Abstract
Remodeling or deregulation of the calcium signaling pathway is a relevant hallmark of cancer including cutaneous melanoma (CM). In this study, using data from a published genome-wide association study (GWAS) from The University of Texas M.D. Anderson Cancer Center, we assessed the role of 41,377 common single-nucleotide polymorphisms (SNPs) of 167 calcium signaling pathway genes in CM survival. We used another GWAS from Harvard University as the validation dataset. In the single-locus analysis, 1830 SNPs were found to be significantly associated with CM-specific survival (CMSS; P ≤ 0.050 and false-positive report probability ≤ 0.2), of which 9 SNPs were validated in the Harvard study (P ≤ 0.050). Among these, three independent SNPs (i.e. PDE1A rs6750552 T>C, ITPR1 rs6785564 A>G and RYR3 rs2596191 C>A) had a predictive role in CMSS, with a meta-analysis-derived hazards ratio of 1.52 (95% confidence interval = 1.19–1.94, P = 7.21 × 10−4), 0.49 (0.33–0.73, 3.94 × 10−4) and 0.67 (0.53–0.86, 0.0017), respectively. Patients with an increasing number of protective genotypes had remarkably improved CMSS. Additional expression quantitative trait loci analysis showed that these genotypes were also significantly associated with mRNA expression levels of the genes. Taken together, these results may help us to identify prospective biomarkers in the calcium signaling pathway for CM prognosis.
We used genotypes from two genome-wide association studies in a two-stage analysis and found that three SNPs in PDE1A, ITPR1 and RYR3 modulated the survival of cutaneous melanoma patients, suggesting that these genetic variants may be promising predictors of cutaneous melanoma.
Introduction
Cutaneous melanoma (CM) remains a clinical challenge for management worldwide. In 2018, an estimated 91,270 adults will be diagnosed with CM, and 9320 patients will die from this disease in the United States (1). As the most unfavorable and lethal skin cancer, the distant-stage CM generally has a poor prognosis with a 5-year survival of ~20% (2). Thus, it is imperative to understand molecular mechanisms underlying the prognosis of CM.
It is broadly accepted that calcium is ubiquitously involved in nearly every aspect of cellular processes in humans, including cell growth, proliferation and even cell death (3,4), and thus the molecule has been appropriately referred to as the life and death signal (5). In 2000, Hanahan and Weinberg (6) proposed six hallmark capabilities of cancer, and calcium signaling is connected either directly or indirectly to each of these processes; therefore, it has been proposed that cancer is a perversion of some normal calcium-related processes and that calcium is the central control of carcinogenesis (7), in which the calcium influx across different cellular compartments is a key trigger or a regulator of the process (8). In the past decades, a growing number of studies have shown that components of the calcium signaling pathway are remodeled or deregulated in cancer (9,10). For example, as one of transient receptor potential channels, TRPM8 was found to be associated with various types of cancer, such as melanoma and cancers of the pancreas, breasts, colorectum and lungs (11–13).
Cancers differ in the types of calcium channels and pumps that were initially recruited, and several studies have explored the role of the calcium signaling in CM development and progression. For example, Maiques et al. (14) found a significant increase in expression of the T-type channel isoform Cav3.1 in primary and malignant melanoma, compared with normal skin and nevi, and the expression levels of another isoform Cav3.2 were significantly higher in metastatic melanoma than in primary melanoma; furthermore, the store-operated Ca2+ entry was found to contribute to melanoma progression and cell migration (15). However, the role of the calcium signaling pathway in the prognosis of melanoma remains unknown.
Genome-wide association study (GWAS) provides a broad approach to identify genes involved in carcinogenesis and tumor progression. An increasing number of genetic variants, such as single-nucleotide polymorphisms (SNPs), have been found to be associated with CM risk or survival (16), in which the two-step gene-set or pathway analysis has been applied to understanding of the effects of genes and their biological pathways on CM development and progression (17). In this study, we performed a gene-set-based pathway analysis of two existing CM GWAS datasets to assess the associations between genetic variants in the calcium signaling pathway genes and CM-specific survival (CMSS).
Materials and methods
Discovery dataset
We used a published GWAS study from The University of Texas MD Anderson Cancer Center (MDACC) as the discovery dataset (18), in which 858 cases who had both complete questionnaire data and detailed clinical information were included in the final analysis. Genotyping data were obtained from the existing GWAS genotyping data generated by Illumina HumanOmni-Quad_v1_0_B array and made available at the National Center for Biotechnology Information Database of Genotypes and Phenotypes (dbGaP Study Accession: phs000187.v1.p1) (19,20), in which the genome-wide imputation was performed by the MACH software based on the 1000 Genomes project phase I v2 CEU (Northern Europeans from Utah; March 2010 release) (21), the 1000 Genomes project reference: 26432245.
Validation dataset
The significant SNPs obtained from the discovery dataset were further validated by using the Harvard GWAS study that was described previously elsewhere (22), in which 409 non-Hispanic white subjects with survival data were included in the final analysis. The Harvard GWAS genotyping was performed with Illumina HumanHap610 array, and the genome-wide imputation was also performed using the MACH software based on the 1000 Genomes Project CEU (Northern Europeans from Utah) data (phase I v3, March 2012) (23), the 1000 Genomes project publication. PMID: 26432245.
All subjects provided a written informed consent at both MDACC and Brigham and Women’s Hospital that had been approved by the local institutional review boards.
Gene and SNP extraction
For the gene-set pathway to be analyzed, 178 genes were selected from the category of ‘calcium signaling pathway’ in the Molecular Signatures Database (http://software.broadinstitute.org/gsea/msigdb/index.jsp). Because there are no standard statistics established for the sex-specific analysis as females carry two copies of chromosome X and males are hemizygous for this chromosome, 10 genes on X chromosome as well as 1 pseudogene were excluded; the remaining 167 genes located on autosomes were used as the candidate genes (Supplementary Table 1, available at Carcinogenesis Online). We then mapped all the SNPs located within 2 kb up- and downstreams of those selected genes and extracted their summary SNP data from the MDACC GWAS dataset. The quality control of the genotyping data included minor allele frequency ≥ 0.05, genotyping rate ≥ 95% and Hardy–Weinberg equilibrium P value ≥ 1×10−5.
Statistical analysis
CMSS was calculated from the date of diagnosis with CM to the CM-related death or the date of the last follow-up. Adjusted hazards ratios (HRs) from the multivariate Cox proportional hazards regression models were conducted using an additive genetic model for both MDACC and Harvard GWAS datasets with the GenABEL package of R software (version 3.3.3). As a result of the imputation that provided the majority of SNPs to be analyzed, there was a high level of correlations among SNPs used in the final analysis, for which the false-positive report probability method was preferably chosen for the multiple testing correction (24), we assigned a prior probability of 0.10 to detect an HR of 2.0 for an association with variant genotypes or minor alleles of the SNPs with P ≤ 0.05. Only SNPs with a false-positive report probability value ≤ 0.2 were chosen for validation in the Harvard GWAS dataset.
Meta-analysis of SNPs from both discovery and validation datasets was also performed using a fixed-effects model. If the Cochran’s Q test P-value ≤ 0.100 or the heterogeneity statistic (I2) ≥ 25%, a random-effects model was employed. Validated SNPs and clinical variables were then put into the multivariable stepwise Cox model to select the independent SNPs, with both the entry and stay points for the models set to 0.05. We summarized the number of genetic variants to evaluate the combined effect of all independent or representative SNPs on CMSS. Receiver operating characteristic (ROC) curve was used to illustrate the ability of area under the curve (AUC) in predicting CMSS. A time-dependent ROC analysis was also performed with timeROC package of R software to assess the accuracy of genetic variants’ continuing effect over the time.
We also performed in silico functional validation of the significant SNPs to further explore the molecular mechanisms underlying the observed CM-death associations with the genotypes. Specifically, we conducted expression quantitative trait loci (eQTL) analysis with data from The Cancer Genome Atlas database (dbGaP Study Accession: phs000178.v9.p8) (25).
All other analyses were performed with SAS (version 9.3.3; SAS Institute, Cary, NC), if not specified otherwise.
Results
Patient characteristics
The final analyses included 858 patients from the MDACC GWAS study and 409 patients from the Harvard GWAS study (Supplementary Table 2, available at Carcinogenesis Online). Because MDACC patients were from a tertiary care center, the patient population tended to be enriched for late-stage and younger patients, compared with the patients from the general population that was captured by the Harvard cohort studies. The use of cases from a cohort also resulted in fewer clinical variables available in the Harvard GWAS Study. In the MDACC study, ages of the patients at diagnosis were between 17 and 94 years (52.4 ± 14.4 years), with a percentage of 57.8% and 42.2% for men and women, respectively, and patients with stages I/II (82.6%) were more than those with stages III/IV (17.4%), which were also defined as regional/distant metastasis. Univariate analysis showed that age, sex, regional/distant metastasis, Breslow thickness, ulceration and mitotic rate were significantly associated with CMSS. In the Harvard study, however, only age, sex, survival outcome and genotype data were available for analysis; the ages of eligible cases at diagnosis were between 34 to 87 years (61.1 ± 10.8 years), and 82.4% of these patients were older than 50 years; there were more women than men, with a percentage of 66.3% and 33.7%, respectively; and only age was significantly associated with CMSS in the univariate analysis. In comparison with patients from the MDACC study that had a median follow-up time of 81.1 months, patients from the Harvard study had a relatively longer median follow-up time (179.0 months), but the death rates during the follow-up period were similar between the MDACC (95/858, 11.1%) and the Harvard studies (48/409, 11.5%).
Associations between SNPs in the calcium signaling pathway genes and CMSS
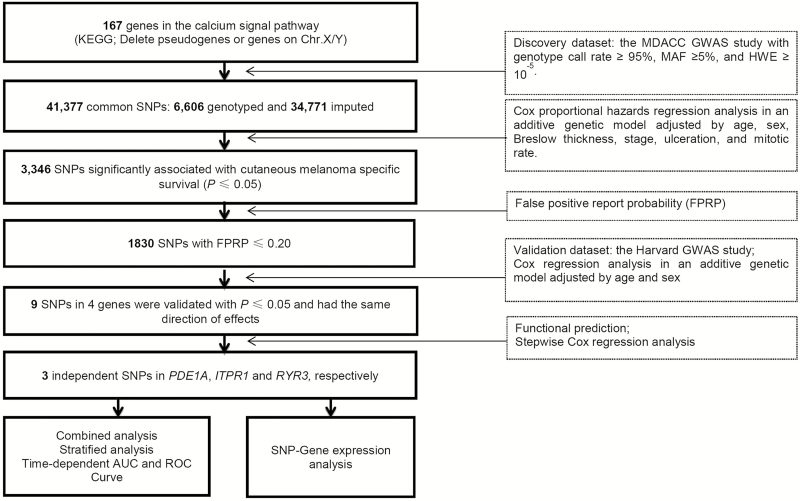
We extracted 41,377 SNPs (6606 genotyped and 34,771 imputed) in the relevant genes with 2 kb flanking regions from the MDACC GWAS dataset, and the study flowchart is presented in Figure 1. The Manhattan plot of associations between these SNPs and CMSS is also presented in Supplementary Figure 1, available at Carcinogenesis Online. In the single-locus analysis of the MDACC discovery dataset, 3346 SNPs were found to be significantly associated with CMSS (P ≤ 0.050), of which 1830 SNPs were worthy of being further explored after the correction by a false-positive report probability ≤ 0.2. The effects of these SNPs on CMSS were then validated in the Harvard GWAS dataset. As a result, nine SNPs in four genes remained significant, of which rs2623439, rs1430157, rs6750552 and rs10931014 in phosphodiesterase 1A (PDE1A) were associated with a poorer survival, whereas rs485412, rs1104370 and rs2841038 in CHRM3, rs6785564 in inositol 1,4,5-trisphosphate receptor type 1 (ITPR1), and rs2596191 in ryanodine receptor 3 (RYR3) were associated with a better survival. Meta-analysis of these nine SNPs from the two GWAS datasets confirmed the same associations (Table 1), and no significant heterogeneity was observed in the effects of these SNPs across the two datasets. Linkage disequilibrium (LD) plots showed that both the three SNPs in CHRM3 and the four SNPs in PDE1A were in LD (Supplementary Figure 2, available at Carcinogenesis Online).
Figure 1.
Study flowchart. AUC, area under the curve; FPRP, false-positive report probability; GWAS, genome-wide association study; HWE, Hardy–Weinberg equilibrium; MAF, minor allele frequency; MDACC, The University of Texas M.D. Anderson Cancer Center; ROC, receiver operating characteristic; SNP, single nucleotide polymorphism.
Table 1.
Meta-analysis of nine validated SNPs using two published melanoma GWAS datasets
| The MDACC Study (n = 858) | The Harvard Study (n = 409) | Meta-analysis | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Allelea | Gene | Chr | EAF | HR (95% CI)b | P b | FDR | FPRP | EAF | HR (95% CI)c | P c | P het | I 2 | HR (95% CI)d | P d |
| rs485412 | T/C | CHRM3 | 1q43 | 0.24 | 0.66 (0.46–0.95) | 0.026 | 0.533 | 0.200 | 0.25 | 0.57 (0.33–0.97) | 0.039 | 0.660 | 0.000 | 0.63 (0.47–0.85) | 2.90 × 10−3 |
| rs1104370 | A/G | CHRM3 | 1q43 | 0.24 | 0.66 (0.46–0.95) | 0.026 | 0.533 | 0.200 | 0.25 | 0.57 (0.33–0.97) | 0.039 | 0.660 | 0.000 | 0.63 (0.47–0.85) | 2.90 × 10−3 |
| rs2841038 | G/T | CHRM3 | 1q43 | 0.24 | 0.65 (0.45–0.94) | 0.024 | 0.513 | 0.187 | 0.26 | 0.56 (0.33–0.96) | 0.036 | 0.650 | 0.000 | 0.62 (0.46–0.84) | 2.10 × 10−3 |
| rs2623439e | A/G | PDE1A | 2q32.1 | 0.30 | 1.44 (1.06–1.95) | 0.020 | 0.479 | 0.154 | 0.28 | 1.51 (1.01–2.28) | 0.047 | 0.860 | 0.000 | 1.46 (1.15–1.87) | 2.22 × 10−3 |
| rs1430157 | C/T | PDE1A | 2q32.1 | 0.31 | 1.41 (1.04–1.90) | 0.026 | 0.530 | 0.188 | 0.29 | 1.61 (1.07–2.41) | 0.016 | 0.610 | 0.000 | 1.48 (1.16–1.88) | 1.52 × 10−3 |
| rs6750552 | T/C | PDE1A | 2q32.1 | 0.33 | 1.47 (1.08–1.98) | 0.013 | 0.446 | 0.106 | 0.30 | 1.62 (1.07–2.45) | 0.021 | 0.710 | 0.000 | 1.52 (1.19–1.94) | 7.21 × 10−4 |
| rs10931014 | T/C | PDE1A | 2q32.1 | 0.33 | 1.39 (1.04–1.87) | 0.027 | 0.535 | 0.200 | 0.30 | 1.65 (1.09–2.49) | 0.018 | 0.510 | 0.000 | 1.47 (1.16–1.87) | 1.59 × 10−3 |
| rs6785564 | A/G | ITPR1 | 3p26.1 | 0.14 | 0.51 (0.32–0.81) | 0.004 | 0.382 | 0.062 | 0.14 | 0.44 (0.20–0.96) | 0.040 | 0.750 | 0.000 | 0.49 (0.33–0.73) | 3.94 × 10−4 |
| rs2596191 | C/A | RYR3 | 15q13.3-q14 | 0.44 | 0.69 (0.51–0.93) | 0.016 | 0.460 | 0.130 | 0.42 | 0.64 (0.41–0.98) | 0.039 | 0.780 | 0.000 | 0.67 (0.53–0.86) | 1.71 × 10−3 |
EAF, effect allele frequency; FDR: false discovery rate; FPRP: false-positive report probability; Phet, P value for heterogeneity by Cochrane’s Q test.
aReference allele/effect allele.
bAdjusted for age, sex, Breslow thickness, distant/regional metastasis, ulceration and mitotic rate in the MDACC study.
cAdjusted for age and sex in the Harvard study.
dMeta-analysis in a fix-effects model.
eSNP genotyped, or else SNP imputed.
Genetic variants in the calcium signaling pathway genes as independent survival predictors
To identify independent genetic predictors of CMSS, the nine validated SNPs together with selected clinical variables from the MDACC study were all included in a multivariable stepwise Cox regression model. As a result, age, metastasis, Breslow thickness, ulceration, mitotic rate and three SNPs (i.e. PDE1A rs6750552, ITPR1 rs6785564 and RYR3 rs2596191), but not sex, were significantly and independently associated with CMSS (Table 2). Therefore, we selected these three SNPs as independent and representative SNPs for further analyses.
Table 2.
Predictors of CMSS obtained from stepwise Cox regression analysis in the MDACC study
| Parametera | Categoryb | Frequency | HR (95% CI) | P |
|---|---|---|---|---|
| Age | ≤50/>50 | 371/487 | 1.02 (1.01–1.04) | 0.0090 |
| Sex | Female/Male | 362/496 | 1.55 (0.97–2.47) | 0.0677 |
| Regional/distant metastasis | No/Yes | 709/149 | 4.52 (2.92–6.99) | <0.0001 |
| Breslow thickness (mm) | ≤1/>1 | 347/511 | 1.16 (1.10–1.22) | <0.0001 |
| Ulceration | No/Yes | 681/155 | 2.79 (1.81–4.29) | <0.0001 |
| Mitotic rate (mm2) | ≤1/>1 | 275/583 | 2.53 (1.21–5.29) | 0.0137 |
| PDE1A rs6750552 T>C | TT/TC/CC | 388/376/94 | 1.48 (1.10–1.99) | 0.0104 |
| ITPR1 rs6785564 A>G | AA/AG/GG | 636/205/17 | 0.51 (0.32–0.80) | 0.0038 |
| RYR3 rs2596191 C>A | CC/CA/AA | 271/411/176 | 0.69 (0.51–0.93) | 0.0166 |
aStepwise analysis included age, sex, regional/distant metastasis, Breslow thickness, ulceration, mitotic rate and nine SNPs in four genes (rs485412, rs1104370, rs2841038 in CHRM3; rs6785564 in ITPR1; rs2596191 in RYR3 and rs2623439, rs1430157, rs6750552, rs10931014 in PDE1A).
bThe leftmost was used as the reference.
As shown in Table 3, in the MDACC study, the risk effect of PDE1A rs6750552 C allele as well as protective effects of ITPR1 rs6785564 G and RYR3 rs2596191 A alleles on CM survival were statistically significant (trend test: P = 0.013, 0.004 and 0.016, respectively), and similar results were observed in the Harvard study (trend test: P = 0.022, 0.040 and 0.039, respectively). When we combined MDACC and Harvard datasets into one dataset (n = 1267), consistent results were observed (trend test: P = 0.007, 0.019, and 0.003, respectively). For illustrative purposes, each SNP in its gene with 20 kb flanking region is shown in a regional association plot (Supplementary Figure 3, available at Carcinogenesis Online).
Table 3.
Associations between three independent SNPs and CMSS of patients in the MDACC study, the Harvard study and the combined dataset
| The MDACC Study (n = 858) | The Harvard Study (n = 409) | MDACC + Harvard (n = 1267) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | All | Death (%) | HR (95% CI)a | P a | All | Death (%) | HR (95% CI)b | P b | All | Death (%) | HR (95% CI)c | P c |
| PDE1A rs6750552 T>C | ||||||||||||
| TT | 388 | 37 (9.54) | 1.00 | 201 | 15 (7.46) | 1.00 | 589 | 52 (8.83) | 1.00 | |||
| TC | 376 | 45 (11.97) | 1.56 (0.99–2.46) | 0.053 | 169 | 28 (16.57) | 2.30 (1.23–4.30) | 0.010 | 545 | 73 (13.39) | 1.59 (1.12–2.27) | 0.010 |
| CC | 94 | 12 (13.83) | 2.06 (1.08–3.94) | 0.029 | 39 | 5 (12.82) | 2.03 (0.74–5.61) | 0.171 | 133 | 18 (13.53) | 1.74 (1.02–2.98) | 0.043 |
| Trend test | 0.013 | 0.022 | 0.007 | |||||||||
| TC+CC | 470 | 58 (12.34) | 1.65 (1.07–2.54) | 0.023 | 208 | 33 (15.87) | 2.25 (1.22–4.15) | 0.009 | 678 | 91 (13.42) | 1.62 (1.15–2.28) | 0.006 |
| ITPR1 rs6785564 A>G | ||||||||||||
| AA | 636 | 75 (11.79) | 1.00 | 299 | 41 (13.71) | 1.00 | 935 | 116 (12.41) | 1.00 | |||
| AG | 205 | 19 (9.27) | 0.58 (0.34–0.99) | 0.047 | 102 | 7 (6.86) | 0.47 (0.21–1.05) | 0.067 | 307 | 26 (8.47) | 0.65 (0.43–1.00) | 0.047 |
| GG | 17 | 1 (5.88) | 0.13 (0.02–0.98) | 0.048 | 8 | 0 (0.0) | — | — | 25 | 1 (4.00) | 0.28 (0.04–1.97) | 0.200 |
| Trend test | 0.004 | 0.040 | 0.019 | |||||||||
| AG+GG | 222 | 20 (9.01) | 0.50 (0.29–0.84) | 0.009 | 110 | 7 (6.36) | 0.44 (0.20–0.99) | 0.046 | 332 | 27 (8.13) | 0.62 (0.41–0.94) | 0.025 |
| RYR3 rs2596191 C>A | ||||||||||||
| CC | 271 | 38 (14.02) | 1.00 | 133 | 22 (16.54) | 1.00 | 404 | 60 (14.85) | 1.00 | |||
| CA | 411 | 45 (10.95) | 0.76 (0.48–1.19) | 0.227 | 205 | 21 (10.24) | 0.64 (0.35–1.16) | 0.137 | 616 | 66 (10.71) | 0.73 (0.51–1.03) | 0.071 |
| AA | 176 | 12 (6.82) | 0.44 (0.23–0.87) | 0.019 | 71 | 5 (7.04) | 0.41 (0.15–1.08) | 0.070 | 247 | 17 (6.88) | 0.46 (0.27–0.79) | 0.005 |
| Trend test | 0.016 | 0.039 | 0.003 | |||||||||
| CA+AA | 587 | 57 (9.71) | 0.66 (0.43–1.01) | 0.054 | 276 | 26 (9.42) | 0.57 (0.33–1.01) | 0.055 | 863 | 83 (9.62) | 0.65 (0.46–0.90) | 0.010 |
| Number of protective genotypesd | ||||||||||||
| 0 | 108 | 20 (18.52) | 1.00 | 51 | 16 (31.37) | 1.00 | 159 | 36 (22.64) | 1.00 | |||
| 1 | 367 | 41 (11.17) | 0.58 (0.34–1.00) | 0.050 | 173 | 18 (10.40) | 0.30 (0.15–0.58) | 0.0004 | 540 | 59 (10.93) | 0.44 (0.29–0.67) | 0.0001 |
| 2 | 319 | 29 (9.09) | 0.37 (0.21–0.68) | 0.001 | 141 | 12 (8.51) | 0.24 (0.11–0.51) | 0.0002 | 460 | 41 (8.91) | 0.37 (0.24–0.58) | <0.0001 |
| 3 | 64 | 5 (7.81) | 0.15 (0.05–0.52) | 0.003 | 44 | 2 (4.55) | 0.13 (0.03–0.56) | 0.006 | 108 | 7 (6.48) | 0.23 (0.10–0.52) | 0.0004 |
| Trend test | <0.001 | 0.0002 | <0.0001 | |||||||||
| 0–1 | 475 | 61 (12.84) | 1.00 | 224 | 34 (15.18) | 1.00 | 699 | 95 (13.59) | 1.00 | |||
| 2–3 | 383 | 34 (8.88) | 0.49 (0.31–0.76) | 0.002 | 185 | 14 (7.57) | 0.48 (0.26–0.90) | 0.021 | 568 | 48 (8.45) | 0.61 (0.43–0.86) | 0.0049 |
aAdjusted for age, sex, Breslow thickness, distant/regional metastasis, ulceration and mitotic rate in Cox models of SNPs and CMSS in the MDACC study.
bAdjusted for age and sex in the Harvard study.
cAdjusted for age and sex in MDACC and Harvard combined dataset.
dProtective genotypes were rs6750552 TT, rs6785564 AG+GG, rs2596191 CA+AA.
Combined analyses of the three independent and representative SNPs
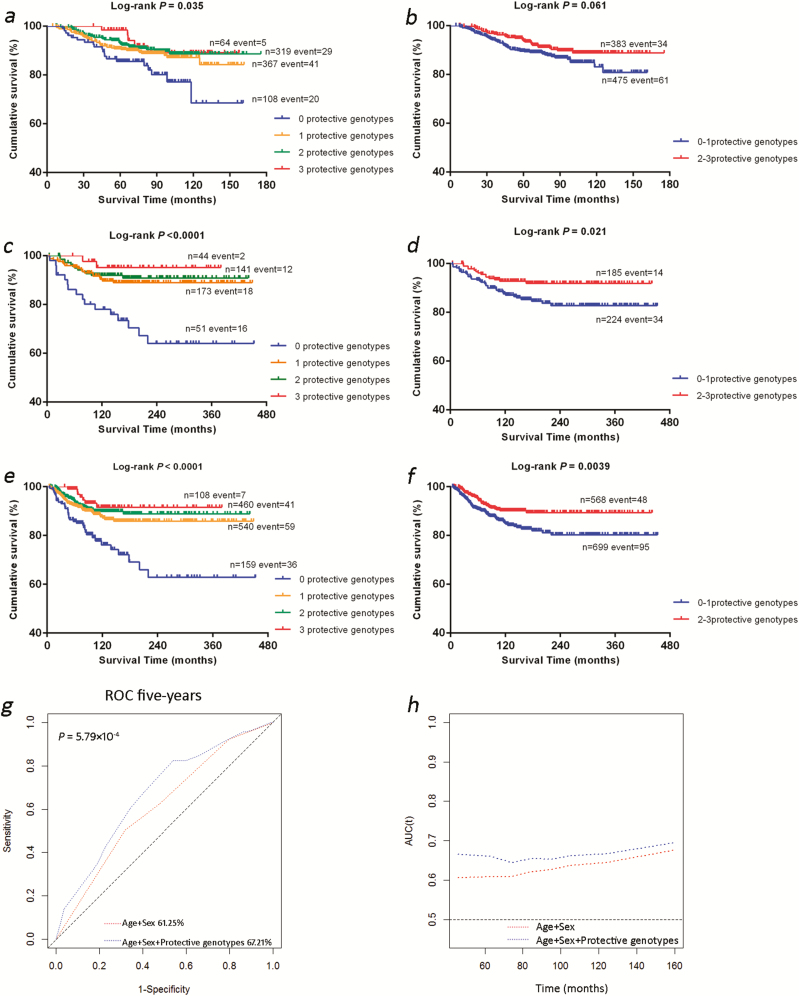
To better interpret the joint effect of the three independent and representative SNPs on risk of death, we combined protective genotypes of rs6750552 TT, rs6785564 AG+GG and rs2596191 CA+AA into one variable of the number of protective genotypes as a genetic score. As shown in Table 3, the trend test demonstrated that an increased genetic score was associated with an improved survival in the MDACC study (P < 0.001), the Harvard study (P = 0.0002) and the combined dataset (P < 0.0001). Compared with those who had no protective genotypes, patients with three protective genotypes had the best survival [MDACC: HR = 0.15, 95% confidence interval (CI) = 0.05–0.52 and P = 0.003; Harvard: HR = 0.13, 95% CI = 0.03–0.56 and P = 0.0063; the combined dataset: HR = 0.23, 95% CI = 0.10–0.52 and P = 0.0004]. Next, we dichotomized all patients into a group with 0–1 protective genotypes and a group with 2–3 protective genotypes, and compared with the former group, the latter group had a significantly better survival (MDACC: HR = 0.49, 95% CI = 0.31–0.76 and P = 0.002; Harvard: HR = 0.48, 95% CI = 0.26–0.90 and P = 0.021; combined dataset: HR = 0.61, 95% CI = 0.43–0.86 and P = 0.0049). Finally, we used Kaplan–Meier curves to visualize associations between the number of protective genotypes and CMSS (Figure 2a–f).
Figure 2.
The independent SNPs and CMSS. (a–f) Kaplan–Meier survival curves of the protective genotypes: the exact numbers of protective genotypes in the (a) MDACC study, (c) Harvard study and (e) combination of these two datasets; dichotomized groups of protective genotypes in the (b) MDACC study, (d) Harvard study and (f) combined dataset. (g and h) Receiver operating characteristic (ROC) curve and time-dependent area under the curve (AUC) estimation for prediction of melanoma-specific survival in the combined dataset. (g) Five-year melanoma-specific survival prediction by ROC curve; (h) time-dependent AUC estimation based on age, sex and the protective genotypes of the three genes.
Stratified analyses for the combined protective genotypes on CMSS
We further conducted stratified analyses to assess whether the combined effect of protective genotypes on CMSS was modified by clinicopathologic variables. In the MDACC study, patients with 2–3 protective genotypes, compared with those with 0–1 protective genotypes, had a significantly reduced risk of CM death in the subgroups of age >50 years, male, patients with regional/distant metastasis, and patients with Breslow thickness >1 mm. The difference was also obvious between subgroups of patients with and without ulceration and mitotic rate ≤1mm2 and >1 mm2. In the Harvard study, although only age and sex were available for the analysis, a similar trend was observed in the subgroup of age >50 years. However, no heterogeneity was observed among all the subgroups of the two studies (Supplementary Table 3, available at Carcinogenesis Online).
In silico functional validation
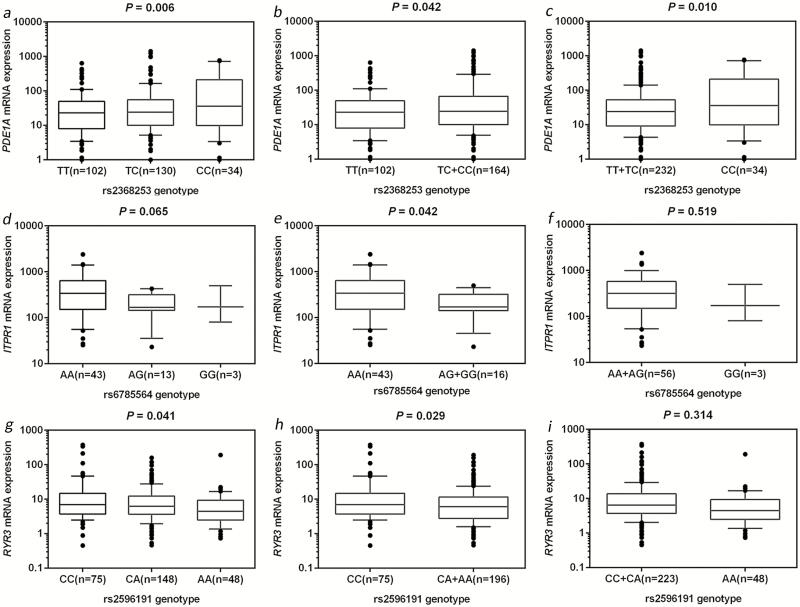
The eQTL analysis with data from the The Cancer Genome Atlas database was performed in two groups: primary and metastatic CM tissues. As genotyping data for PDE1A rs6750552 were not available in the The Cancer Genome Atlas database, we chose PDE1A rs2368253 that is in a high LD with rs6750552 (r2 = 0.84) as an alternative SNP. As shown in Figure 3a–c, the minor rs2368253 C allele had a significant correlation with an increased mRNA expression level of PDE1A in metastatic CM tissue (P value was 0.006, 0.042 and 0.010 in additive, dominant and recessive models, respectively), but no significant difference was observed in primary CM tissue (Supplementary Figure 4a–c, available at Carcinogenesis Online). We also found a significant correlation between the minor ITPR1 rs6785564 G allele and a decreased mRNA expression level of ITPR1 in primary CM tumor tissue in a dominant model (P = 0.042; Figure 3e), but not in other models nor in metastatic CM tissue (Figure 3d and f; Supplementary Figure 4d–f, available at Carcinogenesis Online). A significant correlation between the minor RYR3 rs2596191 A allele and a decreased mRNA expression level of RYR3 was also noticed in metastatic CM tissue in both additive and dominant models (P = 0.041 and 0.029, respectively; Figure 3g and h), but not in primary CM tissue (Supplementary Figure 4g–i, available at Carcinogenesis Online). These results suggest that PDE1A rs6750552 C, ITPR1 rs6785564 G and RYR3 rs2596191 A alleles have an independent effect on their gene expression at the transcription level, which are consistent with their effects on survival of CM patients.
Figure 3.
The eQTLs analysis from The Cancer Genome Atlas database. Correlation between PDE1A mRNA expression and rs2368253 genotypes in metastatic cutaneous melanoma tumor tissue in the (a) additive model, (b) dominant model, (c) recessive model. Correlation between ITPR1 mRNA expression and rs6785564 genotype in primary cutaneous melanoma tumor tissue in the (d) additive model, (e) dominant model and (f) recessive model. Correlation between RYR3 mRNA expression and rs2596191 genotype in metastatic cutaneous melanoma tumor tissue in the (g) additive model, (h) dominant model and (i) recessive model.
We furthermore explored potential functions of these SNPs by using data from the ENCODE Project. PDE1A rs6750552 SNP is located in a DNase I hypersensitive site, and RYR3 rs2596191 SNP is located at the intron region with considerable levels of the H3K4Me1 enrichment, but nothing was found for ITPR1 rs6785564; however, ITPR1 rs7642352, which is in a high LD with rs6785564 (r2 = 0.87), is located in a DNase I hypersensitive site with considerable levels of the H3K4Me1 enrichment (Supplementary Figure 5a–c, available at Carcinogenesis Online).
The ROC curve and time-dependent AUC for CMSS prediction
We further assessed prediction effect of the genotypes of PDE1A rs6750552 C, ITPR1 rs6785564 G and RYR3 rs2596191 A in the same model with age and sex by using the ROC curve and time-dependent AUC in the combined MDACC and Harvard dataset. From the ROC curve, we observed a significant improvement for these protective genotypes in combination with age and sex in prediction performance of the 5-year CMSS, compared with the model with age and sex only (AUC = 61.25–67.21%, P = 5.79×10−4), and the time-dependent AUC curve showed this significant effect continuously through the entire follow-up period (Figure 2g–h).
Discussion
In recent years, it has been recognized that the alternations in the calcium signaling are involved in carcinogenesis and tumor progression. The core components of the calcium signaling system are referred as the ‘calcium toolkit’ (10), and remodeling or deregulation of the calcium signaling pathway, as a cause or consequence of different cancer-related proteins with altered functions, is particularly relevant to the hallmarks of cancer cells (26). Therefore, the key calcium signaling molecules are likely to be promising biomarkers for cancer development and prognosis, even a novel and prospective target for cancer treatment (27,28). However, few studies have investigated the roles of genetic variants in calcium signaling pathway genes in predicting the survival of CM patients. In this study of 167 genes involved in the calcium signaling pathway, we showed that PDE1A rs6750552, ITPR1 rs6785564 and RYR3 rs2596191 were independently or jointly associated the survival of CM patients, suggesting that these genetic variants may be promising prognostic predictors of CM. Therefore, this study highlights the possible role of the calcium signaling pathway in CM progression.
PDE1A, located on chromosome 2q32.1, encodes a member of Ca2+/calmodulin-dependent cyclic nucleotide phosphodiesterase (PDE1), which is one of the key enzymes involved in the complex interactions between the cyclic nucleotide and Ca2+ second messenger systems (29,30). Although associations between the PDE genes and genetic diseases have been investigated for several years, specific members of the PDE family have recently been implicated in carcinogenesis and tumor progression (31,32). For example, one study found a high PDE1A mRNA expression in several malignant tumor cells, including human oral melanoma cell lines (33). Another study showed that inhibition of selective PDE isoforms induced apoptosis and cell cycle arrest of tumor cells and regulated the tumor microenvironment (34). In this study, the rs6750552 C variant genotypes were found to be associated with a decreased CMSS. From the eQTL analyses results of another SNP rs2368253 in a high LD with rs6750552, we inferred that the variant C genotypes were also associated with an increased PDE1A mRNA expression level in metastatic CM tissue. In addition, according to the ENCODE Project data, the rs6750552 SNP is located in a DNase I hypersensitive site; therefore, it is likely that this SNP could affect PDE1A expression by modifying the accessibility of chromatin during transcription, further influencing the gene function. More recently, curcumin, a new candidate for melanoma therapy, has been reported to play an antiproliferative effect on melanoma cells by inhibiting PDE1A, and it was also reported that PDE1A expression was positively correlated with UHRF1 (ubiquitin-like containing PHD and Ring Finger domains 1) expression, which may be a key factor in DNA methylation and histone modification implicated in cell cycle progression (35). Meanwhile, another recent study demonstrated that the elevated expression of UHRF1 played an important role in melanoma cell proliferation and progression, clinically related to high TNM classification and Breslow’s thickness, and that high UHRF1 was positively associated with a shorter overall survival of melanoma patients (36). These may partly explain the potential biology and molecular mechanism of PDE1A underlying the observed association.
ITPR1, located on chromosome 3p26.1, encodes an intracellular receptor for inositol 1,4,5-trisphosphate that is a ligand-gated calcium channel, which modulates intracellular calcium signaling following stimulation by inositol 1,4,5-trisphosphate and mediates calcium release from the endoplasmic reticulum (37). Results from Riker Melanoma in the cancer microarray database (Oncomine) showed a higher expression level of ITPR1 in CM tissue than in normal skin tissues (38), suggesting an oncogenic role of the gene. Recent evidence shows that ITPRs play a crucial role in the regulation of autophagy (39,40), which is involved in regulating the NK-mediated immune response in many tumor cells; for example, ITPR1 was recognized as an autophagy sensor, the overexpression of ITPR1 impaired NK cell-mediated antitumor immune response in clear renal cell carcinoma (41). These results suggest that inhibiting ITPR1/autophagy in tumors may improve their elimination by NK cells in vivo. In this study, the rs6785564 G allele was consistently found to be associated with a decreased mRNA expression level of ITPR1 in primary CM tissue and a better survival of CM patients. Also from the ENCODE Project data, rs7642352 that is in a high LD with rs6785564 is located in a DNase I hypersensitive site with H3K4Me1 enrichment; thus, SNPs in this gene region probably influence the gene function, probably by mediating gene expression at the mRNA transcription level. Taken together, these may explain the possible mechanisms underlying the association between ITPR1 rs6785564 and CMSS.
RYR3, located on chromosome 15q13.3-q14, encodes the third isoform of the RYR family. RYR3 is a Ca2+-induced Ca2+ release channel protein located in the sarcoplasmic reticulum, and it plays a key role in controlling cytosolic calcium levels. Previous studies demonstrated that a genetic variant, rs1044129 A→G, which was present in the microRNA-367 binding site in the 3′untranslated region of RYR3, had an effect on breast cancer progression-free survival, and it was also reported to be relevant to relapse-free survival in Korean patients with resected colonic cancer and postoperative survival in Chinese patients with hepatocellular carcinoma (42–44). In this study, patients with the rs2596191 A allele had a better survival, and eQTL analyses of this genotype showed a correlation with a decreased mRNA expression level of RYR3. In addition, the rs2596191 SNP is located at the intron region with considerable levels of the H3K4Me1 enrichment, according to the ENCODE Project data, which is likely associated with enhancers and transcription starts, and thus SNPs in this region may act as enhancers to affect gene expression by modifying the transcriptional activities. Unfortunately, there is lack of studies of the molecular mechanisms underlying altered RYR3 mRNA expression on CMSS; therefore, further functional investigation is needed.
Limitations of this study should be noticed. First, clinical variables of the discovery and validation datasets were not matched, and only age and sex were available from the Harvard cohort studies; however, no heterogeneity was observed in their comparisons or combined analysis. Second, some valuable clinical information, such as performance status and treatment, were also absent in both studies; thus, we were restricted from an extended CM survival analysis. Third, the exact biological mechanisms about how those variants affect DNA methylation or mRNA expression remains unclear. Therefore, the results of this study should be considered preliminary, and it is necessary to replicate the results in studies with other larger and independent populations with different races or geographic regions, in which functional experiments should also be conducted for further exploration. Once validated, these genetic variants may help us to identify key calcium toolkit molecules that may lead to the development of novel and prospective biomarkers for CM prognosis.
Funding
The MD Anderson Study was supported by National Institutes of Health (NIH)/National Cancer Institute (NCI) (R01 CA100264, 2P50CA093459 and R01CA133996) as well as by The University of Texas MD Anderson Cancer Center Various Donors Melanoma and Skin Cancers Priority Program Fund; the Miriam and Jim Mulva Research Fund; the McCarthy Skin Cancer Research Fund and the Marit Peterson Fund for Melanoma Research. The Harvard Study was in part supported by (NIH/NCI) (R01 CA49449, P01 CA87969, UM1 CA186107 and UM1 CA167552). Q.W. was supported by the start-up funds from Duke Cancer Institute, Duke University Medical Center, and also in part supported by the Duke Cancer Institute as part of the P30 Cancer Center Support Grant (NIH/NCI CA014236). X.W. was supported by the Scholarship from The First Hospital of Jilin University, Changchun, China.
Conflict of Interest Statement:
None declared.
Supplementary Material
Acknowledgements
We thank all of the investigators and funding agencies that enabled the deposition of data in dbGaP that we used in this study (dbGaP Study Accession: phs000187.v1.p1). We also thank the John Hopkins University Center for Inherited Disease Research for conducting high-throughput genotyping for this study. We thank all the participants and staff of the Nurses’ Health Study (NHS) and Health Professionals Follow-up Study (HPFS) for their valuable contributions. The eQTL results published here are in whole or part based upon data generated by The Cancer Genome Atlas pilot project established by the NCI and National Human Genome Research Institute (NHGRI) (dbGaP Study Accession: phs000178.v9.p8). The authors assume full responsibility for analyses and interpretation of these data.
Glossary
Abbreviations
- AUC
area under the curve
- CI
confidence interval
- CM
cutaneous melanoma
- CMSS
cutaneous melanoma-specific survival
- eQTL
expression quantitative trait loci
- GWAS
genome-wide association study
- HR
hazards ratio
- ITPR1
inositol 1,4,5-trisphosphate receptor type 1
- LD
linkage disequilibrium
- MDACC
The University of Texas MD Anderson Cancer Center
- PDE1A
phosphodiesterase 1A
- ROC
receiver operating characteristic
- RYR3
ryanodine receptor 3
- SNP
single-nucleotide polymorphisms
References
- 1. Siegel R.L., et al. (2018) Cancer statistics, 2018. CA. Cancer J. Clin., 68, 7–30. [DOI] [PubMed] [Google Scholar]
- 2. American Cancer Society. (2018) Cancer Facts & Figures 2018 American Cancer Society, Atlanta, GA: http://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-statistics/annual-cancer-facts-and-figures/2018/cancer-facts-and-figures-2018.pdf. (19 June 2018, date last accessed). [Google Scholar]
- 3. Clapham D.E. (2007) Calcium signaling. Cell, 131, 1047–1058. [DOI] [PubMed] [Google Scholar]
- 4. Humeau J., et al. (2018) Calcium signaling and cell cycle: progression or death. Cell Calcium, 70, 3–15. [DOI] [PubMed] [Google Scholar]
- 5. Berridge M.J., et al. (1998) Calcium–a life and death signal. Nature, 395, 645–648. [DOI] [PubMed] [Google Scholar]
- 6. Hanahan D., et al. (2000) The hallmarks of cancer. Cell, 100, 57–70. [DOI] [PubMed] [Google Scholar]
- 7. Kadio B., et al. (2016) Calcium role in human carcinogenesis: a comprehensive analysis and critical review of literature. Cancer Metastasis Rev., 35, 391–411. [DOI] [PubMed] [Google Scholar]
- 8. Monteith G.R., et al. (2012) Calcium channels and pumps in cancer: changes and consequences. J. Biol. Chem., 287, 31666–31673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Stewart T.A., et al. (2015) Altered calcium signaling in cancer cells. Biochim. Biophys. Acta., 1848(Pt B), 2502–2511. [DOI] [PubMed] [Google Scholar]
- 10. Prevarskaya N., et al. (2014) Remodelling of Ca2+ transport in cancer: how it contributes to cancer hallmarks? Philos. Trans. R. Soc. Lond. B. Biol. Sci., 369, 20130097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tsavaler L., et al. (2001) Trp-p8, a novel prostate-specific gene, is up-regulated in prostate cancer and other malignancies and shares high homology with transient receptor potential calcium channel proteins. Cancer Res., 61, 3760–3769. [PubMed] [Google Scholar]
- 12. Liu Z., et al. (2016) TRPM8: a potential target for cancer treatment. J. Cancer Res. Clin. Oncol., 142, 1871–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yee N.S. (2015) Roles of TRPM8 ion channels in cancer: proliferation, survival, and invasion. Cancers (Basel)., 7, 2134–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Maiques O., et al. (2017) Immunohistochemical analysis of T-type calcium channels in acquired melanocytic naevi and melanoma. Br. J. Dermatol., 176, 1247–1258. [DOI] [PubMed] [Google Scholar]
- 15. Umemura M., et al. (2014) Store-operated Ca2+ entry (SOCE) regulates melanoma proliferation and cell migration. PLoS One, 9, e89292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Law M.H., et al. (2015) Genome-wide meta-analysis identifies five new susceptibility loci for cutaneous malignant melanoma. Nat. Genet., 47, 987–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yuan H., et al. (2015) Genetic variants in Hippo pathway genes YAP1, TEAD1 and TEAD4 are associated with melanoma-specific survival. Int. J. Cancer, 137, 638–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Amos C.I., et al. ; GenoMEL Investigators; Q-Mega Investigators; AMFS Investigators (2011) Genome-wide association study identifies novel loci predisposing to cutaneous melanoma. Hum. Mol. Genet., 20, 5012–5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mailman M.D., et al. (2007) The NCBI dbGaP database of genotypes and phenotypes. Nat. Genet., 39, 1181–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tryka K.A., et al. (2014) NCBI’s database of genotypes and phenotypes: dbGaP. Nucleic Acids Res., 42(Database issue), D975–D979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li Y., et al. (2010) MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol., 34, 816–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Song F., et al. (2012) Exonuclease 1 (EXO1) gene variation and melanoma risk. DNA Repair (Amst)., 11, 304–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lappalainen T., et al. ; Geuvadis Consortium (2013) Transcriptome and genome sequencing uncovers functional variation in humans. Nature, 501, 506–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wacholder S., et al. (2004) Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J. Natl. Cancer Inst., 96, 434–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Network T.C.G.A. (2015) Genomic classification of cutaneous melanoma. Cell, 161, 1681–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Roderick H.L., et al. (2008) Ca2+ signalling checkpoints in cancer: remodelling Ca2+ for cancer cell proliferation and survival. Nat. Rev. Cancer, 8, 361–375. [DOI] [PubMed] [Google Scholar]
- 27. Cui C., et al. (2017) Targeting calcium signaling in cancer therapy. Acta Pharm. Sin. B, 7, 3–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Prevarskaya N., et al. (2013) Targeting Ca²⁺ transport in cancer: close reality or long perspective? Expert Opin. Ther. Targets, 17, 225–241. [DOI] [PubMed] [Google Scholar]
- 29. Kakkar R., et al. (1999) Calmodulin-dependent cyclic nucleotide phosphodiesterase (PDE1). Cell. Mol. Life Sci., 55, 1164–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Michibata H., et al. (2001) Human Ca2+/calmodulin-dependent phosphodiesterase PDE1A: novel splice variants, their specific expression, genomic organization, and chromosomal localization. Biochim. Biophys. Acta., 1517, 278–287. [DOI] [PubMed] [Google Scholar]
- 31. Levy I., et al. (2011) Phosphodiesterase function and endocrine cells: links to human disease and roles in tumor development and treatment. Curr. Opin. Pharmacol., 11, 689–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Keravis T., et al. (2012) Cyclic nucleotide phosphodiesterase (PDE) isozymes as targets of the intracellular signalling network: benefits of PDE inhibitors in various diseases and perspectives for future therapeutic developments. Br. J. Pharmacol., 165, 1288–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shimizu K., et al. (2005) Calmodulin-dependent phosphodiesterase (PDE) 1 in human oral melanoma cell lines. Int. J. Oral Surg., 34, 4. [Google Scholar]
- 34. Savai R., et al. (2010) Targeting cancer with phosphodiesterase inhibitors. Expert Opin. Investig. Drugs, 19, 117–131. [DOI] [PubMed] [Google Scholar]
- 35. Abusnina A., et al. (2011) Anti-proliferative effect of curcumin on melanoma cells is mediated by PDE1A inhibition that regulates the epigenetic integrator UHRF1. Mol. Nutr. Food Res., 55, 1677–1689. [DOI] [PubMed] [Google Scholar]
- 36. Wei C., et al. (2018) Upregulation of UHRF1 promotes the progression of melanoma by inducing cell proliferation. Oncol. Rep., 39, 2553–2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Foskett J.K., et al. (2007) Inositol trisphosphate receptor Ca2+ release channels. Physiol. Rev., 87, 593–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Riker A.I., et al. (2008) The gene expression profiles of primary and metastatic melanoma yields a transition point of tumor progression and metastasis. BMC Med. Genomics, 1, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ivanova H., et al. (2014) Inositol 1,4,5-trisphosphate receptor-isoform diversity in cell death and survival. Biochim. Biophys. Acta., 1843, 2164–2183. [DOI] [PubMed] [Google Scholar]
- 40. Decuypere J.P., et al. (2015) ITPRs/inositol 1,4,5-trisphosphate receptors in autophagy: from enemy to ally. Autophagy, 11, 1944–1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Messai Y., et al. (2014) ITPR1 protects renal cancer cells against natural killer cells by inducing autophagy. Cancer Res., 74, 6820–6832. [DOI] [PubMed] [Google Scholar]
- 42. Zhang L., et al. (2011) Functional SNP in the microRNA-367 binding site in the 3’UTR of the calcium channel ryanodine receptor gene 3 (RYR3) affects breast cancer risk and calcification. Proc. Natl. Acad. Sci. USA, 108, 13653–13658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chae Y.S., et al. (2013) Functional polymorphism in the MicroRNA-367 binding site as a prognostic factor for colonic cancer. Anticancer Res., 33, 513–519. [PubMed] [Google Scholar]
- 44. Peng C., et al. (2015) A polymorphism at the microRNA binding site in the 3’ untranslated region of RYR3 is associated with outcome in hepatocellular carcinoma. Onco. Targets. Ther., 8, 2075–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.