Abstract
Adipose tissue represents an abundant and easily accessible source of multipotent cells, which may serve as excellent building blocks for tissue engineering. This article presents a newly described protocol for isolating adipose-derived stromal cells (ASCs) from human lipoaspirate, compared to the standard protocol for harvesting ASCs established in 2001. Human ASC isolation is performed using two methods, and resultant cells are compared through cell yield, cell viability, cell proliferation and regenerative potential. The osteogenic and adipogenic potential of ASCs isolated using both protocols are assessed in vitro and gene expression analysis is performed. The focus of this series of protocols is the regenerative potential of both cell populations in vivo. As such, the two in vivo animal models described are fat graft retention (soft tissue reconstruction) and calvarial defect healing (bone regeneration). The techniques described comprise fat grafting with cell assisted lipotransfer, and calvarial defect creation healed with cell-seeded scaffolds.
Keywords: adipose derived stromal cell, calvarial defect, digest, fat graft, liposuction
INTRODUCTION
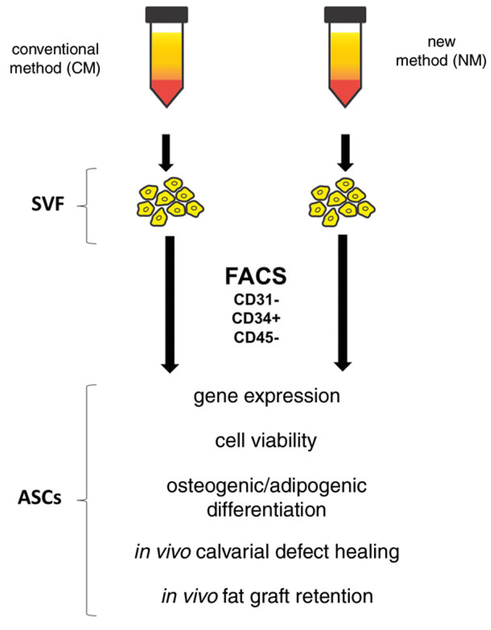
One of the most significant surgical challenges is represented by large bone defects resulting from trauma, tumor resection, and nonunion of fractures. Stem cell based therapies have been extensively studied as a method of bone regeneration. Ten years of both animal and human research has lead the way for promising osteogenic studies (Korbling & Estrov, 2003). Naturally, the healing of bones has been well characterized using bone marrow derived mesenchymal stem cells (BM-MSCs), and has been validated in multiple animal models (Chung et al., 2013; Koob et al., 2011; Levi et al., 2010; Schantz et al., 2003; Zuk et al., 2002). However, the harvesting of BM-MSCs requires collection of cancellous bone found in the medullary cavity. Adipose tissue is a practical alternative to bone marrow, given its abundance, ease of harvest, and lack of donor site morbidity. In 2001, Zuk described the adipose derived stromal cell (ASC) and it’s harvest method (Zuk et al., 2002; Zuk et al., 2001). ASCs possess similar properties to BM-MSCs, including similar cell surface marker profiles and differentiation potential (Uzbas et al., 2014). A key difference is the possible cellular yield from each of these tissues; 100 grams of lipoaspirate will yield 300-fold more cells than 100 ml bone marrow aspirate (Oedayrajsingh-Varma et al., 2006; Pittenger et al., 1999). The use of ASCs as building blocks for osseous regeneration has been researched in multiple different models also, proving important as a potential regenerative medicine therapy (Carvalho et al., 2013; Chung et al., 2013; Kim et al., 2012; Mizuno et al., 2012). Additionally, ASCs are a cell population which does not present any ethical concerns in the harvesting or use thereof, unlike the restrictions associated with embryonic or induced pluripotent stem cells. Our laboratory has shown that it is possible to isolate a subpopulation of ASCs with enhanced osteogenic potential (Chung et al., 2013; McArdle et al., 2014). Considering the hypothesis that fat graft survival and volume retention correlate directly with the amount of viable ASCs in the fat graft (Peer, 1955; Rieck & Schlaak, 2003), the present set of protocols aims to preserve ASC viability during harvest, isolation and transplantation in vivo. There are two processing methods detailed in this protocol; “conventional” method (CM) referring to the standard procedure described by Zuk, and the “new” method (NM) is one which was designed by our laboratory. We have observed a large increase in cell yield and viability using the new method; both critical parameters for downstream success of transplantation. Figure 2H.1.1 shows a schematic representation of the experiments performed in both methods.
Figure 2H.1.1.
Schematic of experimental flow in this protocol; using two different harvest methods for stromal vascular fraction (SVF), flow cytometry to separate adipose-derived stromal cells (ASCs), and downstream analytical in vitro and in vivo experiments.
BASIC PROTOCOL 1
FAT PROCESSING AND CELL HARVEST
This protocol describes two different processes of isolating the stromal vascular fraction (SVF) cells from adipose tissue. The endpoint of this protocol is a dense cell suspension in fully supplemented media.
Materials
Human lipoaspirate samples (biohazard, obtained using appropriate IRB and associated consent form)
Ice
Medium 199 (Gibco, cat. no. 11150059)
Type I collagenase 2.2 mg/ml (Sigma Aldrich)
Collagenase, from Clostridium histolyticum (Sigma Aldrich, cat. no. C6885)
DNase I (Roche, cat. no. 10104159001)
Calcium Chloride dehydrate (Sigma Aldrich, cat. no. C3306)
Bovine Serum Albumin (Sigma Aldrich, cat. no. A2058)
P188 (Sigma Aldrich)
50× HEPES (Life Technologies,)
500 ml sterile FACS buffer [1 × phosphate-buffered saline (PBS; pH 7.4, 1 × Gibco, 10010023), 2% fetal bovine serum, 1% P188, 1% penicillin-streptomycin]
Histopaque, a commercially available density gradient separation medium (SigmaAldrich, cat. no. 10771)
Hank’s balanced salt solution (Cellgro, cat. no. 55022PB)
Sterile serological pipettes (5, 10 and 25 ml; Corning, 357543, 357551, 357525)
Sterile plastic bottles for centrifuging (250 ml; Corning, 430776)
0.22-μm filter system
500-ml sterile PTEG medium bottle
Parafilm®
37°C water bath
Orbital shaker
Centrifuge
100-μm cell filter
Sterile polypropylene centrifuge tubes (50-ml; Fisher Scientific, cat. no. 1443222)
New method (NM)
1a. Place lipoaspirate on ice for 1 hour to allow the fat to congeal and to separate out the fat and blood. Prepare fresh collagenase digestion buffer using M199 medium, Type I collagenase 2.2 mg/ml, 1,000 units/ml DNase, 1000× 1mM calcium chloride, 10% bovine serum albumin, 100× P188, and 50× HEPES and filter using a 0.22-μm filter system.
2a. Transfer congealed fat to a 500-ml sterile PTEG medium bottle and add an equal volume of collagenase digestion buffer. Close and seal the lid with Parafilm®.
3a. Incubate the fat/collagenase mixture at 37°C in a water bath for 10 min to activate the collagenase. Then transfer this mixture to the orbital shaker for 20 min.
4a. Using sterile serological pipettes, neutralize collagenase activity by addition of an equal volume of fluorescent activated cell sorting (FACS) buffer (1 × PBS, 2% fetal bovine serum, 1% P188, 1% penicillin-streptomycin).
5a. Centrifuge the solution for 10 min at 1500 rpm, room temperature. Aspirate the supernatant, and resuspend the stromal vascular fraction (SVF) pellet in 15 ml of room temperature FACS buffer. Strain the suspension through a 100-μm cell filter.
6a. Add 15 ml histopaque, a commercially available density gradient separation medium, to a new 50-ml conical, and gently pour the strained cell solution on top of the histopaque in a 1:1 ratio.
7a. Centrifuge the solution for 15 min at 1450 rpm, room temperature, with acceleration set to low and deceleration settings inactivated.
8a. Transfer the resultant cloudy interface (buffy layer) to a new 50-ml conical, and make up the final volume to 30 ml with FACS buffer. Centrifuge the solution for 5 min at 1300 rpm, 4°C. Aspirate the supernatant and resuspend the pellet in 500 μl FACS buffer in preparation for FACS.
Conventional method (CM)
In the CM, SVF is isolated as previously described by Zuk et al. (2002). The procedure is briefly described below.
1b. Wash the raw lipoaspirate with PBS by adding an equal volume of PBS to the tissue and allow to separate by gravity at room temperature.
2b. Add an equal volume of 0.075% collagenase type I in Hank’s balanced salt solution, and shake for 1 hr at 37°C with gentle agitation (120 rpm).
3b. Treat the cellular pellet with Histopaque, a density gradient separation medium, and then resuspended in 500 μl of FACS buffer in preparation for FACS.
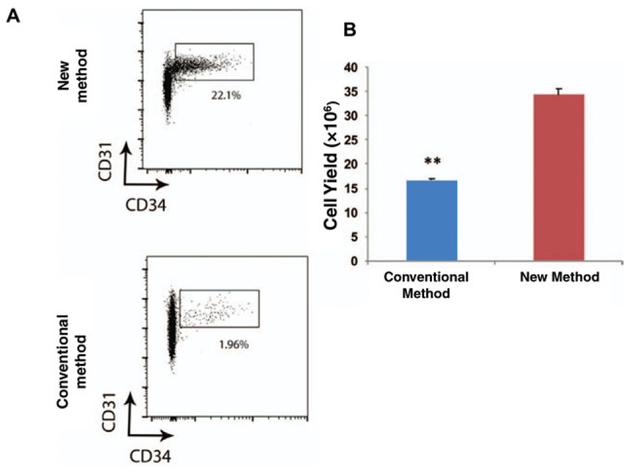
The NM and CM differ in two key areas: the constituents of the collagenase digestion buffer and the use of an orbital shaker. While NM is more labor intensive, we find that it yields a greater number of cells which have higher viability when directly compared to cells isolated from an identical volume of lipoaspirate using the CM. The two methods described above, for ASC isolation, yield statistically different quantities of cells as seen in FACS data output plots (Fig. 2H.1.2a,b)
Figure 2H.1.2.
(A) FACS plots demonstrating the frequency of CD45−CD31−CD34+ cells from the SVF. Top: cells isolated using the CM; bottom: cells isolated using the NM. (B) Graph representing the cell yield following FACS following CM (blue) versus NM (red). (**P 0.0017, t-test) (Used with permission from Plastic and Reconstructive Surgery) (Tevlin et al., 2016).
BASIC PROTOCOL 2
FACS SORTING
Flow cytometry is used here as a method to obtain ASCs from the isolated SVF cell population.
Materials
SVF cells (see Basic Protocol 1)
Human anti-CD45, anti-CD31, and anti-CD34 (BD Biosciences)
500 ml sterile FACS buffer [1×phosphate-buffered saline (PBS; pH 7.4, 1 × Gibco, 10010023), 2% fetal bovine serum, 1% P188, 1% penicillin-streptomycin]
BD FACSDiva™ software
FACS Aria II (BD Biosciences)
100-μm cell strainer (Corning, cat. no. 352360)
15-ml tubes
Incubate SVF cells for 30 min in a FACS buffer containing antibodies to human CD34, CD31, and CD45. Use BD FACSDiva™ software to run Accudrop Delay prior to physical sorting of ASCs using pre-calibrated streams.
Perform a sort on a FACS Aria II, using a 100-μm nozzle.
Sort cells into 15-ml tubes half filled with full medium (DMEM containing 10% FBS and 1% antibiotic) as a suitable buffer to enhance cell viability post-sort.
BASIC PROTOCOL 3
IN VITRO ASSAYS
This protocol details the in vitro tests of viability, adipogenic, and osteogenic potential of obtained ASCs.
Materials
SVF cells
Growth medium (DMEM, 10% FBS, and 1% penicillin/streptomycin) supplemented with recombinant FGF-2
BrdU kit (BrdU Cell Proliferation Kit, Abcam, ab12556)
Trypan blue
XTT-based assay: Cell Proliferation Kit II XTT (Roche Applied Science)
Osteogenic differentiation medium [Sigma Aldrich, L-ascorbic acid (A4403), glycerol phosphate disodium salt hydrate (G6501)]
Adipogenic differentiation medium [Sigma Aldrich, Indomethacin (cat. no. I7378),Dexamethasone (cat. no. D4902), IBMX (cat. no. I7018), Insulin (cat. no. 90177C)]
Dulbecco’s modified Eagle’s medium
Fetal bovine serum (FBS)
Penicillin/streptomycin
96-well plate/6-well plate, standard, untreated (Corning, cat. nos. 3898/CLS3516, respectively)
Spectrophotometer (Roche Applied Science)
GraphPad Prism
Light microscope
Additional reagents and solutions for alkaline phosphatase quantification (Levi et al., 2010), Oil Red O staining (Chung et al., 2013), and quantitative real-time polymerase chain reaction (qRT-PCR; see Basic Protocol 4)
Bromodeoxyuridine (BrdU) assay
Proliferation of NM-ASCs versus the CM-ASCs was assessed by BrdU incorporation, as previously described (James, Xu, Wang, & Longaker, 2008).
-
1
Seed SVF cells onto 96-well plates at a density of 1000 cells per well. After overnight attachment, treat cells with growth medium supplemented with recombinant FGF-2 (5, 10, 50, and 100 ng/ml).
-
2
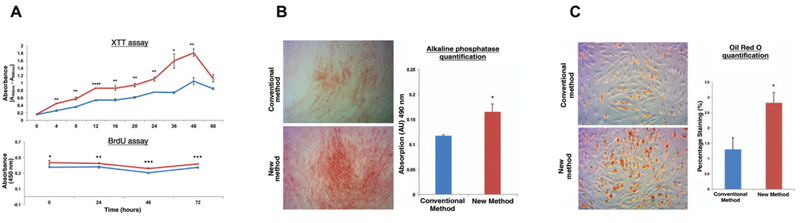
Change the medium every other day. At days 3 and 7, perform spectrophotometric assays per manufacturer protocol. Calculate means and standard deviations using GraphPad Prism statistical software (data output shown in Fig. 2H.1.3a, lower panel)
Figure 2H.1.3.
(A) Graphic representation of XTT assay results demonstrates greater viability of the ASCs isolated using the NM (red) versus CM (blue). Graphic representation of BrdU assay results demonstrate a significantly higher rate of cellular proliferation at all examined time points from 0–72 hr in the cells harvested with the NM (red) relative to the CM (blue) (t0 *p = 0.0487; t24 **p = 0.0086; t48 ***p 0.0003; t72 ***p = 0.0001). B) Representative Brightfield micrographs illustrating alkaline phosphatase staining of ASCs isolated using the CM (top) versus NM (bottom). Graphical representation of the alkaline phosphatase quantification post osteogenic differentiation in vitro demonstrates significantly greater alkaline phosphatase staining of the ASCs isolated using NM (red) versus CM (blue) (*p < 0.05, t-test). (C) Brightfield microscopy illustrating oil red O staining of ASCs isolated using the CM (top) versus the NM (bottom). Graphical representation of the oil red O staining quantification post adipogenic differentiation in vitro assay demonstrates significantly greater oil red O staining of the ASCs isolated using the NM (red) versus the CM (blue) (*p < 0.05, t-test). (Used with permission from Plastic and Reconstructive Surgery) (Tevlin et al., 2016).
XTT (2,3-Bis-(2-Methoxy-4-Nitro-5-Sulfophenyl)-2H-Tetrazolium-5-Carboxanilide) assay
-
3
Use 10 μl cell suspension mixed with 10 μl trypan blue to assess the viability of CM-ASCs versus NM-ASCs. Count viable, large, bright (non-blue) cells using a hemacytometer and observe the using using 20 × magnification on a light microscope. After the cells are harvested (see Basic Protocol 1), seed the cells into 96-well plates. The viable cells will adhere, and the dead cells will remain in suspension, and this will be removed with the first medium change of the plate.
-
4
Measure cell proliferation with an XTT-based assay to compare CM-ASCs versus NM-ASCs (Cell Proliferation Kit II XTT) (Chung et al., 2013) (data output shown in Fig. 2H.1.3a, upper panel).
In vitro osteogenic differentiation assay
-
3
For osteogenic differentiation, seed cells from the two harvest experimental groups at equal densities (100,000 cells per well) in 6-well culture plates using full growth medium (10% FBS, 1% penicillin/streptomycin).
-
4
After attachment, grow cells to at least 80% confluence before culturing in osteogenic differentiation medium (10% FBS, 1% penicillin/streptomycin, 250 μM ascorbate-2-phosphate, 10 mM glycerophosphate) (R&D Systems) (Levi et al., 2010).
-
5
Perform alkaline phosphatase quantification at 7 days, as previously described (Levi et al., 2010).
Data output shown in Fig. 2H.1.3B.
-
6
Analyze gene expression following 0, 7, and 14 days of differentiation by quantitative real-time polymerase chain reaction (qRT-PCR) (see Basic protocol 4).
In vitro adipogenesis
-
3
Seed freshly sorted cells onto 6-well plates (100,000 cells per well) using full growth medium (10% FBS, 1% penicillin/streptomycin).
-
4
Add 5 ml adipogenic differentiation medium (ADM), consisting of Dulbecco’s modified Eagle’s medium, 10% FBS, 1% penicillin/streptomycin, 10 ng/ml insulin, 1 nM dexamethasone, 0.5 mM methylxanthine, and 200 nM indomethacin, is added after cell attachment.
-
5
Perform Oil Red O staining at 7 days of differentiation (Chung et al., 2013).
Data output shown in Fig. 2H.1.3C.
-
6
Finally, examine specific gene expression after 7 days using qRT-PCR (see Basic Protocol 4 for methodology).
BASIC PROTOCOL 4
RNA HARVEST AND qRT-PCR
Below is a description of how to successfully harvest uncontaminated RNA from cells and briefly details the set up for a quantitative real time PCR (qRT-PCR).
Materials
Phosphate-buffered saline (PBS; Gibco, cat. no. 10010023)
TRIzol RNA Isolation Reagent (Thermo Fisher, cat. no. 10296010)
RNA Isolation: RNEasy Mini Kit (Qiagen, cat. no. 74104)
Omniscript RT Kit (Qiagen, cat. no. 205111)
Specific gene primer sequences were obtained from PrimerBank(https://pga.mgh.harvard.edu/primerbank/)
HotStarTaq DNA Polymerase (Qiagen)
Fast SYBR Green Master Mix (Thermo Fisher, cat. no. 4385610)
Pipettes and pipette tips, sterile
Cell scraper, sterile
1.5-ml microcentrifuge tubes (Fisher Brand, catalog no. 05-408-129)
Applied Biosystems Prism 7900HT Sequence Detection System (Applied Biosystems)
LightCycler software
Additional reagents and equipment for harvesting RNA (Levi et al., 2010)
NOTE: Steps 1 to 3 should be performed in a sterile cell culture hood.
While cells are still in 6-well plates, wash the plates three times, each time with 10 ml PBS to ensure total removal of medium. Any remaining FBS in the well will negatively impact the TRIzol™.
Aspirate PBS and pipette 200 μl of TRIzol™ onto each well. Use a sterile cell scraper to forcefully scrape the base of each plate, freeing the adherent cellular material from the polystyrene and releasing the RNA into the TRIzol™.
Use a sterile pipette tip to collect the RNA-saturated TRIzol™ and transfer into a sterile 1.5-microcentrifuge tube. Store indefinitely at −80°C until ready for RNA clean up and processing.
Harvest RNA at 0, 7, and 14 days post-commencement of ODM/ADM to determine specific gene relative expression, as previously described (Levi et al., 2010). Once all the time points are gathered, perform RNA clean up with Qiagen RNEasy Mini kit, per the manufacturer’s protocol.
Perform reverse transcription based on the quantified RNA per sample, per the manufacturer’s protocol. (Heat cycle sequence: 10 min 25°C, for 60 min 48°C, for 5 min at 95°C. Store up to 4 hr at 4°C until ready to use in qRT-PCR).
Perform qRT-PCR with the Applied Biosystems Prism 7900HT Sequence Detection System. Calculate the amount of PCR product using an external GAPDH standard curve and LightCycler software.
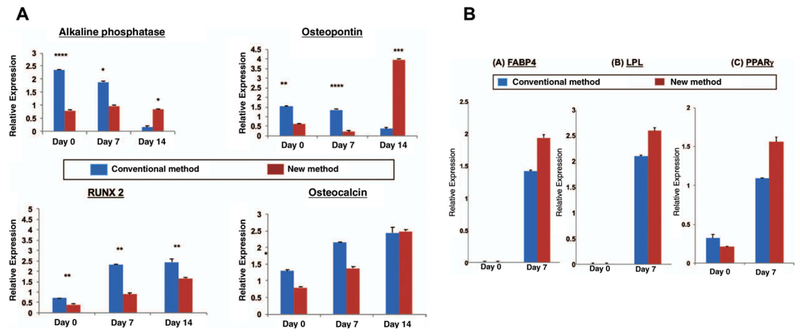
All values should be normalized against GAPDH expression (or alternate housekeeping gene, e.g.,β-actin) in the corresponding samples. Data output should be presented as values normalized to the housekeeping gene (Fig. 2H.1.4). Specific gene primer sequences were obtained from PrimerBank (https://pga.mgh.harvard.edu/primerbank/) (Panetta et al., 2009).
Figure 2H.1.4.
(A) Relative transcriptional expression of osteogenic genes; Alkaline phosphatase, Runx2, Osteopontin and Osteocalcin as determined by qRT-PCR. (B) Relative expression of adipogenic genes by FABP4, LPL, PPAR-g. (Used with permission from Plastic and Reconstructive Surgery) (Tevlin et al., 2016).
Briefly, we discovered that there is significantly higher osteogenic gene expression of cells obtained through the CM when compared to NM (Fig. 2H.1.4a). However, we observed a higher but non-significant expression of adipogenic genes in the ASCs harvested using the NM compared to CM (Fig. 2H.1.4b).
BASIC PROTOCOL 5
IN VIVO MOUSE CALVARIAL DEFECT MODEL
This protocol describes the surgical creation of a 4 mm circular defect in a mouse calvarium using a circular knife and dissection microscope.
Materials
CD-1 nude mice, 8 to10 weeks old (Charles River Laboratories)
Isothesia (Isoflurane, USP, Butler-Schein)
ASCs (see Basic Protocol 1)
(HA)-coated poly(lactic-co-glycolic acid) (PLGA) scaffold, 4 mm diameter (see Cowan et al., 2014)
UV light
96-well culture plates
NSK Z500 drill (Brassler)
4.0 mm circular knife (Xemax, cat. no. CK40)
NOTE: All research must be conducted through APLAC committee–approved protocols. Using your university guidelines, apply for ethical approval by writing a proposed animal protocol.
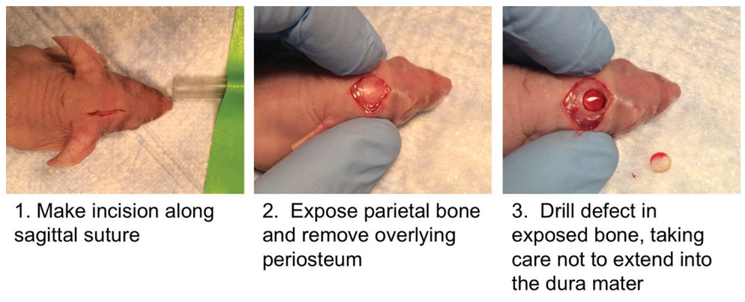
Create 4-mm calvarial defects in the right parietal bone of an 8-week-old male mice (Fig. 2H.1.5)
Figure 2H.1.5.
Macroscopic view of calvarial defect drilling in CD-1 nude mouse.
Athymic nude mice [Crl:NU(NCr)-Foxn1nu] were used for experiments in this study to minimize effects of an immune reaction to retention of human ASCs. This strain of mouse lacks a thymus and, therefore, all associated T cells.
Sterilize prefabricated, sterile hydroxyapatite (HA)-coated poly(lactic-co-glycolic acid) (PLGA) cast from 85/15 PLGA by solvent casting and a particulate leaching process, as previously illustrated (Cowan et al., 2004), by exposure to UV light.
In preparation for cell engraftment, seed scaffolds with 150,000 ASCs in 125 μl of traditional medium in 96-well culture plates 24-hr prior to implantation, as reported previously (Chung et al., 2013).
Divide the animals into test groups, received scaffolds seeded with CM or NM ASCs, including a negative control group with no ASCs.
Animals should be fully anesthetized by initial exposure in a chamber, and then operated upon while the mouse head sits in a nose cone, which is dispensing breathable isoflurane and 2% oxygen. Create the defect using the 4.0 mm circular knife, rotating at 40,000 rpm. Be careful to account for the vaulted surface of the skull, rotating the head and drill bit to evenly cut through the entirety of the skull (Fig. 2H.1.5).
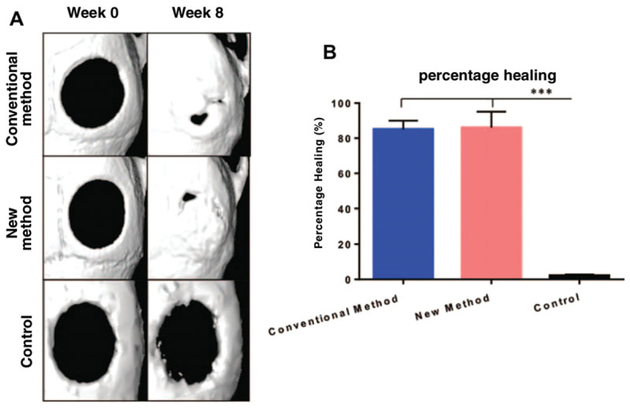
Use small animal micro-computed tomography (μ-CT) scanning to follow calvarial defect healing over 8 weeks (Fig. 2H.1.6a) (Chung et al., 2013).
Evaluate the area of the calvarial defect by quantifying pixels in the defect. Percentage healing is calculated by dividing the defect area by the size of the defect, as recorded on the immediate post-operative μ-CT scan (Fig. 2H.1.6b) (Chung et al., 2013).
Figure 2H.1.6.
(A) Micro-CT reconstructed images demonstrating the critical calvarial bone defects at week 0 (left) and week 8. (top: CM-ASCs; middle: NM-ASCs; bottom: Control scaffold, no cells added). (B) Graphical representation of the percentage healing at 8 weeks of calvarial defects treated with CM-ASCs (blue) versus NM-ASCs (red) method demonstrates equivocal results. These results are significantly different from the acellular control scaffold (black), which exhibits a non-healing critical sized defect (***p < 0.001). (Used with permission from Plastic and Reconstructive Surgery) (Tevlin et al., 2016).
BASIC PROTOCOL 6
IN VIVO FAT GRAFTING MODEL
This protocol describes a mouse model of fat grafting for soft tissue reconstruction.
Materials
Human lipoaspirate samples (biohazard, obtained using appropriate IRB and associated consent form)
Freshly harvested NM- or CM-ASCS (see Basic Protocol 1)
CD-1 nude mice (Charles River, Crl:CD1-Foxn1nu)
Isothesia (Isoflurane, USP, Butler-Schein)
Centrifuge
Small scissors
Absorbent pad
1-ml Luer-lock syringe (BD Biosciences, cat. no. 309628)
14-G blunt-tipped cannula (Tulip Medical, cat. no. INJ_LL)
6–0 Vicryl suture (Ethicon)
NOTE: All research must be conducted through APLAC committee–approved protocols.
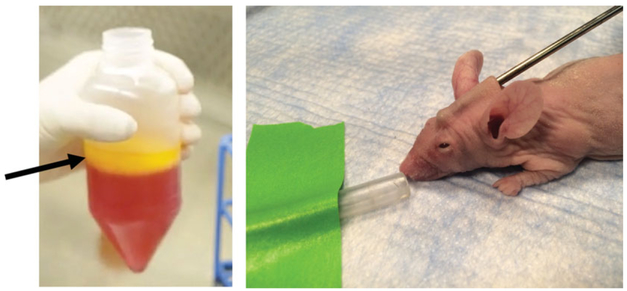
Minimally process whole lipoaspirate for grafting by centrifuging for 6 min at 1300 rpm, 4°C. Aspirate the liquid blood/debris and oil layers, and invert the tube on an absorbent pad for 5 min to remove any remaining oil.
Add freshly harvested NM- or CM-ASCs to the fat stromal layer at a ratio of 10,000 ASCs per 200 μl of fat. Transfer ASC-supplemented fat per group (NM/CM) to 1-ml syringes for injection into mouse scalps.
Animals should be fully anesthetized by initial exposure in a chamber, and then operated upon while the mouse head sits in a nose cone, which is dispensing breathable isoflurane and 2% oxygen. Using scissors, create a minimal incision in the scalp at the base of the skull, and inject a volume of 200 μl of fat into the subcutaneous plane beneath the scalp using a 14-G blunt-tipped cannula (Fig. 2H.1.7).
Close incisions with 6–0 Vicryl suture. Use small animal micro-computed tomography (μ-CT) scanning to follow calvarial defect healing.
Following a baseline volume measurement, perform serial imaging every 2 weeks over a total of 8 weeks.
Figure 2H.1.7.
(Left) Macroscopic view of fat graft model, involving image of raw fat, pre-digest. The layer to be grafted is indicated by the black arrow. (Right) Make small incision at the base of the neck and use a 14 g cannula to undermine the skin overlying the skull. With the beveled tip of the cannula facing upwards, inject 200 μl retrograde of prepared lipoaspirate into the space created by undermining. Retrograde injection means inject as you pull the needle out.
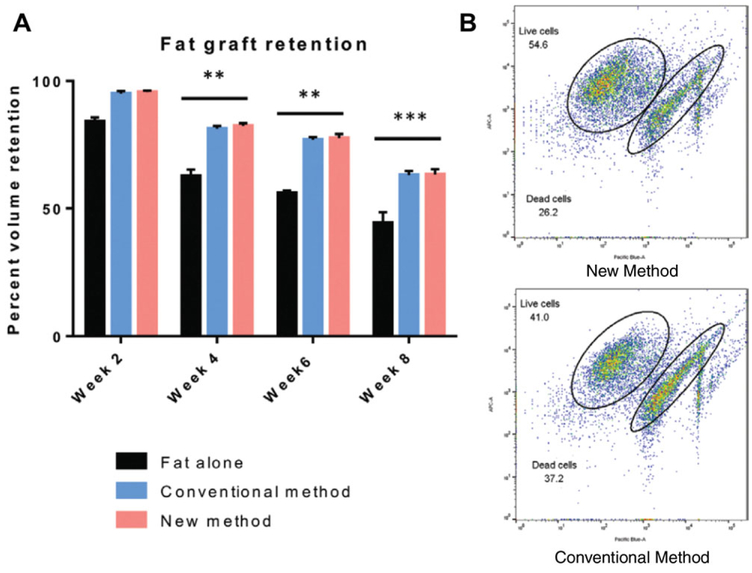
Images were reconstructed as a three-dimensional surface with cubic-spline interpolation using the Inveon Acquisition Workplace (n = 3) (Siemens) (Fig. 2H.1.8a).
Figure 2H.1.8.
(A) Graphical representation of fat graft retention showing an unsupplemented fat graft (black, Fat alone) compared to grafts supplemented with cells obtained using either the CM (blue) or NM (red). While significant differences exist between fat alone control group and groups supplied with ASCs, the graft retention shown with either population of harvested ASCs is equivocal. Data are normalized to Week 0. (**p < 0.01). (B) FACS plots show viability of implanted cells ex-vivo. Labeled cells are identified by presence of Q-dot label detected using APC, and analyzed for viability with DAPI, detected using Pacific Blue. Implanted cell viability was greater in NM-ASCs versus CM-ASCs (CM: 41% viability, NM: 54.6% viability). (Used with permission from Plastic and Reconstructive Surgery) (Tevlin et al., 2016).
BASIC PROTOCOL 7
IN VIVO CELL HARVEST FROM FAT GRAFTS
This protocol describes how to harvest a labeled cell population from fat grafts one week post-op.
Materials
Qtracker 655 Cell Labeling Kit (Thermo fisher Scientific, cat. no. Q25021MP)
500 ml sterile FACS buffer (1×PBS, 2% fetal bovine serum, 1% P188, 1% penicillin-streptomycin)
4′,6-Diamidine-2′-phenylindole dihydrochloride (DAPI)
Medium 199 (Gibco, cat. no. 11150059)
Type I collagenase, 2.2 mg/ml (Sigma Aldrich)
Full medium (DMEM, 10% FBS, 1% penicillin/streptomycin)
NOTE: All research must be conducted through APLAC committee–approved protocols.
Following the FACS detailed in Basic Protocol 2, label NM-ASCs and CM-ASCs separately with Qtracker 655 quantum dots for the purpose of identifying the cells ex vivo 48 hr later. Qtracker 655 is a particle, which is endocytosed and can be excited and analyzed using FACS (using channel APC-Cy7).
At a time point 48 hr post-implantation, explant the fat grafts and digest the tissue using collagenase dissolved in M199, full medium, and a 100-μm cell strainer, consistent with a previously described fat harvest protocol (Garza et al., 2014).
Resuspend the pellet of harvested cells using FACS buffer, with DAPI. Use DAPI as the viability stain (dead cells DAPI positive, live cells DAPI negative), which can be detected using the Pacific Blue channel on BD FACS Aria.
Analyze cells using the gates shown in Figure 2H.1.8b. Percentages of cells in each gate can be calculated using FlowJo software.
COMMENTARY
Background Information
There are two clinically relevant models detailed in this protocol series. While both pertain to plastic surgical and reconstructive procedures, they are highly versatile and allow for myriad hypotheses to be tested. For instance, the fat grafting model can be used in a cancer reconstruction setting. As many breast cancer patients opt for mastectomies, the next common step is reconstruction of the breast after adjuvant radiotherapy, which may be accomplished through fat grafting. It can be very useful to study the effect of irradiated soft tissue recipient sites on mouse fat grafts, with a lens on volume retention of the graft (Garza et al., 2014). Similarly, a model of fat grafting that is gaining traction for its elegance in patient-specific treatment is cell assisted lipotransfer (CAL); adding patient-derived ASCs to fat which is then grafted together. Using CAL in a model of fat grafting and irradiation helps us understand what affect the transplanted ASCs have within the adipose tissue. Previous work shows the capacity of the ASCs to rescue overlying skin in a model of irradiation; reducing fibrotic collagen content and returning normal skin mechanical strength (Luan et al., 2016).
In this situation, is it critical to know the parameters of the cells you are transplanting, and their in vitro capacity, which can allow accurate estimation of their effects in vivo.
Similarly, the model of calvarial defects may be extended to different bone healing hypotheses. It is important to remember the wealth of informative stains available for bone tissue. These can indicate endochondral or intramembranous bone formation, young or old bone, types of bone, and cartilaginous interfaces. Included are Movat’s pentachrome, Trichrome, Aniline blue staining, Alcian blue staining, Alkaline phosphatase, and Alizarin red staining. With more options for tissue analysis come more options for experimental groups. Transgenic mouse models are useful here for simple questioning of bone healing in a knockout model, e.g.: FAK knockout (Kim et al., 2007). Adding cellular components is not a new phenomenon in bone healing regenerative medicine strategies. Specifically studied are the pathologic mechanisms of bony non-unions from a cell therapy standpoint (hypertrophic and atrophic non-unions) (Gómez-Barrena et al., 2015). REBORNE is a multi-center European set of five clinical trials, all focused on the addition of cells to aid bone regeneration. While this consortium initially studied mesenchymal stem cells, there is a recent shift in focus to ASCs, specifically their differentiation to osteoblasts in decellularized bone tissue (Fisher, Peretti, & Scotti, 2016).
Critical Parameters
The cell harvest, FACS sort, and surgical models listed herein can be amalgamated in one day of experimental work. As such, an important consideration prior to embarking is ensuring all reagents are pre-prepared, surgical equipment sterilized, and appropriate facilities booked. The benefit of doing these experiments in the same day is the freshly harvested cells are not cultured in vitro prior to in vivo grafting; adding clinical applicability to the experiment.
While the fat grafting mouse model is relatively low risk, the drilling of the calvarial defect requires skill, practice, can be lethal to the mouse if not performed properly. The dura mater is a much vascularized meningeal layer and must not be disrupted by the drill bit. Breaching this layer will be immediately apparent to the surgeon by extensive bleeding. This injury does not lead to a successful transplant of a cell-laden scaffold, considering the amount of fluid in the operative zone. Meningitis or fatal intracranial bleeding is highly likely to result.
Troubleshooting
Having poor viability of FACS sorted ASCs is a common issue, considering the delicate nature of fresh cell populations. This can be solved by culturing in fully supplemented medium with 20% FBS as opposed to the standard 10%. This higher percentage of nutrients serves to create a “friendlier” environment, leading to increased cell adhesion and survival. Similarly, half-filling the sorting tubes with media containing 20% FBS means the sorting streams will flow into media; maximizing the survival of cells immediately post-sort.
A separate issue which people may face is a lower cell yield than expected. This may be the fault of many steps throughout the harvest procedure. However, making the collagenase digestion mixture involves using chilled M199 and frozen collagenase (a cold solution). Even though the digestion solution and fat mixture is put in an orbital shaker set at 37°C, the mixture itself may not actually reach 37°C; therefore, not activating the collagenase to its full enzymatic activity. Boosting the temperature of the orbital shaker to 40°C ensures a thorough thermal activation of the collagenase, and therefore reliable, high cell yields.
Anticipated Results
If the protocols are performed correctly throughout this procedure, the experimental team should have the means of collecting reliable data to question hypotheses based on ASC yield, survival, lineage commitment, and in vivo potential. With regards to cell yield per volume of adipose tissue, we have found that this varies greatly from donor to donor. There seems to be no fixed example of cell number to expect from a fixed volume of fat. Donor age, gender, smoking status, and comorbidities all appear to play a role in the presence of ASCs in fat. Similarly, another confounding factor may be the location from which the fat is harvested (e.g., abdomen, flank, arm, and thigh). The value of the two protocols described herein is that we have described a technique, which appears to yield the highest amount of viable cells from adipose tissue. If it is the aim of the research team to obtain as many cells as possible from the sample of adipose tissue (>20 million viable ASCs from approximately 300 ml of raw adipose tissue), then use of the NM protocol is advised. Alternatively, if the goal is to harvest under 20 million cells, the CM is slightly more user-friendly and does not require as much digest solution preparation, or expensive reagents (e.g., DNase).
Time Considerations
The series of protocols listed within are relatively straight forward with regard to timing. Having the solutions pre-prepared (media, FACS buffer, red cell lysis buffer, etc.) will lead to smooth, seamless steps in both in vitro and in vivo experimental stages. It is massively advantageous to have multiple teams of personnel if the goal is to complete the cell harvest, FACS sort, and surgical models all in one day. Division of tasks and use of waiting steps allows for a lot of work to run in parallel; for instance, using the 30 minute digest time in the NM, or 60 min digest in the CM allows for the operator to set up the flow cytometer.
Keeping reagents and cells on ice throughout the experimental day will help preserve cell survival and the longevity of reagents involved.
Acknowledgments
M.T.L. was supported by NIH grants, U01 HL099776, R01 DE021683, the Oak Foundation, Hagey Laboratory for Pediatric Regenerative Medicine, and the Gunn/Olivier Fund. D.C.W. was supported by the Hagey Laboratory for Pediatric Regenerative Medicine, NIH grant K08 DE024269, and the Stanford University Child Health Research Institute Faculty Scholar Award.
Literature Cited
- Carvalho PP, Leonor IB, Smith BJ, Dias IR, Reis RL, & Gimble JM (2013). Undifferentiated human adipose-derived stromal/stem cells loaded onto wet-spun starch-polycaprolactone scaffolds enhance bone regeneration: Nude mice calvarial defect in vivo study. Journal of Biomedical Materials Research Part A, 102(9), 3102–3111. doi: 10.1002/jbm.a.34983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung MT, Liu C, Hyun JS, Lo DD, Montoro DT, & Hasegawa M (2013). CD90 (Thy-1)-positive selection enhances osteogenic capacity of human adipose-derived stromal cells. Tissue Engineering Part A, 19(7–8), 989–997. doi: 10.1089/ten.tea.2012.0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan CM, Shi YY, Aalami OO, Chou YF, Mari C, & Thomas R (2004). Adipose-derived adult stromal cells heal critical-size mouse calvarial defects. Nature Biotechnology, 2004;22(5), 560–567. doi: 10.1038/nbt958. [DOI] [PubMed] [Google Scholar]
- Fisher JN, Peretti GM, & Scotti C (2016). Stem cells for bone regeneration: From cell-based therapies to decellularised engineered extracellular matrices. Stem Cells International, 9352598. doi: 10.1155/2016/9352598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garza RM, Paik KJ, Chung MT, Duscher D, Gurtner GC, & Longaker MT (2014). Studies in fat grafting: Part III. Fat grafting irradiated tissue–improved skin quality and de creased fat graft retention. Plastic and Reconstructive Surgery, 134(2), 249–257. doi: 10.1097/PRS.0000000000000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Barrena E, Rosset P, Lozano D, Stanovici J, Ermthaller C, & Gerbhard F (2015). Bone fracture healing: Cell therapy in delayed unions and nonunions. Bone, 70, 93–101. doi: 10.1016/j.bone.2014.07.033. [DOI] [PubMed] [Google Scholar]
- James AW, Xu Y, Wang R, & Longaker MT (2008). Proliferation, osteogenic differentiation, and fgf-2 modulation of posterofrontal/sagittal suture-derived mesenchymal cells in vitro. Plastic and Reconstructive Surgery, 122(1), 53–63. doi: 10.1097/PRS.0b013e31817747b5. [DOI] [PubMed] [Google Scholar]
- Kim HP, Ji YH, Rhee SC, Dhong ES, Park SH, & Yoon ES (2012). Enhancement of bone regeneration using osteogenic-induced adipose-derived stem cells combined with demineralized bone matrix in a rat critically-sized calvarial defect model. Current Stem Cell Research & Therapy, 7(3), 165–172. doi: 10.2174/157488812799859847. [DOI] [PubMed] [Google Scholar]
- Kim JB, Leucht P, Luppen CA, Park YJ, Beggs HE, & Damsky CH (2007). Reconciling the roles of FAK in osteoblast differentiation, osteoclast remodeling, and bone regeneration. Bone, 41(1), 39–51. doi: 10.1016/j.bone.2007.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob S, Torio-Padron N, Stark GB, Hannig C, Stankovic Z, & Finkenzeller G (2011). Bone formation and neovascularization mediated by mesenchymal stem cells and endothelial cells in critical-sized calvarial defects. Tissue Engineering Part A, 17(3–4), 311–321. doi: 10.1089/ten.tea.2010.0338. [DOI] [PubMed] [Google Scholar]
- Korbling M, & Estrov Z (2003). Adult stem cells for tissue repair - a new therapeutic concept? The New England Journal of Medicine, 349(6), 570–582. doi: 10.1056/NEJMra022361. [DOI] [PubMed] [Google Scholar]
- Levi B, James AW, Nelson ER, Vistnes D, Wu B, & Lee M (2010). Human adipose derived stromal cells heal critical size mouse calvarial defects. PLoS One, 5(6) doi: 10.1371/journal.pone.0011177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan A, Duscher D, Whittam AJ, Paik KJ, Zielins ER, Brett EA, … Wan DC (2016). Cell-Assisted lipotransfer improves volume retention in irradiated recipient sites and rescues radiation-induced skin changes. Stem Cells, 34(3), 668–673. doi: 10.1002/stem.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArdle A, Chung MT, Paik KJ, Duldulao C, Chan C, Rennert R, … Longaker MT (2014). Positive selection for bone morphogenetic protein receptor type-IB promotes differentiation and specification of human adipose-derived stromal cells toward an osteogenic lineage. Tissue Engineering Part A, 20(21–22), 3031–3040. doi: 10.1089/ten.tea.2014.0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno H, Tobita M, & Uysal AC (2012). Concise review: Adipose-derived stem cells as a novel tool for future regenerative medicine. Stem Cells (Dayton, Ohio), 30(5), 804–810. doi: 10.1002/stem.1076. [DOI] [PubMed] [Google Scholar]
- Oedayrajsingh-Varma MJ, van Ham SM, Knippenberg M, Helder MN, Klein-Nulend J, Schouten TE, … van Milligen FJ (2006). Adipose tissue-derived mesenchymal stem cell yield and growth characteristics are affected by the tissue-harvesting procedure. Cytotherapy, 8(2), 166–177. doi: 10.1080/14653240600621125. [DOI] [PubMed] [Google Scholar]
- Panetta NJ, Gupta DM, Kwan MD, Wan DC, Commons GW, & Longaker MT (2009). Tissue harvest by means of suction-assisted or third-generation ultrasound-assisted lipoaspiration has no effect on osteogenic potential of human adipose-derived stromal cells. Plastic and Reconstructive Surgery, 124(1), 65–73. doi: 10.1097/PRS.0b013e3181ab10cd. [DOI] [PubMed] [Google Scholar]
- Peer LA (1955). Cell survival theory versus replacement theory. Plastic and Reconstructive Surgery, (1946);16(3), 161–168. doi: 10.1097/00006534-195509000-00001. [DOI] [PubMed] [Google Scholar]
- Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, … Marshak DR (1999). Multilineage potential of adult human mesenchymal stem cells. Science (New York, NY), 284(5411), 143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- Rieck B, & Schlaak S (2003). Measurement in vivo of the survival rate in autologous adipocyte transplantation. Plastic and Reconstructive Surgery, 111(7), 2315–2323. doi: 10.1097/01.PRS.0000060797.59958.55. [DOI] [PubMed] [Google Scholar]
- Schantz JT, Hutmacher DW, Lam CX, Brinkmann M, Wong KM, Lim TC, … Teoh SH (2003). Repair of calvarial defects with customised tissue-engineered bone grafts II. Evaluation of cellular efficiency and efficacy in vivo. Tissue Engineering, 9(Suppl 1), S127–139. doi: 10.1089/10763270360697030. [DOI] [PubMed] [Google Scholar]
- Tevlin R, McArdle A, Brett E, Paik K, Seo EY, Walmsley GG, … Longaker MT (2016). A novel method of human adipose derived stem cell isolation with resultant increased cell yield. Plastic and Reconstructive Surgery, 138(6), 983e–996e. doi: 10.1097/PRS.0000000000002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzbas F, May ID, Parisi AM, Thompson SK, Kaya A, Perkins AD, & Memili E (2014). Molecular physiognomies and applications of adipose-derived stem cells. Stem Cell Reviews, 11(2), 298–308. doi: 10.1007/s12015-014-9578-0. [DOI] [PubMed] [Google Scholar]
- Zuk PA, Zhu M, Ashjian P, De Ugarte DA, Huang JI, Mizuno H, … Hedrick MH (2002). Human adipose tissue is a source of multipotent stem cells. Molecular Biology of the Cell, 13(12), 4279–4295. doi: 10.1091/mbc.E02-02-0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuk PA, Zhu M, Mizuno H, Huang J, Futrell JW, Katz AJ, … Hedrick MH (2001). Multilineage cells from human adipose tissue: Implications for cell-based therapies. Tissue Engineering, 7(2), 211–228. doi: 10.1089/107632701300062859. [DOI] [PubMed] [Google Scholar]