Abstract
Identification of factors associated with human papillomavirus (HPV) cervical histopathology or recurrence/relapse following loop electrosurgical excision procedure (LEEP) would allow for better management of the disease. We investigated whether gene signatures could (i) associate with HPV cervical histopathology and (ii) identify women with post-LEEP disease recurrence/relapse. Gene array analysis was performed on paraffin-embedded cervical tissue-isolated RNA from two cross-sectional cohorts of antiretroviral therapy (ART)-suppressed HIV+HPV+ coinfected women: (i) 55 women in South Africa recruited into three groups: high risk (HR) (−) (n = 16) and HR (+) (n = 15) HPV without cervical histopathology and HR (+) HPV with cervical intraepithelial neoplasia (CIN) grade 1/2/3 (n = 24), (ii) 28 women in Botswana with CIN2/3 treated with LEEP 12-month prior to recruitment and presenting with (n = 13) and without (n = 15) lesion recurrence/relapse (tissue was analyzed at first LEEP). Three distinct gene expression signatures identified were able to segregate: (i) HR+ HPV and CIN1/2/3, (ii) HR HPV-free and cervical histopathology-free and (iii) HR+ HPV and cervical histopathology-free. Immune activation and neoplasia-associated genes (n = 272 genes; e.g. IL-1A, IL-8, TCAM1, POU4F1, MCM2, SMC1B, CXCL6, MMP12) were a feature of cancer precursor dysplasia within HR HPV infection. No difference in LEEP tissue gene expression was detected between women with or without recurrence/relapse. In conclusion, distinctive gene signatures were associated with presence of cervical histopathology in tissues from ART-suppressed HIV+/HPV+ coinfected women. Lack of detection of LEEP tissue gene signature able to segregate subsequent post-LEEP disease recurrence/relapse indicates additional factors independent of local gene expression as determinants of recurrence/relapse.
In this study of cervical tissue gene expression in ART-treated HIV+HPV+ co-infected women, we report the presence of distinct gene signatures in tissue biopsies able to segregate between tissues with and without cervical histopathology within HR HPV infection, as well as the lack of a specific gene signature in LEEP-excised tissues that could inform post-LEEP disease recurrence/relapse.
Introduction
Genital human papillomaviruses (HPVs) are characterized based on the strength of their association with cervical cancer, as oncogenic [high risk (HR)] types which act as carcinogens in the development of cervical cancer (1,2) and non-oncogenic types (low risk) (3). Approximately, 80% of new HPV infections are cleared within 12–18 months (4,5). In a small proportion where the immune response fails to clear or control the infection, a persistent infection is established, often with locally high levels of HR HPV DNA replication and true cancer precursor dysplasia [cervical intraepithelial neoplasia (CIN), divided into grades 1, 2 and 3] (6). As a standard cancer prevention strategy, in cases of confirmed high-grade lesions with a histological result of CIN2 or CIN3, treatment of the lesion is indicated by either ablative or excisional methods [i.e. loop electrosurgical excision procedure of the transition zone of cervix (LEEP)].
Human immunodeficiency virus 1 (HIV-1) infection alters the natural history of HPV-associated oncogenesis. HIV+ women with invasive cervical cancer have different frequency of HPV types compared with non-HIV women (7) and an increased risk of progression from subclinical HPV infection to disease (8). The degree of immunosuppression (CD4+ T-cell count <200 cells/mm3) has already been positively associated with increased risk of persistent HPV infection and progression of disease irrespective of viral load, CIN prevalence and severity (9). Treatment failure rates (defined as incomplete ectocervical and/or endocervical margins on pathology specimens irrespective of clear margins) leading to lesion relapse after LEEP are between 10 and 15% in immunocompetent women (10) and up to 50% worldwide in HIV+ women.
Previous studies suggest that local tissue gene expression could serve as a pathogenesis differentiator relative to lesion grade within HR HPV types (11–14) with several genes proposed as diagnostic markers for the detection of cervical neoplasia. In addition, although the local immune reaction to HPV is likely to play a significant role toward progression to cancer, there is a need for elucidation of the factors that could distinguish recurrent/relapsing HPV-associated premalignant lesions following LEEP from non-recurrent/relapsing lesions. It remains to be determined whether there is a specific HPV-associated gene signature that could predict disease recurrence/relapse after LEEP or whether recurrence/relapse is independent of local gene expression. Identification of markers associated with cervical histopathology and/or recurrence/relapse of HPV-associated premalignant lesions following LEEP could lead to the development of a screening method that would allow for better diagnosis, follow-up and evaluation of interventions to prevent recurrence/relapse.
We performed a cross-sectional study of antiretroviral therapy (ART)-suppressed HIV+/HPV+ coinfected women to test the hypothesis that cervical tissue gene signatures could (i) associate with cervical histopathology and (ii) identify women with post-LEEP disease recurrence/relapse.
Subjects, materials and methods
Participants
Paraffin-embedded cervical tissue samples were collected from two cross-sectional cohorts of ART-treated HIV+HPV+ coinfected women.
The first cohort consisted of 55 women identified from populations of patients of the Themba Lethu clinic and Clinical HIV Research Unit at the Helen Joseph hospital in South Africa. All women were negative for pregnancy and sexually transmitted infections tests at screening, with no clinical evidence of an inflammatory disease, and confirmed CD4+ T-cell count >200 cells/mm3 and HIV-1 viral load <50 copies for >6 months and at screening. These women participated in a study assessing the relationship between HR HPV type infection and immune activation/exhaustion. The main findings for this study are described in our prior publication (15). As described in our prior publication (15), HPV genotyping was performed in DNA isolated from a cytobrush sample by using the qualitative Roche Linear Array HPV genotyping test (Roche Molecular Systems, NJ) according to the manufacturer’s instructions. This qualitative test allowed for the detection of 37 HPV types. Individual HPV types were divided into 14 HR types: 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, 68, and 23 LR types: HPV 6, 11, 26, 40, 42, 53, 54, 55, 61, 62, 64, 67, 69, 70, 71, 72, 73, 81, 82, 83, 84, IS39 and CP6108. Women were divided into the following three groups: (i) negative for HR HPV [HR (−) HPV] with negative cervical histopathology (group A, n = 16), (ii) positive for HR HPV [HR (+) HPV] with negative cervical histopathology (group B, n = 15) and (iii) positive for HR HPV [HR (+) HPV] with CIN1/2/3 (group C, n = 24). Fixed colposcopic punch biopsies from these women were used for gene expression assessment.
The second cohort consisted of 28 women in Botswana with CIN2/3 lesion stage who underwent LEEP with clear margins noted 12 months prior to recruitment. All women were negative for pregnancy test, >18 years with CD4+ T-cell count >200 cells/mm3 and <50 copies/ml HIV viral load. Tissue block for resection, clinical case history and information on whether clear endomargins and ectomargins were noted were available for all women. The fixed excised tissue was available and used for gene expression assessment. In addition, data on recurrence/relapse of the precancer lesion within a period of 12 months were available for these women, allowing their division at recruitment into the following two groups: (i) women who showed recurrence/relapse of the precancer lesion within 12 months after excision date [group R (recurrent), n = 13], and (ii) women who did not show recurrence/relapse of the precancer lesion for at least 12 months after excision date [group NR (non-recurrent), n = 15]. Disease recurrence/relapse was defined by histopathologically proven CIN2/3 lesion in women treated for such lesions within the prior year, in which margins of the resected tissue were clear from cancerous of precancerous cells. Samples from this cohort were obtained from the specimen archive at the National Health Laboratory, Gaborone in Botswana as part of the Botswana-UPenn partnership.
The study protocol was approved by the Institutional Review Boards of the authors’ institutions and by the Human Research Committee of the Botswana Government.
Tissue gene expression assessment in cervical biopsies by microarray
Cervical tissue biopsies from all women meeting study inclusion criteria were collected in South Africa and Botswana as part of the clinical standard of care (no minimum threshold for dysplastic content was set as criteria). Eight unstained slices (10 µm thickness each) were scrapped from one (South African cohort) and two (Botswana cohort) formalin-embedded cervical tissue biopsies and combined for RNA isolation using the Tri-reagent (Sigma Aldrich, St Louis, MO) according to the manufacturer’s instructions. For the gene expression microarrays, amplified cRNA was generated from 100 ng RNA using the TargetAmp Nano-g Biotin-aRNA Labeling Kit (Epicentre, Madison WI) and then hybridized to the HumanHT-12 v4 Expression BeadChips (Illumina 44k bead array, Illumina, San Diego, CA).
Data preprocessing
Raw gene expression microarray data were quantile normalized. Non-informative probes, which were either expressed at background level or showed <1.2-fold change between all samples pairs, were removed prior to further analysis. Data preprocessing was performed in MATLAB R201a.
Statistical analysis
Single-point differences in the baseline mean mRNA expression between groups were tested for each gene using two-sided, paired t-tests. Significant gene results were further filtered to satisfy fold enrichment >1.5 criteria. False discovery rate (FDR)-based q values were reported with a threshold of 15% considered meaningful (16). A list of differential ranked genes was derived using linear kernel SVM-RFE with 10-fold re-sampling cross-validation (17,18). Cross-validation iterations started with 1000 top significant genes (based on t-test), reducing the number by 10% at each elimination step based on gene SVM-scores. Final ranking of the genes was calculated using the Borda count procedure. To directly assess the impact of gene expression as a predictor of lesion recurrence/relapse, heat maps for total versus specific clusters of genes were composed using two-way hierarchical clustering using Euclidean distance to cluster samples/conditions and Spearman correlation distance to cluster genes. To better define predictor mechanisms, pathway and functional analysis was performed using Ingenuity pathways analysis software (Ingenuity, CA) using Ingenuity core analysis (IPA 6.0, Ingenuity® Systems) with Benjamini–Hochberg multiple testing corrected P value of <0.05 as a significance threshold. Where appropriate, enrichments of gene ontology terms, KEGG and BIOCARTA pathways along with Swiss-Prot, INTERPRO and SMART keywords in a gene list were performed using DAVID software.
Results
Study subjects characteristics
Patient characteristics of the South African and the Botswana cohort are described in Table 1. No statistical difference was observed among study groups of each cohort for any of the clinical or demographic parameters described.
Table 1.
Characteristics of the South African and the Botswana cohort
| South African cohort | Botswana cohort | ||||
|---|---|---|---|---|---|
| Variable | Negative for high- risk HPV, negative histopathology (group A, n = 16) | Positive for high- risk HPV, negative histopathology (group B, n = 15) | Positive for high- risk HPV, CIN1/2/3 (group C, n = 24) | Recurrent (group R, n = 13) | Non-recurrent (group NR, n = 15) |
| Age (years), median (25th, 75th percentile) | 35 (31.5, 39.75) | 37 (29, 41) | 34.5 (28.25, 39.75) | 35 (29, 35.5) | 33 (30, 35) |
| CD4+ T-cell count at recruitment (cells/mm3), median (25th, 75th percentile) | 568.5 (395.7, 754) | 517 (399.5, 602) | 435.5 (308, 673.75) | 321 (238.5, 458.5) | 341 (282, 477) |
| Number of sexual partners, median (25th, 75th percentile) | 4.5 (3, 6) | 5 (3, 9) | 5.5 (4, 98) | 5 (3, 8.5) | 4.5 (3, 10) |
| Number of pregnancies, median (25th, 75th percentile) | 2 (2, 4) | 3 (1, 3) | 2 (1, 3) | 2 (1.5, 3) | 2 (1, 3) |
| Absolute number of women smoking cigarettes or chewing tobacco | 2 | 1 | 5 | 1 | 1 |
Increased expression of immune and neoplasia-associated genes in women with HR HPV type and CIN1/2/3 when compared with women without cervical histopathology irrespective of absence or presence of HR HPV
Cervical tissue biopsies from the South African cohort were used to determine whether distinctive gene signatures are associated with cervical histopathology.
In the absence of cervical histopathology, tissue gene expression analysis by the absence or presence of HR HPV (groups A and B, respectively) did not detect gene signatures distinguishing the two groups. On the other hand, three gene signatures were identified after comparisons between tissues with HR HPV and cervical histopathology (CIN1/2/3, group C) and tissues without cervical histopathology irrespective of the absence or presence of HR HPV (groups A and B, respectively).
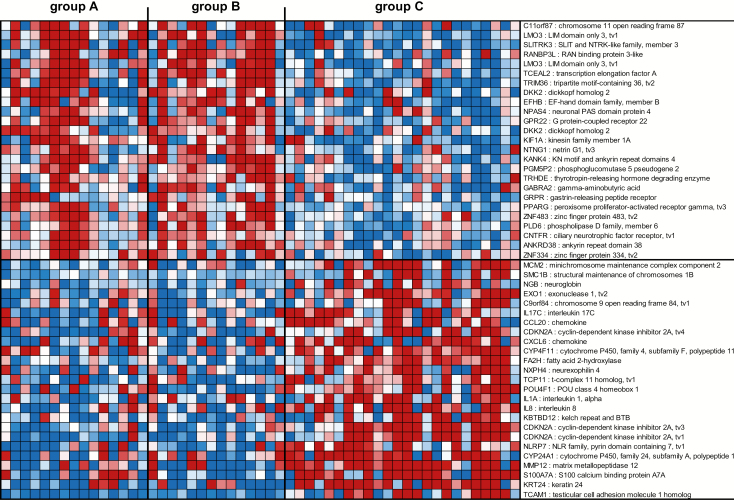
The first gene signature consisted of 272 genes (Supplementary Table 1, available at Carcinogenesis Online) and was able to segregate tissues with HR HPV and cervical histopathology (group C) from tissues without cervical histopathology irrespective of the absence or presence of HR HPV (groups A and B, respectively). The top 50 genes in this HR+ HPV and CIN1/2/3 signature are shown in Figure 1 and include genes associated with cell adhesion, cancer, metastasis or immune response. Briefly, LIM domain only 3 (LMO3) (19), SLIT and NTRK like family member 3 (SLITRK3) (20), tripartite motif containing 36 (TRIM36) (21), dickkopf WNT signaling pathway inhibitor 2 (DKK2) (22), gastrin releasing peptide receptor (GRPR) (23) and peroxisome proliferator activated receptor gamma (PPARG) (24) were downregulated in tissues with HR HPV and CIN1/2/3 (group C), while minichromosome maintenance complex component 2 (MCM2) (25), exonuclease 1 (EXO1) (26), interleukin-17C (IL-17C) (27), C-C motif chemokine ligand 20 (CCL20) (28), cyclin-dependent kinase inhibitor 2A (CDKN2A) (29), C-X-C motif chemokine ligand 6 (CXCL6) (30), POU class 4 homeobox 1 (POU4F1) (31), IL-1A (32), IL-8 (33), NLR family pyrin domain containing 7 (NLRP7) (34), matrix metallopeptidase 12 (MMP12) (35) and S100 calcium-binding protein A7A (S100A7A) (36) were upregulated. These results were further confirmed by ingenuity analysis showing an over-representation in tissues with HR HPV and CIN1/2/3 of molecular mechanisms of cancer, p53 signaling, DNA damage and repair, cell adhesion and role of cytokines in mediating cell communication (Supplementary Table 2, available at Carcinogenesis Online).
Figure 1.
Profiles of the top 50 mRNA probes of the first gene signature identified in the South African cohort. Expression ratios for the top 50 mRNA probes of the first gene signature (HR+ HPV and CIN1/2/3 gene signature) able to differentiate group C from groups A and B. The first gene signature consisted of 272 mRNA probes with FDR <15% and fold change >1.5 in the comparison of group A + B with group C and P < 0.05 in the comparisons of groups A, B and A + B with group C. Shown are the top 25 mRNA probes that displayed <−1.66-fold change and the top 25 mRNA probes that displayed >1.98-fold change in expression ratios.
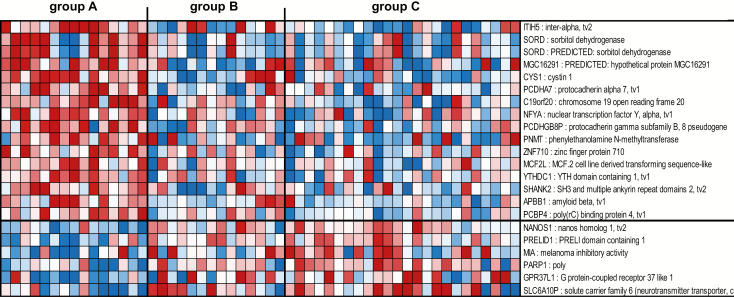
The second gene signature consisted of 22 genes not represented in the 272 gene signature described above able to segregate tissues without HR HPV and without cervical histopathology (group A), from tissues with HR HPV irrespective of the absence or presence of cervical histopathology (groups B and C, respectively) (Figure 2, Supplementary Table 3, available at Carcinogenesis Online). This HR HPV-free and cervical histopathology-free gene signature included an upregulation of genes associated with prevention of tumor metastasis [inter-alpha-trypsin inhibitor heavy chain family member 5 (ITIH5)] (37), or induction of apoptosis and inhibition of proliferation [poly(RC) binding protein 4 (PCBP4)] (38) in tissues without HR HPV and without cervical histopathology (group A), in contrast to a downregulation of these genes in tissues with HR HPV irrespective of the absence or presence of cervical histopathology (groups B and C, respectively). On the other hand, genes associated with tumor invasion [nanos C2HC-type zinc finger 1 (NANOS1)] (39) and transformation [poly(ADP-ribose) polymerase 1 (PARP1)] (40) were downregulated in tissues without HR HPV and without cervical histopathology (group A) and upregulated in tissues with HR HPV irrespective of the absence or presence of cervical histopathology (groups B and C, respectively) (Figure 2, Supplementary Table 3, available at Carcinogenesis Online). No significant pathways were identified by ingenuity analysis due to the low number of genes (n = 22) in this HR HPV-free and cervical histopathology-free signature.
Figure 2.
Profiles of the 22 mRNA probes of the second gene signature identified in the South African cohort. Expression ratios for the 22 mRNA probes of the second gene signature (HR HPV-free and cervical histopathology-free) able to differentiate group A from groups B and C. The second gene signature consisted of 22 genes not represented in the first gene signature (n = 272 genes), with FDR <15% and fold change >1.5 in the comparison of group A with group C and with P < 0.05 in the comparison of group A with group B. Shown are all 22 genes of the signature.
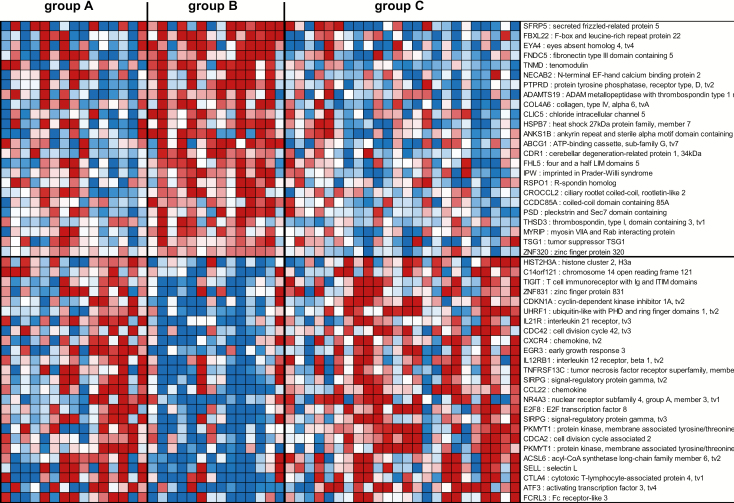
Finally, the third gene signature consisted of 81 genes not represented in the 272 gene signature described above able to segregate tissues with HR HPV and without cervical histopathology (group B) from tissues with HR HPV and CIN1/2/3 (group C) as well as from tissues without HR HPV and without cervical histopathology (group A) (Figure 3, Supplementary Table 4, available at Carcinogenesis Online). Among the genes identified in this HR+ HPV and cervical histopathology-free gene signature were genes associated with oncogene regulation [protein tyrosine phosphatase receptor type D (PTPRD), EYA transcriptional coactivator and phosphatase 4 (EYA4)] (41,42) and inhibition of angiogenesis [tenomodulin (TNMD)] (43) that were upregulated in tissues with HR HPV and without cervical histopathology (group B), while genes associated with cell cycle, cell migration, cancer, T-cell response and proliferation [e.g. T-cell immunoreceptor with Ig and ITIM domains (TIGIT) (44), CDKN1A (45), ubiquitin like with PHD and ring finger domains 1 (UHRF1) (46), IL-21R (47), cell division cycle 42 (CDC42) (48), early growth response 3 (EGR3) (49), signal regulatory protein gamma (SIRPG) (50), CCL22 (51), nuclear receptor subfamily 4 group A member 3 (NR4A3) (52), E2F transcription factor 8 (E2F8) (53), cytotoxic T lymphocyte associated protein 4 (CTLA4) (54) and activating transcription factor 3 (ATF3) (55)] were downregulated (Figure 3). No significant pathways were identified by ingenuity analysis due to the low numbers of genes (n = 81) in this HR+ HPV and cervical histopathology-free gene signature.
Figure 3.
Profiles of the 81 mRNA probes of the third gene signature identified in the South African cohort. Expression ratios for the 81 mRNA probes of the third gene signature (HR+ HPV and cervical histopathology-free) able to differentiate group B from groups A and C. The third gene signature identified 81 genes not represented in the first gene signature (n = 272 genes), with FDR <15% and fold change >1.5 in the comparison of group B with group C and with P < 0.05 in the comparison of group A with group B. Shown are the top 25 mRNA probes that displayed <−1.66-fold change and the top 24 mRNA probes that displayed >1.98-fold change in expression ratios.
Taken together, these findings suggest the presence of distinct cervical biopsy tissue-based gene signatures in ART-treated HIV+HPV+ coinfected women in South Africa that were associated with (i) the presence of cervical histopathology (CIN1/2/3) in the presence of HR HPV infection (HR+ HPV and CIN1/2/3 gene signature), (ii) the absence of cervical histopathology in the absence of HR HPV infection (HR HPV-free and cervical histopathology-free gene signature) or (iii) the absence of cervical histopathology in the presence of HR HPV infection (HR+ HPV and cervical histopathology-free gene signature).
Lack of significant difference in LEEP-excised cervical tissue gene expression between women with and without post-LEEP disease recurrence/relapse
LEEP-excised cervical tissue from the Botswana cohort was used to determine whether distinctive gene signatures are predictive of post-LEEP disease recurrence/relapse.
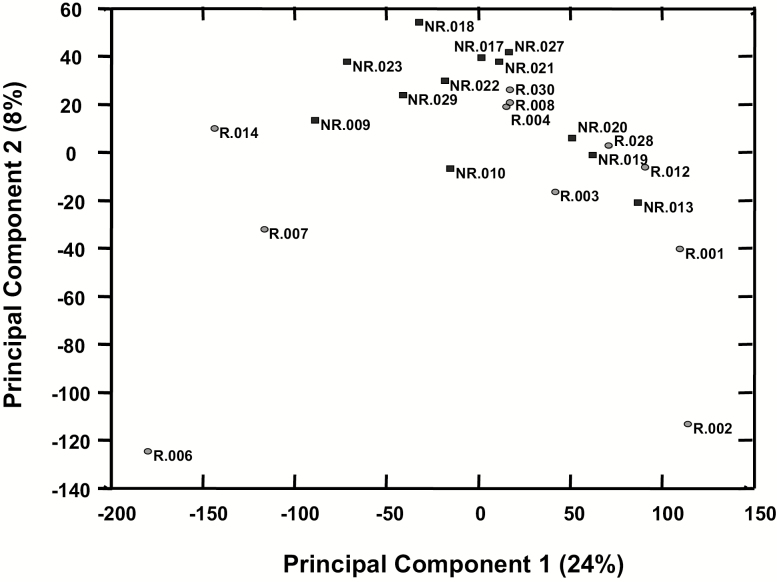
Microarray data preprocessing consisted of exclusion of five outliers (i.e. group R: n = 2; group NR: n = 3), quantile normalization and filtering out genes that had poor detection in all samples (i.e. detection P > 0.05 in all samples) or genes that have <1.2-fold change between any two samples. This resulted in 26 643 informative genes. Using these genes, a principal component analysis did not segregate groups defined by presence/absence of post-LEEP disease recurrence/relapse (Figure 4). Furthermore, t-test comparison between the two groups showed 964 genes with P < 0.05 but with FDR 84%. Although the FDR limits the ability to establish differences between the two groups, it is of interest to note for future investigation that of the 964 genes identified in the LEEP-excised cervical tissues, a total of 27 genes showed a fold change >3, with 14/27 genes being upregulated in LEEP-excised cervical tissues from women with post-LEEP disease recurrence/relapse when compared with LEEP-excised cervical tissues from women without post-LEEP disease recurrence/relapse and 13/27 genes being downregulated (Supplementary Figure 1A, available at Carcinogenesis Online). As shown in Supplementary Figure 1B, available at Carcinogenesis Online, among the genes upregulated in LEEP-excised cervical tissues from women with post-LEEP disease recurrence/relapse were genes that are associated with tumorigenesis [PDZ binding kinase (PBK)] (56), cell growth and differentiation [metallothionein-Like 5, testis-specific (MTL5), alcohol dehydrogenase 7 (ADH7)] (57,58) and apoptosis [deoxyribonuclease 1 like 3 (DNASE1L3)] (59).
Figure 4.
Principal component analysis of mRNA probes in the Botswana cohort. Principal component analysis using the 26 643 informative mRNA probes [i.e. genes with good detection in all samples (i.e. detection P < 0.05 in all samples), >1.2-fold change between any two samples and background level of expression about 82]. Data are shown for each patient from group R (i.e. women with post-LEEP disease recurrence/relapse, indicated with black circles) and group NR (i.e. women without post-LEEP disease recurrence/relapse, indicated with white squares) of the Botswana cohort.
Taken together, these findings support a lack of detection of a cervical gene expression signature at time of LEEP excision that could inform subsequent 12-month disease recurrence/relapse.
Discussion
In this study of cervical tissue gene expression in ART-treated HIV+HPV+ coinfected women, we report the presence of distinct gene signatures in tissue biopsies able to segregate between tissues with and without cervical histopathology within HR HPV infection, as well as the lack of a specific gene signature in LEEP-excised tissues that could inform post-LEEP disease recurrence/relapse.
The lack of differences in gene expression between LEEP-excised tissues with subsequent recurrence/relapse and LEEP-excised tissues without subsequent recurrence/relapse despite LEEP with clear margin suggests that added factors, apart from possible remains of dysplastic residual tissue after LEEP, could determine post-LEEP disease recurrence/relapse. For example, the HPV-infected cells that could possibly remain outside the LEEP-excised lesion site could continue to progress after LEEP. Furthermore, post-LEEP disease recurrence/relapse could also be explained by possible post-LEEP re-infection, a scenario that cannot be ruled out in our study. Moreover, prior reports indicate that host or viral factors may also play a role in the observed disease recurrence/relapse in HIV+ women where CD4+ count <200 cells/mm3, detectable HIV viremia and lack of adequate ART (10,60,61) are present. Although our cohort was stable with regards to CD4+ T-cell count >200 cells/mm3 and undetectable HIV, it is possible that the re-emergence of a cervical lesion may signal a state of immune activation or dysfunction in spite of suppressive ART (62,63).
Importantly, within biopsies of tissues with HR HPV and cervical histopathology (CIN1/2/3), we did find an enrichment in the expression of genes that were associated with oncogenesis and immune responses. These data add to previous reports highlighting that cervical gene expression can serve as a pathogenesis differentiator after HR HPV infection (11–14,64–67). Our data are also informed by usage of the same tissue blocks as used in our prior published report (15) on the same cohort of ART-suppressed HIV+/HPV+ coinfected South African women. Specifically, having previously established in the same cohort that the presence of HR HPV infection could be associated with greater immune activation in the blood irrespective of absence or presence of cervical histopathology (15), it was unexpected that the predominant gene signature (i.e. HR+ HPV and CIN1/2/3 gene signature) identified in tissue segregated the presence of cervical histopathology from the absence of cervical histopathology within HR HPV infection. The latter supports a compartmentalization of tissue microenvironment changes that are distinct from blood after HR HPV infection of ART-suppressed HIV+ women. The presence of sustained systemic immune activation in the blood of women coinfected with HR HPV and HIV aligns with data showing increased immune activation in hepatitis C virus/HIV coinfected subjects on ART as compared with hepatitis C virus mono-infected subjects (68–72). We now expand on these data by showing an association between tissue gene expression and cervical histopathology within HR HPV types. More precisely, as suggested by the predominant gene signature (i.e. HR+ HPV and CIN1/2/3 gene signature) identified, we show in cervical tissues with CIN1/2/3 an increased gene expression of markers of inflammation (i.e. IL-1A, IL-8, CCL20, S100A7A) (28,32,33,36). Among these genes, inflammatory molecules IL-1A (73,74), and IL-8 (74,75) have been described to be associated in high-grade lesions. Furthermore, and besides immune-associated genes, we also show in cervical tissues with CIN1/2/3 an increased gene expression of molecules associated with DNA replication and cell division [i.e. MCM2, structural maintenance of chromosomes 1B (SMC1B)] (25,76), chemotaxis and angiogenesis (i.e. CXCL6) (30), metastasis (i.e. MMP12) (35) and cancer (i.e. POU4F1) (31). Of interest, POU4F1 (11,12,64,65), MCM2 (13,66) and MMP12 (14,67) have been described previously to be associated with high-grade cervical lesions and were proposed to be used as diagnostic markers for the detection of cervical neoplasia. Furthermore, as suggested by the other two signatures (i.e. HR HPV-free and cervical histopathology-free gene signature, and HR+ HPV and cervical histopathology-free gene signature) identified, we also confirm upregulation of PARP1, UHRF1, CDC42 and CCL22 in cervical tissues with CIN1/2/3 in agreement with previous studies showing an association between lesion stage, cervical cancer or tumor invasion and increased expression of these genes (77–80).
Our study supports and further expands studies in HIV− subjects reporting gene signatures able to distinguish HPV-associated cancers (e.g in the head and neck) from normal tissue or HPV− cancers (81–87). More precisely, we now show that in HIV+ women, several of these cancer-associated genes involved in DNA replication, cell cycle and cell proliferation [e.g. CDKN2A, MCM2, cell division cycle 7 (CDC7), cyclin E2 (CCNE2), proliferating cell nuclear antigen (PCNA), centromere protein K (CENPK), enhancer of Zeste homologue 2 (EZH2), stathmin 1 (STMN1)] distinguish HR+ HPV and cervical histopathology (CIN1/2/3) from HR− and HR+ HPV with negative cervical histopathology. As a result, our data suggest that many of these cancer-associated signature genes that are dysregulated in cervical neoplasia are common to other HPV-associated neoplasms arising in HIV+ and HIV− subjects.
This study has limitations. First, genes were identified by microarray analysis and require validation by an independent cohort and other methods. Second, collection of samples was performed as clinical standard of care in a limited-resources setting, and as a result, the possible presence of normal tissue along with dysplastic lesions in the analyzed samples cannot be ruled out. Third, in the South African cohort, due to the limited number of subjects with CIN1 (n = 9), all subjects with cervical dysplasia (group C: CIN1/2/3 n = 24) were analyzed together rather than segregated in groups with low (CIN1) and high (CIN2/3) grade cervical histopathology. Fourth, in the Botswana cohort, the limited sample size, the relatively short follow-up period (i.e. at least 12 months) for assessment of post-LEEP disease recurrence/relapse and the lack of methods that could exclude the possibility of post-LEEP re-infection could account for the lack of a gene signature able to distinguish between the two groups in our study and should be addressed in future studies. Finally, as a cross-sectional study, the stability of gene signatures as predictive of lesion regression or cancer remains to be confirmed by future studies.
In conclusion, our data suggest that in contrast to changes in systemic immune activation that were associated with HR HPV infection and not with cervical histopathology (15), cervical gene expression can serve as a differentiator to identify the presence of cervical histopathology with HR HPV infection. Furthermore, our data showing that post-LEEP disease recurrence/relapse is not associated with a distinct cervical tissue gene expression at the time of excision indicate that additional factors independent of local gene expression could serve as determinants of recurrence/relapse. It will be important to design future larger studies to determine whether the genes described here could serve as (i) potential biomarkers for grade of dysplasia, lesion progression or clinical outcome following excision of cervical cancer-associated lesions with clear margins or (ii) aid in identifying the source for the dysregulated state in HIV infection in spite of suppressive ART.
Funding
This work was partially supported by National Institutes of Health/National Institute of Allergy and Infectious Diseases grant UO1AI51986 to L.J.M. Additional support was provided by The Philadelphia Foundation (Robert I. Jacobs Fund), Ken Nimblett and the Summerhill Trust, AIDS funds from the Commonwealth of Pennsylvania and from the Commonwealth Universal Research Enhancement Program, The Pennsylvania Department of Health, as well as by the Wistar Cancer Center Support Grant (CCSG) CA010815. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript. There are no potential conflicts of interest. The content of this publication does not necessarily reflect the views or policies of NIAID, nor does mention of trade names, commercial projects or organizations imply endorsement by the US Government.
Supplementary Material
Acknowledgement
We would like to thank subjects who participated in the study and their providers.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- ART
antiretroviral therapy
- CIN
cervical intraepithelial neoplasia
- FDR
false discovery rate
- HIV
human immunodeficiency virus
- HPV
human papillomavirus
- HR
high risk
- LEEP
loop electrosurgical excision procedure
- MCM2
minichromosome maintenance complex component 2
- MMP12
matrix metallopeptidase 12
- POU4F1
POU class 4 homeobox 1
References
- 1. de Sanjose S. et al.; Retrospective International Survey and HPV Time Trends Study Group (2010) Human papillomavirus genotype attribution in invasive cervical cancer: a retrospective cross-sectional worldwide study. Lancet. Oncol., 11, 1048–1056. [DOI] [PubMed] [Google Scholar]
- 2. Walboomers J.M. et al. (1999) Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol., 189, 12–19. [DOI] [PubMed] [Google Scholar]
- 3. Muñoz N. et al.; International Agency for Research on Cancer Multicenter Cervical Cancer Study Group (2003) Epidemiologic classification of human papillomavirus types associated with cervical cancer. N. Engl. J. Med., 348, 518–527. [DOI] [PubMed] [Google Scholar]
- 4. Palefsky J.M. et al. (1998) High incidence of anal high-grade squamous intra-epithelial lesions among HIV-positive and HIV-negative homosexual and bisexual men. AIDS, 12, 495–503. [DOI] [PubMed] [Google Scholar]
- 5. Moscicki A.B. et al. (2001) Risks for incident human papillomavirus infection and low-grade squamous intraepithelial lesion development in young females. JAMA, 285, 2995–3002. [DOI] [PubMed] [Google Scholar]
- 6. Palefsky J. (2006) Human papillomavirus-related tumors in HIV. Curr. Opin. Oncol., 18, 463–468. [DOI] [PubMed] [Google Scholar]
- 7. Clifford G.M. et al. (2016) Effect of HIV infection on human papillomavirus types causing invasive cervical cancer in Africa. J. Acquir. Immune Defic. Syndr., 73, 332–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chirgwin K.D. et al. (1995) Incidence of venereal warts in human immunodeficiency virus-infected and uninfected women. J. Infect. Dis., 172, 235–238. [DOI] [PubMed] [Google Scholar]
- 9. Strickler H.D. et al. (2003) Human papillomavirus type 16 and immune status in human immunodeficiency virus-seropositive women. J. Natl. Cancer Inst., 95, 1062–1071. [DOI] [PubMed] [Google Scholar]
- 10. Cox J.T. (2002) Management of precursor lesions of cervical carcinoma: history, host defense, and a survey of modalities. Obstet. Gynecol. Clin. North Am., 29, 751–785. [DOI] [PubMed] [Google Scholar]
- 11. Ndisdang D. et al. (1998) The HPV-activating cellular transcription factor Brn-3a is overexpressed in CIN3 cervical lesions. J. Clin. Invest., 101, 1687–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sindos M. et al. (2003) Detection of cervical neoplasia using measurement of Brn-3a in cervical smears with persistent minor abnormality. Int. J. Gynecol. Cancer, 13, 515–517. [DOI] [PubMed] [Google Scholar]
- 13. Zheng J. (2015) Diagnostic value of MCM2 immunocytochemical staining in cervical lesions and its relationship with HPV infection. Int. J. Clin. Exp. Pathol., 8, 875–880. [PMC free article] [PubMed] [Google Scholar]
- 14. Valdivia A. et al. (2011) Co-expression of metalloproteinases 11 and 12 in cervical scrapes cells from cervical precursor lesions. Int. J. Clin. Exp. Pathol., 4, 674–682. [PMC free article] [PubMed] [Google Scholar]
- 15. Papasavvas E. et al. (2016) High-risk oncogenic HPV genotype infection associates with increased immune activation and T cell exhaustion in ART-suppressed HIV-1-infected women. Oncoimmunology, 5, e1128612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Storey J.D. et al. (2003) Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA, 100, 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Forthal D.N. et al. (2009) Fc receptor-mediated antiviral antibodies. Curr. Opin. HIV AIDS, 4, 388–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li Q. et al. (2009) Visualizing antigen-specific and infected cells in situ predicts outcomes in early viral infection. Science, 323, 1726–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Song Y.F. et al. (2015) miR-630 targets LMO3 to regulate cell growth and metastasis in lung cancer. Am. J. Transl. Res., 7, 1271–1279. [PMC free article] [PubMed] [Google Scholar]
- 20. Aruga J. et al. (2003) Human SLITRK family genes: genomic organization and expression profiling in normal brain and brain tumor tissue. Gene, 315, 87–94. [DOI] [PubMed] [Google Scholar]
- 21. Balint I. et al. (2004) Cloning and characterisation of the RBCC728/TRIM36 zinc-binding protein from the tumor suppressor gene region at chromosome 5q22.3. Gene, 332, 45–50. [DOI] [PubMed] [Google Scholar]
- 22. Park H. et al. (2014) Distinct roles of DKK1 and DKK2 in tumor angiogenesis. Angiogenesis, 17, 221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jensen R.T. et al. (2008) International Union of Pharmacology. LXVIII. Mammalian bombesin receptors: nomenclature, distribution, pharmacology, signaling, and functions in normal and disease states. Pharmacol. Rev., 60, 1–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xu W. et al. (2010) PPARgamma polymorphisms and cancer risk: a meta-analysis involving 32,138 subjects. Oncol. Rep., 24, 579–585. [PubMed] [Google Scholar]
- 25. Simon N.E. et al. (2014) The Mcm2-7 replicative helicase: a promising chemotherapeutic target. Biomed Res. Int., 2014, 549719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sperka T. et al. (2012) DNA damage checkpoints in stem cells, ageing and cancer. Nat. Rev. Mol. Cell Biol., 13, 579–590. [DOI] [PubMed] [Google Scholar]
- 27. Niu Y.M. et al. (2014) Interleukin-17 gene polymorphisms contribute to cancer risk. Mediators Inflamm., 2014, 128490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schutyser E. et al. (2003) The CC chemokine CCL20 and its receptor CCR6. Cytokine Growth Factor Rev., 14, 409–426. [DOI] [PubMed] [Google Scholar]
- 29. Rayess H. et al. (2012) Cellular senescence and tumor suppressor gene p16. Int. J. Cancer, 130, 1715–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Strieter R.M. et al. (2006) Cancer CXC chemokine networks and tumour angiogenesis. Eur. J. Cancer, 42, 768–778. [DOI] [PubMed] [Google Scholar]
- 31. Hohenauer T. et al. (2013) The neural crest transcription factor Brn3a is expressed in melanoma and required for cell cycle progression and survival. EMBO Mol. Med., 5, 919–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rider P. et al. (2013) Interleukin-1α. Semin. Immunol., 25, 430–438. [DOI] [PubMed] [Google Scholar]
- 33. Harada A. et al. (1996) Interleukin 8 as a novel target for intervention therapy in acute inflammatory diseases. Mol. Med. Today, 2, 482–489. [DOI] [PubMed] [Google Scholar]
- 34. Radian A.D. et al. (2013) NLRP7 and related inflammasome activating pattern recognition receptors and their function in host defense and disease. Microbes Infect., 15, 630–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Egeblad M. et al. (2002) New functions for the matrix metalloproteinases in cancer progression. Nat. Rev. Cancer, 2, 161–174. [DOI] [PubMed] [Google Scholar]
- 36. Kulski J.K. et al. (2003) Genomic and phylogenetic analysis of the S100A7 (Psoriasin) gene duplications within the region of the S100 gene cluster on human chromosome 1q21. J. Mol. Evol., 56, 397–406. [DOI] [PubMed] [Google Scholar]
- 37. Dötsch M.M. et al. (2015) Low expression of ITIH5 in adenocarcinoma of the lung is associated with unfavorable patients’ outcome. Epigenetics, 10, 903–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhu J. et al. (2000) MCG10, a novel p53 target gene that encodes a KH domain RNA-binding protein, is capable of inducing apoptosis and cell cycle arrest in G(2)-M. Mol. Cell. Biol., 20, 5602–5618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Strumane K. et al. (2006) E-cadherin regulates human Nanos1, which interacts with p120ctn and induces tumor cell migration and invasion. Cancer Res., 66, 10007–10015. [DOI] [PubMed] [Google Scholar]
- 40. Schiewer M.J. et al. (2014) Transcriptional roles of PARP1 in cancer. Mol. Cancer Res., 12, 1069–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ostman A. et al. (2006) Protein-tyrosine phosphatases and cancer. Nat. Rev. Cancer, 6, 307–320. [DOI] [PubMed] [Google Scholar]
- 42. Mo S.J. et al. (2016) EYA4 functions as tumor suppressor gene and prognostic marker in pancreatic ductal adenocarcinoma through β-catenin/ID2 pathway. Cancer Lett., 380, 403–412. [DOI] [PubMed] [Google Scholar]
- 43. Shukunami C. et al. (2007) Chondromodulin-I and tenomodulin: the negative control of angiogenesis in connective tissue. Curr. Pharm. Des., 13, 2101–2112. [DOI] [PubMed] [Google Scholar]
- 44. Blake S.J. et al. (2016) Molecular pathways: targeting CD96 and TIGIT for cancer immunotherapy. Clin. Cancer Res., 22, 5183–5188. [DOI] [PubMed] [Google Scholar]
- 45. Parveen A. et al. (2016) Dual role of p21 in the progression of cancer and its treatment. Crit. Rev. Eukaryot. Gene Expr., 26, 49–62. [DOI] [PubMed] [Google Scholar]
- 46. Alhosin M. et al. (2016) Signalling pathways in UHRF1-dependent regulation of tumor suppressor genes in cancer. J. Exp. Clin. Cancer Res., 35, 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Davis M.R. et al. (2015) The role of IL-21 in immunity and cancer. Cancer Lett., 358, 107–114. [DOI] [PubMed] [Google Scholar]
- 48. Qadir M.I. et al. (2015) Cdc42: role in cancer management. Chem. Biol. Drug Des., 86, 432–439. [DOI] [PubMed] [Google Scholar]
- 49. Taefehshokr S. et al. (2017) Early growth response 2 and Egr3 are unique regulators in immune system. Cent. Eur. J. Immunol., 42, 205–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Weiskopf K. (2017) Cancer immunotherapy targeting the CD47/SIRPα axis. Eur. J. Cancer, 76, 100–109. [DOI] [PubMed] [Google Scholar]
- 51. Gobert M. et al. (2009) Regulatory T cells recruited through CCL22/CCR4 are selectively activated in lymphoid infiltrates surrounding primary breast tumors and lead to an adverse clinical outcome. Cancer Res., 69, 2000–2009. [DOI] [PubMed] [Google Scholar]
- 52. Wenzl K. et al. (2015) The nuclear orphan receptor NR4A1 and NR4A3 as tumor suppressors in hematologic neoplasms. Curr. Drug Targets, 16, 38–46. [DOI] [PubMed] [Google Scholar]
- 53. DeGregori J. et al. (2006) Distinct and overlapping roles for E2F family members in transcription, proliferation and apoptosis. Curr. Mol. Med., 6, 739–748. [DOI] [PubMed] [Google Scholar]
- 54. Hodi F.S. et al. (2003) Biologic activity of cytotoxic T lymphocyte-associated antigen 4 antibody blockade in previously vaccinated metastatic melanoma and ovarian carcinoma patients. Proc. Natl. Acad. Sci. USA, 100, 4712–4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Thompson M.R. et al. (2009) ATF3 transcription factor and its emerging roles in immunity and cancer. J. Mol. Med. (Berl)., 87, 1053–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Stricker T.P. et al. (2013) Expression profiling of 519 kinase genes in matched malignant peripheral nerve sheath tumor/plexiform neurofibroma samples is discriminatory and identifies mitotic regulators BUB1B, PBK and NEK2 as overexpressed with transformation. Mod. Pathol., 26, 930–943. [DOI] [PubMed] [Google Scholar]
- 57. Sugihara T. et al. (1999) A novel testis-specific metallothionein-like protein, tesmin, is an early marker of male germ cell differentiation. Genomics, 57, 130–136. [DOI] [PubMed] [Google Scholar]
- 58. Chen X. et al. (2016) Differentiation-inducing and anti-proliferative activities of isoliquiritigenin and all-trans-retinoic acid on B16F0 melanoma cells: mechanisms profiling by RNA-seq. Gene, 592, 86–98. [DOI] [PubMed] [Google Scholar]
- 59. Sisirak V. et al. (2016) Digestion of chromatin in apoptotic cell microparticles prevents autoimmunity. Cell, 166, 88–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Adam Y. et al. (2008) Predictors of persistent cytologic abnormalities after treatment of cervical intraepithelial neoplasia in Soweto, South Africa: a cohort study in a HIV high prevalence population. BMC Cancer, 8, 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tebeu P.M. et al. (2006) The recurrence of cervical intraepithelial neoplasia in HIV-positive women: a review of the literature. Int. J. STD AIDS, 17, 507–511. [DOI] [PubMed] [Google Scholar]
- 62. Sandler N.G. et al.; INSIGHT SMART Study Group (2011) Plasma levels of soluble CD14 independently predict mortality in HIV infection. J. Infect. Dis., 203, 780–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sunil M. et al. (2016) Unchanged levels of soluble CD14 and IL-6 over time predict serious non-AIDS events in HIV-1-infected people. AIDS Res. Hum. Retroviruses, 32, 1205–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ndisang D. et al. (2009) The HPV cellular transactivator Brn-3a can be used to predict cervical adenocarcinoma and squamous carcinoma precancer lesions in the developed and developing worlds. Obstet. Gynecol. Int., 2009, 359457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sindos M. et al. (2003) Measurement of Brn-3a levels in Pap smears provides a novel diagnostic marker for the detection of cervical neoplasia. Gynecol. Oncol., 90, 366–371. [DOI] [PubMed] [Google Scholar]
- 66. Saritha V.N. et al. (2018) Significance of DNA replication licensing proteins (MCM2, MCM5 and CDC6), p16 and p63 as markers of premalignant lesions of the uterine cervix: its usefulness to predict malignant potential. Asian Pac. J. Cancer Prev., 19, 141–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Vazquez-Ortiz G. et al. (2005) Overexpression of cathepsin F, matrix metalloproteinases 11 and 12 in cervical cancer. BMC Cancer, 5, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Papasavvas E. et al. (2017) HCV viraemia associates with NK cell activation and dysfunction in antiretroviral therapy-treated HIV/HCV-co-infected subjects. J. Viral Hepat., 24, 865–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Gonzalez V.D. et al. (2009) High levels of chronic immune activation in the T-cell compartments of patients coinfected with hepatitis C virus and human immunodeficiency virus type 1 and on highly active antiretroviral therapy are reverted by alpha interferon and ribavirin treatment. J. Virol., 83, 11407–11411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kottilil S. et al. (2009) Human immunodeficiency virus and hepatitis C infections induce distinct immunologic imprints in peripheral mononuclear cells. Hepatology, 50, 34–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kovacs A. et al. (2008) CD8(+) T cell activation in women coinfected with human immunodeficiency virus type 1 and hepatitis C virus. J. Infect. Dis., 197, 1402–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Rempel H. et al. (2013) Monocyte activation in HIV/HCV coinfection correlates with cognitive impairment. PLoS One, 8, e55776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Iglesias M. et al. (1998) Human papillomavirus type 16 E7 protein sensitizes cervical keratinocytes to apoptosis and release of interleukin-1alpha. Oncogene, 17, 1195–1205. [DOI] [PubMed] [Google Scholar]
- 74. Mhatre M. et al. (2012) Cervical intraepithelial neoplasia is associated with genital tract mucosal inflammation. Sex. Transm. Dis., 39, 591–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Iwata T. et al. (2015) Cytokine profile in cervical mucosa of Japanese patients with cervical intraepithelial neoplasia. Int. J. Clin. Oncol., 20, 126–133. [DOI] [PubMed] [Google Scholar]
- 76. Mannini L. et al. (2015) SMC1B is present in mammalian somatic cells and interacts with mitotic cohesin proteins. Sci. Rep., 5, 18472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Hassumi-Fukasawa M.K. et al. (2012) Expression of BAG-1 and PARP-1 in precursor lesions and invasive cervical cancer associated with human papillomavirus (HPV). Pathol. Oncol. Res., 18, 929–937. [DOI] [PubMed] [Google Scholar]
- 78. Oliver A.W. et al. (2011) The HPV16 E6 binding protein Tip-1 interacts with ARHGEF16, which activates Cdc42. Br. J. Cancer, 104, 324–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Zhang Q. et al. (2018) UHRF1 epigenetically down-regulates UbcH8 to inhibit apoptosis in cervical cancer cells. Cell Cycle, 17, 300–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Zhao M. et al. (2017) Negative immune factors might predominate local tumor immune status and promote carcinogenesis in cervical carcinoma. Virol. J., 14, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Chakravarthy A. et al. (2016) Human papillomavirus drives tumor development throughout the head and neck: improved prognosis is associated with an immune response largely restricted to the oropharynx. J. Clin. Oncol., 34, 4132–4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Jung A.C. et al. (2010) Biological and clinical relevance of transcriptionally active human papillomavirus (HPV) infection in oropharynx squamous cell carcinoma. Int. J. Cancer, 126, 1882–1894. [DOI] [PubMed] [Google Scholar]
- 83. Lohavanichbutr P. et al. (2009) Genomewide gene expression profiles of HPV-positive and HPV-negative oropharyngeal cancer: potential implications for treatment choices. Arch. Otolaryngol. Head. Neck Surg., 135, 180–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Pyeon D. et al. (2007) Fundamental differences in cell cycle deregulation in human papillomavirus-positive and human papillomavirus-negative head/neck and cervical cancers. Cancer Res., 67, 4605–4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Schlecht N.F. et al. (2007) Gene expression profiles in HPV-infected head and neck cancer. J. Pathol., 213, 283–293. [DOI] [PubMed] [Google Scholar]
- 86. Slebos R.J. et al. (2006) Gene expression differences associated with human papillomavirus status in head and neck squamous cell carcinoma. Clin. Cancer Res., 12 (3 Pt 1), 701–709. [DOI] [PubMed] [Google Scholar]
- 87. Tang K.W. et al. (2013) The landscape of viral expression and host gene fusion and adaptation in human cancer. Nat. Commun., 4, 2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.