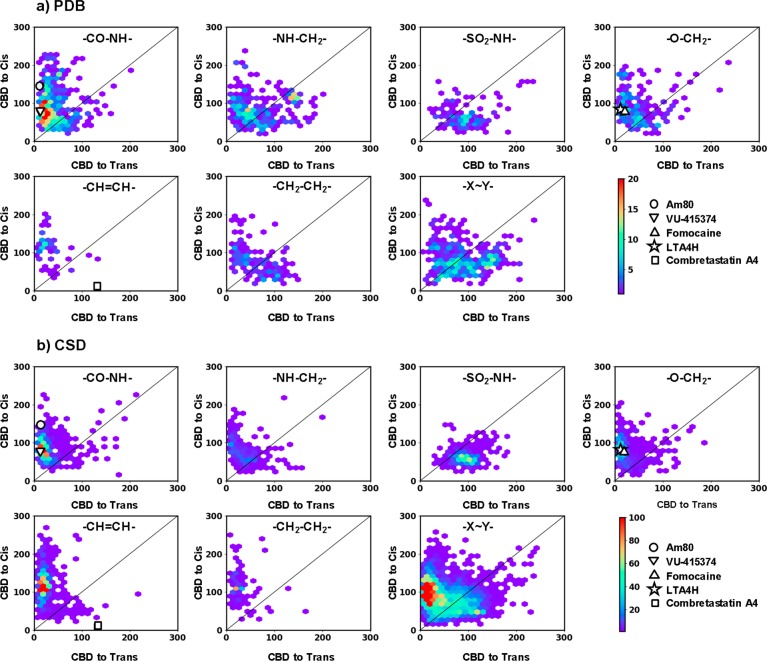
Next, we computationally generated cis- and trans-azologs for all PDB/CSD structures identified in our screen using the 3D builder CORINA.23 We compared the shape similarity of the parent compound to the computed cis- and trans-azologs using the 3D atom pair fingerprint 3DAPfp, which is a 16-dimensional fingerprint counting the number of atom pairs at exponentially increasing distances in a molecule, and which encodes molecular shape.18 3D structural homology between parent drug and either azolog isomer was quantified using city block distance (CBD) metric, which is the sum of the absolute differences between the 16 pairs of bit values in the respective 3DAPfp fingerprints, such that a low CBD3DAPfp value indicates a high shape similarity between two molecules. We subsequently analyzed the entire azolog space comparing the CBD values of parent structures with the respective cis/trans azologs. The results from the analysis were scatter plotted by linker type and database (Figure 3) and are individually discussed for the major compound classes below. In addition, we investigated for each class whether the linker engages in hydrogen bonding, which would be partially or fully lost upon azologization. We arbitrarily selected 30 benzyl anilines, N-alkyl benzyl anilines, N-aryl benzamides, and biaryl sulfonamides from the PDB ligands and analyzed the crystal structures for potential target engagement (SI Table 1).
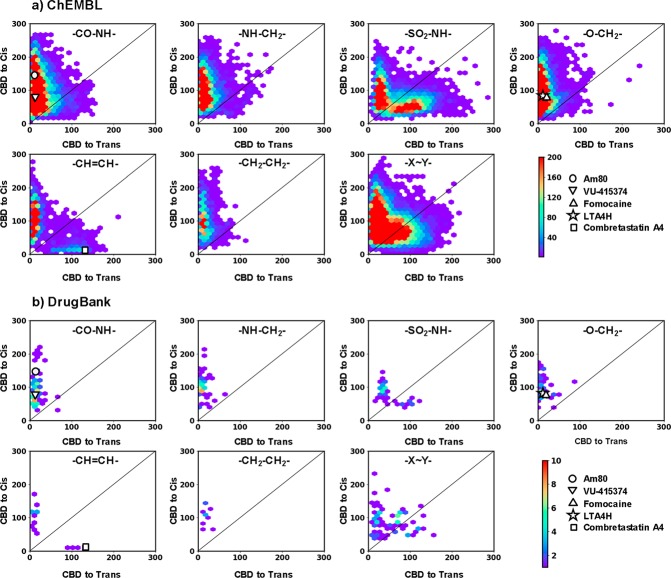
Figure 3.
Scatter plots showing the 3D-shape similarities of potential azologable compounds toward their cis-azolog (vertical axis) and trans-azolog (horizontal axis), for compounds from PDB (a) and CSD (b). 3D Shape similarities are reported as the city block distance (CBD) in the 3DAPfp, with the lowest value indicating highest similarity. Scatter plots are shown for six highly populated linker groups and rest of the groups merged together (−X–Y−). Each of the scatter plots is marked with the structure of respective linker type. Color scale shows the density of molecules. The compounds further investigated in this study are highlighted on the scatter plots.
Azobenzenes
In addition to the azologable targets analyzed above, PDB and CSD also contained a large number of azobenzenes themselves. Of course, these compounds would be the most straightforward photoswitches to use. In our search for bioactive azobenzenes we found 14 molecules in Drugbank, 45 in PDB, and 1602 in ChEMBL. Similar to the analysis performed above, we analyzed their experimentally determined 3D structures as reported in the PDB and CSD compared the dihedral angles across the C–N=N–C moiety (Figure 2). The majority of the azobenzenes were trans isomers with angles close to 180°, whereas a few examples of cis-azobenzenes existed that had dihedral angles close to 0° (examples: PDB 2H4B, 2M0Z, and 2CLX). None of the diazene units appear to engage in hydrogen bonding, although azobenzenes are protonated at this site under strongly acidic conditions (the pKa of methyl orange is 3.4).
Stilbenes
Stilbenes provide the most obvious azologization motif since the dihedral angles of trans-stilbenes match those of trans-azobenzenes, while cis-stilbenes would be expected to give cis-active azologs. However, surprisingly few cis-stilbenes have been found in our analysis. Combretastatin A-4 is a notable exception (Figure 3).
1,2-Diarylethanes
To the best of our knowledge, 1,2-diarylethanes have not yet been azologed for use in photopharmacology. Their dihedral angle analysis shows that approximately 50% of the PDB ligands bind their biological target with angles closely matching those of trans-azobenzenes. Like stilbenes, they cannot engage in hydrogen bonding at the linker region.
N-Aryl Benzamides
The most abundant azologization motifs found are N-aryl benzamides. The dihedral angle and 3D structural homology analyses confirm that these molecules should be prime targets for azologization and are expected to give trans-active compounds. Their linker amide can function as hydrogen bond donor or acceptor. Azologization could therefore lead to a loss of the bioactivity. However, amide linkers are often employed in medicinal chemistry to connect two fragments with an easily formed bond. Indeed, we found that approximately one-third of carboxamide linkers (11/30) do not engage in hydrogen bonding, increasing the chances that the corresponding azolog has comparable potency to the parent compound.
Benzyl Phenyl Ethers
Benzyl phenyl ethers have been successfully employed on a few occasions. Our study suggests that this linker class is highly amenable to azologization. The majority of benzyl phenyl ethers possesses dihedral angles around 180°, which primes them as potential trans-azologs. A few benzyl phenyl ethers have gauche angles making them less suitable for azologization. Only 2 of the 30 benzyl phenyl ethers analyzed showed hydrogen bonding to the linker, which is significantly less compared to N-aryl benzamides or benzyl anilines.
N-Benzyl Anilines
While benzyl anilines have been successfully azologed previously, our analysis suggests that only a few of these compounds have suitable dihedral angles. A surprisingly large proportion of these compounds exhibit gauche-like dihedral angles. Approximately one-third of secondary amine linkers (11/30) and tertiary amine linkers (11/30) show hydrogen bonding in the linker region, which is significantly less than in N-aryl benzamides but more than in benzyl phenyl ethers.
Biaryl Sulfonamides
Our analysis suggests that biaryl sulfonamides are generally challenging targets for azologization. They almost exclusively exhibit gauche conformations in their bioactive form and do not show a pronounced tendency for structural overlap with either trans- or cis-azologs in our 3D structural homology analysis. Roughly one-third of the biaryl sulfonamide analyzed (11/30) also showed hydrogen bonding in the linker region.
Analysis of Drugbank and ChEMBL
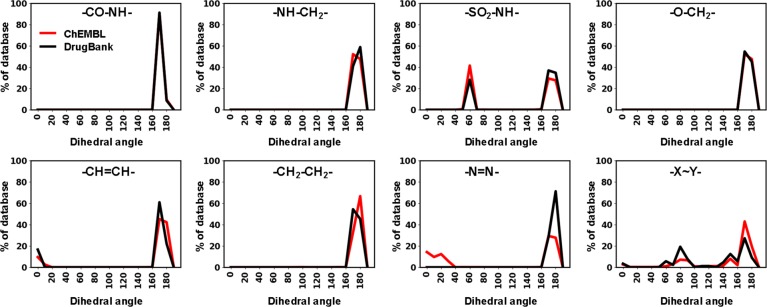
Next, we repeated the analysis for Drugbank and ChEMBL using the computationally generated 3D structures (CORINA calculation). With the exception of sulfonamides, all linker types overwhelmingly showed preferred dihedral angles close to 180° (Figure 4).
Figure 4.
Dihedral angle analysis of ChEMBL and DrugBank compounds with azologable linkers.
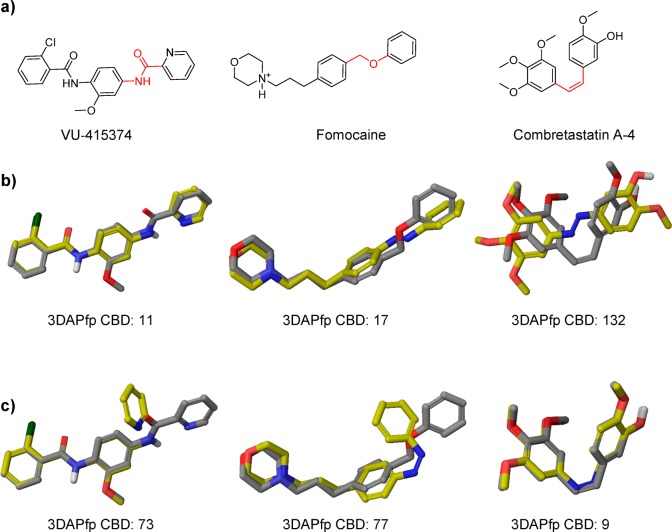
The scatter plots show similar distributions to that of PDB and CSD compounds, suggesting that the CBD values could indeed be indicative of isomer activity and guide the choice of compounds for azologization (Figure 5). We calibrated this analysis with three known photoswitches, which had previously been developed using an azologization strategy (Figure 6). These include VU-415374, a positive allosteric modulator of metabotropic glutamate receptor type 4, the ion channel blocker fomocaine, and the microtubule inhibitor combretastatin A-4. For VU-415374 and fomocaine, the 3D structure overlay with the respective trans-azolog showed more overlap and significantly lower CBD scores compared to that of the cis-azolog. For combretastatin A-4 we obtained the reversed result and significantly better overlap between parent drug and cis-azolog. These results matched the experimental results and demonstrate that low CBD scores give good predictions about which isomer of an azolog is active.
Figure 5.
Scatter plots showing the 3D shape similarities of potential azologable compounds toward their cis-azolog (vertical axis) and trans-azolog (horizontal axis), for compounds from PDB (a) and CSD (b). 3D Shape similarities are reported as the city block distance (CBD) in the 3DAPfp, with lowest value indicating highest similarity. Scatter plots are shown for six highly populated linker groups and rest of the groups merged together (−X–Y−). Each of the scatter plots is marked with the structure of respective linker type. Color scale shows the density of molecules. The compounds further investigated in this study are highlighted on the scatter plots.
Figure 6.
(a) Structures of previously reported azologable compounds. The two atom linkers which were replaced by the diazo group are highlighted in red. (b) 3D-overlays of parent azologable compounds (gray) with corresponding trans-azologs (yellow). (c) 3D-overlays of parent azologable compounds (gray) with corresponding cis-azologs (yellow).
Azologs and Their Biological Targets
From the analysis above, we selected all molecules from ChEMBL, DrugBank, and PDB having two-atom linkers with experimental or predicted linker dihedral angle falling within the range 0–20° (cis-azologization targets) or within the range 160–180° (trans-azologization targets; Table 2).
Table 2. Azologable Compounds.
| no.
of cmpds. as per top five linker types |
||||||
|---|---|---|---|---|---|---|
| database | no. of azologable cmpds | –CO–NH– | –NH–CH2– | –SO2–NH– | –O–CH2– | –CH=CH- |
| PDB Ligand | 949 | 577 | 37 | 5 | 117 | 70 |
| DrugBank | 281 | 91 | 54 | 32 | 40 | 16 |
| ChEMBL | 40,719 | 16,097 | 4,746 | 3,915 | 7,785 | 1,186 |
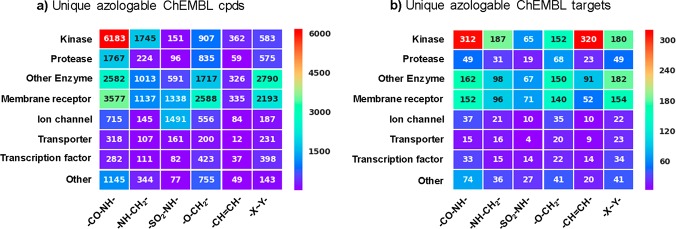
We analyzed these hits according to biological targets. In total, more than 1200 biological targets in the CHEMBL database could be amenable to azologization. Multiple hits were found for most, which increases the chances to find a useful photopharmaceutical. Strikingly, only a small fraction of these biological targets has been previously addressed by photopharmacology. For example, relatively few enzymes found were put under photocontrol. Kinases present the largest class of biological targets that were identified in our screen (Figure 7). While some efforts in optogenetics have been geared toward the optical control of kinases, only a few studies in photopharmacology have addressed them. Other target classes that could benefit from the spatiotemporal resolution of photopharmacology include transcription factors, GPCRs, ion channels, and transporters.
Figure 7.
Heatmaps showing (a) number of unique ChEMBL compounds and (b) number of unique ChEMBL targets as a function of linker type and protein target family. Only azologable compounds within the correct predicted dihedral angle windows having IC50 and EC50 values of <10 μM (∼41 K compounds) were considered for this analysis.
Optical Control of RARα Receptor
To demonstrate the usefulness of our analysis, we synthesized and evaluated a number of hits. To this end, we chose molecules that are easily synthetically accessible and could modulate biological targets that have not been previously addressed with photopharmacology.
Nuclear hormone receptors (NHRs) are hormone targeted transcription factors which bind to DNA and regulate various biological processes.24,25 Apart from the modulation of transcription levels on a relatively slow time scale, several NHRs are known to mediate rapid nongenomic effects which are thought to occur through protein–protein interactions.26 Nongenomic functions of NHRs include the regulation of kinases, phosphatase, and ion channels.27 Optical control of NHRs could enable the dissection between genomic and nongenomic mechanisms with the resolution of single cells in complex cellular networks.
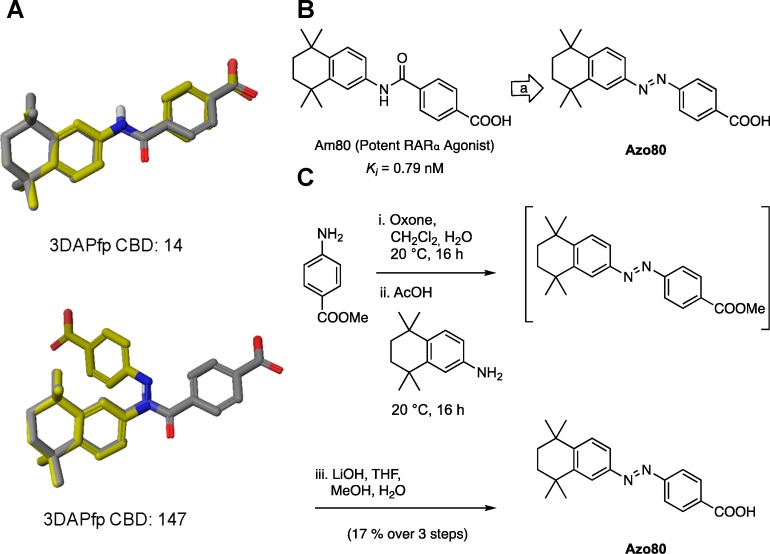
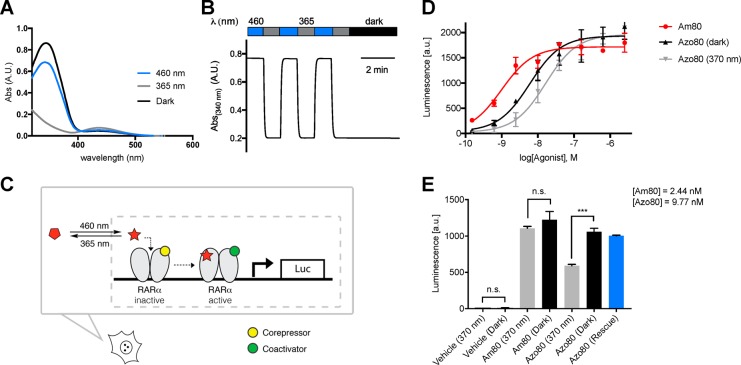
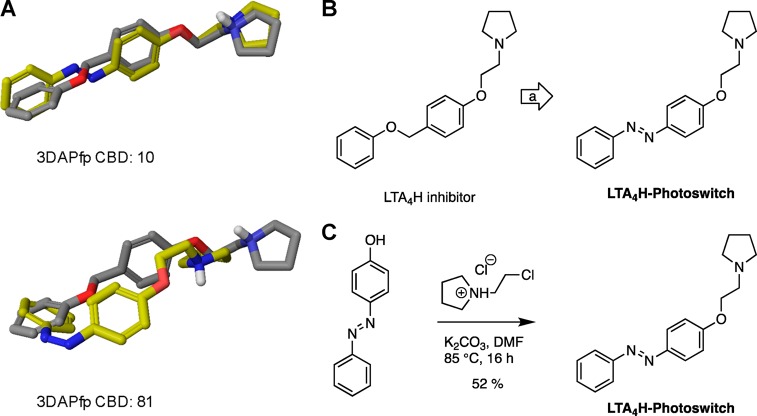
Our computational screen for azologization motifs led to the identification of several hits for a number of nuclear hormone receptors, including retinoic X receptor, retinoic acid receptor, thyroid hormone receptor, peroxisome proliferator-activated receptor, liver X receptor, and estrogen receptor. We decided to synthesize the azolog of one of these compounds targeting retinoic acid receptor α (RARα). The potent RARα agonist Am80 is commercially available and widely used in biological research. At the same time our screen suggests that Am80 is an ideal target for azologization (3DAPfp CBD trans: 14; 3DAPfp cis: 147; Figure 8 A). Interestingly, the azolog of Am80 was already synthesized in 1989 and evaluated in SAR studies (CHEMBL 13150).28 However, its potential for optical control has never been evaluated, and the compound was exclusively tested in its nonirradiated form. We resynthesized the azolog of Am80, termed Azo80 (Figure 8B), using Baeyer-Mills conditions with subsequent ester hydrolysis (Figure 8C).
Figure 8.
Computational prediction, design, and synthesis of Azo80. (A) 3D overlays of parent azologable compounds (gray) with corresponding cis- and trans- azologs (yellow) and 3DAPfp scores of 3D shape similarity comparison. (B) Design of Azo80 based on the azologization of the N-aryl benzamides Am80. (C) Chemical synthesis of Azo80.
The photophysical properties of Azo80 are similar to those of a classical nonsubstituted azobenzene. Azo80 can be efficiently switched between cis/trans using 365/460 nm light and is bistable (Figure 9A,B). To test Azo80 for the ability to photocontrol RARα, we used a reporter gene assay in which the activation of RARα induces transcription of luciferase (Figure 9C). Upon addition of luciferase substrate after 24 h incubation, a luminescent signal proportional to luciferase transcription and RARα activation was quantified. We were pleased to find that the EC50 of cis-Azo80 (1.6 × 10–8 M) is significantly lower than that of trans-Azo80 (6.0 × 10–9 M) (Figure 9D). In a subsequent control experiment (Figure 9E), we demonstrated that light does not mediate or alter transcription levels in the absence of Azo80, while reversible optical control is achieved with Azo80. To demonstrate reversibility, Azo80 was added to cells as 365 nm-adapted cis-Azo80 and after 5 min the cells were illuminated with 460 nm light for 2 min to reactivate Azo80. Similar levels of transcription were observed in this rescue experiment compared to the experiment with dark-adapted trans-Azo80. The trans-activity of Azo80 is coherent with the computational prediction, which suggested better 3D homology of Am80 with its trans-azolog.
Figure 9.
Photophysical and biological evaluation of Azo80. (A) The UV–vis spectrum of Azo80 in the dark-adapted (black, trans), 365 nm adapted (gray, cis), and 460 nm adapted (blue, trans) photostationary states. (B) Reversible cycling between isomers with alternating illumination at 365/460 nm. (C) Schematic depiction of RARα activation with a small molecule photoswitch leading to corepressor/coactivator exchange and transcription of target genes (here: luciferase–luc). (D) Dose responses of Am80, cis-Azo80, and trans-Azo80 in a luminescent reporter cell line after 22 h. Samples were run in duplicates and in two independent experiments. Error bars represent mean ± SD (E) Control and rescue (reversibility) experiments. Samples were run in triplicates. Error bars represent SEM *** p < 0.001, n.s., not significant, student’s t-test.
Optical Control of Leuktriene-A4 Hydrolase
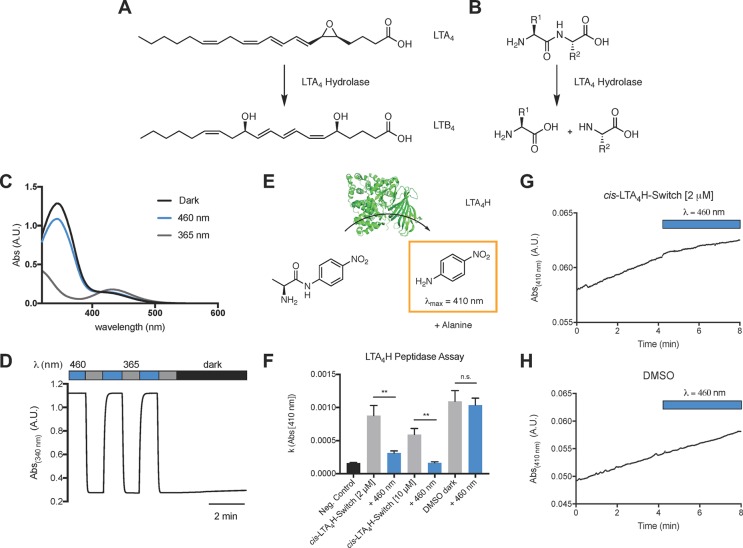
Lipid photopharmacology is a rapidly growing field, and photoswitchable lipids have enabled the control of a wide range of biological pathways.29−31 In this context, the optical modulation of lipid metabolic networks could provide important insights. Our computational screen identified a number of small molecules that target various lipid metabolizing enzymes, including fatty acid amide hydrolases, lipoxygenases, phospholipases, leukotriene hydrolases, lipid kinases, and phosphorylases. Many of these enzymes could be interesting targets for photopharmacology. We decided to synthesize and test a photoswitchable inhibitor of the enzyme Leukotriene A4 hydrolase (LTA4H). LTA4H is a dual enzyme that functions both as a hydrolase and as aminopeptidase (Figure 11).32 It catalyzes the conversion of LTA4 to LTB4 and the degradation of chemoattractant tripeptide molecules. Multiple inhibitors for LTA4H have been in development as anti-inflammatory drugs.33 The CHEMBL database contained several benzyl phenyl ether based LTA4H inhibitors, and we chose a relatively potent candidate of this compound family with straightforward synthesis for azologization (Figure 10). The CBD values suggest that this inhibitor exhibits more structural homology with its trans-azolog (3DAPfp CBD trans: 10; 3DAPfp cis: 81; Figure 10 A). LTA4h-Photoswitch was synthesized from 4-phenylazophenol through Williamson ether synthesis (Figure 10C).
Figure 11.
Photophysical evaluation and LTA4-hydrolase peptidase assay with LTA4H-Photoswitch. (A, B) Enzymatic reactions catalyzed by LTA4-hydrolase. (C) The UV–vis spectrum of LTA4H-Photoswitch in the dark-adapted (black, trans), 365 nm adapted (gray, cis), and 460 nm adapted (blue, trans) photostationary states. (D) Reversible cycling between isomers with alternating illumination at 365/460 nm. (E) Schematic depiction of l-alanine 4-nitroanilide cleavage by LTA4H (PDB: 2VJ8(34)). (F) LTA4H peptidase assay with LTA4 h (1.1 μg) and l-alanine 4-nitroanilide (1 mM) in the presence and absence of cis-LTA4H-Photoswitch at different concentrations. Samples were irradiated with 460 nm light after 4 min to yield trans-LTA4H-Photoswitch. The slope of 4-nitroaniline absorption (λ = 410 nm) was plotted. (G, H) Representative traces of 4-nitroaniline absorption (λ = 410 nm) before and after application of 460 nm light. Samples were run in triplicates. Error bars represent SEM ** p < 0.01, n.s., not significant, student’s t-test.
Figure 10.
Synthesis and isomerization of LTA4h-Photoswitch. (A) 3D-overlays of parent azologable compounds (gray) with corresponding cis- and trans- azologs (yellow) and 3DAPfp scores of 3D shape similarity comparison. (B) Design of LTA4h-Photoswitch based on the azologization of a benzyl phenyl ethers. (C) Chemical synthesis of LTA4h-Photoswitch.
We used a colorimetric LTA4H peptidase assay to evaluate the potential of LTA4H-Photoswitch for the optical control of LTA4H aminopeptidase activity using l-alanine 4-nitroanilide which is converted to the strongly absorbing 4-nitroaniline (λ = 410). This assay is most commonly used to screen for LTA4H inhibitors. In good agreement with the computational prediction, trans-LTA4H-Photoswitch was a more potent inhibitor of LTA4H peptidase than the cis-isomer (Figure 11).
Azo80 and LTA4H-Photoswitch are two new photoswitches obtained through the azologization of a N-aryl benzamide and a benzyl phenyl ether which were identified through the mapping of the photopharmacological azolog space in CHEMBL. These examples demonstrate the utility of the database for the development of bioactive photoswitches for the modulation of new photopharmacological targets from two different azologization motifs.
Concluding Remarks
Our study sheds light on the scope and limitations of azologization in photopharmacology. The azolog space was found to be surprisingly large, comprising more than 40 000 bioactive molecules, which modulate more than 1200 biological targets. Relatively few of those have been put under optical control to date. The identification of functional azologs for two completely different new targets classes (LTA4H and RARα) illustrates the usefulness of our systematic approach.
3DAPfp shape similarity scores and analysis of dihedral angles gave good predictions of the active isomer for the examples explored and for the newly developed photoswitches reported. These scores could be used to predict which isomer is bioactive. Additional insights for the design and prediction can often be drawn from crystal structures and structure activity relationship studies.
Our results suggest that stilbenes, 1,2-diarylethanes, N-aryl benzamides, and benzyl phenyl ethers are excellent candidates for azologization. Benzylanilines and especially sulfonamides appear to be less suited for this approach. The analysis of both dihedral angles and shape similarity indicates that the vast majority of azologs are likely to be more bioactive in their trans-form. Usually cis-active photoswitches are more desirable as they are inactive in the dark-adapted state and can be activated with light. However, trans-active compounds can still be highly useful (e.g., as tonic ion channel blockers) and bistable photoswitches that can be easily kept in the inactive cis-state. In addition, cis-stable azobenzene photoswitches are emerging that could be adapted to photopharmacology.35
The large number of potential molecular targets (>1200) that should be addressable with photopharmacology suggests that this approach to optical control is very versatile. It should be noted, however, that methods other than azologization further increase the reach of photopharmacology. Many photoswitches were designed through extension of the core with azobenzenes instead of replacement (“azo-extension” approach). An n-alkyl chain has been replaced with an azobenzene in the optical control of glycerophospho- and sphingolipids. In addition, photoswitches have been successfully installed in the backbone or side chain of biopolymers, such as nucleic acids and peptides. While “azologization” is the most straightforward design strategy, all existing strategies complement each other to provide an exceptionally versatile photopharmacological toolbox.
Experimental Section
Processing of Database and Azologs Generation
DrugBank (version 5, https://www.drugbank.ca/), PDB ligands (http://ligand-expo.rcsb.org/ld-download.html), and ChEMBL (version 22, https://www.ebi.ac.uk/chembl/) databases were downloaded in SDF format from the respective database Web site in year 2017. CSD was copied from a licensed CD to Dr. Jürg Hauser, University of Bern. Molecules were processed using an in-house developed Java program utilizing the JChem chemistry library from ChemAxon Pvt. Ltd. (https://www.chemaxon.com/). Counter ions were removed, valence errors were checked, and molecules were ionized at pH 7.4. Molecules containing ≥6 and ≤50 heavy atoms, ≤4 stereocenters, 1 Ar-(two atom linker)-Ar, and no Ar-(diazo linker)-Ar were retained in the database. “Ar” stands for aromatic carbon. The two atoms in linker are acyclic atoms and may or may not be substituted. For each database, duplicate molecules were removed based on unique smiles comparisons. The resulting molecules in these processed databases are considered as potential azologable compounds. Afterward for each of the azologable molecules, Ar-(two atom linker)-Ar was replaced by Ar-(diazo linker)-Ar, and corresponding trans- and cis-azologs were generated. It should be noted that, whenever the difference in number of atoms between parent azologable molecules and corresponding azologs were more than six atoms, the corresponding molecules were not considered in the study. This was because some of the two atom linkers in parent molecules were substituted by large groups.
Similarity Calculation
For each drug and its two isomeric azolog, we generated the lowest energy 3D conformer using the CORINA program available from Molecular Networks Pvt. Ltd. For the parent azologable compounds in PDB and CSD databases experimental 3D coordinates were used. To compare molecules, we used an in-house developed 3D atom pair fingerprint as a measure of overall shape similarity.36 For each parent drug and its corresponding cis and trans azolog 3D atom pair fingerprints were computed, and similarities between them were quantified using city block distance metric.
Chemical Synthesis
All reagents and solvents were purchased from commercial sources (Sigma-Aldrich, TCI Europe N.V., Strem Chemicals, etc.) and were used without further purification. Solvents were obtained from Fisher Scientific. Reactions were monitored by TLC on precoated, Merck Silica gel 60 F254 glass backed plates, and the chromatograms were first visualized by UV irradiation at λ = 254 nm. Flash silica gel chromatography was performed using silica gel (SiO2, particle size 40–63 μm) purchased from SiliCycle. NMR spectra were measured on a BRUKER Avance III HD 400 (equipped with a CryoProbe). Multiplicities in the following experimental procedures are abbreviated as follows: s = singlet, d = doublet, t = triplet, q = quartet, m = multiplet. Proton chemical shifts are expressed in parts per million (ppm, δ scale) and are referenced to the residual protium in the NMR solvent (CDCl3 = 7.26; MeOD: δ = 3.31). Carbon chemical shifts are expressed in ppm (δ scale) and are referenced to the carbon resonance of the NMR solvent ((CDCl3: δ = 77.16; MeOD: δ = 49.00). NOTE: Due to the trans/cis isomerization of some compounds containing an azobenzene functionality, more signals were observed in the 1H and 13C spectra than would be expected for the pure trans-isomer. Only signals for the major trans-isomer are reported.
Photophysical Evaluation
UV–vis spectra were recorded using a Varian Cary 50 Bio UV–visible spectrophotometer with Hellma SUPRASIL precision cuvettes (10 mm light path). Switching was achieved using 365 or 460 nm LED light sources. The LEDs were pointed directly into the top of the sample cuvette. An initial spectrum was recorded (dark-adapted state, black) and then again following illumination at λ = 365 nm for 30 s (cis-adapted state, gray). A third spectrum was recorded after irradiation at λ = 470 nm for 30 s (trans-adapted state, blue).
Azo80
A solution of Oxone (378 mg, 0.615 mmol, 2.5 equiv) in water (2 mL) was added to a solution of 4-aminobenzoate (37.2 mg, 0.246 mmol, 1.0 equiv) in CH2Cl2 (2 mL), and the biphasic mixture was stirred for 16 h at rt. The organic layer was washed with 1 M HCl, sat. NaHCO3, water, dried over sodium sulfate, filtered, and concentrated in vacuo. 5,6,7,8-Tetrahydro-5,5,8,8-tetramethyl-2-naphthylamine (50.0 mg, 0.246 mmol, 1.0 equiv) and acetic acid (2.0 mL) were added and stirred for 24 h at rt. Acetic acid was removed in vacuo, and the residue was dissolved in CH2Cl2, washed with sat. NaHCO3, water, dried over sodium sulfate, filtered, and concentrated in vacuo. The residue was dissolved in THF/MeOH (2 mL/2 mL), treated with 1 M LiOH (1 mL), stirred for 2 h at rt, and acidified with 2 M HCl. It was diluted with EtOAc, washed with water, dried over sodium sulfate, filtered, and concentrated in vacuo. Purification by flash column chromatography (CH2Cl2 + 1% AcOH) afforded Azo80 (13.8 mg, 0.041 mmol, 17%) as an orange solid. A small amount of methanol was added to obtain a more concentrated solution for the 13C NMR measurement. 1H NMR (400 MHz, CDCl3) δ 8.32–8.04 (m, 3H), 7.89 (d, J = 1.8 Hz, 1H), 7.86 (d, J = 7.7 Hz, 2H), 7.63 (dd, J = 8.5, 1.9 Hz, 1H), 7.40 (d, J = 8.5 Hz, 1H), 1.69 (s, 4H), 1.33 (s, 6H), 1.28 (s, 6H). 13C NMR (100 MHz, CDCl3) δ 155.5, 150.7, 149.5, 146.2, 131.0, 127.6, 123.6, 123.4, 122.4, 118.5, 35.0, 34.9, 34.8, 34.6, 31.8, 31.7. HRMS: m/z calcd. for C21H23N2O2– ([M + H]−): 335.1765, found: 335.1758.
LTA4H-Photoswitch
A solution of 1-(2-chloroethyl)pyrrolidine·HCl (51.4 mg, 0.30 mmol, 1.2 equiv), 4-phenylazophenol (50.0 mg, 0.25 mmol, 1.0 equiv), and K2CO3 (104 mg, 0.76 mmol, 3.0 equiv) in DMF (3 mL) was stirred at 85 °C for 16 h. The solution was cooled, water was added, and extracted with EtOAc. The combined organic phase was dried over Na2SO4, and concentrated in vacuo. Purification by flash column chromatography with CH2Cl2 → 10% MeOH in CH2Cl2 yielded LTA4H-Photoswitch (38.6 mg, 0.13 mmol, 52%) as an orange liquid. 1H NMR (400 MHz, MeOD): δ 7.93–7.81 (m, 4H), 7.55–7.39 (m, 3H), 7.11–7.04 (m, 2H), 4.19 (t, J = 5.6 Hz, 2H), 2.94 (t, J = 5.6 Hz, 2H), 2.68 (t, J = 6.6 Hz, 4H), 1.86–1.78 (m, 4H). 13C NMR (100 MHz, MeOD): δ 162.8, 154.0, 148.3, 131.6, 130.2, 125.8, 123.5, 115.9, 67.9, 55.5, 24.2. HRMS: m/z calcd. for C18H22N3O ([M + H]+): 296.1757, found: 296.1757.
RARα Reporter Gene Assay
A cell-based human RARα (NR1B1) driven luciferase reporter assay from INDIGO Bioscience (State College, PA) was adapted and used for the biological evaluation of Azo80. In brief, a 10 mM stock solution of Am80 or Azo80 was diluted with the provided cell screening medium to a final concentration of 2.5 μM. A 4-fold dilution series was prepared using this initial concentration and cell screening medium. The dilutions were added to reporter cells in white-bottom 96-well plates. For trans-Azo80, dilutions were added in the dark and cells were incubated for 22 h in the dark. For cis-Azo80, dilutions were irradiated at 365 nm for 3 min, and cells were incubated for 22 h in the presence of a 370 nm LED Cell Disco with light pulses for 75 ms/15 s. To minimize variations in the dilutions, the same dilutions were used for both experiments before and after irradiation. For the rescue experiment, Azo80 was added to cells as 365 nm adapted cis-Azo80 and cells were illuminated after 5 min with 460 nm light for 2 min to reactivate Azo80. After 22 h medium was ejected, and the supplied luciferase detection reagents were added and quantified using a BMG Labtech FLUOstar Omega plate reader.
LTA4H-Peptidase Assay
Recombinant LTA4H was purchased from Cayman Chemicals and stored at −80 °C. LTA4H (1.1 μg) was incubated with l-alanine-p-nitroanilide (1 mM), in 50 mM HEPES (pH = 7.5), 100 mM KCl, 1 mg/mL BSA, 1% DMSO in the presence and absence of LTA4H-Photoswitch. LTA4H-Photoswitch was illuminated with 365 nm light for 3 min before addition of l-alanine-p-nitroanilide, and the reaction was initiated with LTA4H added last. The absorption at 410 nm was recorded for 4 min. After 4 min a 460 nm LED was used to illuminate the sample in the cuvette, and the absorption was recorded for 4 min under constant illumination.
Safety Statement
No unexpected or unusually high safety hazards were encountered in this line of research.
Acknowledgments
We thank New York University for financial support. NMR spectra were acquired using the TCI cryoprobe supported by the NIH (OD016343). This work was supported financially by the Swiss National Science Foundation, NCCR TransCure. J.M. thanks the German Academic Scholarship Foundation for a Ph.D. fellowship and New York University for a MacCracken Ph.D. fellowship. Dr. Bryan Matsuura, Christopher Arp, Martin Reynders, Konstantin Hinnah, Dr. Andrej Shemet, and Dr. Christian Fischer are acknowledged for insightful discussion and critical review of the manuscript.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acscentsci.8b00881.
Analysis of linker–protein contacts, analysis of correlation between similarity scatter plot and dihedral angle, compound characterization by NMR and HRMS (PDF)
Author Contributions
§ J.M. and M.A. contributed equally.
The authors declare no competing financial interest.
Supplementary Material
References
- Hüll K.; Morstein J.; Trauner D. In Vivo Photopharmacology. Chem. Rev. 2018, 118, 10710–10747. 10.1021/acs.chemrev.8b00037. [DOI] [PubMed] [Google Scholar]
- Beharry A. A.; Woolley G. A. Azobenzene Photoswitches for Biomolecules. Chem. Soc. Rev. 2011, 40, 4422–4437. 10.1039/c1cs15023e. [DOI] [PubMed] [Google Scholar]
- Velema W. A.; Szymanski W.; Feringa B. L. Photopharmacology: Beyond Proof of Principle. J. Am. Chem. Soc. 2014, 136, 2178–2191. 10.1021/ja413063e. [DOI] [PubMed] [Google Scholar]
- Broichhagen J.; Frank J. A.; Trauner D. A Roadmap to Success in Photopharmacology. Acc. Chem. Res. 2015, 48, 1947–1960. 10.1021/acs.accounts.5b00129. [DOI] [PubMed] [Google Scholar]
- Schoenberger M.; Damijonaitis A.; Zhang Z.; Nagel D.; Trauner D. Development of a New Photochromic Ion Channel Blocker via Azologization of Fomocaine. ACS Chem. Neurosci. 2014, 5, 514–518. 10.1021/cn500070w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowiak M.; Nahaboo W.; Reynders M.; Nekolla K.; Jalinot P.; Hasserodt J.; Rehberg M.; Delattre M.; Zahler S.; Vollmar A.; et al. Photoswitchable Inhibitors of Microtubule Dynamics Optically Control Mitosis and Cell Death. Cell 2015, 162, 403–411. 10.1016/j.cell.2015.06.049. [DOI] [PubMed] [Google Scholar]
- Engdahl A. J.; Torres E. A.; Lock S. E.; Engdahl T. B.; Mertz P. S.; Streu C. N. Synthesis, Characterization, and Bioactivity of the Photoisomerizable Tubulin Polymerization Inhibitor Azo-Combretastatin A4. Org. Lett. 2015, 17, 4546–4549. 10.1021/acs.orglett.5b02262. [DOI] [PubMed] [Google Scholar]
- Sheldon E.; Dcona M. M.; Lyons C. E.; Hackett J. C.; Hartman M. C. T. Photoswitchable Anticancer Activity via Trans – Cis Isomerization of a Combretastatin A-4 Analog. Org. Biomol. Chem. 2016, 14, 40–49. 10.1039/C5OB02005K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damijonaitis A.; Broichhagen J.; Urushima T.; Hull K.; Nagpal J.; Laprell L.; Schonberger M.; Woodmansee D. H.; Rafiq A.; Sumser M. P.; et al. AzoCholine Enables Optical Control of Alpha 7 Nicotinic Acetylcholine Receptors in Neural Networks. ACS Chem. Neurosci. 2015, 6, 701–707. 10.1021/acschemneuro.5b00030. [DOI] [PubMed] [Google Scholar]
- Broichhagen J.; Jurastow I.; Iwan K.; Kummer W.; Trauner D. Optical Control of Acetylcholinesterase with a Tacrine Switch. Angew. Chem., Int. Ed. 2014, 53, 7657–7660. 10.1002/anie.201403666. [DOI] [PubMed] [Google Scholar]
- Rastogi S. K.; Zhao Z.; Barrett S. L.; Shelton S. D.; Zafferani M.; Anderson H. E.; Blumenthal M. O.; Jones L. R.; Wang L.; Li X.; et al. Photoresponsive Azo-Combretastatin A-4 Analogues. Eur. J. Med. Chem. 2018, 143, 1–7. 10.1016/j.ejmech.2017.11.012. [DOI] [PubMed] [Google Scholar]
- Ferreira R.; Nilsson J. R.; Solano C.; Andréasson J.; Grøtli M. Design, Synthesis and Inhibitory Activity of Photoswitchable RET Kinase Inhibitors. Sci. Rep. 2015, 5, 9769. 10.1038/srep09769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittolo S.; Gómez-Santacana X.; Eckelt K.; Rovira X.; Dalton J.; Goudet C.; Pin J.-P.; Llobet A.; Giraldo J.; Llebaria A.; et al. An Allosteric Modulator to Control Endogenous G Protein-Coupled Receptors with Light. Nat. Chem. Biol. 2014, 10, 813–815. 10.1038/nchembio.1612. [DOI] [PubMed] [Google Scholar]
- Cheng B.; Trauner D.; Shchepakin D.; Kavanaugh M. P. A Photoswitchable Inhibitor of a Glutamate Transporter. ACS Chem. Neurosci. 2017, 8, 1668–1672. 10.1021/acschemneuro.7b00072. [DOI] [PubMed] [Google Scholar]
- Schoenberger M.; Damijonaitis A.; Zhang Z.; Nagel D.; Trauner D. Development of a New Photochromic Ion Channel Blocker via Azologization of Fomocaine. ACS Chem. Neurosci. 2014, 5, 514–518. 10.1021/cn500070w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broichhagen J.; Johnston N. R.; von Ohlen Y.; Meyer-Berg H.; Jones B. J.; Bloom S. R.; Rutter G. A.; Trauner D.; Hodson D. J. Allosteric Optical Control of a Class B G-Protein-Coupled Receptor. Angew. Chem., Int. Ed. 2016, 55, 5865–5868. 10.1002/anie.201600957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J. A.; Yushchenko D.; Fine N. H. F.; Duca M.; Citir M.; Broichhagen J.; Hodson D. J.; Schultz C.; Trauner D. Optical Control of GPR40 Signalling in Pancreatic β-Cells. Chem. Sci. 2017, 8, 7604–7610. 10.1039/C7SC01475A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awale M.; Jin X.; Reymond J.-L. Stereoselective Virtual Screening of the ZINC Database Using Atom Pair 3D-Fingerprints. J. Cheminf. 2015, 7, 3. 10.1186/s13321-014-0051-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burley S. K.; Berman H. M.; Kleywegt G. J.; Markley J. L.; Nakamura H.; Velankar S.. Protein Data Bank (PDB): The Single Global Macromolecular Structure Archive. In Protein Crystallography: Methods and Protocols; Wlodawer A., Dauter Z., Jaskolski M., Eds.; Methods in Molecular Biology; Springer New York: New York, NY, 2017; pp 627–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law V.; Knox C.; Djoumbou Y.; Jewison T.; Guo A. C.; Liu Y.; Maciejewski A.; Arndt D.; Wilson M.; Neveu V.; et al. DrugBank 4.0: Shedding New Light on Drug Metabolism. Nucleic Acids Res. 2014, 42, D1091–D1097. 10.1093/nar/gkt1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bento A. P.; Gaulton A.; Hersey A.; Bellis L. J.; Chambers J.; Davies M.; Krüger F. A.; Light Y.; Mak L.; McGlinchey S.; et al. The ChEMBL Bioactivity Database: An Update. Nucleic Acids Res. 2014, 42, D1083–D1090. 10.1093/nar/gkt1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groom C. R.; Bruno I. J.; Lightfoot M. P.; Ward S. C. The Cambridge Structural Database. Acta Crystallogr., Sect. B: Struct. Sci., Cryst. Eng. Mater. 2016, 72, 171–179. 10.1107/S2052520616003954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski J.; Gasteiger J. From Atoms and Bonds to Three-Dimensional Atomic Coordinates: Automatic Model Builders. Chem. Rev. 1993, 93, 2567–2581. 10.1021/cr00023a012. [DOI] [Google Scholar]
- da Silva S. L.; Burbach J. P. H. The Nuclear Hormone-Receptor Family in the Brain: Classics and Orphans. Trends Neurosci. 1995, 18, 542–548. 10.1016/0166-2236(95)98376-A. [DOI] [PubMed] [Google Scholar]
- Liu S.; Downes M.; Evans R. M.. Metabolic Regulation by Nuclear Receptors. In Innovative Medicine: Basic Research and Development; Nakao K., Minato N., Uemoto S., Eds.; Springer: Tokyo, 2015. [PubMed] [Google Scholar]
- Ordóñez-Morán P.; Muñoz A. Nuclear Receptors: Genomic and Non-Genomic Effects Converge. Cell Cycle 2009, 8, 1675–1680. 10.4161/cc.8.11.8579. [DOI] [PubMed] [Google Scholar]
- Unsworth A. J.; Flora G. D.; Gibbins J. M. Non-Genomic Effects of Nuclear Receptors: Insights from the Anucleate Platelet. Cardiovasc. Res. 2018, 114, 645–655. 10.1093/cvr/cvy044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagechika H.; Himi T.; Namikawa K.; Kawachi E.; Hashimoto Y.; Shudo K. Retinobenzoic Acids. 3. Structure-Activity Relationships of Retinoidal Azobenzene-4-Carboxylic Acids and Stilbene-4-Carboxylic Acids. J. Med. Chem. 1989, 32, 1098–1108. 10.1021/jm00125a027. [DOI] [PubMed] [Google Scholar]
- Frank J. A.; Yushchenko D. A.; Hodson D. J.; Lipstein N.; Nagpal J.; Rutter G. A.; Rhee J.-S.; Gottschalk A.; Brose N.; Schultz C.; et al. Photoswitchable Diacylglycerols Enable Optical Control of Protein Kinase C. Nat. Chem. Biol. 2016, 12, 755–762. 10.1038/nchembio.2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J. A.; Moroni M.; Moshourab R.; Sumser M.; Lewin G. R.; Trauner D. Nat. Commun. 2015, 6, 7118. 10.1038/ncomms8118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J. A.; Franquelim H. G.; Schwille P.; Trauner D. Optical Control of Lipid Rafts with Photoswitchable Ceramides. J. Am. Chem. Soc. 2016, 138, 12981–12986. 10.1021/jacs.6b07278. [DOI] [PubMed] [Google Scholar]
- Haeggström J. Z. Leukotriene A4 Hydrolase/Aminopeptidase, the Gatekeeper of Chemotactic Leukotriene B4 Biosynthesis. J. Biol. Chem. 2004, 279, 50639–50642. 10.1074/jbc.R400027200. [DOI] [PubMed] [Google Scholar]
- Çalıskan B.; Banoglu E. Overview of Recent Drug Discovery Approaches for New Generation Leukotriene A4 Hydrolase Inhibitors. Expert Opin. Drug Discovery 2013, 8, 49–63. 10.1517/17460441.2013.735228. [DOI] [PubMed] [Google Scholar]
- Thunnissen M. M. G. M.; Andersson B.; Samuelsson B.; Wong C.-H.; Haeggström J. Z. Crystal Structures of Leukotriene A4 Hydrolase in Complex with Captopril and Two Competitive Tight-Binding Inhibitors. FASEB J. 2002, 16, 1648–1650. 10.1096/fj.01-1017fje. [DOI] [PubMed] [Google Scholar]
- Siewertsen R.; Neumann H.; Buchheim-Stehn B.; Herges R.; Näther C.; Renth F.; Temps F. Highly Efficient Reversible Z–E Photoisomerization of a Bridged Azobenzene with Visible Light through Resolved S1(Nπ*) Absorption Bands. J. Am. Chem. Soc. 2009, 131, 15594–15595. 10.1021/ja906547d. [DOI] [PubMed] [Google Scholar]
- Nicholls A.; McGaughey G. B.; Sheridan R. P.; Good A. C.; Warren G.; Mathieu M.; Muchmore S. W.; Brown S. P.; Grant J. A.; Haigh J. A.; et al. Molecular Shape and Medicinal Chemistry: A Perspective. J. Med. Chem. 2010, 53, 3862–3886. 10.1021/jm900818s. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.