Abstract
Introduction:
Increasing evidence suggests a role for the gut microbiome in central nervous system disorders and specific role for the gut-brain axis in neurodegeneration. Bile acids (BA), products of cholesterol metabolism and clearance, are produced in the liver and are further metabolized by gut bacteria. They have major regulatory and signaling functions and seem dysregulated in Alzheimer disease (AD).
Methods:
Serum levels of 15 primary and secondary BAs and their conjugated forms were measured in 1,464 subjects including 370 cognitively normal older adults (CN), 284 with early mild cognitive impairment (MCI), 505 with late MCI, and 305 AD cases enrolled in the AD Neuroimaging Initiative. We assessed associations of BA profiles including selected ratios with diagnosis, cognition, and AD-related genetic variants, adjusting for cofounders and multiple testing.
Results:
In AD compared to CN, we observed significantly lower serum concentrations of a primary BA (cholic acid CA) and increased levels of the bacterially produced, secondary BA, deoxycholic acid (DCA), and its glycine and taurine conjugated forms. An increased ratio of DCA:CA, which reflects 7α-dehydroxylation of CA by gut bacteria, strongly associated with cognitive decline, a finding replicated in serum and brain samples in the Rush Religious Orders and Memory and Aging Project. Several genetic variants in immune response related genes implicated in AD showed associations with BA profiles.
Conclusion:
We report for the first time an association between altered BA profile, genetic variants implicated in AD and cognitive changes in disease using a large multicenter study. These findings warrant further investigation of gut dysbiosis and possible role of gut liver brain axis in the pathogenesis of AD.
Keywords: Metabolomics, metabolome, lipidomics, Alzheimer’s disease, gut microbiome, gut liver brain axis, atlas for Alzheimer, genetic variants, immunity, inflammation
1. Introduction
Alzheimer’s disease (AD), a progressive neurodegenerative disorder, is the leading cause of dementia in old age affecting over 40 million people worldwide[1]. There are currently no therapies to prevent or slow AD progression, highlighting our incomplete knowledge of disease mechanisms and the need for new drug targets. A large number of biochemical processes are affected in AD and genes implicated in AD highlight the possible roles for lipid processing, immune function, phagocytosis, (innate) immunity and neurotransmitter function, biological pathways that may affect metabolism[2, 3]. Recent AD hypotheses implicate viral and bacterial contributions to disease pathogenesis[4-6].
Bidirectional biochemical communication between the brain and the gut contribute to a variety of neurodegenerative and psychiatric diseases[7-10]. The gut microbiome and the host collaboratively produce a large array of small molecules that impacts human health[11, 12]. Recently, a role for the gut microbiome in motor dysfunction in Parkinson’s disease has been highlighted[13] and several animal models of AD showed a possible role of gut bacteria in amyloid-beta (Aβ) pathology[14, 15]. The APP transgenic mouse model of AD had a drastically altered gut microbiome composition compared to wild-type mice[15]. Other studies linked pro-inflammatory bacteria, such as gram-negative producers of neurotoxic lipopolysaccharides, to brain amyloidosis and systemic inflammation, a central feature of AD[16, 17]. These studies suggest microbial dysbiosis or imbalance could potentially contribute to AD pathogenesis.
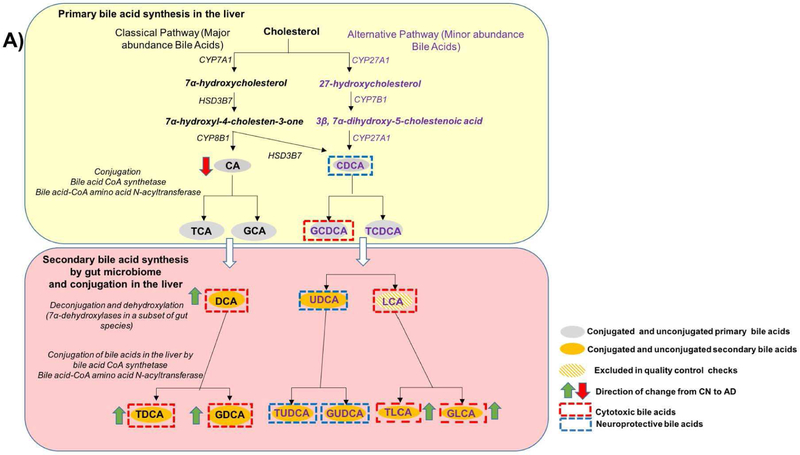
Cholesterol metabolism in the liver is thought to play a key role in AD[18]. In fact, many cholesterol metabolism related genes (e.g., BIN1, CLU, PICALM, ABCA7, ABCG1, and SORL1) are among the top AD susceptibility loci identified by genome-wide association studies[2, 19]. Cholesterol is cleared through production of bile acids (BAs). Primary BAs, chenodeoxycholic acid (CDCA) and cholic acid (CA), are synthesized from cholesterol in the liver, conjugated with glycine or taurine, secreted into the gallbladder via the bile salt export pump (BSEP), and transported to the intestine to be metabolized by gut bacteria (Fig. 1). Intestinal anaerobic bacteria deconjugate the liver-derived BAs through the action of bile salt hydrolases (BSH) to their respective free BAs. Subsequently, anaerobe bacteria convert primary BAs to the secondary BAs. That is, CA is converted to deoxycholic acid (DCA). CDCA is converted to lithocholic acid (LCA) and ursodeoxycholic (UDCA) through 7α or 7β –dehydroxylation, respectively[20, 21]. In the terminal ileum and colon, BAs are reabsorbed by the enterocytes and released into the portal vein for return to the liver where they are conjugated to produce their glycine and taurine forms.
Fig. 1. Bile acid synthesis and cholesterol clearance pathway.
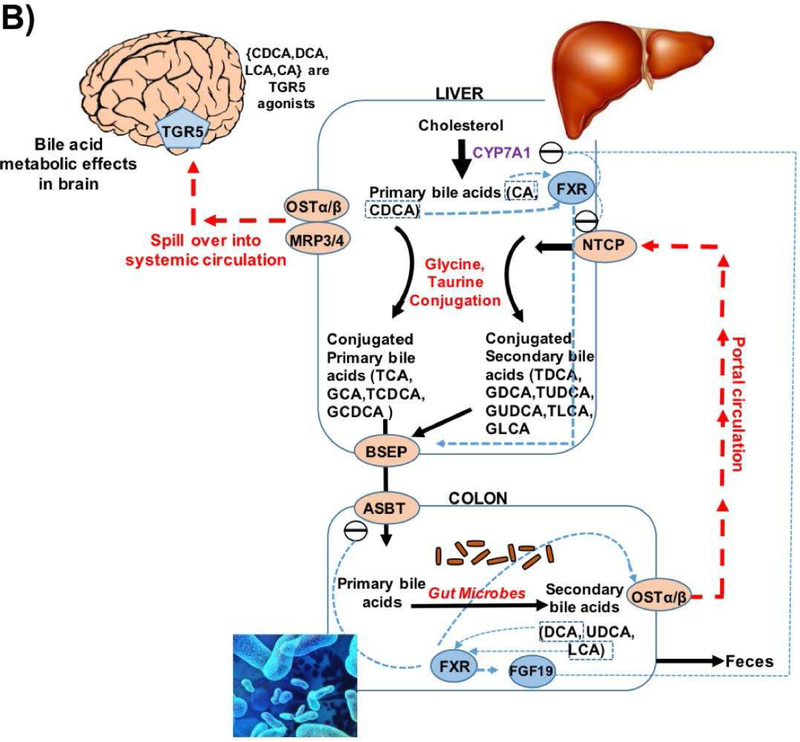
Regulation of bile acid synthesis by feedback mechanism and bile acid transport through enterohepatic circulation. In the liver the bile acids (CDCA, DCA, LCA, CA) activate FXR that inhibits (via a repressor SHP, not shown here) the rate-limiting enzyme CYP7A1. The bile acids via FXR/SHP also inhibit the influx transporter NTCP; induce BSEP and canalicular bile acid secretion. In the intestine, bile acids, via FXR, inhibit the uptake transporter ASBT, decreasing absorption and increasing basolateral secretion into portal circulation by inducing OSTα & β. Bile acid activated FXR in the intestine also exerts inhibition on CYP7A1 in the liver via FGF19 pathway. At the basolateral membrane of hepatocytes, transporters OSTα & β, and also MRP3 and MRP4, secrete bile acids into the systemic circulation.
Abbreviations: ASBT: Apical Sodium-dependent Bile acid Transporters; BSEP: Bile Salt Export Pump; FXR: Farnesoid X Receptor; NTCP: Sodium/Taurocholate Co-transporting Polypeptide; SHP: Small heterodimer partner.
Beyond BAs role in cholesterol clearance, BAs are major regulators for maintaining energy homeostasis through binding to nuclear receptors, including FXR and LXR among others. BAs also modulate the gut microbiome[22, 23] and seem to be indicators of gut dysbiosis. Both primary and secondary BAs are present in the brains of mice and possibly humans with evidence that they cross the blood-brain barrier[24-29]. Some BAs such UDCA exert beneficial effects while others are known to be cytotoxic[30-34]. In particular, DCA’s toxicity has been associated with modulating apoptosis involving mitochondrial pathways in a variety of tissues and cell types[35-38].
In recent pilot human studies, BA profiles were shown to be affected in AD[26, 39-42]. Here, we used a targeted metabolomics approach to evaluate BA profiles in a large cohort of 1,464 individuals enrolled in the AD Neuroimaging Initiative (ADNI) where rich clinical, imaging, and genetic data exist. A schematic representation of study design is shown in Fig. 2. We used this data to address the following:
Fig.2.
Schematic representation of study design.
Investigate if BA profiles are altered in MCI and AD patients and if these differences are related to cognitive decline.
Use ratios of BAs to pinpoint possible enzymatic alterations in the liver and in the gut microbiome that directly contribute to altered BA profile.
Investigate whether immune related AD genome-wide significant genes affect levels of BAs in circulation as markers for altered gut microbiome function.
In a subsequent paper we evaluated correlations between BAs and ATN (amyloid, tau, and neurodegenerative) biomarkers of AD including cerebrospinal fluid (CSF) biomarkers, brain atrophy, and brain glucose metabolism.
2. Methods
2.1. Study cohorts and samples:
2.1.1. ADNI baseline samples
Data used in the preparation of this article were downloaded from the ADNI database (http://adni.loni.usc.edu/). The ADNI studies have recruited over 1,500 adults, ages 55 to 90, consisting of cognitively normal older individuals (CN), individuals with subjective memory concerns (SMC), subjects with early (EMCI) or late mild cognitive impairment (LMCI), and patients with early probable AD dementia. Subjects categorized as SMC were excluded in this study. For key clinical and demographic variables of ADNI participants included in this study, see Table 1.
Table 1.A.
Demographics of ADNI participants stratified by baseline diagnosisa.
| Variable | N | CN (N=370) |
EMCI (N=284) |
LMCI (N=505) |
AD (N=305) |
p-valueb |
|---|---|---|---|---|---|---|
| Age | 1464 | 74.58(5.71) | 71.12(7.51) | 73.95(7.59) | 74.70(7.79) | P<0.001 |
| Sex: Female, No. (%) | 1464 | 190(51%) | 130(46%) | 197(39%) | 139(46%) | P=0.004 |
| Education, years | 1464 | 16.28(2.92) | 15.95(2.66) | 15.87(2.90) | 15.16(3.00) | P<0.001 |
| BMI (Kg/M2) | 1461 | 27.05(4.46) | 28.06(5.41) | 26.54(4.25) | 25.83(4.69) | P<0.001 |
| ≥1APOE ε4 allele, No.(%) | 1464 | 104(28%) | 121(43%) | 273(54%) | 202(66%) | P<0.001 |
| ADAS-Cog13c | 1455 | 9.19(4.17) | 12.64(5.40) | 18.67(6.62) | 29.67(8.20) | P<0.001 |
Data are reported as mean(SD) unless otherwise indicated. Bolded values indicate statistical significance. SD: Standard deviation.
Based on 2-sample t tests, or Pearson χ2 tests.
Score explanations: ADAS-Cog13 range, 0 (best) to 85 (worst).
Abbreviations: AD: Alzheimer’s disease; BMI: Body mass index; CN: Cognitively normal; EMCI: Early mild cognitive impairment; LMCI: Late mild cognitive impairment; ADAS-Cog13, Alzheimer Disease Assessment Scale 13-item cognitive subscale.
2.1.2. The Religious Orders Study and the Rush Memory and Aging Project (ROS/MAP) for replication of key finding
The ROS/MAP studies are both longitudinal cohort studies of aging and AD at Rush University, and are designed to be used in joint analyses to maximize sample size. ROS enrolled individuals from religious orders (nuns, priests, brothers) across the United States[43]. MAP was designed to complement the ROS study by using a similar structure and design as ROS, but enrolling participants with a wider range of life experiences and socioeconomic status from the Chicago, IL metropolitan area[44]. The entire ROS/MAP cohort consists of approximately 3,300 participants, more than 1,500 of whom have come to autopsy (www.radc.rush.edu). We measured a subset of serum BAs in 566 subjects (446 CN, 109 MCI, and 11 AD), as well as a subset of BAs in postmortem brain samples from the dorsolateral pre-frontal cortex of 111 subjects with brain pathology measured (51 CN, 31 MCI, and 27 AD at time of death), of whom 93 had serum BA measurements. Key demographic characteristics of the ROS/MAP cohort are in Supplementary Table 1.
2.1.3. Rotterdam study (RS)
RS was used to examine the association of BAs with AD genetic variants. RS is a prospective population based study[45]. At the baseline examination in 1990-93, 7983 subjects ≥ 55 years of age were recruited from the Ommoord district of Rotterdam (RS-I). All the study participants were extensively interviewed and physically examined at baseline and after every 3 to 4 years. During 2000 to 2001, the baseline cohort (RS-I) was expanded with 3011 subjects ≥55 years of age, who were not yet part of RS-I (RS-II). In this analysis, fasting serum BAs were measured for 488 dementia-free subjects with mean(SD) age of 73.1(6.3) from RS-I using Metabolon platform (Durham, North Carolina, USA) as described previously[46] (see Supplementary Table 2 for demographics).
2.2. Sample collection and quantification of BAs
Targeted metabolomics profiling was performed to measure concentrations of 20 BA metabolites in serum samples of the ADNI cohorts. Morning fasting serum samples from the baseline visit were collected and aliquoted as described in the ADNI standard operating procedures. BA quantification was performed by liquid chromatography tandem mass spectrometry using the Biocrates® Life Sciences Bile Acids Kit (BIOCRATES Life Science AG, Innsbruck, Austria) according to manufacturer’s instructions (see Table 2 for list of BAs, abbreviations and their levels across diagnosis groups).
Table 2.
Levels of primary and secondary bile acids measured in the ADNI cohort stratified by clinical diagnosisa.
| Bile Acid | Category | Nb | CN (N=370) |
EMCI (N=284) |
LMCI (N=505) |
AD (N=305) |
|---|---|---|---|---|---|---|
| CA, Cholic | Primary | 1446 | 0.221(0.024) | 0.155(0.021) | 0.192(0.021) | 0.135(0.025) |
| CDCA, Chenodeoxycholic | Primary | 1357 | 0.285(0.042) | 0.241(0.034) | 0.288(0.033) | 0.216(0.033) |
| GCA, Glycocholic | Primary Conjugated | 1463 | 0.236(0.019) | 0.234(0.021) | 0.239(0.014) | 0.297(0.037) |
| GCDCA, Glycochenodeoxycholic | Primary Conjugated | 1464 | 0.658(0.035) | 0.724(0.059) | 0.710(0.037) | 0.806(0.049) |
| TCA, Taurocholic | Primary Conjugated | 1020 | 0.068(0.008) | 0.057(0.006) | 0.068(0.006) | 0.066(0.009) |
| TCDCA, Taurochenodeoxycholic | Primary Conjugated | 1426 | 0.090(0.006) | 0.088(0.007) | 0.091(0.006) | 0.097(0.008) |
| TMCA, Tauromuricholic | Primary Conjugated | 1146 | 0.012(0.001) | 0.011(0.001) | 0.014(0.002) | 0.014(0.002) |
| DCA, Deoxycholic | Secondary | 1445 | 0.526(0.041) | 0.574(0.043) | 0.529(0.026) | 0.627(0.045) |
| UDCA, Ursodeoxycholic | Secondary | 1111 | 0.065(0.007) | 0.072(0.011) | 0.091(0.010) | 0.087(0.012) |
| GDCA, Glycodeoxycholic | Secondary Conjugated | 1439 | 0.440(0.034) | 0.488(0.038) | 0.502(0.031) | 0.672(0.054) |
| TDCA, Taurodeoxycholic | Secondary Conjugated | 1430 | 0.058(0.006) | 0.059(0.005) | 0.065(0.005) | 0.077(0.006) |
| GLCA, Glycolithocholic | Secondary Conjugated | 1037 | 0.027(0.002) | 0.034(0.003) | 0.030(0.002) | 0.039(0.003) |
| TLCA, Taurolithocholic | Secondary Conjugated | 1008 | 0.005(0.0002) | 0.005(0.0003) | 0.005(0.0003) | 0.006(0.0005) |
| GUDCA, Glycoursodeoxycholic | Secondary Conjugated | 1401 | 0.115(0.010) | 0.114(0.012) | 0.129(0.012) | 0.136(0.015) |
| TUDCA, Tauroursodeoxycholic | Secondary Conjugated | 1369 | 0.008(0.001) | 0.008(0.001) | 0.008(0.001) | 0.008(0.001) |
Values represent μM in mean (standard error of the mean).
Number of non-missing measurements.
In the ROS/MAP, quantification of BA concentrations in 566 serum samples and 111 postmortem brain samples was performed at the University of Hawaii cancer center using ultraperformance liquid chromatography coupled to a tandem mass spectrometry (UPLC-MS/MS) system (ACQUITY UPLC-Xevo TQ-S, Waters Corp., Milford, MA)[47].
In the RS study, serum BAs were measured in 488 serum samples using the non-targeted Metabolon platform (Durham, North Carolina, USA).
2.3. Quality control of BA profiles
Metabolomics lab staff were blinded to diagnosis and pathological data in all the studies. In ADNI, after unblinding and data release, metabolite profiles went through quality-control (QC) checks and data preprocessing including batch-effect adjustment, missing value imputation, and log-transformation (Supplementary Methods and Supplementary Table 3). After QC correction, the dataset included 15 BAs (5 BAs did not pass QC criteria) for a total of 1,464 subjects (after excluding 99 SMC). The preprocessed BA values after QC were used for subsequent association analyses directly or were adjusted to take into account the effect of medications on BA levels[48]. The list of medications selected for adjustment for each BA is shown in Supplementary Table 7. We performed all analyses using both medication adjusted and unadjusted BA levels, results derived from medication-adjusted data and the adjustment process are described in Supplementary Methods and its accompanying tables.
In both RS and ROS/MAP, missing metabolite levels were imputed using half of the limit of detection. Log-transformed values were used in subsequent analyses.
2.4. Clinical Outcomes
For ADNI data, continuous response variables included the modified Alzheimer Disease Assessment Scale 13-item cognitive subscale (ADAS-Cog13; range, 0 [best] to 85 [worst] points), an index of general cognitive functioning. Categorical response variables included clinical diagnosis at baseline and MCI conversion (MCI-NonConverter, MCI-Converter). For the ROS/MAP cohort, cognition was measured using a battery of tests (details are published[49-52]). A composite measure of global cognition was created by averaging the z-scores of all tests as previously described[52]. Mean and standard deviation at baseline were used to compute z-scores. A negative z-score means that an individual has an overall score that is lower than the average of the entire sample at baseline. Cognitive tests were used from the same cycle as serum, and proximate to death for brain.
2.5. Genotype and whole genome sequencing data
Whole genome sequencing:
For 817 ADNI participants, whole-genome sequencing (WGS) was performed on blood-derived genomic DNA. Samples were sequenced on the Illumina HiSeq2000 using paired-end read chemistry and read-length of 100bp at 30–40X coverage. For data processing and QC, an established analysis pipeline based on GATK was used. The QC steps included participant sex check, participant identity check, and variant quality check of the Illumina-generated VCF files (see Saykin et al., 2015 for details[53]).
DNA genotyping in the participants of the RS cohort was performed using 550K, 550K duo, or 610K Illumina arrays at the internal genotyping facility of the Erasmus Medical Center, Rotterdam. Study samples with excess autosomal heterozygosity, call rate < 97.5%, ethnic outliers, and duplicate or family relationships were excluded during quality control analysis. Genotype exclusion criteria further included call rate < 95%, Hardy-Weinberg equilibrium p < 1.0×10−6 and Minor Allele Frequency (MAF) < 1%. Genetic variants were imputed to the Haplotype Reference Consortium (HRC) reference panel (version 1.0)[54] using the Michigan imputation server[55].
Reference genetic associations with BA profiles in healthy individuals were obtained from supplementary data of the atlas of genetic influences on blood metabolites[46]. To obtain genome-wide genetic associations with DCA, we considered all suggestive significant results with P < 1.0 × 10−5. Gene and complex trait annotations of the 13 resulting genetic loci were performed using the SNiPA tool v3.2[56] and the NHGRI-EBI Catalog of published genome-wide association studies (www.ebi.ac.uk/gwas; accessed 02/01/2018, version 1.0)[57]. Lookup of AD genetic associations for DCA candidate variants was performed using the IGAP repository[2].
2.6. Statistical analysis
Differences of demographic, clinical, and cognitive measurements among the clinical diagnostic groups were evaluated using 2-sample t-test (for continuous variables) and Pearson Chi-squared test (for categorical variables). All analyses were performed in a metabolite-wise manner and Bonferroni-adjusted critical values were used to assess statistical significance. All models included age at baseline, sex, APOE ε4, and log10-transformed body mass index (BMI). For cognition, number of years of education was added as an additional covariate.
Separate binary logistic regression models were conducted to examine cross-sectional association of each metabolite with baseline diagnosis (6 models per metabolite). We performed logistic regression models to compare BA levels between the MCI-NonConverter and MCI-Converter groups. Cox proportional hazard models were used to evaluate the association of metabolite levels with progression from MCI (combined EMCI and LMCI subjects) to AD. The cross-sectional association of ADAS-Cog13 with BAs was assessed using linear regression models with square root of ADAS-Cog13 as the dependent variable.
In ROS/MAP, one sample per individual was used. Linear regression models with global cognition score as dependent variable and metabolites as independent variables were used to assess the association of serum BAs with cognition, while adjusting for sex, age, APOE ε4, and years of education. Similar analyses were conducted for brain BAs separately.
We restricted our genetic variant analysis to single nucleotide polymorphisms (SNPs) in genes involved in the immune response pathway that were significantly associated with AD genome-wide[2, 58-60]. Selected genetic variant included rs616338-T(ABI3), rs143332484-T(TREM2), rs72824905-C(PLCG2), rs9331896-T(CLU), rs6656401-A(CR1), rs35349669-T(INPP5D), rs11771145-G(EPHA1), rs983392-A(MS4A6A), and rs190982 -A(MEF2C). Associations of AD risk variants in immune-related genes with selected metabolic traits in ADNI and RS were computed using sex, age, and BMI as covariates.
3. Results
Characteristics of ADNI participants are depicted in Table 1. Baseline cognitive measurements were significantly different among diagnostic groups, as expected. AD patients were more often carriers of at least one APOE ε4 allele. In addition, ADAS-Cog13 scores were not significantly different between the MCI-converter and Non-Converter groups. However, the proportion of APOE ε4 carriers was higher in the MCI-Converter group.
3.1. Serum BA profiles are significantly altered in AD
The Bonferroni-corrected threshold for statistical significance was determined as P < 4.76 × 10−4 (0.05 divided by 15 metabolites times 7 phenotypes including cognition). When we compared BA profile in AD to CN, we detected a significant decrease in levels of the primary BA, CA (P = 1.56 × 10−4). In contrast, a significant increase of bacterially produced secondary BA, DCA was noted (P = 1.61 × 10−4) along with several secondary conjugated BAs, GDCA, TDCA, and GLCA (Table 3). GDCA and GLCA were significantly associated with ADAS-Cog13 where higher levels indicate worse cognition. Comparing BA levels between AD and both MCI groups yielded similar results, while the comparison of BA levels between the CN and MCI groups did not reach statistical significance (Supplementary Table 4).
Table 3.
Cross-sectional association of bile acids with clinical diagnosis and cognition in the ADNI studya.
| Bile Acid | CN vs. AD (n=673) OR (95% CI); P-valueb |
ADAS-Cog13 (n=1453) β(95% CI); P-value c |
|---|---|---|
| CA | 0.85(0.78,0.92);1.56E-04 | −0.04(−0.07,−0.01);2.81E-03 |
| CDCA | 0.94(0.87,1.01);7.19E-02 | −0.02(−0.04,0.00);1.07E-01 |
| GCA | 1.07(0.96,1.18);2.03E-01 | 0.01(−0.02,0.05);4.36E-01 |
| GCDCA | 1.15(1.02,1.29);2.07E-02 | 0.06(0.02,0.09);4.60E-03 |
| TCA | 1.03(0.94,1.12);5.32E-01 | −0.01(−0.03,0.03);7.92E-01 |
| TCDCA | 1.04(0.94,1.15);4.29E-01 | 0.02(−0.02,0.05);3.39E-01 |
| TMCA | 1.09(1.00,1.18);4.46E-02 | 0.029(0.00,0.06);4.21E-02 |
| DCA | 1.24(1.11,1.39);1.61E-04 | 0.05(0.01,0.08);9.26E-03 |
| UDCA | 0.96(0.90,1.03);2.41E-01 | −0.01(−0.03,0.01);2.44E-01 |
| GDCA | 1.30(1.17,1.43);4.20E-07 | 0.07(0.04,0.10);1.05E-05 |
| TDCA | 1.19(1.08,1.30);3.26E-04 | 0.05(0.02,0.08);2.39E-03 |
| GLCA | 1.33(1.20,1.48);9.21E-08 | 0.07(0.04,0.11);1.97E-05 |
| TLCA | 1.19(1.07,1.31);9.53E-04 | 0.06(0.03,0.1);3.18E-04 |
| GUDCA | 1.09(1.00,1.19);5.39E-02 | 0.03(−0.00,0.06);6.04E-02 |
| TUDCA | 1.08(0.96,1.20);1.86E-01 | 0.01(−0.02,0.05);4.85E-01 |
Statistically significant associations that passed Bonferroni correction are bolded.
Odds ratios and p-values were obtained from logistic regressions. Models were corrected for age, sex, body mass index, and APOE ε4 status; Bonferroni-adjusted critical value was set to 5.76 × 10−4 (0.05 divided by 15 metabolites times 7 phenotypes including cognition)
Outcome: Square root of ADASCog-13 (0 [best] to 85 [worst]); Models were corrected for age, sex, years of education, body mass index and APOE ε4 status; Bonferroni-adjusted critical value was set to 2.17E-03.
3.2. Ratios reflective of conversion of BAs by gut microbiome are significantly associated with AD and cognitive performance
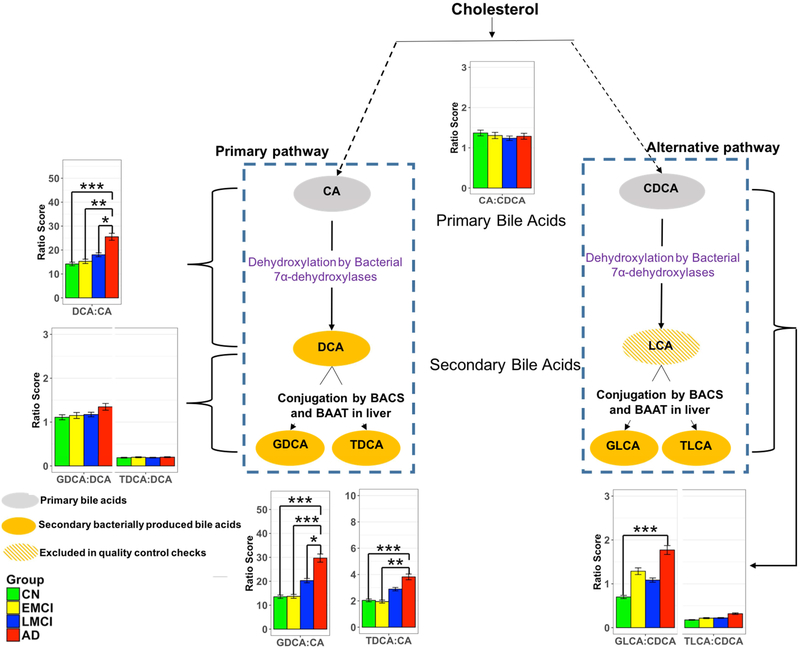
To determine which enzymatic processes in BA metabolism may underlie the differences noted in AD, we investigated eight selected ratios reflective of enzymatic activities in the liver and the gut microbiome. These ratios included:
The CA:CDCA ratio was selected to test if a possible shift in BA synthesis from the primary to the alternative BA pathway occurs in the liver.
Ratios of secondary to primary BAs (DCA:CA, GLCA:CDCA, and TLCA:CDCA) to investigate differences in gut microbiome enzymatic activity leading to altered production of secondary BAs. Since LCA was excluded in QC steps, the GLCA:CDCA and TLCA:CDCA ratios were used as proxies for LCA:CDCA ratio.
GDCA:DCA and TDCA:DCA ratios were used to test if the observed secondary BA dysregulation is related to enzymatic differences related to their taurine and glycine conjugation.
Here, we considered associations as significant at a Bonferroni-corrected P < 3.11 × 10−4 (0.05 divided by all 23 metabolic traits times 7 phenotypes, which include cognition). The ratio of the primary BAs (CA:CDCA) showed no significant association with AD. Yet, for the ratio of DCA:CA (i.e. the conversion of unconjugated primary to unconjugated secondary BA) we observed a highly significant association with AD diagnosis (P=1.53 × 10−8). Ratios between primary and secondary conjugated BAs showed the same effect and direction, including GDCA:CA (P=8.53 × 10−10), TDCA:CA (P=9.83 × 10−7), and GLCA:CDCA (P=3.61 × 10−6). Ratios modeling the glycine and taurine conjugation step of DCA (i.e. GDCA:DCA, TDCA:DCA) were not significantly associated with diagnosis (Fig. 3 and Table 4).
Fig. 3.
Ratios of bile acids reflective of liver and gut microbiome enzymatic activities in CN, Early MCI, Late MCI and AD patients.
Three types of ratios were calculated to inform about possible enzymatic activity changes in Alzheimer’s patients. These ratios reflect one of the following: (1) Shift in bile acid metabolism from primary to alternative pathway. (2) Changes in gut microbiome correlated with production of secondary bile acids. (3) Changes in glycine and taurine conjugation of secondary bile acids. Color code: Green: cognitively normal; Yellow: EMCI; Blue: LMCI; Red: AD. Composition of selected ratios stratified by clinical diagnosis. Error bars indicate standard error of the means; Asterisks indicate statistical significance (*P<10−03, ** P< 10−04, and ***P< 10−05). P-values were estimated from logistic regression models and adjusted for age, sex, body mass index, and APOE ε4 status. The significance level was adjusted for multiple testing according to Bonferroni method to 0.05/138 = 3.62E-4; LCA was excluded in the quality control steps.
Table 4.
Ratios of bile acids reflective of gut microbiome and liver enzymatic activities and their correlation with disease status and cognitive functiona.
| Ratios informative about metabolic processes |
Ratios calculated | CN vs. AD (n=673) OR (95% CI); P-valueb |
ADAS-Cog13 (n=1453) β(95% CI); P-value c |
|---|---|---|---|
| Bile acid synthesis: primary vs. alternative pathway | CA:CDCA | 0.87(0.77,0.97);1.67E-02 | −0.03(−0.07,0.01);1.27E-01 |
| Conversion from primary to secondary BA by the gut microbiome | DCA:CA | 1.25(1.16,1.35);1.53E-08 | 0.05(0.03,0.08);1.05E-05 |
| GDCA:CA | 1.24(1.16,1.33);8.53E-10 | 0.06(0.04,0.08);1.20E-07 | |
| TDCA:CA | 1.16(1.10,1.24);9.83E-07 | 0.04(0.02,0.06);5.40E-05 | |
| GLCA:CDCA | 1.16(1.09,1.23);3.61E-06 | 0.04(0.02,0.06);9.15E-05 | |
| TLCA:CDCA | 1.09(1.03,1.16); 1.60E-03 | 0.03(0.01,0.05);1.50E-03 | |
| Glycine or Taurine conjugation of secondary bile acids by liver enzymes | GDCA:DCA | 1.16(1.02,1.31);2.41E-02 | 0.05(0.02,0.10);5.49E-03 |
| TDCA:DCA | 1.02(0.93,1.11);7.40E-01 | 0.01(−0.02,0.04);4.15E-01 |
Several ratios were calculated to inform about possible enzymatic activity changes in Alzheimer’s patients. These ratios reflect: (1) Shift in bile acid metabolism from primary to alternative pathway. (2) Changes in gut microbiome correlated with production of secondary bile acids. (3) Changes in glycine and taurine conjugation of secondary bile acids.
Outcome: Baseline diagnosis; Odds ratios and p-values were obtained from logistics regressions. Models were corrected for age, sex, body mass index, and APOE ε4 status; Bonferroni-adjusted critical value was set to 1.04E-03 based on 6 possible pairwise comparison of diagnosis groups (CN, EMCI, LMCI, and AD) for 8 ratios.
Outcome: Square root of ADASCog-13 (0 [best] to 85 [worst]); Models were corrected for age, sex, years of education, body mass index, and APOE ε4 status; Bonferroni-adjusted critical value was set to .11 × 10−4 (0.05 divided by all 23 metabolic traits times 7 phenotypes, which include cognitive function).
Four ratios (including DCA:CA and GLCA:CDCA) were significantly associated with ADAS-Cog13. For the ratios we observed the same pattern as AD diagnosis, with higher ratios of secondary to primary BAs being highly significantly associated with worse cognitive performance, while neither conjugation, nor a shift between primary and alternative BA pathways in the liver were significantly linked to cognition (Table 4).
3.3. Serum BA levels were associated with progression from MCI to AD in ADNI
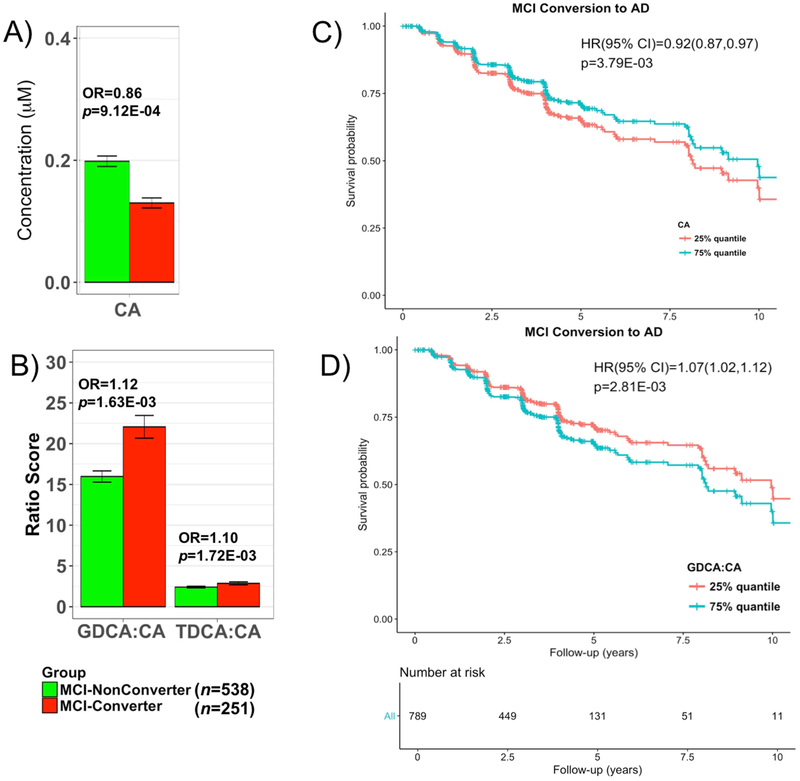
The 9 metabolites and ratios associated with diagnosis were further investigated to assess their relationship with progression from MCI to AD. Out of 789 MCI (EMCI and LMCI) patients with mean (SD) follow-up 3.94 (2.35), 32.2% progressed to AD dementia in four years (labeled as MCI-Converter (n=251) vs. those that did not progressed MCI Non-Converter (n=538)). BA profiles were compared between the two groups using logistic regression models with conversion status as dependent variable and metabolite as independent variable. Models were adjusted for age, sex, BMI, baseline ADAS-Cog13 score, and APOE ε4. The Bonferroni-corrected threshold for statistical significance was determined as P < 5.56 × 10−3 (0.05 divided by 9 metabolites and ratios). We noted a decrease in CA levels (P=9.12 × 10−4) and an increase in ratios of GDCA:CA (P=1.63 × 10−3) and TDCA:CA (P=1.72 × 10−3) in MCI-Converters (Fig. 4 and Supplementary Table 5). Further survival analysis also revealed that levels of CA (hazard ratio (HR), 0.92; P =3.79 × −3), GDCA:CA (HR, 1.07; P=2.81 × −3), and TDCA:CA (HR, 1.06; P=3.19 × 10−3) ratios predicted MCI progression (Fig. 4).
Fig. 4.
Comparison of bile levels in MCI subjects who convert and those who did not convert to AD dementia.
A and B. Lower levels of CA and higher levels of two secondary to primary ratios were significantly associated with higher odds of converting from MCI to AD. EMCI and LMCI patients that converted to AD dementia in 4 years after baseline were labeled as MCI-Converter; 9 bile acids and ratios that were significantly dysregulated between CN to AD were assessed; P-values were estimated from logistic regression models and adjusted for age, sex, body mass index, and APOE ε4 status; the significance level was adjusted for multiple testing according to Bonferroni 0.05/9 = 5.56 × 10−3. C and D. Cox hazards model of the association of conversion from MCI to AD. Red line: 1st quantile, Red line: 3rd quantile. Analysis was conducted using quantitative values and stratification by quantiles was used only for graphical representation.
3.4. Replication of association between cognition and DCA:CA ratio in serum and brain from ROS/MAP
In order to confirm the associations observed in ADNI, we used an independent cohort of older adults (ROS/MAP) with measures of BAs in serum and brain to replicate our findings. Since the sample sizes in ROS/MAP were smaller than ADNI and AD cases were strongly underrepresented (566 serum samples 11 of which were AD and 111 brains 27 of which were AD), we focused on replicating our key findings related to the association between cognition and the DCA:CA ratio (as proxy for BA processing by the gut microbiome). Here we had to use global cognition scores where higher values indicate better cognition. Separate linear regression models were used for brain and serum samples. Pearson’s correlation coefficient between serum DCA:CA and DCA:CA in 93 matching brain samples was 0.303 (P=0.003). In both serum and brain samples, higher levels of DCA:CA were associated with worse cognition (serum: β = −0.06; P = 0.011; brain: β = −0.21; P = 0.032), confirming our ADNI finding.
3.5. Genetic risk variants for AD in genes related to immune function are associated with bile acid levels
To further evaluate that altered BA profiles in AD are related to processes in the gut microbiome, we investigated if BA profiles were associated with immune-related AD risk genes which may contribute to differences in gut microbiome composition. Using the ADNI (n=817 with WGS data) and RS (n=488) cohorts, association of selected BAs in the primary BA pathway (CA, DCA, GDCA, and TDCA) as well as the DCA:CA ratio with the selected genetic risk variants in 9 candidate genes with immune-related functions was assessed. In addition, we included associations from a published large cohort-based study [46] to increase sample size. With the exception of rs983392 inMS4A6A, we found nominally significant associations for the candidate variants in all of these genes (Supplementary Table 10). Three associations were significant after Bonferroni-correction (P < 1.1 × 10−3) in at least one of the studies: rs616338 (ABI3) and rs190982 (MEF2C) were significantly associated with the DCA:CA ratio and rs11771145 (EPHA1) was significantly linked to both DCA and TDCA.
3.6. Genetic loci associated with DCA may influence susceptibility for AD
To follow up on the hypothesis that elevated DCA levels in AD which are linked to gut dysbiosis are relevant in the pathogenesis of AD, we collected (suggestive) significant genetic associations with DCA levels (P < 1.0 × 10−5) from a previous study of genetic influences on blood metabolite levels in large population-based cohorts (n~7,800)[46]. We then annotated the resulting 13 loci with genetic trait associations, including AD associations from the IGAP study[2], and tried to replicate associations with DCA in ADNI (Supplementary Table 11). Two of the 13 genes, CYP7A1 and IMPA2, also showed association with DCA levels in ADNI subjects. Notably, six of the 13 genes have been previously linked via genetic studies to AD (ABCA7) or AD phenotypes, including cognitive decline and CSF protein levels (LRRC7, CYCS, GPC6, FOXN3 and CNTNAP4).
4. Discussion
In this study, we interrogated a possible role for BA end products of cholesterol metabolism and clearance in cognitive changes in AD. Using stored blood samples from ADNI studies we established that BA profile is significantly altered in AD patients. We noted a significant decrease in serum levels of a liver-derived primary BA (CA) and an increase in levels of a bacterially produced secondary BAs and their conjugated forms (DCA, GDCA and TDCA, GLCA) in AD patients compared to CN subjects (Table 3, Fig. 1A). Higher levels of secondary conjugated BAs (GDCA, GLCA, and TLCA) were significantly associated with worse cognitive function (ADAS-Cog13; Table 3). In a follow up paper, we illustrate that these changes are also correlated with changes in CSF markers of disease and with brain imaging changes.
To inform about enzymatic activity changes in the liver and the gut, three types of metabolite ratios were evaluated to inform about mechanisms leading to the noted altered BA profile in AD. We found no shift in metabolism between primary and alternative pathways (Fig. 3; no change in CA:CDCA); a significant change in production of secondary BAs via enzymatic activities in the gut microbiome (increased DCA:CA as well as GLCA:CDCA and TLCA:CDCA as proxies for LCA:CDCA) and no change in processes involved in glycine and taurine conjugation of secondary BAs in the liver (no change in GDCA:DCA or TDCA:DCA). The significant increase in ratios of secondary to primary BAs (e.g. DCA:CA; Fig. 3), suggest altered activity of bacterial 7β-dehydroxylases leading to excess production of secondary BAs many of which are cytotoxic[34, 61-63]. This indicates potential gut dysbiosis in AD patients possibly caused by enhanced colonization of the large and possibly the small intestine with anaerobic bacteria capable of CA and CDCA 7α-dehydroxylation. Increases in these ratios also significantly correlated with poorer cognition (Table 4). Together, these findings suggest that enzymatic steps in conversion of primary to secondary BAs in the gut might contribute to disease.
We also evaluated effects of BA levels on risk of progression to AD among 789 MCI patients. Lower levels of CA and higher ratio of secondary to primary BAs, GDCA:CA, and TDCA:CA were significantly associated with risk of developing AD dementia (Fig. 4, Supplementary Table 6).
The increased production of bacterially produced DCA from CA modeled by ratio DCA:CA and its link to cognition was replicated in the independent ROS/MAP cohort. Association of the DCA:CA ratio with disease severity was evaluated separately in 566 serum and 111 brain samples. Due to the small number of AD patients (n=11, serum n=27, brains), we used global cognitive scores as an index of disease severity. Most of the BAs primary and bacterially produced secondary were found in the brain. Similar to ADNI findings, an increase in the DCA:CA ratio in both serum and brain were significantly associated with worse cognition. This finding suggests that downstream effects of the gut-directed dysregulation of primary vs. secondary BAs are not limited to the periphery, but also might affect metabolic homeostasis and/or signaling functions in the human brain.
Earlier smaller studies suggested differences in BA levels in AD [26, 38-42]. For example, in a study of 495 plasma metabolites comparing MCI (n=58) and AD (n = 100) with those of cognitively normal controls (n=93), levels of DCA, LCA, and GLCA were significantly elevated in the disease state [41]. Mapstone and colleagues[39] identified increased levels of glycoursodeoxycholic acid (GUDCA) in subjects likely to develop amnestic MCI or AD within 2 to 3 years compared to controls. In a small pilot study Marksteiner and colleagues [42] reported increased levels of LCA, GDCA, and GLCA in AD (n=30) relative to MCI (n=20). We replicated these findings with the exception of LCA (excluded during QC) and GUDCA which showed only a non-significant trend of upregulation in the AD group (P = 0.054). Marksteiner [42] did not report a significant increase in DCA or decrease in CA which we observed in the ADNI cohort. However, there is a trend in their data to suggest that DCA levels are increased in AD relative to CN. Our analyses build upon these pilot studies to include a large well-characterized cohort with rich clinical, neuroimaging, and genetics data. Our analyses include links to innate immunity related genes which was not possible in smaller studies. In addition, we controlled for medication use which is known to significantly affect the gut microbiome and bile acids. In our follow-up paper we explore the association of serum BAs with CSF and neuroimaging biomarkers of AD.
Composition and functional changes of the gut microbiome have been implicated in several diseases. Microbiome GWAS revealed that variants in many human genes involved in immunity and gut architecture are associated with an altered gut microbiome composition [64]. Although many factors such as diet can affect the microbial organisms residing in the gut, emerging data support the hypothesis that certain host genetic variants predispose an individual towards microbiome dysbiosis and this can be linked to disorders of metabolism and immunity such as type 2 diabetes mellitus, obesity, and autism[64].
Accumulating evidence links dysregulation of the immune system to AD pathology. In particular, genetic association studies in AD have robustly identified several genetic risk variants in immune-related genes[2, 60]. Using the ADNI and RS cohorts, we investigated the association of BA profiles of CN subjects with genetic variants in nine AD-related and innate immunity genes. Eight genetic variants were associated with selected BA levels at nominal significance (Supplementary Table 10). Three of these associations were significant after Bonferroni-correction, with rs616338 (ABI3) and rs190982 (MEF2C) associated with the DCA:CA ratio, and rs11771145 (EPHA1) linked to both DCA and TDCA. The association of the BAs to AD genes suggest that these immune related genes may influence the risk of AD through BA metabolism or changes in the gut microbiome. Interestingly, both ABI3 and MEF2C are thought to be involved in immune reactions to pro-inflammatory stimuli that are partially secreted by microbes[65, 66]. The link to the DCA:CA ratio may thus mirror differences in gut microbiome composition due to altered immune response in AD, providing a mechanistic hypothesis for our findings. The function of EPHA1 is not well understood but it has been hypothesized that when activated, this receptor may affect the integrity of the blood brain barrier (BBB)[67]. Its association with levels of DCA is intriguing as DCA is known to be cytotoxic and can disrupt the BBB and then enter the brain[28]. rs11771145 is associated with gene expression levels of EPHA1[56], and as DCA is not known to be produced by human metabolism, changed expression and activity of EPHA1 may be related to DCA-mediated cytotoxic effects.
Using an established atlas of genetic influences on human blood metabolites[46], we further investigated a potential cytotoxic role of DCA. For almost half of the 13 identified loci, we found genetic evidence for involvement in AD-linked complex traits (Supplementary Table 11). In particular, ABCA7 is an AD risk gene replicated in several genetic studies[68, 69]. Five additional genes (LRRC7, CYCS, GPC6, FOXN3, and CNTNAP4) genetically influence AD phenotypes, including cognitive decline and CSF markers. While it remains speculative if and how these genes interact with DCA to contribute to AD risk, it is intriguing that we identified ABCA7 by screening for associations with DCA levels. ABCA7 is highly expressed in the brain, and functions in the efflux of lipids, including cholesterol, from cells. Due to the structural similarity of DCA and cholesterol, we hypothesize that ABCA7 may be able to also transport this BA, reconciling metabolomics findings via a functional hypothesis to a risk gene for AD. The findings that BA levels are regulated by AD related genes might provide new mechanistic insights.
There is growing support for strong connections between the intestinal environment, with its diverse microbial composition and activity, and the functions of the central nervous system. The “gut-brain metabolic axis” facilitates bidirectional chemical communication between the central and enteric nervous systems through mechanisms just starting to be defined[7-9]. Such a metabolic axis is thought to be involved in the regulation of multiple host metabolic pathways in which levels of hormones, neurotransmitters, amines, GABA, short-chain fatty acids (SCFA), lipid metabolites, and others are regulated by gut microbiome activity[12]. Changes in the composition of intestinal bacterial populations are associated with a wide array of neurological and neurodevelopmental disorders including multiple sclerosis, autism, depression, schizophrenia, and Parkinson’s disease[70-72]. In addition, increasing evidence suggest that liver disease may impact cognitive functions and contribute to AD[73].
Our findings suggest novel metabolic links in AD where BAs represent a component of the gut-liver-brain axis that relates to cognition. We hypothesize that interconnected immune and gut microbiome dysregulation leads to increase in production of cytotoxic secondary bile acids like DCA and its derivatives and these can module the BBB and build up in the brain leading to impaired metabolic functions mediated by their receptors and targets. Such dysregulation includes cholesterol and glucose homeostasis.
It is of interest that BAs are ligands for nuclear receptors including FXR, LXRs among others and they acts synergistically as metabolic sensors to regulate energy homeostasis[74, 75] peripherally and might also propagate their effects to the brain. Interestingly, levels of four BAs produced by the gut microbiome and that we show to be significantly correlated with disease status and cognition (DCA, GLCA, TLCA, TDCA) are hydrophobic and cytotoxic[34, 35, 76, 77]. Cell lines, animal models, and human studies suggest that levels of such BAs, particularly DCA, lead to a disruption of mitochondrial membranes resulting in increased reactive oxygen species, markers of inflammation, and apoptosis as well as decreases in cell viability and DNA synthesis[34, 35, 78]. Studies in rodents with deuterium labelled DCA, demonstrated DCA crosses the BBB and increases its permeability[27, 29]. Increased amounts of secondary BAs in blood may enter the brain through induced permeability of the BBB, affecting brain physiology and metabolism[28]. Several studies in human and animal brains also revealed that the full panel of BAs are found in the brain[24-27], but it is unclear whether this is due to transport from the periphery, from local synthesis, or both. The function of these BAs in the brain remains poorly defined with some support for them acting as neurosteroids[79].
BA levels and the gut microbiome influence each other, where BSH-rich bacteria readily modify the BA profile while, on the other hand, intestinal BAs control the growth and maintenance of commensal bacteria, maintain barrier integrity, and modulate the immune system[80-83]. Such changes might impact brain functions. Significant data support a role for cholesterol metabolism in the pathogenies of AD including large genetic studies. Cholesterol homeostasis is regulated in part by the gut microbiome suggesting that cholesterol intermediates including those produced by gut might present as one gut brain axis of communication that needs to be further investigated in human and animal studies.
4.1. Limitations
This is an observational study, the results of which may contain confounding biases. For example, diet, lifestyle, exposome and other factors may contribute to changes in the gut. It remains unclear how these important factors are related to AD pathogenesis and whether the observed differences we note are causes or consequences of disease. Further studies of metabolic changes in normal aging are required to help define which aspects of BA metabolism might be related to disease vs normal aging. Fecal material was not collected in the ADNI cohorts and most large studied therefore precluding a direct analysis of microbiota changes across the trajectory of disease. Such studies have just been initiated. Use of medications was extensively evaluated as a possible confound (Supplementary methods and Tables 7-9) and our key findings remained after controlling for medication use but larger studies need to further evaluate the effect of these medications. Additional experimental studies are needed to more fully define the expression of BAs and their receptors in the brain and mechanistic roles of BAs in the development of AD. The impact of BAs on FXR, TGR5, vitamin, and hormone receptors in the brain and the signaling pathways impacted are currently unclear. It is important to evaluate in other large community studies the generalizability of our findings. The genetic links need to be tested in large populations.
Longitudinal studies covering pre-symptomatic stages are needed to establish the influence of immune changes on gut microbiome composition and activity in AD patients and the impact of this on BAs and cholesterol homeostasis. Tracking earliest changes in BA and other gut derived metabolites might provide insights into causality. Labeling studies are needed to evaluate if BAs cross the BBB and build up in brain with further elucidation of their signaling and regulatory functions centrally. However, we cannot exclude the possibility that changes in the brain during disease can also impact the gut and liver, and hence some of our findings might be brain derived.
5. Conclusions
In summary, there is evidence of a relationship among intestinal BA profile, gut microbial composition and/or activity, innate immunity, and genetic variants implicated in AD. When disrupted, BAs may contribute to cognitive changes, highlighting the importance of cholesterol clearance and its regulation in AD. Disorders in BA metabolism cause cholestatic liver diseases, dyslipidemia, fatty liver diseases, cardiovascular diseases, and diabetes, which are all associated with risk of cognitive decline, directly or indirectly. Our results lend support to this relationship in the context of AD and cohorts at risk for AD. Our evolving understanding of the gut microbiome’s role in aging and in central nervous system diseases and their progression could open potential new hypotheses in the field, regardless of whether the role is ultimately found to be causative, consequence, or contributory. The role of the gut microbiome in AD needs to be further investigated along with the emerging links between central and peripheral metabolic failures that might contribute to brain health and disease during aging.
Supplementary Material
Table 1.B.
Demographics of ADNI participants stratified by MCI progression to ADa.
| Variable | N | MCI-NonConverter (N=538) |
MCI-Converter (N=251) |
p-valueb |
|---|---|---|---|---|
| Age | 789 | 72.47(7.90) | 73.91(7.08) | P=0.01 |
| Sex: Female, No. (%) | 789 | 41% (223) | 41% (104) | P=1 |
| Education, years | 789 | 15.95(2.85) | 15.79(2.76) | P=0.43 |
| BMI (Kg/M2) | 788 | 27.37(4.80) | 26.47(4.61) | P=0.005 |
| ≥1APOE ε4 allele, No.(%) | 789 | 41%(223) | 68%(171) | P<0.001 |
| ADAS-Cog13 | 786 | 14.26(6.04) | 21.31(5.94) | P=0.29 |
MCI Subjects that converted to AD dementia in 4 years after baseline were labeled as MCI Converter.
Research in Context.
Systematic review: The authors reviewed the literature using PubMed, Google, Web of Science, Medline, Research Gate and through meeting abstracts and presentations. While the role of the gut microbiome in AD is not yet as widely studied, there are several recent publications implicating the gut's role in other neuropsychiatric diseases. These relevant citations are appropriately cited.
Interpretation: We report correlations between secondary gut microbiome produced BA and cognitive decline in AD with innate immunity genes contributing to altered BA profile. Our findings highlight a possible role for cholesterol clearance and gut microbiome in disease mechanism.
Future directions: Our evolving understanding of the gut microbiome's role in aging and related diseases could open new hypotheses for AD, regardless of whether the role is ultimately found to be causative, consequence, or contributory. These findings warrant further investigation of the possible role of gut liver brain axis in AD pathogenesis.
Acknowledgments
Funding/Support:
Funding for ADMC (Alzheimer’s Disease Metabolomics Consortium, led by Dr R.K.-D. at Duke University) was provided by the National Institute on Aging grant R01AG046171, a component of the Accelerated Medicines Partnership for AD (AMP-AD) Target Discovery and Preclinical Validation Project (https://www.nia.nih.gov/research/dn/amp-ad-target-discovery-and-preclinical-validation-project) and the National Institute on Aging grant RF1 AG0151550, a component of the M2OVE-AD Consortium (Molecular Mechanisms of the Vascular Etiology of AD – Consortium https://www.nia.nih.gov/news/decoding-molecular-ties-between-vascular-disease-and-alzheimers).
Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (A.D.N.I.) (National Institutes of Health Grant U01 AG024904) and DOD A.D.N.I. (Department of Defense award number W81XWH-12-2-0012). A.D.N.I. is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support A.D.N.I. clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Disease Cooperative Study at the University of California, San Diego. A.D.N.I. data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
The work of various Consortium Investigators are also supported by various NIA grants [U01AG024904-09S4, P50NS053488, R01AG19771, P30AG10133, P30AG10124, K01AG049050], the National Library of Medicine [R01LM011360, R00LM011384], and the National Institute of Biomedical Imaging and Bioengineering [R01EB022574]. Additional support came from Helmholtz Zentrum, the Alzheimer’s Association, the Indiana Clinical and Translational Science Institute, and the Indiana University-IU Health Strategic Neuroscience Research Initiative.
DISCLOSURES
J.B.T. reports investigator-initiated research support from Eli Lilly unrelated to the work reported here. S.S., S.K, and T.K. are employed by Biocrates Life Sciences AG. These authors have no other financial relationships relevant to this article to disclose. J.Q.T. may accrue revenue in the future on patents submitted by the University of Pennsylvania wherein he is a coinventor and he received revenue from the sale of Avid to Eli Lily as a co-inventor on imaging-related patents submitted by the University of Pennsylvania. L.M.S. receives research funding from NIH (U01 AG024904; R01 MH 098260; R01 AG 046171; 1RF AG 051550); MJFox Foundation for PD Research and is a consultant for Eli Lilly; Novartis; Roche; he provides QC over-sight for the Roche Elecsys immunoassay as part of responsibilities for the ADNI3 study. A.J.S. reports investigator-initiated research support from Eli Lilly unrelated to the work reported here. He has received consulting fees and travel expenses from Eli Lilly and Siemens Healthcare and is a consultant to Arkley BioTek. He also receives support from Springer publishing as an editor in chief of Brain Imaging and Behavior. M.W.W. reports stock/stock options from Elan, Synarc, travel expenses from Novartis, Tohoku University, Fundacio Ace, Travel eDreams, MCI Group, NSAS, Danone Trading, ANT Congress, NeuroVigil, CHRU-Hopital Roger Salengro, Siemens, AstraZeneca, Geneva University Hospitals, Lilly, University of California, San Diego–ADNI, Paris University, Institut Catala de Neurociencies Aplicades, University of New Mexico School of Medicine, Ipsen, Clinical Trials on Alzheimer’s Disease, Pfizer, AD PD meeting. PMD has received research grants and advisory/speaking fees from several companies for other projects, and he owns stock in several companies. Full disclosures will be made through the IJCME form. R.K.D. is inventor on key patents in the field of metabolomics including applications for Alzheimer disease. All other authors report no disclosures.
Role of the Funder/Sponsor: [Funders listed above] had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Abbreviations:
- AD
Alzheimer’s disease
- ADAS-Cog13
13-item Alzheimer’s Disease Assessment Scale-Cognitive subscale
- ADMC
Alzheimer’s Disease Metabolomics Consortium
- ADNI
Alzheimer’s Disease Neuroimaging Initiative
- APOE
apolipoprotein E
- ASBT
Apical sodium-dependent bile acid transporters
- BBB
blood-brain barrier
- BSEP
Bile salt export pump
- BSH
Bile salt hydrolases
- BA
bile acid
- CA
Cholic acid
- cAMP
Cyclic adenosine monophosphate
- CDCA
Chenodeoxycholic acid
- CN
Cognitively normal older control
- DCA
Deoxycholic acid
- EMCI
Early mild cognitive impairment
- FDR
False discovery rate
- FXR
Farnesoid X Receptor
- GCA
Glycocholic acid
- GCDCA
Glycochenodeoxycholic acid
- GDCA
Glycodeoxycholic acid
- GLCA
Glycolithocholic acid
- GUDCA
Glycoursodeoxycholic acid
- LCA
Lithocholic acid
- LMCI
Late mild cognitive impairment
- MCI
Mild cognitive impairment
- NTCP
Sodium/Taurocholate co-transporting polypeptide
- OST-α
Organic solute transporter alpha
- OST-β
Organic solute transporter beta
- ROS/MAP
The Religious Orders Study (ROS) and the Memory and Aging Project (MAP)
- RS
Rotterdam Study
- SHP
Small heterodimer partner
- SMC
Subjective memory concern
- TCA
Taurocholic acid
- TCDCA
taurochenodeoxycholic acid
- TDCA
Taurodeoxycholic acid
- TLCA
Taurolithocholic acid
- TMCA
Trimethoxycinnamic acid
- TUDCA
Tauroursodeoxycholic acid
- UDCA
Ursodeoxycholic acid
Footnotes
Statistical analyses also included: Toledo, Arnold, Bhattacharyya, Jia, Rotroff, Jack, Kastenmüller
Data management and medication term mapping: Tenenbaum and Blach
Concept and design: Kaddurah-Daouk lead concept and design team that included all coauthors
Data analysis and quality control of metabolomic data: Schafferer, Klein, Koal, St. John Williams, Thompson, Moseley
Acquisition, quality control and processing of metabolomic data: St. John Williams, Thompson, Moseley
Drafting of the manuscript: MahmoudianDehkordi, Nho, Arnold, Louie, Kastenmüller, Kueider-Paisley, Kaddurah-Daouk
Biochemical, genomics and medications integration: Kastenmüller, Baillie, Han, Risacher, Arnold; Nho
Data deposition: Alzheimer’s Disease Neuroimaging Initiative (see note)
Harmonization of methods: Alzheimer’s Disease Metabolomics Consortium
Technical, bibliographic research and/or material support: Louie
Biochemical interpretation: Baillie, Han, Kaddurah-Daouk, Jia, Bhattacharyya, Arnold
Critical revision of the manuscript for important intellectual content: Saykin, Doraiswamy, Kastenmüller, Kaddurah-Daouk Obtained funding: Kaddurah-Daouk
Supervision: Motsinger-Reif, Trojanowski, Shaw, Weiner, Doraiswamy, Saykin, Kastenmüller, Kaddurah-Daouk
The Alzheimer’s Disease Metabolomics Consortium (ADMC): A complete listing of ADMC investigators can be found at: https://sites.duke.edu/adnimetab/who-we-are/
The Alzheimer’s Disease Neuroimaging Initiative (ADNI): Data used in the preparation of this article were obtained from the ADNI database (http://adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf.
Additional Contributions: The authors are grateful to Lisa Howerton for administrative support and the numerous ADNI study volunteers and their families.
An email with links to the Authorship Form will be sent to authors for completion after manuscripts have been submitted
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf.
REFERENCES
- [1].Alzheimer's A. 2017 Alzheimer's disease facts and figures. Alzheimer's & Dementia: The Journal of the Alzheimer's Association. 2017;13:325–73. [Google Scholar]
- [2].Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat Genet. 2013;45:1452–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Jones L, Lambert J-C, Wang L-S, Choi S-H, Harold D, Vedernikov A, et al. Convergent genetic and expression data implicate immunity in Alzheimer's disease. Alzheimer's & dementia: the journal of the Alzheimer's Association. 2015;11:658–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Sampson TR, Mazmanian SK. Control of brain development, function, and behavior by the microbiome. Cell Host Microbe. 2015;17:565–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Wu S-C, Cao Z-S, Chang K-M, Juang J-L. Intestinal microbial dysbiosis aggravates the progression of Alzheimer’s disease in Drosophila. Nature Communications. 2017;8:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kumar DK, Choi SH, Washicosky KJ, Eimer WA, Tucker S, Ghofrani J, et al. Amyloid-beta peptide protects against microbial infection in mouse and worm models of Alzheimer's disease. Sci Transl Med. 2016;8:340ra72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ghaisas S, Maher J, Kanthasamy A. Gut microbiome in health and disease: linking the microbiome-gut-brain axis and environmental factors in the pathogenesis of systemic and neurodegenerative diseases. Pharmacology & therapeutics. 2016;158:52–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Yarandi SS, Peterson DA, Treisman GJ, Moran TH, Pasricha PJ. Modulatory Effects of Gut Microbiota on the Central Nervous System: How Gut Could Play a Role in Neuropsychiatric Health and Diseases. Journal of Neurogastroenterology and Motility. 2016;22:201–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Tognini P. Gut Microbiota: A Potential Regulator of Neurodevelopment. Frontiers in Cellular Neuroscience. 2017;11:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Tremlett H, Bauer KC, Appel-Cresswell S, Finlay BB, Waubant E. The gut microbiome in human neurological disease: A review. Annals of Neurology. 2017;81:369–82. [DOI] [PubMed] [Google Scholar]
- [11].Holmes E, Li JV, Marchesi JR, Nicholson JK. Gut microbiota composition and activity in relation to host metabolic phenotype and disease risk. Cell Metab. 2012;16:559–64. [DOI] [PubMed] [Google Scholar]
- [12].Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, et al. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–7. [DOI] [PubMed] [Google Scholar]
- [13].Sampson TR, Debelius JW, Thron T, Janssen S, Shastri GG, Ilhan ZE, et al. Gut Microbiota Regulate Motor Deficits and Neuroinflammation in a Model of Parkinson's Disease. Cell. 2016;167:1469–80 e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Minter MR, Hinterleitner R, Meisel M, Zhang C, Leone V, Zhang X, et al. Antibiotic-induced perturbations in microbial diversity during post-natal development alters amyloid pathology in an aged APPSWE/PS1DeltaE9 murine model of Alzheimer's disease. Sci Rep. 2017;7:10411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Harach T, Marungruang N, Duthilleul N, Cheatham V, Mc Coy KD, Frisoni G, et al. Reduction of Abeta amyloid pathology in APPPS1 transgenic mice in the absence of gut microbiota. Sci Rep. 2017;7:41802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Cattaneo A, Cattane N, Galluzzi S, Provasi S, Lopizzo N, Festari C, et al. Association of brain amyloidosis with pro-inflammatory gut bacterial taxa and peripheral inflammation markers in cognitively impaired elderly. Neurobiol Aging. 2017;49:60–8. [DOI] [PubMed] [Google Scholar]
- [17].Bester J, Soma P, Kell DB, Pretorius E. Viscoelastic and ultrastructural characteristics of whole blood and plasma in Alzheimer-type dementia, and the possible role of bacterial lipopolysaccharides (LPS). Oncotarget. 2015;6:35284–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Di Paolo G, Kim TW. Linking lipids to Alzheimer's disease: cholesterol and beyond. Nature reviews Neuroscience. 2011;12:284–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Beecham GW, Hamilton K, Naj AC, Martin ER, Huentelman M, Myers AJ, et al. Genome-Wide Association Meta-analysis of Neuropathologic Features of Alzheimer's Disease and Related Dementias. PLOS Genetics. 2014;10:e1004606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47:241–59. [DOI] [PubMed] [Google Scholar]
- [21].Donova MV. [Transformation of steroids by actinobacteria: a review]. Prikladnaia biokhimiia i mikrobiologiia. 2007;43:5–18. [PubMed] [Google Scholar]
- [22].Islam KBMS, Fukiya S, Hagio M, Fujii N, Ishizuka S, Ooka T, et al. Bile Acid Is a Host Factor That Regulates the Composition of the Cecal Microbiota in Rats. Gastroenterology. 2011;141:1773–81. [DOI] [PubMed] [Google Scholar]
- [23].Camilleri M, Gores GJ. Therapeutic targeting of bile acids. Am J Physiol Gastrointest Liver Physiol. 2015;309:G209–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Mano N, Goto T, Uchida M, Nishimura K, Ando M, Kobayashi N, et al. Presence of protein-bound unconjugated bile acids in the cytoplasmic fraction of rat brain. Journal of lipid research. 2004;45:295–300. [DOI] [PubMed] [Google Scholar]
- [25].Naqvi SH, Herndon BL, Kelley MT, Bleisch V, Aexel RT, Nicholas HJ. Detection of monohydroxy "bile" acids in the brains of guinea pigs afflicted with experimental allergic encephalomyelitis. Journal of lipid research. 1969;10:115–20. [PubMed] [Google Scholar]
- [26].Pan X, Elliott CT, McGuinness B, Passmore P, Kehoe PG, Holscher C, et al. Metabolomic Profiling of Bile Acids in Clinical and Experimental Samples of Alzheimer's Disease. Metabolites. 2017;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Higashi T, Watanabe S, Tomaru K, Yamazaki W, Yoshizawa K, Ogawa S, et al. Unconjugated bile acids in rat brain: Analytical method based on LC/ESI-MS/MS with chemical derivatization and estimation of their origin by comparison to serum levels. Steroids. 2017;125:107–13. [DOI] [PubMed] [Google Scholar]
- [28].Quinn M, McMillin M, Galindo C, Frampton G, Pae HY, DeMorrow S. Bile acids permeabilize the blood brain barrier after bile duct ligation in rats via Rac1-dependent mechanisms. Digest Liver Dis. 2014;46:527–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Lalic-Popovic M, Vasovic V, Milijasevic B, Golocorbin-Kon S, Al-Salami H, Mikov M. Deoxycholic Acid as a Modifier of the Permeation of Gliclazide through the Blood Brain Barrier of a Rat. J Diabetes Res. 2013;2013:598603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Cecilia MPR, Stephen RS, Susana S, Andrew WG, Cheryle L-S, Walter CL, et al. Neuroprotection by a Bile Acid in an Acute Stroke Model in the Rat. Journal of Cerebral Blood Flow & Metabolism. 2002;22:463–71. [DOI] [PubMed] [Google Scholar]
- [31].Dionisio PA, Amaral JD, Ribeiro MF, Lo AC, D'Hooge R, Rodrigues CMP. Amyloid-beta pathology is attenuated by tauroursodeoxycholic acid treatment in APP/PS1 mice after disease onset. Neurobiol Aging. 2015;36:228–40. [DOI] [PubMed] [Google Scholar]
- [32].Nunes AF, Amaral JD, Lo AC, Fonseca MB, Viana RJ, Callaerts-Vegh Z, et al. TUDCA, a bile acid, attenuates amyloid precursor protein processing and amyloid-beta deposition in APP/PS1 mice. Mol Neurobiol. 2012;45:440–54. [DOI] [PubMed] [Google Scholar]
- [33].Paolini M, Pozzetti L, Montagnani M, Potenza G, Sabatini L, Antelli A, et al. Ursodeoxycholic acid (UDCA) prevents DCA effects on male mouse liver via up-regulation of CYP [correction of CXP] and preservation of BSEP activities. Hepatology. 2002;36:305–14. [DOI] [PubMed] [Google Scholar]
- [34].Ignacio Barrasa J, Olmo N, Perez-Ramos P, Santiago-Gomez A, Lecona E, Turnay J, et al. Deoxycholic and chenodeoxycholic bile acids induce apoptosis via oxidative stress in human colon adenocarcinoma cells. Apoptosis. 2011;16:1054–67. [DOI] [PubMed] [Google Scholar]
- [35].Martinez-Diez MC, Serrano MA, Monte MJ, Marin JJ. Comparison of the effects of bile acids on cell viability and DNA synthesis by rat hepatocytes in primary culture. Biochim Biophys Acta. 2000;1500:153–60. [DOI] [PubMed] [Google Scholar]
- [36].Ramalho RM, Viana RJS, Low WC, Steer CJ, Rodrigues CMP. Bile acids and apoptosis modulation: an emerging role in experimental Alzheimer's disease. Trends in molecular medicine. 2008;14:54–62. [DOI] [PubMed] [Google Scholar]
- [37].Schulz S, Schmitt S, Wimmer R, Aichler M, Eisenhofer S, Lichtmannegger J, et al. Progressive stages of mitochondrial destruction caused by cell toxic bile salts. Biochimica et biophysica acta. 2013;1828:2121–33. [DOI] [PubMed] [Google Scholar]
- [38].Marksteiner J, Blasko I, Kemmler G, Koal T, Humpel C. Bile acid quantification of 20 plasma metabolites identifies lithocholic acid as a putative biomarker in Alzheimer's disease. Metabolomics. 2018;14:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Mapstone M, Cheema AK, Fiandaca MS, Zhong X, Mhyre TR, MacArthur LH, et al. Plasma phospholipids identify antecedent memory impairment in older adults. Nat Med. 2014;20:415–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Greenberg N, Grassano A, Thambisetty M, Lovestone S, Legido-Quigley C. A proposed metabolic strategy for monitoring disease progression in Alzheimer's disease. Electrophoresis. 2009;30:1235–9. [DOI] [PubMed] [Google Scholar]
- [41].Olazaran J, Gil-de-Gomez L, Rodriguez-Martin A, Valenti-Soler M, Frades-Payo B, Marin-Munoz J, et al. A blood-based, 7-metabolite signature for the early diagnosis of Alzheimer's disease. Journal of Alzheimer's disease : JAD. 2015;45:1157–73. [DOI] [PubMed] [Google Scholar]
- [42].Marksteiner J, Blasko I, Kemmler G, Koal T, Humpel C. Bile acid quantification of 20 plasma metabolites identifies lithocholic acid as a putative biomarker in Alzheimer’s disease. Metabolomics. 2018;14:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Bennett DA, Schneider JA, Arvanitakis Z, Wilson RS. Overview and findings from the religious orders study. Current Alzheimer research. 2012;9:628–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Bennett DA, Schneider JA, Buchman AS, Barnes LL, Boyle PA, Wilson RS. Overview and findings from the rush Memory and Aging Project. Current Alzheimer research. 2012;9:646–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Hofman A, Brusselle GG, Darwish Murad S, van Duijn CM, Franco OH, Goedegebure A, et al. The Rotterdam Study: 2016 objectives and design update. Eur J Epidemiol. 2015;30:661–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Shin SY, Fauman EB, Petersen AK, Krumsiek J, Santos R, Huang J, et al. An atlas of genetic influences on human blood metabolites. Nat Genet. 2014;46:543–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Xie G, Zhong W, Li H, Li Q, Qiu Y, Zheng X, et al. Alteration of bile acid metabolism in the rat induced by chronic ethanol consumption. The FASEB Journal. 2013;27:3583–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].St John-Williams L, Blach C, Toledo JB, Rotroff DM, Kim S, Klavins K, et al. Targeted metabolomics and medication classification data from participants in the ADNI1 cohort. Sci Data. 2017;4:170140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Wilson R, Evans D, Bienias J, De Leon CM, Schneider J, Bennett D. Proneness to psychological distress is associated with risk of Alzheimer’s disease. Neurology. 2003;61:1479–85. [DOI] [PubMed] [Google Scholar]
- [50].Wilson RS, De Leon CFM, Barnes LL, Schneider JA, Bienias JL, Evans DA, et al. Participation in cognitively stimulating activities and risk of incident Alzheimer disease. Jama. 2002;287:742–8. [DOI] [PubMed] [Google Scholar]
- [51].Wilson RS, Bienias JL, Evans DA, Bennett DA. Religious Orders Study: overview and change in cognitive and motor speed. Aging Neuropsychology and Cognition. 2004;11:280–303. [Google Scholar]
- [52].Wilson RS, Barnes LL, Krueger KR, Hoganson G, Bienias JL, Bennett DA. Early and late life cognitive activity and cognitive systems in old age. Journal of the International Neuropsychological Society. 2005;11:400–7. [PubMed] [Google Scholar]
- [53].Saykin AJ, Shen L, Yao X, Kim S, Nho K, Risacher SL, et al. Genetic studies of quantitative MCI and AD phenotypes in ADNI: Progress, opportunities, and plans. Alzheimer's & dementia: the journal of the Alzheimer's Association. 2015;11:792–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016;48:1279–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Das S, Forer L, Schonherr S, Sidore C, Locke AE, Kwong A, et al. Next-generation genotype imputation service and methods. Nat Genet. 2016;48:1284–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Arnold M, Raffler J, Pfeufer A, Suhre K, Kastenmüller G. SNiPA: an interactive, genetic variant-centered annotation browser. Bioinformatics. 2015;31:1334–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].MacArthur J, Bowler E, Cerezo M, Gil L, Hall P, Hastings E, et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 2017;45:D896–D901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Guerreiro R, Bras J, Hardy J. Snapshot: genetics of Alzheimer's disease. Cell. 2013;155:968–e1. [DOI] [PubMed] [Google Scholar]
- [59].Villegas-Llerena C, Phillips A, Garcia-Reitboeck P, Hardy J, Pocock JM. Microglial genes regulating neuroinflammation in the progression of Alzheimer's disease. Current opinion in neurobiology. 2016;36:74–81. [DOI] [PubMed] [Google Scholar]
- [60].Sims R, van der Lee SJ, Naj AC, Bellenguez C, Badarinarayan N, Jakobsdottir J, et al. Rare coding variants in PLCG2, ABI3, and TREM2 implicate microglial-mediated innate immunity in Alzheimer's disease. Nat Genet. 2017;49:1373–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Rolo AP, Palmeira CM, Wallace KB. Mitochondrially mediated synergistic cell killing by bile acids. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease. 2003;1637:127–32. [DOI] [PubMed] [Google Scholar]
- [62].Shivaram KN, Winklhofer-Roob BM, Straka MS, Devereaux MW, Everson G, Mierau GW, et al. The effect of idebenone, a coenzyme Q analogue, on hydrophobic bile acid toxicity to isolated rat hepatocytes and hepatic mitochondria. Free radical biology & medicine. 1998;25:480–92. [DOI] [PubMed] [Google Scholar]
- [63].Rodrigues CM, Fan G, Ma X, Kren BT, Steer CJ. A novel role for ursodeoxycholic acid in inhibiting apoptosis by modulating mitochondrial membrane perturbation. J Clin Invest. 1998;101:2790–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Hall AB, Tolonen AC, Xavier RJ. Human genetic variation and the gut microbiome in disease. Nature Reviews Genetics. 2017;18:690. [DOI] [PubMed] [Google Scholar]
- [65].Han J, Jiang Y, Li Z, Kravchenko VV, Ulevitch RJ. Activation of the transcription factor MEF2C by the MAP kinase p38 in inflammation. Nature. 1997;386:296–9. [DOI] [PubMed] [Google Scholar]
- [66].Fairfax BP, Humburg P, Makino S, Naranbhai V, Wong D, Lau E, et al. Innate immune activity conditions the effect of regulatory variants upon monocyte gene expression. Science. 2014;343:1246949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Shoemark DK, Allen SJ. The microbiome and disease: reviewing the links between the oral microbiome, aging, and Alzheimer's disease. J Alzheimers Dis. 2015;43:725–38. [DOI] [PubMed] [Google Scholar]
- [68].Hollingworth P, Harold D, Sims R, Gerrish A, Lambert JC, Carrasquillo MM, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nat Genet. 2011;43:429–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Le Guennec K, Nicolas G, Quenez O, Charbonnier C, Wallon D, Bellenguez C, et al. ABCA7 rare variants and Alzheimer disease risk. Neurology. 2016;86:2134–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Holmqvist S, Chutna O, Bousset L, Aldrin-Kirk P, Li W, Bjorklund T, et al. Direct evidence of Parkinson pathology spread from the gastrointestinal tract to the brain in rats. Acta Neuropathol. 2014;128:805–20. [DOI] [PubMed] [Google Scholar]
- [71].Jangi S, Gandhi R, Cox LM, Li N, von Glehn F, Yan R, et al. Alterations of the human gut microbiome in multiple sclerosis. Nature communications. 2016;7:12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Dinan TG, Cryan JF. Gut instincts: microbiota as a key regulator of brain development, ageing and neurodegeneration. J Physiol. 2017;595:489–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Kim DG, Krenz A, Toussaint LE, Maurer KJ, Robinson SA, Yan A, et al. Non-alcoholic fatty liver disease induces signs of Alzheimer's disease (AD) in wild-type mice and accelerates pathological signs of AD in an AD model. J Neuroinflammation. 2016;13:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Calkin AC, Tontonoz P. Transcriptional integration of metabolism by the nuclear sterol-activated receptors LXR and FXR. Nat Rev Mol Cell Biol. 2012;13:213–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Schaap FG, Trauner M, Jansen PLM. Bile acid receptors as targets for drug development. Nat Rev Gastroenterol Hepatol. 2014;11:55–67. [DOI] [PubMed] [Google Scholar]
- [76].Faubion WA, Guicciardi ME, Miyoshi H, Bronk SF, Roberts PJ, Svingen PA, et al. Toxic bile salts induce rodent hepatocyte apoptosis via direct activation of Fas. The Journal of clinical investigation. 1999;103:137–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Perez MJ, Briz O. Bile-acid-induced cell injury and protection. World J Gastroenterol. 2009;15:1677–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Sousa T, Castro RE, Pinto SN, Coutinho A, Lucas SD, Moreira R, et al. Deoxycholic acid modulates cell death signaling through changes in mitochondrial membrane properties. Journal of lipid research. 2015;56:2158–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Keitel V, Gorg B, Bidmon HJ, Zemtsova I, Spomer L, Zilles K, et al. The bile acid receptor TGR5 (Gpbar-1) acts as a neurosteroid receptor in brain. Glia. 2010;58:1794–805. [DOI] [PubMed] [Google Scholar]
- [80].Stenman LK, Holma R, Eggert A, Korpela R. A novel mechanism for gut barrier dysfunction by dietary fat: epithelial disruption by hydrophobic bile acids. Am J Physiol Gastrointest Liver Physiol. 2013;304:29. [DOI] [PubMed] [Google Scholar]
- [81].Shapiro H, Thaiss CA, Levy M, Elinav E. The cross talk between microbiota and the immune system: metabolites take center stage. Current Opinion in Immunology. 2014;30:54–62. [DOI] [PubMed] [Google Scholar]
- [82].Baptissart M, Vega A, Maqdasy S, Caira F, Baron S, Lobaccaro JM, et al. Bile acids: from digestion to cancers. Biochimie. 2013;95:504–17. [DOI] [PubMed] [Google Scholar]
- [83].Allegretti JR, Kearney S, Li N, Bogart E, Bullock K, Gerber GK, et al. Recurrent Clostridium difficile infection associates with distinct bile acid and microbiome profiles. Aliment Pharmacol Ther. 2016;43:1142–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.