Abstract
Since wild-type p53 is central for maintaining genomic stability and preventing oncogenesis, its coding gene TP53 is highly mutated in ~50% of human cancers, and its activity is almost abrogated in the rest of cancers. Approximately 80% of p53 mutations are single point mutations with several hotspot mutations. Besides loss of function and dominant-negative effect on the wild-type p53 activity, the hotspot p53 mutants also acquire new oncogenic functions, so-called ‘gain-of-functions’ (GOF). Because the GOF of mutant p53 is highly associated with late-stage malignance and drug resistance, these p53 mutants have become hot targets for developing novel cancer therapies. In this essay, we review some recent progresses in better understanding of the role of mutant p53 GOF in chemoresistance and the underlying mechanisms, and discuss the pros and cons of targeting mutant p53 for the development of anti-cancer therapies.
Keywords: mutant p53, gain-of-function, cancer therapy, chemoresistance, synthetic lethality
Introduction
The tumor suppressor p53 guards the genome and prevents tumorigenesis by promoting cell growth arrest, DNA repair, senescence, and cell death, as well as modulating autophagy and cancer metabolism (Levine and Oren, 2009; Kastenhuber and Lowe, 2017). However, to boost their own survival and growth, cancer cells exploit multiple tactics to disarm p53. The most straight and effective way to inactivate p53 is to mutate the p53-encoding gene TP53 (Freed-Pastor and Prives, 2012; Muller and Vousden, 2014). Indeed, TP53 is the most frequently mutated gene in human cancers. The cancer-derived p53 mutants include missense, frameshift, truncation, and deletion mutations, most of which are missense mutations (~74%), a single substitution of the original amino acid with a different one. Among the missense mutations, ~80% of them occur in the p53 DNA-binding domain (DBD), and remarkably, several hotspot mutations have been identified in this domain, such as Arg-175, Tyr-220, Gly-245, Arg-248, Arg-249, Arg-273, and Arg-282. Generally speaking, p53 mutants can be grouped into two categories, DNA-contact mutants that replace the amino acids critical for DNA binding, such as Arg-248-Gln (R248Q), Arg-273-His (R273H), and Arg-282-Trp (R282W), and conformational mutants that cause unfolded structure or altered conformation, such as Arg-175-His (R175H), Tyr-220-Cys (Y220C), Gly-245-Ser (G245S), and Arg-249-Ser (R249S). While the vast majority of p53 mutants lose the wild-type function or exert a ‘dominant-negative’ effect on the wild-type allele, some of them, such as R248Q, R273H, R175H, and R249S, have been shown to acquire ‘gain-of-functions’ (GOF) that further promote cancer malignance and confer chemoresistance (Figure 1). This essay is thus composed to review the recent progresses in better understanding of the role of mutant p53 in cancer development and drug resistance and to discuss about the potential of developing anti-cancer treatment by targeting these hotspot p53 mutants. For further reading, readers are referred to several recent reviews on the related subjects (Freed-Pastor and Prives, 2012; Muller and Vousden, 2014; Bykov et al., 2018; Kim and Lozano, 2018; Sabapathy and Lane, 2018).
Figure 1.
p53 mutants promote cancer development and therapeutic resistance through their GOF.
Gain-of-functions of mutant p53
The wild-type p53 forms a homotetramer to act as a nuclear transcription factor that regulates the expression of a myriad of genes in response to various stressors. Although it can also interact with cytosolic proteins, e.g. BCL family proteins, leading to mitochondrial outer membrane permeabilization (MOMP) and augmented apoptosis (Riley et al., 2008; Green and Kroemer, 2009), it is mostly through its transcriptional activity that p53 executes its cancer cell killing functions. This is also part of the reasons for why 80% of mutations are clustered in the DBD of p53. These hotspot p53 mutants are not only unable to specifically bind to the canonical p53-responsive DNA elements (p53RE) but also obtain a new GOF activity essential for cancer cell survival and growth. Though incompletely understood, they generally execute their GOF activity via distinct molecular mechanisms. For instance, p53 mutants can interact with other transcription factors or cofactors, resulting in increased gene transcription and expression. Also, they can prohibit gene transcription by associating with and blocking the DNA-binding activity of other transcription factors or cofactors. In some cases, they could directly bind to DNA, particularly with specifically structured DNA, such as matrix attachment regions (MARs), and regulate transcription. Furthermore, they could bind to proteins that are not related to transcription, modulating their cellular functions. Through these mechanisms, hotspot p53 mutants implement their GOF, promote cancer malignance, and cause drug resistance (Figures 1 and 2). The oncogenic activity of mutant p53 was actually reported in the early 1980s when the first p53 cDNA was cloned from cancer cells that harbor a hotspot mutation (Eliyahu et al., 1984; Jenkins et al., 1984; Parada et al., 1984). Later on and particularly recently, the GOF of mutant p53 has caught more attention from cancer researchers. Hence, a remarkable progress has been made in dissecting the biochemical and molecular mechanisms underlying the GOF activity of some of the hotspot p53 mutants and in understanding of its role in cancer development and progression as well as drug resistance (Dittmer et al., 1993), which will be further discussed below.
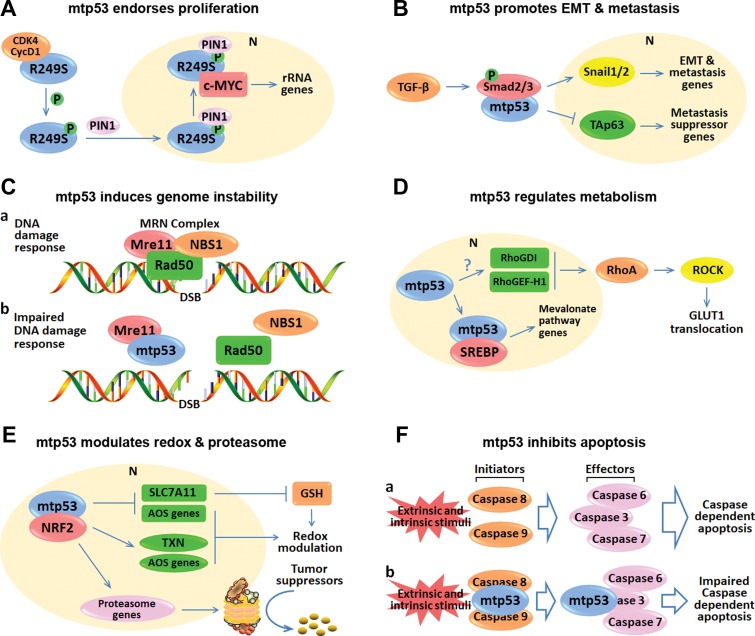
Figure 2.
Representative working modes of mutant p53 GOF. (A) p53-R249S is phosphorylated by CDK4/cyclin D1 and translocated into the nucleus as mediated by PIN1. The nuclear R249S binds to and augments c-Myc activity, leading to enhanced ribosome biogenesis and proliferation. (B) Mutant p53 prompts TGF-β-mediated EMT and metastasis by binding to Smad2/3. The interaction between mutant p53 and Smads leads to elevation of the downstream EMT and metastasis genes. Also, mutant p53, Smads, and TAp63 form a ternary complex that suppresses the anti-metastasis activity of TAp63. (C) Mutant p53 interferes with the assembly of the Mre11–Rad50–NBS1 complex on DNA double-stranded breaks, leading to genome instability. (D) Mutant p53 transcriptionally induces RhoGD1 and RhoGEF-H1 (Mizuarai et al., 2006; Bossi et al., 2008) through mechanisms to be identified, which elicits RhoA–ROCK–GLUT1 cascade, leading to increased aerobic glycolysis. Additionally, mutant p53 binds to the transcription factor SREBP, leading to the activated mevalonate pathway responsible for the regulation of lipid metabolism. (E) Mutant p53 modulates redox and proteasome by interacting with NRF2. Mutant p53 can induce and repress NRF2 target genes to control the glutathione (GSH) level and the redox balance. Moreover, mutant p53 promotes the expression of NRF2 target genes involved in proteasome machinery, leading to increased proteasomal degradation of tumor suppressors. (F) Mutant p53 suppresses apoptosis by directly binding to caspases. Upon external and/or internal stimuli, the initiator caspases, caspase-8, and caspase-9, can proteolytically activate the effector caspases, caspase-3, caspase-6, and caspase-7, thus triggering the caspase-dependent apoptosis. However, mutant p53 can interact with and inhibit caspase-8, caspase-9, and caspase-3 to compromise the caspase-dependent apoptosis. In conclusion, p53 mutants execute their oncogenic GOF by interacting with transcription factors or cofactors to promote (A, B, D, E) or repress (B, E) gene expression, or with proteins irrelevant to transcription (C, F). mtp53 indicates mutant p53.
Mutant p53 endorses proliferation
One of the cellular outcomes owing to the GOF activity of mutant p53 is ‘uncontrolled’ cell proliferation. It has been shown that the p53 mutants, such as R175H, R273H, and D281G, are able to form a ternary complex with the transcription factor NF-Y and the co-factor p300. This NF-Y, p300, and mutant p53 complex mediates histone acetylation and induces transcriptional activation of NF-Y target genes that promotes DNA synthesis and cell cycle progression. These target genes include a panel of cell cycle-regulated genes CCNA2, CCNB1, CCNB2, CDK1, and CDC25C (Di Agostino et al., 2006). The other oncogenes, such as c-MYC, MAP2K3, CXCL1, and CCNE2, have also been shown to be induced by p53 mutants via different mechanisms and to enhance cancer cell proliferation (Frazier et al., 1998; Yan and Chen, 2009; Gurtner et al., 2010; Girardini et al., 2011). Very recently, we unveiled a unique mechanism for the GOF activity of a hepatocellular carcinoma (HCC)-specific p53 mutant, Arg-249 to Ser (R249S) (Figure 2A) (Liao et al., 2017). Interestingly, this mechanism recruited several other oncoproteins, such as CDK4/Cyclin D1, PIN1 and c-MYC to ‘materialize’ the GOF activity of mutant p53. Specifically, p53-R249S is phosphorylated by CDK4/Cyclin D1 at the cancer-derived serine 249 residue, and then interacts with PIN1 that facilitates nuclear import of the phosphorylated p53-R249S. Once in the nucleus, it binds to and stabilizes c-MYC, and the latter becomes more active to boost the transcription of its target genes involved in ribosomal biogenesis, consequently enhancing HCC proliferation and survival (Liao et al., 2017; Wang et al., 2018). Thus, different p53 mutants utilize distinct mechanisms to exhibit their oncogenic GOF activity and promote cancer cell proliferation and survival.
Mutant p53 promotes invasion and metastasis
The GOF also endows mutant p53 with the activity to drive epithelial-mesenchymal transition (EMT) and, subsequently, the more aggressive pathological changes—cancer cell invasion and metastasis (Jiang et al., 2015). The EMT process includes the loss of cell-to-cell and cell-to-ECM (extracellular matrix) adherence and acquirement of mesenchymal characteristics. The first hint that mutant p53 can regulate EMT was found in 1997, when overexpression of the mutant p53-R175H exacerbated the morphological changes of the transformed cells by the oncogenic Ras (Gloushankova et al., 1997). Later on, numerous molecules or signaling pathways, such as TGF-β, EGFR, and cadherins, have been characterized in the EMT and the resultant metastasis. Among these key molecules, TGF-β activates several EMT and metastasis-associated transcription factors, Snail1/2, ZEB1/2, and Slug, and supports cancer metastasis. The p53 mutants, R175H and R280K, were found to prompt TGF-β-mediated cancer cell spread by connecting the TGF-β:Smad cascade with TAp63, a p53 family member (Figure 2B) (Adorno et al., 2009). A recent study also showed that p53-R175H modulates a subset of TGF-β target genes by directly binding to Smad3 (Figure 2B) (Ji et al., 2015). E-cadherin is another crucial regulator that maintains the epithelial status of cells, while loss of its expression in cancer cells suppresses cell-to-cell adhesion. Interestingly, wild-type p53 was shown to induce E-cadherin expression through the miR-192/200 family–ZEB1/2 cascade (Kim et al., 2011), whereas several p53 mutants diminish E-cadherin expression and promote EMT via the miR-130b–ZEB1 axis (Roger et al., 2010; Dong et al., 2013). Furthermore, several hotspot p53 mutants can also trigger constitutive activation of EGFR/integrin signaling, resulting in the detachment of epithelial cells from the ECM (Muller et al., 2009). Hence, together with other lines of evidence (Muller et al., 2011), p53 mutants exert the GOF by enhancing cancer cell EMT and metastasis.
Mutant p53 and genome instability
The GOF activity of mutant p53 is also engaged in the induction of replication stress and genome instability. Triggered by replication stress, genome instability is a causative and characteristic feature of almost all human cancers. There are two hypotheses for interpreting the mechanisms underlying genome instability in tumorigenesis—the mutator hypothesis and the oncogene-induced replication stress model (Negrini et al., 2010). The hereditary cancers are believed to be driven by genome instability as proposed by the mutator hypothesis, in which germline mutations of DNA repair or mitotic checkpoint genes are descended and present in every single cell of patients, leading to accelerating genome instability and early onset of various cancers. For example, p53 mutations, particularly the GOF mutations, in germline are the source of genome instability resulting in Li-Fraumeni syndrome (LFS), a complex inherited cancer predisposition disorder (McBride et al., 2014). However, the mutator hypothesis cannot explain the onsets of sporadic cancers, as very few mutations of DNA repair or mitotic checkpoint genes are identified in these cancers through genome-wide associated studies (Negrini et al., 2010). It was also shown that genome instability can be detected in the precancerous lesions prior to the establishment of mutations of tumor suppressor genes, such as TP53 (Halazonetis et al., 2008). In fact, genome instability is more likely to arise from carcinogen or oncogene-induced replication stress during the formation of sporadic cancers. Oncogene-induced uncontrolled rapid cell proliferation should impose additional burden on DNA replication leading to two distinct but interrelated consequences—DNA damage response and genome instability. This replication stress provokes p53-dependent cell death at the early stage of cancer and, however, causes TP53 gene mutations that further hasten genome instability as the cancer progresses to a later stage (Halazonetis et al., 2008). There are two mechanisms possibly accounting for the acquirement of p53 mutations. First, the induction of p53-mediated apoptosis may lead to selection of cancer cells for p53 mutation. Alternatively, the TP53 gene locus could be associated with the genomic fragile sites that are highly sensitive to genomic instability (Durkin and Glover, 2007). Thus, p53 mutants act as proponents rather than sources of genome instability in the sporadic cancers. Mechanistically, the hotspot p53 mutants, R248W and R273H, were found to interact with the MRN (Mre11-Rad50-NBS1) complex and prevents its association with the DNA double-stranded breaks, consequently leading to replication stress and impaired DNA damage response (Figure 2C) (Song et al., 2007). Therefore, the crosstalk between mutant p53 and genome instability is pivotal to cancer development.
Mutant p53 is involved in cancer metabolism, redox homeostasis, and others
Recently, the mutant p53 GOF has been also attributed to its capability of modulating cancer cell metabolism. Deregulated energy metabolism represents one of the hallmarks of cancer (Hanahan and Weinberg, 2011). The Warburg effect theory, which was postulated in 1956 (Warburg, 1956) and is favorably utilized by cancer cells, depicts the fact that cancer cells reprogram their glucose metabolism by preferentially utilizing glycolysis even under the aerobic condition (Vander Heiden et al., 2009). Intriguingly, p53 mutants enhance the Warburg effect or aerobic glycolysis via the RhoA–ROCK–GLUT1 cascade (Figure 2D) (Zhang et al., 2013) or by inactivating AMPK signaling (Zhou et al., 2014). Moreover, the hotspot p53 mutants, such as R175H, R248Q, R248W, and R273H, have also been shown to activate the mevalonate pathway and elevate sterol biosynthesis by directly binding to and boosting the SREBP transcription factors (Figure 2D) (Freed-Pastor et al., 2012). Recently, the P72R polymorphism of p53 was shown to associate with and bolster PGC-1α function and enhance oxidative phosphorylation in cancer cells (Basu et al., 2018). Taken together, these studies reveal that p53 mutants sustain cancer cell survival through metabolic regulation.
Of note, an interesting study unveiled a potential link between mutant p53 and redox homeostasis through modulation of activity of the master antioxidant transcription factor NRF2 (Kalo et al., 2012). Later on, the p53 mutants, R175H, R248W/Q, and R273H, were found to interact with NRF2 and repress transcription of SLC7A11, leading to reduction of intracellular glutathione, as well as numerous NRF2 target genes required for monitoring cellular redox balance (Liu et al., 2017). Interestingly, another study revealed that the interaction between mutant p53 and NRF2 also leads to enhanced expression of NRF2 target genes involved in redox regulation, which endorses cancer cell survival and is negatively associated with cancer prognosis (Figure 2E) (Lisek et al., 2018). Remarkably, a universal mutant p53 GOF has been demonstrated to globally regulate protein turnover by activating the NRF2-mediated proteasome gene program (Figure 2E) (Walerych et al., 2016).
In addition to the abovementioned GOF, p53 mutants are also found to induce angiogenesis, enable replicative immortality, regulate GTPase activity and so on, which have been elaborately discussed in other review articles (Freed-Pastor and Prives, 2012; Muller and Vousden, 2014; Bykov et al., 2018; Kim and Lozano, 2018; Sabapathy and Lane, 2018).
Mutant p53 fosters cancer development in vivo
Notably, the GOF of mutant p53 is also demonstrated in genetically engineered mouse models and highly associated with clinical outcomes. The first mutant p53 mouse model, manifesting a high incidence of lung tumors, osteosarcomas, and lymphomas, was generated by germline overexpression of mutant Trp53 transgenes derived from murine cancers (Lavigueur et al., 1989). Because of the existence of innate wild-type Trp53 alleles, this transgenic mouse model did not elucidate if those phenotypes are resulted from GOF or the dominant-negative effect (Lavigueur et al., 1989). After that, a number of tissue-specific transgenic mouse models were generated to demonstrate the function of mouse mutant p53-R172H (equivalent to human R175H), R246S (equivalent to human R249S), and human mutant p53-R273H in the development of mammary tumors (Li et al., 1997), hepatocellular carcinoma (Ghebranious and Sell, 1998), and lung adenocarcinoma (Duan et al., 2002), respectively. Interestingly, the p53-R172L (equivalent to human R175L) transgenic mice driven by the mammary gland or prostate-specific promoter manifested normal or even reduced tumor predisposition (Li et al., 1994; Hernandez et al., 2003). These findings are in agreement with a later study showing that the germline p53-R175L variant retains partial tumor suppressive activity to prevent LFS, although leads to the pediatric adrenal cortical carcinoma (West et al., 2006). The oncogenic GOF of mutant p53 was then validated by knock-in mouse models in 2004, when the murine p53-R172H or R270H (equivalent to human R273H) was engineered into the endogenous p53 locus in mice independently by the Lozano group (Lang et al., 2004) and the Jacks group (Olive et al., 2004). In the former study, the p53+/R172H mice exhibited more aggressive or metastatic malignancies than did the p53+/- mice. Consistently, the embryonic fibroblasts from the p53R172H/R172H mice displayed augmented cell proliferation, DNA synthesis and transformation potential compared to those from the p53−/− mice (Lang et al., 2004). The latter study not only demonstrated the oncogenic GOF of mouse p53 mutants R172H and R270H, but also ascertained the differential effects of the DNA-contact (R270H) and conformational (R172H) mutants on tumorigenesis (Olive et al., 2004). Additionally, a recent study has revealed that mutant p53-R172H and R245W (equivalent to human R248W) differentially prompt breast cancer development by generating somatic mutations in murine mammary epithelial cells (Zhang et al., 2018). Altogether, these genetic studies elegantly depict the role of the cancer-associated hotspot mutants in vivo (Donehower and Lozano, 2009).
In the LFS patients, the TP53 missense mutations with GOF in the germline have been shown to induce earlier cancer onset and more aggressive malignancies compared to the deletion or truncation mutations (McBride et al., 2014). The lifetime likelihood for LFS patients of developing malignancies is ~75% in males and nearly 100% in females. Importantly, the missense mutations, particularly the most GOF mutations, result in the first tumor onset at age 22.3–22.6, while patients with other types of mutations begin to develop tumors at age 31.4–37.5 (Bougeard et al., 2008; Zerdoumi et al., 2013). These bench-to-bed findings again verify that mutant p53 GOF promotes human cancer formation and development, which certainly makes it an essential target protein in cancer therapy.
Roles of mutant p53 in cancer therapy
The major principle of cancer therapy is to inhibit cell proliferation and promote cell death. However, to survive, cancer cells can develop adaptation to the therapeutic treatment by, for example, eventually mutating the TP53 gene at their very late stage in most of the cancer cases. Indeed, p53 mutants are the key molecules endowing cancer cells with chemoresistance. The first line of evidence showing that p53 mutation could be associated with drug resistance is the identification of MDR1 (multidrug resistance gene 1) as a mutant p53 target gene (Chin et al., 1992; Zastawny et al., 1993). MDR1, also known as ABCB1, which is found to be substantially overexpressed in cancer, encodes an ATP-dependent drug efflux pump responsible for the induction of chemoresistance with broad spectrum activity. While wild-type p53 suppresses MDR1 expression, several p53 mutants are shown to specifically associate with the core promoter region of MDR1 and stimulate its expression (Chin et al., 1992; Zastawny et al., 1993). The clinical correlation between therapeutic resistance and p53 mutation has been studied since 1990s. Sequencing of the complete coding region of the TP53 gene in breast cancer from a cohort of 316 patients revealed that p53 mutation is associated with significantly worse prognosis and resistance to adjuvant systemic therapy (Bergh et al., 1995). Another study involving an array of 63 patients showed that p53 mutation dampens the response of breast cancer to Doxorubicin treatment (Aas et al., 1996). Ovarian cancer patients harboring mutant p53 were also found to exhibit less sensitivity to Cisplatin therapy (Shelling, 1997). Moreover, the association of p53 mutation with chemoresistance and poor prognosis was also observed in lung cancer (Horio et al., 1993), gastric and colorectal cancers (Hamada et al., 1996), and hematologic malignancies (Wattel et al., 1994; Wilson et al., 1997). Apart from triggering chemoresistance, p53 mutants are able to attenuate cancer cell response to radiotherapy. This was firstly noticed by ectopically introducing a mutant p53 gene, p53-V143A, into the colon cancer RKO cells sustaining endogenous wild-type p53 (Kuerbitz et al., 1992). The study also suggested that the mutant p53-mediated radioresistance was at least partially attributed to its dominant-negative regulation over the wild-type allele. Later, mouse models demonstrated the radioresistance activity of mutant p53 in vivo (Lee and Bernstein, 1993; Lowe et al., 1994). Moreover, the clinical studies revealed that p53 mutants reduce radiosensitivity and worsens prognosis in the treatment of numerous human cancers (Bergh et al., 1995; Hamada et al., 1996; Koch et al., 1996). Next, we further discuss the molecular basis underlying mutant p53-induced therapeutic resistance by attenuating apoptosis, impairing autophagic cell death, and boosting cancer cell stemness (Figure 1).
Mutant p53 confers resistance to apoptosis
To elicit apoptosis is the major way that most therapeutic strategies eliminate cancer cells. The protein products of TP53 target genes play pivotal roles in the process. In response to external stresses induced by cytotoxic agents or irradiation, wild-type p53 can be activated mostly through post-translational modifications, such as acetylation and phosphorylation, resulting in robust apoptosis. The stress-responsive p53 transcriptionally upregulates the expression of BH3-only proteins, including PUMA, NOXA, BID, and NIX, as well as other pro-apoptotic proteins, such as BAX and PIDD. These proteins are involved in the initiation of MOMP and consequently drive caspase activation and apoptosis (Vazquez et al., 2008). Meanwhile, the cytosolic p53, via the DBD or the region adjacent to DBD, can either directly interact with and neutralize the anti-apoptotic activity of BCL-2 and BCL-XL, or associate with BAX and BAK to trigger MOMP by forming supramolecular structures (Green and Kroemer, 2009). Mutation of p53 not only impairs its transcriptional activity in the nucleus but also abrogates p53 interaction with the BCL-2 family proteins in the cytoplasm. Hence, p53 mutation has dual anti-apoptotic effects by inhibiting both nuclear and cytosolic functions of wild-type p53.
Of note, p53 mutants have also been shown to repress apoptosis and bring therapeutic resistance independently of their wild-type counterpart and through their GOF. Although most p53 mutants lose the DNA-binding capability due to the mutations within the DBD, they can still indirectly modulate gene transcription via association with other transcription factors or cofactors (Figure 1). For example, p53 mutants bind to and inactivate the p53 homologs, TAp63 and TAp73, that also induce their target genes shared with wild-type p53 (Di Como et al., 1999; Gaiddon et al., 2001; Liu et al., 2011). Another study demonstrated that p53-R175H or R248W in cooperation with ETS2 associates with the TDP2 promoter and activates its expression, which is responsible for Etoposide resistance (Do et al., 2012). Recently, p53 mutants were also shown to induce apoptosis resistance by transcriptionally upregulating Ephrin-B2 expression and through JNK–c-Jun and Src–ERK pathways (Alam et al., 2016). In addition, p53 mutants have been found to induce drug resistance by orchestrating the microRNA network. For instance, p53-R175H transcriptionally elevates the expression of miRNA-128-2 that targets E2F5 resulting in resistance to Cisplatin, Doxorubicin or 5-Fluorouracil-induced apoptosis (Donzelli et al., 2012). Conversely, it inhibits miR-223 expression via association with the transcriptional repressor ZEB-1. The miR-233 inhibition derepresses the expression of STMN-1 that triggers chemoresistance (Masciarelli et al., 2014). Other major mechanisms also include the physical interaction of mutant p53 with proteins irrelevant to transcription. It has been shown that p53 mutants repress apoptosis through directly crippling the caspase-dependent apoptotic singling cascade. By sensing a variety of stress signals, the initiator caspases, including caspase-2, caspase-8, and caspase-9, can induce MOMP or directly activate the downstream effector caspases, such as caspases-3, caspase-6, and caspase-7, eventually leading to apoptosis (Marino et al., 2014). Markedly, p53 mutants are not only able to hamper the activation of caspase-8 and caspase-9 (Ameyar-Zazoua et al., 2002; Chee et al., 2013), but also impede the cleavage of caspase-3 through directly binding to these proteins (Figure 2F) (Frank et al., 2011). Altogether, p53 mutants evoke tumor cell resistance to cancer therapy by inhibiting apoptosis through multifarious mechanisms.
Mutant p53 suppresses autophagic cell death
Autophagy is a conserved lysosome-associated degradation pathway throughout the eukaryotes. As an important cellular homeostatic mechanism, autophagy is responsible for clearance and recycling of the excessive and dysfunctional molecules as well as the damaged and aged organelles (Marino et al., 2014). On one hand, by doing so, autophagy serves as protective machinery that supports survival of both normal and cancer cells. On the other hand, persistent autophagy caused by therapeutic treatment has been found to stimulate ‘autophagic cell death’, an independent cell death modality that does not involve, but could be accompanied by, apoptosis (Sui et al., 2013). Therefore, autophagy is a double-edged sword in tumorigenesis by either turning on pro-survival signals or driving autophagic cell death.
It has been well documented that many anti-cancer drugs induce autophagic cell death, particularly, in the cancer cells refractory to apoptosis (Kondo and Kondo, 2006). For instance, Sorafenib and its derivative, SC-59, elicit autophagic cell death through a SHP1-STAT3-Mcl1-Beclin1 cascade in hepatocellular carcinoma (Tai et al., 2013). Mono-Pt, a novel monofunctional platinum analog, initiates autophagic cell death by involving the AKT1–mTOR–RPS6KB1 and MAPK1 (ERK2)/MAPK3 (ERK1) pathways in ovarian carcinomas (Guo et al., 2013). Moreover, 5-Fluorouracil has been found to evoke autophagic cell death in the apoptosis-impaired colon cancer cell lines, HCT116 BAX−/− and HCT116 PUMA−/− (Xiong et al., 2010). Several studies have also demonstrated that the potential anti-cancer agents, such as cannabinoids, suppress tumor growth by triggering autophagic cell death in glioma, hepatocellular carcinoma, and pancreatic cancer cells (Salazar et al., 2009; Donadelli et al., 2011; Lorente et al., 2011).
Intriguingly, p53 mutants have been shown to impair autophagy and result in restrained autophagic cell death. The first report to reveal p53 mutants as inhibitors of autophagy was described by Kroemer and colleagues (Morselli et al., 2008). In the study, they found that cytoplasmically localized p53 mutants, such as p53-R273H, p53-R273L, p53-A161T, p53-S227R, and p53-E258K, preferentially repress autophagy (Morselli et al., 2008). Mechanistically, p53 mutants suppress autophagy by regulating mTOR and AMPK signaling pathways. In general, inhibition of the mTOR activity is a strategy to provoke autophagy, but p53 mutants were surprisingly found to antagonize the autophagic machinery by enhancing the mTOR activity (Zhang et al., 2015; Agarwal et al., 2016). AMPK is crucial for cells to sense metabolic stress and often plays an opposed role to that of mTOR (Cordani et al., 2017). p53 mutants are able to compromise the AMPK signaling through direct interaction with AMPKα subunit or by inhibiting Sestrin proteins, the activators of AMPK (Zhou et al., 2014; Cordani et al., 2016). By doing so, p53 mutants suppress autophagy by activating mTOR. Other mechanisms also include mutant p53-mediated repression of ATG12, which is required for the early formation of autophagosome, and enhancement of HIF-1 activity (Cordani et al., 2017). Therefore, p53 mutants can cause the drug resistance at least partially through their inhibitory effect on autophagic cell death triggered by anti-cancer therapy (Figure 1).
Mutant p53 facilitates cancer stemness
Normal stem cells can be generally categorized into two groups: (i) embryonic stem cells (ESCs) that are pluripotent and able to self-renew and differentiate into all different cell lineages; (ii) adult stem cells (ASCs) with multipotency to differentiate into tissue-specific cells. Wild-type p53 was found to suppress self-renewal and promote differentiation of both ESCs and ASCs (Molchadsky and Rotter, 2017). Also, p53 serves as a barrier to the generation of induced pluripotent stem cells (iPSC) (Molchadsky and Rotter, 2017). However, inactivation or mutation of the wild-type p53 enhances the stem-like properties and promotes malignant transformation of stem cells, as thus favoring the formation of cancer stem cells (CSCs) (Shetzer et al., 2016).
CSCs represent a small number of tumorigenic progenitors (also known as tumor-initiating cells) that maintain the potentials of self-renewal, differentiation into cancerous cells, migration to adjacent tissues and resistance to conventional therapeutic response (Pattabiraman and Weinberg, 2014). Mounting evidence implies the involvement of mutant p53 in the regulation of CSCs (Figure 1). For example, p53 mutations have been shown to induce the transformation of hematopoietic progenitors, neural stem cells, ovarian surface epithelial stem cells as well as iPSC (Shounan et al., 1997; Wang et al., 2009; Sarig et al., 2010; Flesken-Nikitin et al., 2013; Bartesaghi et al., 2015). In addition, p53 mutants support CSC survival and proliferation by promoting the stemness-associated gene expression signature (Mizuno et al., 2010). In particular, they raise the expression of stem cell markers, including CD133, CD44, YAP/TAZ, Lgr5, and ALDH (Escoll et al., 2017; Solomon et al., 2018). Interestingly, a cancer-associated isoform of p53 has been shown to induce several key stemness factors, such as SOX2, OCT3/4, and NANOG (Arsic et al., 2015). Hence, stimulation of stem-like features by mutant p53 contributes to the induction of drug resistance, although the molecular mechanism for how p53 mutants endorse the emergence and propagation of CSCs awaits further investigation.
Therapeutic strategies to target mutant p53
p53 mutation is regarded as an obstacle in cancer therapy because of its loss of function, dominant-negative effect and, particularly, the oncogenic GOF. Thus, a multitude of small molecules have been developed to reverse the therapeutic resistance by targeting mutant p53. There are two types of small molecules proved to be potent and effective to antagonize or neutralize the oncogenic functions of mutant p53. They can either accelerate mutant p53 protein turnover or convert it into the wild-type conformation. Some of these molecules in combination with the conventional therapies have been shown to greatly improve cancer treatment. However, recent studies have revealed that mutant p53 could also make cancer more vulnerable to be treated. Wild-type p53 is essential to the G1 checkpoint activation and DNA repair in response to a variety of DNA damage insults. Once the TP53 gene is mutated, cancer cells have to rely on the G2 checkpoint activation as a compensating survival mechanism upon DNA damage stress. Further impairment of the G2 checkpoint is lethal to these cancer cells, which is considered as the ‘synthetic lethality’. Because of these complex outcomes of cancer cells upon chemotherapy pending on the p53 status, mutant p53 therefore can be regarded as both a barrier and a path to cancer therapy. Hereafter, we have discussed some important progresses in the development of anti-cancer molecules by targeting mutant p53 (Figure 3).
Figure 3.
Therapeutic strategies targeting mutant p53. (A) Ablation of mutant p53 in cancer is one of the most effective strategies for developing cancer therapy. HSP90 is highly expressed in cancer and crucial to mutant p53 stabilization. HSP90 inhibitors block HSP90 function and lead to MDM2- or CHIP-mediated proteasomal degradation of mutant p53 proteins. In addition, both Spautin-1 and MCB-613 can induce lysosomal degradation of mutant p53 through HSPA8/LAMP2A-mediated CMA pathway and inhibition of the deubiquitinase USP15, respectively. Moreover, HDAC inhibitors can suppress mutant p53 gene transcription. (B) Another attractive strategy is to restore the mutant p53 to a wild-type-resembling conformation. APR-246, COTI-2, and PK11007 can reactivate mutant p53 and trigger the expression of wild-type p53 target genes. Also, APR-246 and PK11007 can kill cancer cells by elevating the ROS level and inducing oxidative stress. (C) The TP53 gene mutations also impose vulnerability on cancer cells. In response to DNA damage stress, wild-type p53-mediated G1/S arrest and WEE1-mediated G2/M arrest are essential for cells to repair damaged DNA. Nevertheless, the WEE1-mediated G2/M checkpoint renders crucial survival dependency in the mutant p53-sustaining cancer cells with defective G1/S checkpoint. Thus, the WEE1 inhibitor can cause synthetic lethality to these cancer cells by abrogating the remaining G2/M checkpoint. mtp53 indicates mutant p53.
Deprivation of mutant p53
Inhibition of a mutant p53 by promoting its protein degradation has been attested to be an efficient strategy for the treatment of cancers that sustain p53 mutations (Figure 3A). The rationale of depleting mutant p53 is based on the fact that the mutant p53 proteins are inherently unstable in normal cells (Terzian et al., 2008), while it is stabilized by, for example, HSP90 that is usually overexpressed in cancer (Trepel et al., 2010). The discovery of several HSP90 inhibitors has led to the feasibility of ablation of mutant p53 as a potential anti-cancer approach. The first HSP90 inhibitor 17-AAG, an analog of Geldanamycin, was shown to destabilize p53 mutants such as p53-V143A, R175H, S241F, R273C/H, and R280K, by inactivating HSP90 (Blagosklonny et al., 1995, 1996; Egorin et al., 1998) and consequently triggering mutant p53 degradation by the E3 ligases MDM2 or CHIP (Peng et al., 2001; Li et al., 2011b). Notably, another HSP90 inhibitor, Ganetespib, also known as STA-9090, was shown to exhibit >50-fold more potency than 17-AAG in degrading p53-R175H and R248Q using mouse models (Alexandrova et al., 2015). Thus far, more than a dozen of HSP90 inhibitors have been under preclinical or clinical evaluations, which would hopefully lead to a clinically useful therapy for cancer eventually (Trepel et al., 2010). HDAC inhibitors are another group of compounds that can reduce the enrichment of mutant p53. The first line of evidence that HDAC inhibitors, such as trichostatin A and FR901228, prompt ablation of p53 mutants including p53-R175H, P223L, V274F, and R280K was reported by Blagosklonny et al. (2005). Mechanistically, HDAC inhibitors can counteract mutant p53 at both transcriptional and post-translational layers. Inhibition of the class I HDACs, including HDAC1, HDAC2, and HDAC8, by SAHA, NaB, or MS275 restrains transcription of the mutant TP53 gene by abolishing the activity of HOXA5, Yin Yang 1 (YY1), or c-MYC (Yan et al., 2013; Wang et al., 2016; Stojanovic et al., 2017). Also, SAHA can block the HDAC6–HSA90 cascade, thus leading to MDM2- or CHIP-mediated degradation of mutant p53 (Li et al., 2011a). Interestingly, HSP90 inhibitors synergize the effect of SAHA on degradation of mutant p53 and inhibition of tumor cell growth in vitro and in vivo (Alexandrova et al., 2015). In addition, a broad range of mutant p53 proteins including p53-R175H/C/D, S241F, G245C, R248Q/W/L, E258K, R273H/L, R280K, and R282W are also degraded through the chaperone-mediated autophagy (CMA) pathway triggered by Spautin-1, a small molecule designed for inhibition of macroautophagy (Vakifahmetoglu-Norberg et al., 2013). Very recently, a small molecule named MCB-613 has been found to preferentially target p53-R175H for lysosomal degradation by subverting the deubiquitinase USP15-mediated mutant p53 stabilization (Padmanabhan et al., 2018). Hence, targeting mutant p53 for degradation by small molecule compounds could be potentially beneficial to the improvement of therapeutic sensitivity of human cancers that harbor mutant p53.
Restoration of wild-type activity to mutant p53
An attractive strategy by targeting mutant p53 for cancer therapy is to convert it to a version that mimics the wild-type p53 protein (Figure 3B). In 1999, Foster et al. (1999) identified a compound, CP-31398, that could accumulate the active conformational DBDs of p53-V173A, R175S, R249S, and R273H, and thus trigger the expression of p21 as well as a p53-inducible luciferase reporter gene in cancer cells and in tumor. Later on, more compounds that can render p53 mutants into their wild-type versions have been identified, such as, P53R3 (Weinmann et al., 2008), NSC319726 (Yu et al., 2012), PK7088 (Liu et al., 2013), PEITC (Aggarwal et al., 2016), and RITA (Zhao et al., 2010; Burmakin et al., 2013). Noteworthily, two compounds, COTI-2 and APR-246/PRIMA-1MET, are currently undergoing clinical trials (Duffy et al., 2017; Zhao et al., 2017). COTI-2, a thiosemicarbazone-related compound, displays significantly high efficiency in constraining tumor growth by retrieving wild-type p53 activity from a panel of cancer cell lines that contain mutant p53-R175H, Y220C, R248Q, I255N, and R273H, respectively (Salim et al., 2016; Duffy et al., 2017). p53 reactivation and induction of massive apoptosis (PRIMA-1) was identified through an approach designed for screening compounds engaged in repression of mutant p53-dependent cell growth (Bykov et al., 2002). This molecule can restore sequence-specific DNA binding capability to the mutant p53 proteins and present in vivo tumor suppressive activity with no evident toxicity (Bykov et al., 2002). Mechanistically, the decomposition products of PRIMA-1 covalently associate and form adducts with thiols of p53-R173H, R248Q, and R273H (Lambert et al., 2009). APR-246, also known as PRIMA-1MET, is a methylated and improved version of PRIMA-1 and exhibits higher efficacy in reactivating mutant p53 and eliciting apoptosis. Additionally, several studies also uncovered a mutant p53-independent effect of APR-246 by depleting glutathione content and inducing elevated ROS and oxidative damage to cancer cells (Peng et al., 2013; Tessoulin et al., 2014). The diverse mechanisms for APR-246 are reminiscent of another important small 2-sulfonylpyrimidine molecule, PK11007, that reactivates p53 mutants Y220C and V143A, while, more generally, sensitizes mutant p53-containing cancer cells to oxidative stress (Bauer et al., 2016). Of note, APR-246 displays a synergistic inhibitory effect on tumor growth in combination of several commonly used chemotherapeutic drugs (Bykov et al., 2005). It also cooperates with Sulfasalazine, Auranofin or Carfilzomib in eliciting cell death by exploiting the aforementioned mutant p53/NRF2-mediated oxidative or proteasomal signal (Walerych et al., 2016; Liu et al., 2017; Lisek et al., 2018). Recently, a cell-penetrating peptide ReACp53 has been shown to impede aggregation of p53 mutants, particularly R175H and R248Q, and therefore increase the pool of functional and wild-type-like p53 protein in the high-grade serous ovarian carcinomas (HGSOC) (Soragni et al., 2016). These studies demonstrate that reactivation of mutant p53 by converting it to a tumor suppressive conformation is a promising anti-cancer strategy, as the field is awaiting the outcomes of their clinical trials with hope.
Synthetic lethality imposed by mutant p53
The concept of synthetic lethality emerged from genetic studies of the Drosophila model system in which simultaneous mutations of two or multiple separate and non-essential genes result in cell death. Likewise, in the area of cancer biology, synthetic lethality describes a cellular phenomenon that an oncogenic mutation or tumor suppressor defect may offer cancer cells secondary survival dependency (Kaelin, 2005). The cancer cells are vulnerable to attack by further targeting the secondary survival signal. Since cancer cells are genomic instable with frequent gene mutations, amplifications, or deletions, it is critically important to identify potentially druggable signaling pathways that are synthetic lethal to the cancer cells.
Accumulating evidence has revealed that the TP53 gene mutations provide an opportunity for achieving synthetic lethality in cancer cells (Figure 3C). Wild-type p53 has been shown to protect the genome by mainly inducing G1-arrest, offering time for the cells to repair their damaged DNA in response to moderate genotoxic stress. However, p53 mutations impair DNA damage response of the cells by abrogating G1 checkpoint, thus those cells have to rely on other compensating or parallel pathways, such as G2 checkpoint, to react properly with the DNA damage insults. Supporting this notion are several studies showing that inactivation of the G2 or S checkpoint-associated ATR/CHK1, ATM/CHK2, or p38MAPK/MK2 pathway is synthetic lethal to the p53-deficient cancer cells (Gurpinar and Vousden, 2015; Zhao et al., 2017). Thus, it is efficient to eliminate cancer cells by targeting these crucial secondary survival-dependent signals. The most successful example is the clinical application of the WEE1 inhibitor as a synthetic lethal agent in the TP53-mutated human cancers. The WEE1 kinase was shown to arrest cell cycle at the G2 phase in response to DNA damage stress by phosphorylating and inactivating CDK1/Cyclin B complex in the cancer cells with the deficient G1 checkpoint (Matheson et al., 2016). Remarkably, recent phase I and II clinical trials gracefully demonstrated that AZD1775, a potent and selective WEE1 inhibitor, substantially improves advanced solid tumor treatment and enhances Carboplatin efficacy in ovarian cancer patients who are refractory to the first-line therapy and harbor mutant p53 (Leijen et al., 2016a, b). Therefore, p53 mutation also bestows cancer cells in vulnerability that can be efficiently targeted as a strategy for developing cancer therapy.
Although a growing number of strategies targeting mutant p53 for cancer therapy have been developed, the field also faces a number of challenges. First, the compounds inducing mutant p53 degradation should be ineffective in killing cancer cells sustaining the ‘loss of function’ mutants. Second, the cellular response to the mutant p53-targeting compounds may display considerable variation, because most agents are only specifically potent for several p53 mutants with regard to their distinct conformations and in different cellular contexts. Third, reactivation of mutant p53 should also enhance the expression of wild-type p53’s auto-regulators, such as MDM2, NGFR, and PHLDB3, which on the contrary promote degradation of p53 leading to impaired cell death (Chao et al., 2016; Zhou et al., 2016, 2017). Furthermore, like other anti-cancer agents, the mutant p53-targeting agents also have off-target activity that may cause mutant p53-independent adverse effect by triggering cytotoxicity in the normal cells. Finally, there are few compounds targeting the truncation mutants or variants of p53 that could also possess the oncogenic GOF (Arsic et al., 2015). It is anticipated that thorough investigation of the biochemical and molecular basis of mutant p53 GOF actions should shed light on some possible approaches to solve these problems.
Concluding remarks
To date, the TP53 gene has been extensively studied as reflected in more than 90000 publications via PubMed search (https://www.ncbi.nlm.nih.gov/pubmed/?term=p53). However, among them, only 7% of the studies are related to mutant p53. Given that TP53 is mutated in ~50% of human cancers and tends to be much more frequently mutated (>80%) in aggressive or late-stage cancers, more attention needs to be paid to the basic and translational study of mutant p53. Although the past decade has witnessed a tremendous progress in our better understanding of the mutant p53 GOF in cancer, those studies have also raised more outstanding questions than definitive answers. Generally, what is the molecular basis for cellular events that lead to the mutations of the TP53 gene? Is mutant p53 able to serve as a diagnostic and/or prognostic marker by combining with other risk factors? Can the GOF mutants be categorized according to their structures or cancer-driving functions and, eventually, targeted by designing specific strategies? In addition, the TP53 gene displays diverse mutation patterns in the context of different cancers, which suggests cancer-specific regulation may exist. For instance, TP53 exhibits extremely high frequency of mutation in certain types of cancer, such as HGSOC with the mutation rate at ~96%–99% (Cancer Genome Atlas Research Network, 2011; Patch et al., 2015). It is tempting to explore the mechanism underlying the prevalence and function of mutant p53 and to develop clinical approaches by targeting the mutant p53 proteins in this specific type of cancer. Also, as mentioned above, we recently demonstrated a mutant p53-mediated, HCC-specific pathway that is responsive to AFB1 or HBV infection. This newly identified CDK4–PIN1–p53 (R249S)–c-MYC cascade is crucial to HCC proliferation and survival and thus potentially to be targeted for developing anti-HCC therapy (Liao et al., 2017). Hence, in light of the studies described above and elsewhere (Freed-Pastor and Prives, 2012; Muller and Vousden, 2014; Bykov et al., 2018; Kim and Lozano, 2018; Sabapathy and Lane, 2018), more efforts are demanded to decipher the precise roles of p53 mutants and the detailed mechanisms underlying their roles in human cancers. The knowledge gained from these systematic studies would provide more new molecules for future development of target-specific therapies against late stage or more aggressive cancer that harbor mutant p53, which would open one of the boulevards to eventually lead to the triumph of the anti-cancer battle. Finally, we still do not have a high resolution 3D image about the wild type or mutant p53 protein (Okorokov et al., 2006; Tidow et al., 2007), as this atomic and structural information is critically important for moving the p53 research and its drug discovery to the next level.
Funding
X.Z. was supported by the National Natural Science Foundation of China (81672566 and 81874053), Q.H. was supported by the National Natural Science Foundation of China (81702352), and H.L. was in part supported by the National Institutes of Health-National Cancer Institute grants (R01CA095441 and R01CA127724).
Conflict of interest
none declared.
References
- Aas T., Borresen A.L., Geisler S., et al. (1996). Specific P53 mutations are associated with de novo resistance to doxorubicin in breast cancer patients. Nat. Med. 2, 811–814. [DOI] [PubMed] [Google Scholar]
- Adorno M., Cordenonsi M., Montagner M., et al. (2009). A mutant-p53/Smad complex opposes p63 to empower TGFβ-induced metastasis. Cell 137, 87–98. [DOI] [PubMed] [Google Scholar]
- Agarwal S., Bell C.M., Taylor S.M., et al. (2016). p53 deletion or hotspot mutations enhance mTORC1 activity by altering lysosomal dynamics of TSC2 and Rheb. Mol. Cancer Res. 14, 66–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggarwal M., Saxena R., Sinclair E., et al. (2016). Reactivation of mutant p53 by a dietary-related compound phenethyl isothiocyanate inhibits tumor growth. Cell Death Differ. 23, 1615–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam S.K., Yadav V.K., Bajaj S., et al. (2016). DNA damage-induced ephrin-B2 reverse signaling promotes chemoresistance and drives EMT in colorectal carcinoma harboring mutant p53. Cell Death Differ. 23, 707–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrova E.M., Yallowitz A.R., Li D., et al. (2015). Improving survival by exploiting tumour dependence on stabilized mutant p53 for treatment. Nature 523, 352–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameyar-Zazoua M., Larochette N., Dorothee G., et al. (2002). Wild-type p53 induced sensitization of mutant p53 TNF-resistant cells: role of caspase-8 and mitochondria. Cancer Gene Ther. 9, 219–227. [DOI] [PubMed] [Google Scholar]
- Arsic N., Gadea G., Lagerqvist E.L., et al. (2015). The p53 isoform △133p53β promotes cancer stem cell potential. Stem Cell Rep. 4, 531–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartesaghi S., Graziano V., Galavotti S., et al. (2015). Inhibition of oxidative metabolism leads to p53 genetic inactivation and transformation in neural stem cells. Proc. Natl Acad. Sci. USA 112, 1059–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S., Gnanapradeepan K., Barnoud T., et al. (2018). Mutant p53 controls tumor metabolism and metastasis by regulating PGC-1α. Genes Dev. 32, 230–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer M.R., Joerger A.C., and Fersht A.R. (2016). 2-Sulfonylpyrimidines: Mild alkylating agents with anticancer activity toward p53-compromised cells. Proc. Natl Acad. Sci. USA 113, E5271–E5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergh J., Norberg T., Sjogren S., et al. (1995). Complete sequencing of the p53 gene provides prognostic information in breast cancer patients, particularly in relation to adjuvant systemic therapy and radiotherapy. Nat. Med. 1, 1029–1034. [DOI] [PubMed] [Google Scholar]
- Blagosklonny M.V., Toretsky J., and Neckers L. (1995). Geldanamycin selectively destabilizes and conformationally alters mutated p53. Oncogene 11, 933–939. [PubMed] [Google Scholar]
- Blagosklonny M.V., Trostel S., Kayastha G., et al. (2005). Depletion of mutant p53 and cytotoxicity of histone deacetylase inhibitors. Cancer Res. 65, 7386–7392. [DOI] [PubMed] [Google Scholar]
- Blagosklonny M.V., Toretsky J., Bohen S., et al. (1996). Mutant conformation of p53 translated in vitro or in vivo requires functional HSP90. Proc. Natl Acad. Sci. USA 93, 8379–8383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossi G., Marampon F., Maor-Aloni R., et al. (2008). Conditional RNA interference in vivo to study mutant p53 oncogenic gain of function on tumor malignancy. Cell Cycle 7, 1870–1879. [DOI] [PubMed] [Google Scholar]
- Bougeard G., Sesboue R., Baert-Desurmont S., et al. (2008). Molecular basis of the Li-Fraumeni syndrome: an update from the French LFS families. J. Med. Genet. 45, 535–538. [DOI] [PubMed] [Google Scholar]
- Burmakin M., Shi Y., Hedstrom E., et al. (2013). Dual targeting of wild-type and mutant p53 by small molecule RITA results in the inhibition of N-Myc and key survival oncogenes and kills neuroblastoma cells in vivo and in vitro. Clin Cancer Res. 19, 5092–5103. [DOI] [PubMed] [Google Scholar]
- Bykov V.J.N., Eriksson S.E., Bianchi J., et al. (2018). Targeting mutant p53 for efficient cancer therapy. Nat. Rev. Cancer 18, 89–102. [DOI] [PubMed] [Google Scholar]
- Bykov V.J., Issaeva N., Shilov A., et al. (2002). Restoration of the tumor suppressor function to mutant p53 by a low-molecular-weight compound. Nat. Med. 8, 282–288. [DOI] [PubMed] [Google Scholar]
- Bykov V.J., Zache N., Stridh H., et al. (2005). PRIMA-1(MET) synergizes with cisplatin to induce tumor cell apoptosis. Oncogene 24, 3484–3491. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. (2011). Integrated genomic analyses of ovarian carcinoma. Nature 474, 609–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao T., Zhou X., Cao B., et al. (2016). Pleckstrin homology domain-containing protein PHLDB3 supports cancer growth via a negative feedback loop involving p53. Nat. Commun. 7, 13755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chee J.L., Saidin S., Lane D.P., et al. (2013). Wild-type and mutant p53 mediate cisplatin resistance through interaction and inhibition of active caspase-9. Cell Cycle 12, 278–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin K.V., Ueda K., Pastan I., et al. (1992). Modulation of activity of the promoter of the human MDR1 gene by Ras and p53. Science 255, 459–462. [DOI] [PubMed] [Google Scholar]
- Cordani M., Butera G., Pacchiana R., et al. (2017). Molecular interplay between mutant p53 proteins and autophagy in cancer cells. Biochim. Biophys. Acta 1867, 19–28. [DOI] [PubMed] [Google Scholar]
- Cordani M., Oppici E., Dando I., et al. (2016). Mutant p53 proteins counteract autophagic mechanism sensitizing cancer cells to mTOR inhibition. Mol. Oncol. 10, 1008–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Agostino S., Strano S., Emiliozzi V., et al. (2006). Gain of function of mutant p53: the mutant p53/NF-Y protein complex reveals an aberrant transcriptional mechanism of cell cycle regulation. Cancer Cell 10, 191–202. [DOI] [PubMed] [Google Scholar]
- Di Como C.J., Gaiddon C., and Prives C. (1999). p73 function is inhibited by tumor-derived p53 mutants in mammalian cells. Mol. Cell. Biol. 19, 1438–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmer D., Pati S., Zambetti G., et al. (1993). Gain of function mutations in p53. Nat. Genet. 4, 42–46. [DOI] [PubMed] [Google Scholar]
- Do P.M., Varanasi L., Fan S., et al. (2012). Mutant p53 cooperates with ETS2 to promote etoposide resistance. Genes Dev. 26, 830–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donadelli M., Dando I., Zaniboni T., et al. (2011). Gemcitabine/cannabinoid combination triggers autophagy in pancreatic cancer cells through a ROS-mediated mechanism. Cell Death Dis. 2, e152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donehower L.A., and Lozano G. (2009). 20 years studying p53 functions in genetically engineered mice. Nat. Rev. Cancer 9, 831–841. [DOI] [PubMed] [Google Scholar]
- Dong P., Karaayvaz M., Jia N., et al. (2013). Mutant p53 gain-of-function induces epithelial-mesenchymal transition through modulation of the miR-130b-ZEB1 axis. Oncogene 32, 3286–3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donzelli S., Fontemaggi G., Fazi F., et al. (2012). MicroRNA-128-2 targets the transcriptional repressor E2F5 enhancing mutant p53 gain of function. Cell Death Differ. 19, 1038–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan W., Ding H., Subler M.A., et al. (2002). Lung-specific expression of human mutant p53-273H is associated with a high frequency of lung adenocarcinoma in transgenic mice. Oncogene 21, 7831–7838. [DOI] [PubMed] [Google Scholar]
- Duffy M.J., Synnott N.C., and Crown J. (2017). Mutant p53 as a target for cancer treatment. Eur. J. Cancer 83, 258–265. [DOI] [PubMed] [Google Scholar]
- Durkin S.G., and Glover T.W. (2007). Chromosome fragile sites. Annu. Rev. Genet. 41, 169–192. [DOI] [PubMed] [Google Scholar]
- Egorin M.J., Rosen D.M., Wolff J.H., et al. (1998). Metabolism of 17-(allylamino)-17-demethoxygeldanamycin (NSC 330507) by murine and human hepatic preparations. Cancer Res. 58, 2385–2396. [PubMed] [Google Scholar]
- Eliyahu D., Raz A., Gruss P., et al. (1984). Participation of p53 cellular tumour antigen in transformation of normal embryonic cells. Nature 312, 646–649. [DOI] [PubMed] [Google Scholar]
- Escoll M., Gargini R., Cuadrado A., et al. (2017). Mutant p53 oncogenic functions in cancer stem cells are regulated by WIP through YAP/TAZ. Oncogene 36, 3515–3527. [DOI] [PubMed] [Google Scholar]
- Flesken-Nikitin A., Hwang C.I., Cheng C.Y., et al. (2013). Ovarian surface epithelium at the junction area contains a cancer-prone stem cell niche. Nature 495, 241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster B.A., Coffey H.A., Morin M.J., et al. (1999). Pharmacological rescue of mutant p53 conformation and function. Science 286, 2507–2510. [DOI] [PubMed] [Google Scholar]
- Frank A.K., Pietsch E.C., Dumont P., et al. (2011). Wild-type and mutant p53 proteins interact with mitochondrial caspase-3. Cancer Biol. Ther. 11, 740–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazier M.W., He X., Wang J., et al. (1998). Activation of c-myc gene expression by tumor-derived p53 mutants requires a discrete C-terminal domain. Mol. Cell. Biol. 18, 3735–3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed-Pastor W.A., Mizuno H., Zhao X., et al. (2012). Mutant p53 disrupts mammary tissue architecture via the mevalonate pathway. Cell 148, 244–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed-Pastor W.A., and Prives C. (2012). Mutant p53: one name, many proteins. Genes Dev. 26, 1268–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaiddon C., Lokshin M., Ahn J., et al. (2001). A subset of tumor-derived mutant forms of p53 down-regulate p63 and p73 through a direct interaction with the p53 core domain. Mol. Cell. Biol. 21, 1874–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghebranious N., and Sell S. (1998). The mouse equivalent of the human p53ser249 mutation p53ser246 enhances aflatoxin hepatocarcinogenesis in hepatitis B surface antigen transgenic and p53 heterozygous null mice. Hepatology 27, 967–973. [DOI] [PubMed] [Google Scholar]
- Girardini J.E., Napoli M., Piazza S., et al. (2011). A Pin1/mutant p53 axis promotes aggressiveness in breast cancer. Cancer Cell 20, 79–91. [DOI] [PubMed] [Google Scholar]
- Gloushankova N., Ossovskaya V., Vasiliev J., et al. (1997). Changes in p53 expression can modify cell shape of ras-transformed fibroblasts and epitheliocytes. Oncogene 15, 2985–2989. [DOI] [PubMed] [Google Scholar]
- Green D.R., and Kroemer G. (2009). Cytoplasmic functions of the tumour suppressor p53. Nature 458, 1127–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W.J., Zhang Y.M., Zhang L., et al. (2013). Novel monofunctional platinum (II) complex Mono-Pt induces apoptosis-independent autophagic cell death in human ovarian carcinoma cells, distinct from cisplatin. Autophagy 9, 996–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurpinar E., and Vousden K.H. (2015). Hitting cancers’ weak spots: vulnerabilities imposed by p53 mutation. Trends Cell Biol. 25, 486–495. [DOI] [PubMed] [Google Scholar]
- Gurtner A., Starace G., Norelli G., et al. (2010). Mutant p53-induced up-regulation of mitogen-activated protein kinase kinase 3 contributes to gain of function. J. Biol. Chem. 285, 14160–14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halazonetis T.D., Gorgoulis V.G., and Bartek J. (2008). An oncogene-induced DNA damage model for cancer development. Science 319, 1352–1355. [DOI] [PubMed] [Google Scholar]
- Hamada M., Fujiwara T., Hizuta A., et al. (1996). The p53 gene is a potent determinant of chemosensitivity and radiosensitivity in gastric and colorectal cancers. J. Cancer Res. Clin. Oncol. 122, 360–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D., and Weinberg R.A. (2011). Hallmarks of cancer: the next generation. Cell 144, 646–674. [DOI] [PubMed] [Google Scholar]
- Hernandez I., Maddison L.A., Wei Y., et al. (2003). Prostate-specific expression of p53(R172L) differentially regulates p21, Bax, and mdm2 to inhibit prostate cancer progression and prolong survival. Mol. Cancer Res. 1, 1036–1047. [PubMed] [Google Scholar]
- Horio Y., Takahashi T., Kuroishi T., et al. (1993). Prognostic significance of p53 mutations and 3p deletions in primary resected non-small cell lung cancer. Cancer Res. 53, 1–4. [PubMed] [Google Scholar]
- Jenkins J.R., Rudge K., and Currie G.A. (1984). Cellular immortalization by a cDNA clone encoding the transformation-associated phosphoprotein p53. Nature 312, 651–654. [DOI] [PubMed] [Google Scholar]
- Ji L., Xu J., Liu J., et al. (2015). Mutant p53 promotes tumor cell malignancy by both positive and negative regulation of the transforming growth factor β (TGF-β) pathway. J. Biol. Chem. 290, 11729–11740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W.G., Sanders A.J., Katoh M., et al. (2015). Tissue invasion and metastasis: Molecular, biological and clinical perspectives. Semin. Cancer Biol. 35(Suppl), S244–S275. [DOI] [PubMed] [Google Scholar]
- Kaelin W.G., Jr. (2005). The concept of synthetic lethality in the context of anticancer therapy. Nat. Rev. Cancer 5, 689–698. [DOI] [PubMed] [Google Scholar]
- Kalo E., Kogan-Sakin I., Solomon H., et al. (2012). Mutant p53R273H attenuates the expression of phase 2 detoxifying enzymes and promotes the survival of cells with high levels of reactive oxygen species. J. Cell Sci. 125, 5578–5586. [DOI] [PubMed] [Google Scholar]
- Kastenhuber E.R., and Lowe S.W. (2017). Putting p53 in context. Cell 170, 1062–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.P., and Lozano G. (2018). Mutant p53 partners in crime. Cell Death Differ. 25, 161–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T., Veronese A., Pichiorri F., et al. (2011). p53 regulates epithelial-mesenchymal transition through microRNAs targeting ZEB1 and ZEB2. J. Exp. Med. 208, 875–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch W.M., Brennan J.A., Zahurak M., et al. (1996). p53 mutation and locoregional treatment failure in head and neck squamous cell carcinoma. J. Natl Cancer Inst. 88, 1580–1586. [DOI] [PubMed] [Google Scholar]
- Kondo Y., and Kondo S. (2006). Autophagy and cancer therapy. Autophagy 2, 85–90. [DOI] [PubMed] [Google Scholar]
- Kuerbitz S.J., Plunkett B.S., Walsh W.V., et al. (1992). Wild-type p53 is a cell cycle checkpoint determinant following irradiation. Proc. Natl. Acad. Sci. USA 89, 7491–7495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert J.M., Gorzov P., Veprintsev D.B., et al. (2009). PRIMA-1 reactivates mutant p53 by covalent binding to the core domain. Cancer Cell 15, 376–388. [DOI] [PubMed] [Google Scholar]
- Lang G.A., Iwakuma T., Suh Y.A., et al. (2004). Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell 119, 861–872. [DOI] [PubMed] [Google Scholar]
- Lavigueur A., Maltby V., Mock D., et al. (1989). High incidence of lung, bone, and lymphoid tumors in transgenic mice overexpressing mutant alleles of the p53 oncogene. Mol. Cell. Biol. 9, 3982–3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.M., and Bernstein A. (1993). p53 mutations increase resistance to ionizing radiation. Proc. Natl Acad. Sci. USA 90, 5742–5746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leijen S., van Geel R.M., Pavlick A.C., et al. (2016. a). Phase I study evaluating WEE1 inhibitor AZD1775 as monotherapy and in combination with gemcitabine, cisplatin, or carboplatin in patients with advanced solid tumors. J. Clin. Oncol. 34, 4371–4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leijen S., van Geel R.M., Sonke G.S., et al. (2016. b). Phase II study of WEE1 inhibitor AZD1775 plus carboplatin in patients with TP53-mutated ovarian cancer refractory or resistant to first-line therapy within 3 months. J. Clin. Oncol. 34, 4354–4361. [DOI] [PubMed] [Google Scholar]
- Levine A.J., and Oren M. (2009). The first 30 years of p53: growing ever more complex. Nat. Rev. Cancer 9, 749–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Greenberg N., Stephens L.C., et al. (1994). Preferential overexpression of a 172Arg→Leu mutant p53 in the mammary gland of transgenic mice results in altered lobuloalveolar development. Cell Growth Differ. 5, 711–721. [PubMed] [Google Scholar]
- Li D., Marchenko N.D., and Moll U.M. (2011. a). SAHA shows preferential cytotoxicity in mutant p53 cancer cells by destabilizing mutant p53 through inhibition of the HDAC6-Hsp90 chaperone axis. Cell Death Differ. 18, 1904–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Marchenko N.D., Schulz R., et al. (2011. b). Functional inactivation of endogenous MDM2 and CHIP by HSP90 causes aberrant stabilization of mutant p53 in human cancer cells. Mol. Cancer Res. 9, 577–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Rosen J.M., McMenamin-Balano J., et al. (1997). neu/ERBB2 cooperates with p53-172H during mammary tumorigenesis in transgenic mice. Mol. Cell. Biol. 17, 3155–3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao P., Zeng S.X., Zhou X., et al. (2017). Mutant p53 gains its function via c-Myc activation upon CDK4 phosphorylation at serine 249 and consequent PIN1 binding. Mol. Cell 68, 1134–1146 e1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisek K., Campaner E., Ciani Y., et al. (2018). Mutant p53 tunes the NRF2-dependent antioxidant response to support survival of cancer cells. Oncotarget 9, 20508–20523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D.S., Duong C.P., Haupt S., et al. (2017). Inhibiting the system xC− glutathione axis selectively targets cancers with mutant-p53 accumulation. Nat. Commun. 8, 14844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K., Ling S., and Lin W.C. (2011). TopBP1 mediates mutant p53 gain of function through NF-Y and p63/p73. Mol. Cell. Biol. 31, 4464–4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Wilcken R., Joerger A.C., et al. (2013). Small molecule induced reactivation of mutant p53 in cancer cells. Nucleic Acids Res. 41, 6034–6044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorente M., Torres S., Salazar M., et al. (2011). Stimulation of the midkine/ALK axis renders glioma cells resistant to cannabinoid antitumoral action. Cell Death Differ. 18, 959–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe S.W., Bodis S., McClatchey A., et al. (1994). p53 status and the efficacy of cancer therapy in vivo. Science 266, 807–810. [DOI] [PubMed] [Google Scholar]
- Marino G., Niso-Santano M., Baehrecke E.H., et al. (2014). Self-consumption: the interplay of autophagy and apoptosis. Nat. Rev. Mol. Cell Biol. 15, 81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masciarelli S., Fontemaggi G., Di Agostino S., et al. (2014). Gain-of-function mutant p53 downregulates miR-223 contributing to chemoresistance of cultured tumor cells. Oncogene 33, 1601–1608. [DOI] [PubMed] [Google Scholar]
- Matheson C.J., Backos D.S., and Reigan P. (2016). Targeting WEE1 kinase in cancer. Trends Pharmacol. Sci. 37, 872–881. [DOI] [PubMed] [Google Scholar]
- McBride K.A., Ballinger M.L., Killick E., et al. (2014). Li-Fraumeni syndrome: cancer risk assessment and clinical management. Nature reviews. Clin. Oncol. 11, 260–271. [DOI] [PubMed] [Google Scholar]
- Mizuarai S., Yamanaka K., and Kotani H. (2006). Mutant p53 induces the GEF-H1 oncogene, a guanine nucleotide exchange factor-H1 for RhoA, resulting in accelerated cell proliferation in tumor cells. Cancer Res. 66, 6319–6326. [DOI] [PubMed] [Google Scholar]
- Mizuno H., Spike B.T., Wahl G.M., et al. (2010). Inactivation of p53 in breast cancers correlates with stem cell transcriptional signatures. Proc. Natl Acad. Sci. USA 107, 22745–22750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molchadsky A., and Rotter V. (2017). p53 and its mutants on the slippery road from stemness to carcinogenesis. Carcinogenesis 38, 347–358. [DOI] [PubMed] [Google Scholar]
- Morselli E., Tasdemir E., Maiuri M.C., et al. (2008). Mutant p53 protein localized in the cytoplasm inhibits autophagy. Cell Cycle 7, 3056–3061. [DOI] [PubMed] [Google Scholar]
- Muller P.A., Caswell P.T., Doyle B., et al. (2009). Mutant p53 drives invasion by promoting integrin recycling. Cell 139, 1327–1341. [DOI] [PubMed] [Google Scholar]
- Muller P.A., and Vousden K.H. (2014). Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell 25, 304–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller P.A., Vousden K.H., and Norman J.C. (2011). p53 and its mutants in tumor cell migration and invasion. J. Cell Biol. 192, 209–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrini S., Gorgoulis V.G., and Halazonetis T.D. (2010). Genomic instability—an evolving hallmark of cancer. Nat. Rev. Mol. Cell Biol. 11, 220–228. [DOI] [PubMed] [Google Scholar]
- Okorokov A.L., Sherman M.B., Plisson C., et al. (2006). The structure of p53 tumour suppressor protein reveals the basis for its functional plasticity. EMBO J. 25, 5191–5200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive K.P., Tuveson D.A., Ruhe Z.C., et al. (2004). Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell 119, 847–860. [DOI] [PubMed] [Google Scholar]
- Padmanabhan A., Candelaria N., Wong K.K., et al. (2018). USP15-dependent lysosomal pathway controls p53-R175H turnover in ovarian cancer cells. Nat. Commun. 9, 1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parada L.F., Land H., Weinberg R.A., et al. (1984). Cooperation between gene encoding p53 tumour antigen and ras in cellular transformation. Nature 312, 649–651. [DOI] [PubMed] [Google Scholar]
- Patch A.M., Christie E.L., Etemadmoghadam D., et al. (2015). Whole-genome characterization of chemoresistant ovarian cancer. Nature 521, 489–494. [DOI] [PubMed] [Google Scholar]
- Pattabiraman D.R., and Weinberg R.A. (2014). Tackling the cancer stem cells - what challenges do they pose? Nat. Rev. Drug Discov. 13, 497–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y., Chen L., Li C., et al. (2001). Inhibition of MDM2 by hsp90 contributes to mutant p53 stabilization. J. Biol. Chem. 276, 40583–40590. [DOI] [PubMed] [Google Scholar]
- Peng X., Zhang M.Q., Conserva F., et al. (2013). APR-246/PRIMA-1MET inhibits thioredoxin reductase 1 and converts the enzyme to a dedicated NADPH oxidase. Cell Death Dis. 4, e881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley T., Sontag E., Chen P., et al. (2008). Transcriptional control of human p53-regulated genes. Nat. Rev. Mol. Cell Biol. 9, 402–412. [DOI] [PubMed] [Google Scholar]
- Roger L., Jullien L., Gire V., et al. (2010). Gain of oncogenic function of p53 mutants regulates E-cadherin expression uncoupled from cell invasion in colon cancer cells. J. Cell Sci. 123, 1295–1305. [DOI] [PubMed] [Google Scholar]
- Sabapathy K., and Lane D.P. (2018). Therapeutic targeting of p53: all mutants are equal, but some mutants are more equal than others. Nat. Rev. Clin. Oncol. 15, 13–30. [DOI] [PubMed] [Google Scholar]
- Salazar M., Carracedo A., Salanueva I.J., et al. (2009). Cannabinoid action induces autophagy-mediated cell death through stimulation of ER stress in human glioma cells. J. Clin. Invest. 119, 1359–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salim K.Y., Maleki Vareki S., Danter W.R., et al. (2016). COTI-2, a novel small molecule that is active against multiple human cancer cell lines in vitro and in vivo. Oncotarget 7, 41363–41379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarig R., Rivlin N., Brosh R., et al. (2010). Mutant p53 facilitates somatic cell reprogramming and augments the malignant potential of reprogrammed cells. J. Exp. Med. 207, 2127–2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelling A.N. (1997). Role of p53 in drug resistance in ovarian cancer. Lancet 349, 744–745. [DOI] [PubMed] [Google Scholar]
- Shetzer Y., Molchadsky A., and Rotter V. (2016). Oncogenic mutant p53 gain of function nourishes the vicious cycle of tumor development and cancer stem-cell formation. Cold Spring Harb Perspect. Med. 6, pii: a026203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shounan Y., MacKenzie K., Dolnikov A., et al. (1997). Myeloproliferative disease and myelodysplastic syndrome induced by transplantation of bone marrow cells expressing mutant p53. Leukemia 11, 1641–1649. [DOI] [PubMed] [Google Scholar]
- Solomon H., Dinowitz N., Pateras I.S., et al. (2018). Mutant p53 gain of function underlies high expression levels of colorectal cancer stem cells markers. Oncogene 37, 1669–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H., Hollstein M., and Xu Y. (2007). p53 gain-of-function cancer mutants induce genetic instability by inactivating ATM. Nat. Cell Biol. 9, 573–580. [DOI] [PubMed] [Google Scholar]
- Soragni A., Janzen D.M., Johnson L.M., et al. (2016). A designed inhibitor of p53 aggregation rescues p53 tumor suppression in ovarian carcinomas. Cancer Cell 29, 90–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojanovic N., Hassan Z., Wirth M., et al. (2017). HDAC1 and HDAC2 integrate the expression of p53 mutants in pancreatic cancer. Oncogene 36, 1804–1815. [DOI] [PubMed] [Google Scholar]
- Sui X., Chen R., Wang Z., et al. (2013). Autophagy and chemotherapy resistance: a promising therapeutic target for cancer treatment. Cell Death Dis. 4, e838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai W.T., Shiau C.W., Chen H.L., et al. (2013). Mcl-1-dependent activation of Beclin 1 mediates autophagic cell death induced by sorafenib and SC-59 in hepatocellular carcinoma cells. Cell Death Dis. 4, e485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzian T., Suh Y.A., Iwakuma T., et al. (2008). The inherent instability of mutant p53 is alleviated by Mdm2 or p16INK4a loss. Genes Dev. 22, 1337–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessoulin B., Descamps G., Moreau P., et al. (2014). PRIMA-1Met induces myeloma cell death independent of p53 by impairing the GSH/ROS balance. Blood 124, 1626–1636. [DOI] [PubMed] [Google Scholar]
- Tidow H., Melero R., Mylonas E., et al. (2007). Quaternary structures of tumor suppressor p53 and a specific p53 DNA complex. Proc. Natl Acad. Sci. USA 104, 12324–12329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trepel J., Mollapour M., Giaccone G., et al. (2010). Targeting the dynamic HSP90 complex in cancer. Nat. Rev. Cancer 10, 537–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakifahmetoglu-Norberg H., Kim M., Xia H.G., et al. (2013). Chaperone-mediated autophagy degrades mutant p53. Genes Dev. 27, 1718–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden M.G., Cantley L.C., and Thompson C.B. (2009). Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez A., Bond E.E., Levine A.J., et al. (2008). The genetics of the p53 pathway, apoptosis and cancer therapy. Nat. Rev. Drug Discov. 7, 979–987. [DOI] [PubMed] [Google Scholar]
- Walerych D., Lisek K., Sommaggio R., et al. (2016). Proteasome machinery is instrumental in a common gain-of-function program of the p53 missense mutants in cancer. Nat. Cell Biol. 18, 897–909. [DOI] [PubMed] [Google Scholar]
- Wang Z.T., Chen Z.J., Jiang G.M., et al. (2016). Histone deacetylase inhibitors suppress mutant p53 transcription via HDAC8/YY1 signals in triple negative breast cancer cells. Cell. Signal. 28, 506–515. [DOI] [PubMed] [Google Scholar]
- Wang H., Liao P., Zeng S.X., et al. (2018). It takes a team: a gain-of-function story of p53-R249S. J. Mol. Cell Biol. doi:10.1093/jmcb/mjy086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Yang J., Zheng H., et al. (2009). Expression of mutant p53 proteins implicates a lineage relationship between neural stem cells and malignant astrocytic glioma in a murine model. Cancer Cell 15, 514–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. (1956). On respiratory impairment in cancer cells. Science 124, 269–270. [PubMed] [Google Scholar]
- Wattel E., Preudhomme C., Hecquet B., et al. (1994). p53 mutations are associated with resistance to chemotherapy and short survival in hematologic malignancies. Blood 84, 3148–3157. [PubMed] [Google Scholar]
- Weinmann L., Wischhusen J., Demma M.J., et al. (2008). A novel p53 rescue compound induces p53-dependent growth arrest and sensitises glioma cells to Apo2L/TRAIL-induced apoptosis. Cell Death Differ. 15, 718–729. [DOI] [PubMed] [Google Scholar]
- West A.N., Ribeiro R.C., Jenkins J., et al. (2006). Identification of a novel germ line variant hotspot mutant p53-R175L in pediatric adrenal cortical carcinoma. Cancer Res. 66, 5056–5062. [DOI] [PubMed] [Google Scholar]
- Wilson W.H., Teruya-Feldstein J., Fest T., et al. (1997). Relationship of p53, bcl-2, and tumor proliferation to clinical drug resistance in non-Hodgkin’s lymphomas. Blood 89, 601–609. [PubMed] [Google Scholar]
- Xiong H.Y., Guo X.L., Bu X.X., et al. (2010). Autophagic cell death induced by 5-FU in Bax or PUMA deficient human colon cancer cell. Cancer Lett. 288, 68–74. [DOI] [PubMed] [Google Scholar]
- Yan W., and Chen X. (2009). Identification of GRO1 as a critical determinant for mutant p53 gain of function. J. Biol. Chem. 284, 12178–12187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W., Liu S., Xu E., et al. (2013). Histone deacetylase inhibitors suppress mutant p53 transcription via histone deacetylase 8. Oncogene 32, 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Vazquez A., Levine A.J., et al. (2012). Allele-specific p53 mutant reactivation. Cancer Cell 21, 614–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zastawny R.L., Salvino R., Chen J., et al. (1993). The core promoter region of the P-glycoprotein gene is sufficient to confer differential responsiveness to wild-type and mutant p53. Oncogene 8, 1529–1535. [PubMed] [Google Scholar]
- Zerdoumi Y., Aury-Landas J., Bonaiti-Pellie C., et al. (2013). Drastic effect of germline TP53 missense mutations in Li-Fraumeni patients. Hum. Mutat. 34, 453–461. [DOI] [PubMed] [Google Scholar]
- Zhang Z.Y., Hong D., Nam S.H., et al. (2015). SIRT1 regulates oncogenesis via a mutant p53-dependent pathway in hepatocellular carcinoma. J. Hepatol. 62, 121–130. [DOI] [PubMed] [Google Scholar]
- Zhang C., Liu J., Liang Y., et al. (2013). Tumour-associated mutant p53 drives the Warburg effect. Nat. Commun. 4, 2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Xiong S., Liu B., et al. (2018). Somatic Trp53 mutations differentially drive breast cancer and evolution of metastases. Nat. Commun. 9, 3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C.Y., Grinkevich V.V., Nikulenkov F., et al. (2010). Rescue of the apoptotic-inducing function of mutant p53 by small molecule RITA. Cell Cycle 9, 1847–1855. [DOI] [PubMed] [Google Scholar]
- Zhao D., Tahaney W.M., Mazumdar A., et al. (2017). Molecularly targeted therapies for p53-mutant cancers. Cell. Mol. Life Sci. 74, 4171–4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Cao B., and Lu H. (2017). Negative auto-regulators trap p53 in their web. J. Mol. Cell Biol. 9, 62–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Hao Q., Liao P., et al. (2016). Nerve growth factor receptor negates the tumor suppressor p53 as a feedback regulator. eLife 5, e15099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G., Wang J., Zhao M., et al. (2014). Gain-of-function mutant p53 promotes cell growth and cancer cell metabolism via inhibition of AMPK activation. Mol. Cell 54, 960–974. [DOI] [PMC free article] [PubMed] [Google Scholar]