Abstract
A number of studies indicate that rare copy number variations (CNVs) contribute to the risk of schizophrenia (SCZ). Most of these studies have focused on protein-coding genes residing in the CNVs. Here, we investigated long non-coding RNAs (lncRNAs) within ten SCZ risk-associated CNV deletion regions (CNV-lncRNAs) and examined their potential contribution to SCZ risk. We used RNA-Seq transcriptomics data derived from postmortem brain tissue from control individuals without psychiatric disease as part of the PsychENCODE BrainGVEX and Developmental Capstone projects. We carried out weighted gene coexpression network analysis (WGCNA) to identify protein-coding genes coexpressed with CNV-lncRNAs in the human brain. We identified one neuronal function-related coexpression module shared by both data sets. This module contained a lncRNA within the 22q11.2 CNV region called DGCR5, which was identified as a hub gene. Protein-coding genes associated with SCZ GWAS signals, de novo mutations or differential expression were also contained in this neuronal module. Using DGCR5 knockdown and overexpression experiments in human neural progenitor cells derived from human induced pluripotent stem cells, we identified a potential role for DGCR5 in regulating certain SCZ-related genes.
One Sentence Summary:
The long non-coding RNA DGCR5 in the 22q11.2 deletion may regulate expression of certain protein-coding genes that have been associated with schizophrenia.
Accessible Summary:
Copy number variations (CNVs) have strong effects on risk of schizophrenia. However, the potential contribution of long non-coding RNAs (lncRNAs) within CNV regions to SCZ risk is not clear. Through a genome-wide search, we identified that a lncRNA, DGCR5, inside 22q11.2 deletion regulates expression of many protein-coding genes related to schizophrenia GWAS signals, de novo mutation or differential expression. DGCR5 was found to be down-regulated in schizophrenia brains. Thus, DGCR5 may contribute to risk of schizophrenia through regulating expression of protein-coding genes implicated in schizophrenia.
INTRODUCTION
Schizophrenia (SCZ) is a severe psychiatric disorder with a complex genetic basis and high heritability (1, 2). It affects approximately 1% of the world’s population and causes considerable social and economic burden (3). Studies have indicated that rare copy number variations (CNVs) have strong effects on SCZ risk (4, 5). Recently, the largest SCZ CNV study ever, based on 21,094 SCZ cases and 20,227 control samples, identified more than ten SCZ risk-associated CNVs (6). However, the mechanisms by which these CNVs contribute to SCZ risk are not clear. CNVs may increase SCZ risk by changing gene dosage or disrupting the sequences of genes within the deleted regions. Some of these genes encode proteins involved in neurodevelopmental pathways including synaptic long-term potentiation and neuregulin signaling (7). For example, CNV deletions at 2p16.3 and at 22q11 associated with SCZ risk (NRXN1 and COMT, respectively) each contain a protein-coding gene related to synaptic function (8, 9).
Protein-coding genes have been the focus of studies investigating SCZ-associated CNVs. For example, protein-coding genes COMT, PRODH, and ZDHHC8 have been regarded as potential SCZ candidate genes because they are located within the 22q11.2 CNV region (10). However, CNV regions also harbor non-coding RNAs. One common type is long non-coding RNA (lncRNA), defined as RNA of 200 nucleotides or longer in size with little or no protein-coding potential (11, 12). The majority of lncRNAs show spatial-temporal, brain region-specific or cell type-specific expression patterns in the brain (13, 14). LncRNAs can regulate gene expression via both cis and trans regulation (15), and may contribute to synaptic plasticity as well as neuronal development and differentiation (13, 16). This is of particular importance since SCZ is generally considered to be a neurodevelopmental disorder (17). Dysregulation of lncRNAs that are involved in neuronal functions may contribute to the etiology of SCZ. The expression of lncRNAs located in CNV deletion regions (CNV-lncRNAs) could be disrupted by the deletion, which, in turn, may alter the expression of their regulatory targets (18, 19). The potential functions of lncRNAs located within CNV deletion regions associated with SCZ risk are intriguing and likely to be important in the etiology of SCZ. However, the roles of CNV lncRNAs in SCZ have not yet been investigated systematically.
To study the functions of lncRNAs, coexpression analysis among well-annotated protein-coding genes and lncRNAs has been used (20), because coexpression implies some degree of coregulation (21, 22). However, it is important to recognize that coexpression does not necessarily translate into a causal regulatory relationship. Experimental validation of the predicted regulatory relationships between a lncRNA and its coexpressed genes becomes critically important. Thus, identifying lncRNAs that are coexpressed with protein-coding genes implicated in SCZ suggests that these lncRNAs may contribute to SCZ through their regulation of SCZ-related genes.
In this study, we hypothesized that CNV lncRNAs may be involved in the etiology of SCZ through regulation of coexpressed SCZ-related genes. To test this hypothesis, we retrieved annotated lncRNAs mapped to ten SCZ risk-associated CNV deletions and identified protein-coding genes that were coexpressed with CNV lncRNAs in postmortem control brains from individuals without psychiatric disease using WGCNA (23) based on brain transcriptome data from the PsychENCODE BrainGVEX (24) and Developmental Capstone (25) projects. The BrainGVEX data set was used to study the regulatory roles of CNV-lncRNAs in postmortem brain tissue from adult individuals without psychiatric disorders. The Developmental Capstone data set was used to identify the temporal regulation of CNV-lncRNAs during normal human brain development. We then focused on hub CNV-lncRNAs in neuronal modules that were the most interconnected lncRNAs based on their correlations with the eigengene (the first principal module component). Finally, we used knockdown and overexpression experiments in human neural progenitor cells (hNPCs) to validate the predicted regulatory relationships between hub CNV-lncRNAs and SCZ-related protein-coding genes. Through this genome-wide search, we identified one lncRNA, DGCR5 in the 22q11.2 CNV region that served as a hub regulator for the expression of several SCZ-associated protein-coding genes, including those related to common single nucleotide polymorphisms (SNPs).
RESULTS
After sample preparation, RNA sequencing (RNA-Seq) and data processing, data from 259 BrainGVEX control postmortem brain samples and 37 Developmental Capstone postmortem samples from the developing human brain were used to perform WGCNA respectively. LncRNAs mapped to ten SCZ risk-associated CNV deletions (Fig. 1) as reported in a recent SCZ CNV study (6) were retrieved. A total of 124 lncRNAs annotated in GENCODE version19 were retrieved from eight SCZ risk-associated CNV deletion regions: 1q21.1, 3q29, 7p36.3, 8q22.2, 15q11.2, 15q13.3, distal 16p11.2, and 22q11.2 (2p16.3 and 9p24.3 were omitted because they did not harbor annotated lncRNAs) (Data file S1). Eighty-eight CNV-lncRNAs and 17,717 protein-coding genes from the BrainGVEX data set passed filtering and were included in the WGCNA. Eighty-seven CNV-lncRNAs and 17,472 protein-coding genes from the Developmental Capstone data set were included in WGCNA after data filtering.
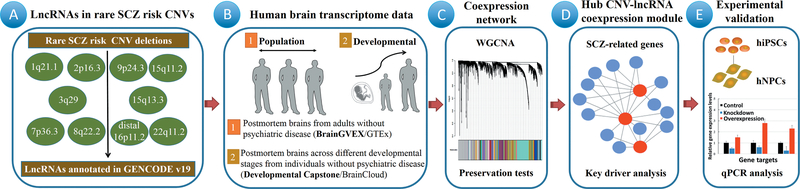
Fig. 1. Study Workflow.
We retrieved annotated lncRNAs mapped to ten SCZ risk-associated CNV deletions (A). We analyzed four human brain transcriptome data sets (BrainGVEX, Developmental Capstone, GTEx, and BrainCloud) from individuals without psychiatric disease. The BrainGVEX (N = 259) and GTEx (N = 101) data sets contained human adult control brains; the Developmental Capstone (N = 37) and BrainCloud (N = 269) data sets contained developing human brains (B). We used WGCNA to identify SCZ-related genes that were coexpressed with the CNV-lncRNAs (C). After identifying hub CNV-lncRNAs from modules related to neuronal functions (D), in vitro experiments were used to validate the predicted regulation of coexpressed SCZ-related genes by hub CNV-lncRNAs (E).
A coexpression module with CNV-lncRNA as a hub gene in adult human control brain
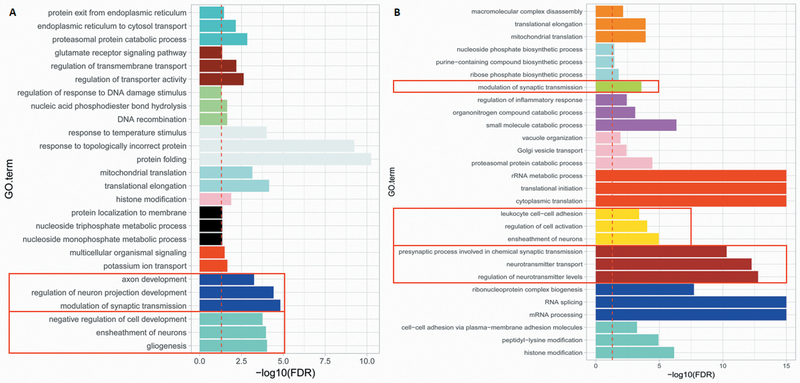
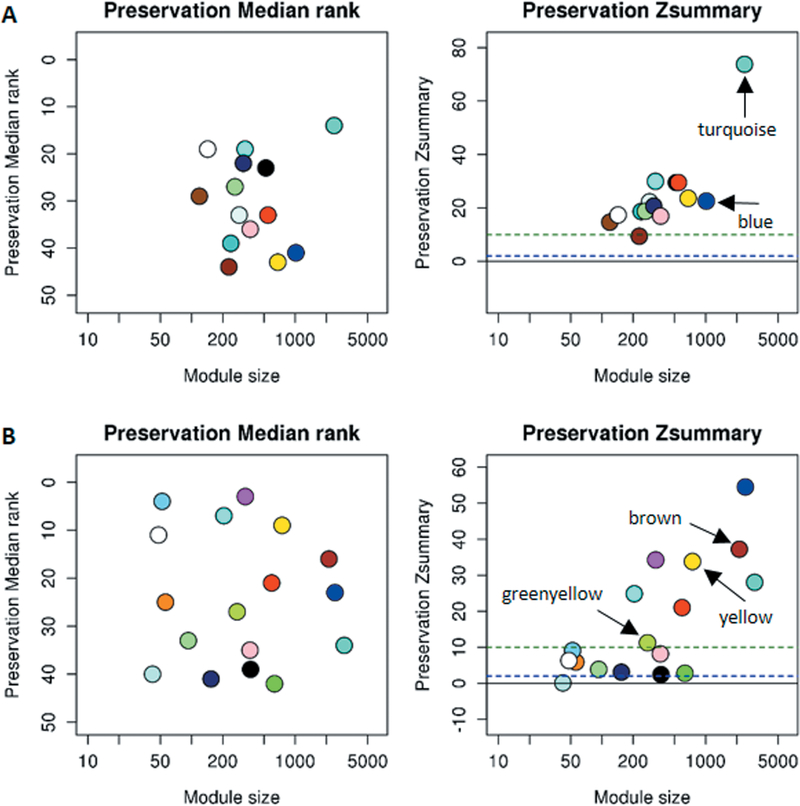
To investigate the regulatory roles of CNV-lncRNAs in the control adult human brain, we performed WGCNA on the RNA-Seq data from 259 BrainGVEX control adult human brain tissue samples and identified a total of 45 coexpression modules. Fourteen of these contained 32 of the 88 CNV-lncRNAs (Data file S2). The remaining 56 CNV-lncRNAs were not clustered into any module. Protein-coding genes in the CNV-lncRNA coexpression modules were used as input for Gene Ontology (GO) and pathway analysis. We then limited our focus to those neuronal function-related modules containing at least one hub CNV-lncRNA (module membership (MM) ≥ 0.8, P < 0.05), in which the hub CNV-lncRNA was predicted to exert important regulatory functions. Of the 14 modules containing CNV-lncRNAs, the largest two modules contained protein-coding genes enriched for neuronal functions (Fig. 2A, turquoise and blue). The turquoise module contained 2,453 protein-coding genes and three non-hub CNV-lncRNAs located within the 15q11.2 and 15q13.3 deletion regions. The blue module included two lncRNAs: RP11–701H24.10 from 15q11.2 and a hub CNV-lncRNA DGCR5 (MM = 0.80, P = 3.82E-60) from 22q11.2, as well as 1,039 protein-coding genes. Protein-coding genes in the blue module were related to modulation of synaptic transmission (false discovery rate (FDR) = 1.50E-05), regulation of neuronal projections during development (FDR = 3.83E-05), and axon development (FDR = 5.60E-04). However, these genes were not enriched in any Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway. A module preservation test, which tests for module similarity, showed that all of tthe CNV-lncRNA coexpression modules were preserved in the brain transcriptome data from the Genotype-Tissue Expression (GTEx) project (26) with Zsummary scores > 2 (a score between 2 and 10 indicates that a module is moderately preserved, whereas a score of 10 or above indicates a module is highly preserved) (23). The two neuronal modules mentioned above were highly preserved with a Zsummary = 73.8 for the turquoise module, and a Zsummary = 22.6 for the blue module (Fig. 3A).
Fig. 2. Functional annotation of CNV-lncRNA coexpression modules.
Modules containing CNV-lncRNAs are represented as horizontal bars with different colors. Neuronal modules (turquoise and blue) from the BrainGVEX data set (human adult control brains, N = 259) (A), and neuronal modules (brown, yellow, and greenyellow) from the Developmental Capstone data set (developing human brains, N = 37) (B) are indicated within the red box. The red dashed line indicates FDR = 0.05. For each module, the top 3 Gene Ontology (GO) terms (FDR < 0.05) are listed on the y axis, and values of -log10(FDR) are shown on the x axis.
Fig. 3. Preservation tests for CNV-lncRNA coexpression modules.
Circles with different colors represent different CNV-lncRNA coexpression modules. The horizontal and vertical axes represent gene number and Zsummary values for each module, respectively. The blue dotted line indicates Zsummary = 2, and the green dotted line indicates Zsummary = 10. Neuronal modules in the BrainGVEX data set (human adult control brains, N = 259) (A) and Developmental Capstone data set (developing human brains, N = 37) (B) are indicated with arrows.
A coexpression module with a hub CNV-lncRNA in the developing human brain
Given that SCZ is thought to be a neurodevelopmental disorder (17), we investigated temporal regulatory roles of CNV-lncRNAs. The ventrolateral frontal cortex (VFC) of the brain, a part of the prefrontal cortex, has been implicated in the etiology of SCZ (27). Thus, we investigated the temporal regulation of CNV-lncRNAs by applying WGCNA to RNA-Seq data from 37 VFC brain tissue samples (across nine stages of human brain development) from the PsychENCODE Developmental Capstone project (25). We identified 71 CNV-lncRNAs clustered in 17 out of 44 coexpression modules (Data file S2). Three CNV-lncRNA coexpression modules (WGCNA color-coded as brown, yellow, and greenyellow) were enriched for neuronal activities (Fig. 2B). A preservation test showed that all of the CNV-lncRNA coexpression modules except the pale turquoise module (Zsummary = 0.11) were preserved in the BrainCloud (28) transcriptome data set (Zsummary > 2). These three neuronal modules were highly preserved with Zsummary > 10 (brown module = 37.2, yellow module = 33.8, and greenyellow module = 11.3) (Fig. 3B). However, only the brown module contained a hub CNV-lncRNA. This module consisted of ten non-hub CNV-lncRNAs from CNVs at 1q21.1, 3q29, 15q11.2, distal 16p11.2, as well as the hub lncRNA DGCR5 (MM = 0.87, P = 1.93E-12) and 2,687 protein-coding genes. Protein-coding genes in this module were enriched for those regulating neurotransmitter production (FDR = 1.69E-13), presynaptic processes involved in chemical synaptic transmission (FDR = 5.05E-11), and potassium ion transport (FDR = 3.21E-10). KEGG pathway analysis demonstrated that the genes in this module were involved in the synaptic vesicle cycle pathway (FDR = 2.50E-07), the neuroactive ligand-receptor interaction pathway (FDR = 5.77E-07), and the calcium signaling pathway (FDR = 5.17E-07).
Protein-coding genes coexpressed with CNV-lncRNA DGCR5 are enriched for SCZ-related genes
The coexpression modules with a hub CNV-lncRNA identified in both the BrainGVEX and Developmental Capstone data sets differed to some extent, given that they captured different spatiotemporal dynamics. Only 187 protein-coding genes and one hub CNV-lncRNA, DGCR5, overlapped between these two modules. DGCR5 was found to have the highest MM value (BrainGVEX data set, MM = 0.80; Developmental Capstone data set, MM = 0.87) among the CNV-lncRNAs in these two modules suggesting that it has an important regulatory function.
To investigate a potential association between DGCR5 and SCZ, we assessed the enrichment of 500 module genes the expression of which highly correlated with DGCR5 expression (Spearman correlation |r| > 0.5, P < 0.05) (Data file S3) against four lists of SCZ-related genes. The first SCZ-related gene set included 343 protein-coding genes adjacent to the 108 SCZ loci identified from the genome-wide association study (GWAS) (denoted GWAS genes) by the Psychiatric Genomics Consortium (PGC) (29). The second list comprised 291 protein-coding genes containing SCZ de novo mutations (DNM) (30). The third list was the published 693 SCZ differentially expressed genes (DEGs) identified from the CommonMind data set (31). We also included 747 SCZ DEGs identified from the BrainGVEX data set (fig. S1 and Data file S4) in the enrichment analysis.
In the BrainGVEX data set, we found that 19 of the top 500 genes coexpressed with DGCR5 were significantly enriched in the SCZ GWAS genes (PH = 1.86E-03, PH represents hypergeometric P value), and that 22 of the top 500 genes were enriched in the SCZ DNM genes (PH = 9.34E-06) (fig. S2A). Considering that gene length can affect enrichment (32), we performed permutation tests (N = 1.00 × 106) by controlling for gene length and found that the enrichment was not due to the length of SCZ GWAS and DNM genes but reflected their coexpression with DGCR5 (fig. S3). However, the top 500 genes coexpressed with DGCR5 were not enriched with the DEGs from either the BrainGVEX or CommonMind data sets. In the Developmental Capstone data set, 28 of the top 500 genes coexpressed with DGCR5 overlapped with the BrainGVEX DEGs (PH = 1.27E-02), and 47 of the 500 genes were enriched in the CommonMind DEGs (PH = 5.66E-10) (fig. S2B). However, enrichment was not significant for the SCZ GWAS genes or DNM genes.
DGCR5 may be a potential regulator of SCZ-related genes in the coexpression module
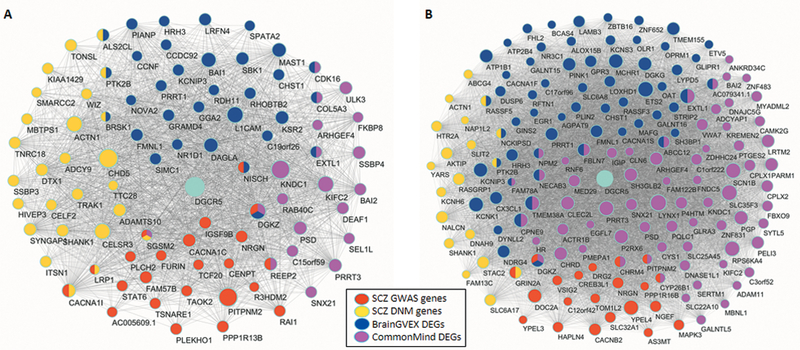
The significant enrichment of genes coexpressed with DGCR5 for SCZ-related genes implied that DGCR5 may be a regulator of these SCZ-related genes. We performed key driver analysis (KDA) for the DGCR5 coexpression module in both the BrainGVEX data set (blue module) and Developmental Capstone data set (brown module) using a previously published method (33). Key drivers are those genes whose neighbors are significantly enriched in the input gene list in contrast to other random genes from the same network. Using the four SCZ-related gene sets mentioned above as input genes in KDA, we identified 135 key driver genes in the DGCR5 coexpression module of the BrainGVEX data set, and 321 key driver genes for the Developmental Capstone data set (Data file S5). DGCR5 was one of the 21 key drivers shared in the coexpression module between the BrainGVEX data set (P = 2.04E-97) and the Developmental Capstone data set (P = 4.05E-164) (Fig. 4). These results suggested that DGCR5 may act as a regulator of SCZ-related genes in the coexpression module.
Fig. 4. The CNV-lncRNA DGCR5 in the coexpression module.
Shown is the key driver analysis for the CNV-lncRNA DGCR5 coexpression module from the BrainGVEX data set (human adult control brains, N = 259) (A) and Developmental Capstone data set (developing human brains, N = 37) (B) are. Subsets of SCZ-related genes are indicated in different colors. Sizes of the nodes represent their weighted correlations with DGCR5.
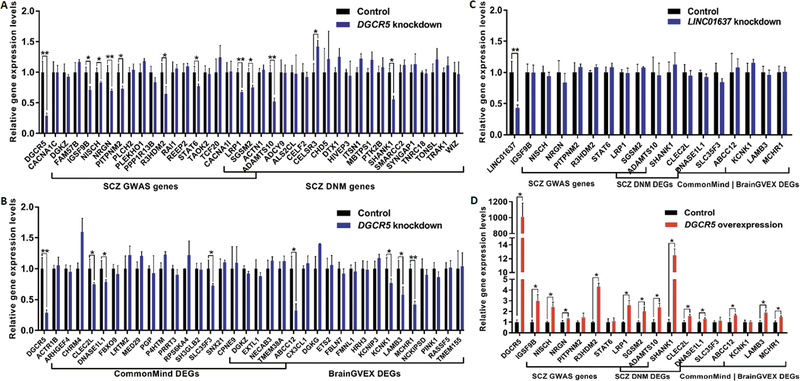
We decided to validate the predicted regulation by DGCR5 of SCZ-related genes in the list of the top 500 DGCR5 coexpressed genes using human neural progenitor cells ( hNPCs) derived from normal human induced pluripotent stem cells (hiPSCs) (fig. S4). Nineteen SCZ GWAS genes and 22 SCZ DNM genes from the BrainGVEX coexpression module, and the top 20 BrainGVEX DEGs and the top 20 CommonMind DEGs ranked by the correlation with DGCR5 in the Developmental Capstone coexpression module, were selected for quantitative PCR (qPCR) analysis (Data file S3). Of the selected genes, two genes (ABCC12 and LAMB3) were not detected in previous RNA-Seq studies of normal control hNPCs derived from hiPSCs or human embryonic stem cells (34, 35), but both of these genes showed low expression in hNPCs from our qPCR analysis. We then observed the effects of DGCR5 knockdown or overexpression in the hNPCs on the expression of these selected genes. Eight out of the 19 SCZ GWAS genes, and four out of the 22 SCZ DNM genes were downregulated in response to DGCR5 knockdown (72% efficiency, Padjust = 8.67E-03) (Fig. 5A. Four BrainGVEX DEGs and three CommonMind DEGs were downregulated in response to DGCR5 knockdown (Fig. 5B). Knocking down LINC01637, a CNV-lncRNA selected from a non-neuronal module that served as a negative control for DGCR5 knockdown, did not perturb expression of any of the genes coexpressed with DGCR5 (Fig. 5C). We next performed a DGCR5 overexpression assay to verify the regulation of DGCR5 on the coexpressed genes. Six of the SCZ GWAS genes, four DNM genes, and five DEG genes were altered in response to DGCR5 overexpression (Fig. 5D). The expression of a total of 13 SCZ-related genes was found to be altered in response to both DGCR5 overexpression and knockdown (table S1).
Fig. 5. DGCR5 may regulate expression of the coexpressed SCZ-related genes.
qPCR analysis was used to detect expression changes of SCZ-related genes that coexpressed with DGCR5 after DGCR5 knockdown or overexpression in hNPCs derived from hiPSCs. The blue and red bars represent the DGCR5 knockdown and overexpression groups, respectively; the black bars represent the group used as either negative control for knockdown of DGCR5 and LINC01637, or the empty vector group for overexpression. qPCR analysis was first used to detect expression changes in selected SCZ GWAS and DNM genes (A) and BrainGVEX DEGs and CommonMind DEGs (B) after knockdown of DGCR5 in hNPCs derived from hiPSCs. Positive results from (A) and (B) were further validated in experiments where the control lncRNA LINC01637 was knocked down (C) and DGCR5 was overexpressed (D) in hNPCs derived from hiPSCs. The knockdown and overexpression experiments were conducted in three biological replicates. Data are shown as means ± SEM. Gene expression in the control group was normalized to 1. Two-tailed t test was used for comparison between two groups. P values were adjusted for multiple testing using the Benjamini-Hochberg method. *Padjust < 0.05, **Padjust < 0.01.
DISCUSSION
CNVs have been implicated in SCZ etiology (36, 37). Previous investigations have focused on how protein-coding genes inside the CNVs rather than non-coding genes might contribute to SCZ risk. Although there have been some investigations into non-coding RNAs (lncRNAs and miRNAs) at CNVs (19, 38, 39), the potential importance of CNV-lncRNAs in SCZ etiology has not been well studied. Here, we investigated the potential regulatory roles for CNV-lncRNAs in the human brain and their possible contributions to SCZ. We found that several CNV-lncRNAs were coexpressed with many neuronal protein-coding genes outside the CNV regions where lncRNAs reside. The coexpression patterns persisted in both the BrainGVEX and Developmental Capstone data sets, and were preserved in other independent transcriptome data sets (GTEx and BrainCloud). Importantly, many of the protein-coding genes coexpressed with CNV-lncRNAs were related to SCZ GWAS signals, de novo mutations or differential expression, implying the involvement of CNV-lncRNAs in SCZ.
Among the CNV-lncRNAs in the neuronal modules, the hub CNV-lncRNA DGCR5 was shared by both the BrainGVEX and Developmental Capstone data sets. We experimentally validated that the CNV-lncRNA DGCR5 could regulate some of the selected coexpressed genes (SCZ GWAS genes, DNM genes or DEGs) in hNPCs derived from hiPSCs. Some of these genes, such as IGSF9B (40) and NRGN (41), have been linked to SCZ in animal model or brain imaging studies. DGCR5 has been found to be downregulated in brain tissue from individuals with SCZ (fold-change = 0.73, FDR = 1.35E-02) in the CommonMind data set (31). In the latest PsychENCODE Capstone RNA-Seq data from 601 SCZ cases and 1,253 controls, Gandal et al. (42) found that the DGCR5 coexpression module was associated with SCZ, and that DGCR5 had significantly lower expression in postmortem brain tissue from SCZ cases compared to control brain tissue (fold-change = 0.94, FDR = 3.21E-03). Intriguingly, in a recently published paper (43), DGCR5 was found to be the only CNV-lncRNA in the neuronal coexpression module (CD1) that was downregulated in postmortem brain tissue from individuals with SCZ, autism and bipolar disorder. Taken together, these findings implicate DGCR5 in an increased risk for SCZ potentially through its regulatory effects on coexpressed SCZ-related genes.
Our study provides evidence for a potential regulatory relationship between the CNV-lncRNA DGCR5 and genes outside of the CNV region. Our study also reveals intriguing connections between SCZ-associated rare CNVs and common SNP variants. Through the CNV-lncRNA DGCR5, rare CNVs and common SNP variants may converge onto common pathways regulating gene expression. Our study suggests that non-coding RNAs within the CNV regions may have regulatory roles that implicate them in SCZ etiology.
There are a number of limitations to our study. First, coexpression does not necessarily mean causality. Not all of the genes coexpressed with DGCR5 were found to be regulated by DGCR5. Future experimental validation of gene regulation by DGCR5 will be critically important. Second, the coexpression modules in this study included some transcription factors and their regulatory targets. It is unlikely that simple pairwise regulator-target analysis can fully describe all of these regulatory relationships. Other important genes could contribute to gene expression as upstream regulators or as part of parallel regulation cascades. Taking advantage of the temporal nature of the Developmental Capstone data set, which covered multiple stages of human brain development, we could study the potential temporal regulation of CNV-lncRNAs in the normal human brain. However, the sample size of the Developmental Capstone data set used in this study was small, which limited the ability to identify temporal regulatory roles for CNV-lncRNAs. Only a few selected genes from the coexpression module of the Developmental Capstone data set were found to be regulated by DGCR5. The human NPC cellular model may also be limited in its ability to capture temporal regulation of gene expression by DGCR5 as these cells represent a very short period in human brain development. Further investigations using a greater number of postmortem human brain tissue samples at different stages of development and better statistical and bioinformatics tools may help to elucidate the exact regulatory roles of DGCR5 and other CNV-lncRNAs.
Finally, lncRNAs have diverse functional mechanisms in the regulation of gene expression. They could interact with protein targets or bind to miRNAs as competing endogenous RNAs (44). For example, several studies (45–49) have demonstrated a role for DGCR5 in the regulation of miRNAs in cancer cell lines. In the Gandal et al. study (43), which was based on human brain transcriptomics data, the DGCR5 coexpression module (CD1) contained one microRNA hsa-miR-1227 and some validated targets of DGCR5. The DGCR5-regulated genes within the CD1 module were also predicted to be targets of hsa-miR-1227 (Data file S6), implying a potential mediator effect for DGCR5 on these target genes. In the future, integration of multi-dimensional data sets will help to reveal the regulatory mechanisms of DGCR5 in SCZ.
In summary, population (PsychENCODE BrainGVEX and GTEx) and developmental (PsychENCODE Developmental Capstone and BrainCloud) brain transcriptomics data sets suggested a potential regulatory relationship between a hub CNV-lncRNA, DGCR5, and SCZ-related genes. DGCR5 potentially could represent a bridge connecting rare CNV deletions and common GWAS findings, given that the 22q11.2 deletion may extend its functional impact beyond the protein-coding genes residing in the deleted regions.
MATERIALS AND METHODS
Study design
The objective of this study was to investigate the potential contribution of CNV-lncRNAs to SCZ risk. First, we retrieved annotated lncRNAs mapped to SCZ risk-associated CNV deletion regions reported in the SCZ CNV study from Psychiatric Genomics Consortium (PGC) (6). We then identified protein-coding genes that were coexpressed with the CNV-lncRNAs in postmortem human brain tissue from individuals without psychiatric disorders (BrainGVEX data set, N = 259) and across different stages of brain development (Developmental Capstone data set, N = 37). Finally, after identifying hub CNV-lncRNAs, we performed knockdown and overexpression experiments in hNPCs derived from hiPSCs. Then we used qPCR analysis to validate the predicted regulation of SCZ-related protein-coding genes by hub CNV-lncRNAs. The knockdown and overexpression experiments were conducted in three biological replicates.
For brain sample collection in the BrainGVEX project, inclusion and exclusion criteria are described below. RNA samples were randomly selected for RNA-sequencing. ComBat was used to correct for the batch effect. For both the BrainGVEX and Developmental Capstone data sets, detailed procedures regarding RNA-Seq quality control and data exclusion criteria are described in the Supplementary Materials. The sample size of both data sets was much larger than the recommended number (N > 15) for WGCNA, which enabled the capture of robust CNV-lncRNA coexpression relationships. We also used other independent brain transcriptome data sets (GTEx, N = 101; BrainCloud, N = 269) to validate the CNV-lncRNA coexpression patterns and to boost the power of our study.
Brain samples and data collection
PsychENCODE BrainGVEX data.
As frontal cortex has been implicated in the etiology of SCZ (27, 50), we chose frontal cortex samples to study the regulatory roles of CNV-lncRNAs in human adult brains. Human post-mortem brain tissues were collected from four collections of the Stanley Medical Research Institute (SMRI): The Neuropathology Consortium, Array Collection, New Collection, and Extra Collection. SCZ diagnosis was based on the DSM-IV criteria and medical records. Diagnosis of unaffected controls was according to the structured interviews with their family member(s). For all brain specimens, the samples will be excluded if they met any one of the exclusion criteria: 1) significant structural brain pathology on postmortem examination; 2) history of significant focal neurological signs pre-mortem; 3) history of a CNS disease that could be expected to alter gene expression in a persistent way; 4) documented IQ < 70; and 5) poor RNA quality. For unaffected controls, two additional exclusion criteria were used: 1) substance abuse within one year of death or significant alcohol-related changes in the liver; 2) age less than 30. Thus, we obtained a total of 96 SCZ cases and 75 control brain samples from SMRI. Additional 185 brain samples of unaffected controls were collected from the Banner Sun Health Research Institute (BSHRI) according to the individual medical records or neuropsychological screening assessment. Total RNA was extracted from the collected brain samples and used for RNA sequencing (Supplementary Materials). This study generated RNA-Seq data from a total of 356 samples. Previous paper published by Gandal et al. used part of this data (43). Metadata for the 356 samples, including sex, ethnicity, the age of death, diagnosis, post-mortem interval (PMI), RNA integrity number (RIN), batch, and sequencing quality matrix are listed in Data file S7. The RNA-Seq data of SCZ cases and controls were used for differential gene expression analysis (Supplementary Materials), and the RNA-Seq data of control samples were used for WGCNA to capture protein-coding genes that were coexpressed with the CNV-lncRNAs in non-psychiatric individuals.
PsychENCODE Developmental Capstone data.
The PsychENCODE Developmental Capstone project (25) was designed to study transcriptional mechanisms involving human brain development. In this project, RNA-Seq data (Illumina GAIIx, polyA+ selection, 76 bp single-end read) were generated from a total of 606 non-psychiatric brain samples, which were collected from 16 brain regions from 41 individuals spanning from 8 post-conceptional weeks to over 40 years of age. As VFC has been found being involved in SCZ (27), we included RNA-Seq data of 40 VFC samples from the PsychENCODE Developmental Capstone project into WGCNA to study the temporal regulation of CNV-lncRNAs. Three VFC samples with no individual information were removed. The remaining 37 samples (15 prenatal and 22 postnatal samples) covered nine different brain developmental stages from early-prenatal to adult (Data file S7). More detailed information about the Developmental Capstone data could be found on the PsychENCODE website (http://www.psychENCODE.org/).
Validation data (GTEx and BrainCloud).
To validate the CNV-lncRNA coexpression patterns in the BrainGVEX data, we used RNA-Seq data of 101 adult frontal cortex samples (RIN > 6) from the GTEx project (26). We also used the BrainCloud transcriptome data (28) to validate the CNV-lncRNA coexpression relationships detected in the Developmental Capstone data. The BrainCloud data are microarray data (Gene Expression Omnibus accession number: GSE30272) of 269 prefrontal cortex samples from fetus to adults. Metadata of the 101 GTEx samples and 269 BrainCloud samples are listed in Data file S7.
CNV-lncRNA retrieval
As CNV-lncRNAs could be directly disrupted by CNV deletions, we chose to focus on the SCZ risk CNV deletion regions but not those protective or duplicated CNVs. Thus, ten SCZ risk-associated CNV deletions identified in the PGC study (6) were included in this study. Eight of the ten CNV regions (1q21.1, 2p16.3, 3q29, 7p36.3, 15q11.2, 15q13.3, distal 16p11.2 and 22q11.2) have been previously implicated in SCZ, and the other two loci (8q22.2 and 9p24.3) are novel SCZ CNVs reported by PGC (6). We obtained the deletion positions of the CNVs by combining the CNV deletion information from the DECIPHER database (51) (https://decipher.sanger.ac.uk/). Annotated lncRNAs mapped to the aforementioned ten CNV deletion regions were retrieved from GENCODE v19.
Weighted gene coexpression network analysis
Raw expression data of the BrainGVEX and Developmental Capstone were processed (Supplementary Materials) before gene coexpression analysis. LncRNAs and mRNAs that had FPKM (fragments per kilobase per million mapped reads) values > 0.1 in at least ten samples were selected for coexpression analysis in the BrainGVEX data. RNAs with FPKM values > 0.1 in at least 10% of the samples were used for coexpression analysis in the Developmental Capstone data. WGCNA (v1.51) in R package was used to identify CNV-lncRNA coexpression modules. Biweight midcorrelation (bicor) was used to calculate pairwise gene correlations in the BrainGVEX data, which is recommended as a robust alternative to outlier measurements. For Developmental Capstone data, Spearman correlation was used to calculate the pairwise gene correlations, which is a non-parametric measure of ranked correlations between variables. It does not depend on the data distribution. For coexpression network construction, the power value was set at 4 and 10 for the BrainGVEX and Developmental Capstone data respectively, using the pickSoftThreshold function. The dynamicTreeCut algorithm was used with the following parameters: deepSplit = 4, pamStage = T, pamRespectsDendro = F, and minClusterSize = 30. CNV-lncRNAs and protein-coding genes with similar expression patterns were clustered into the same module. The CNV-lncRNA coexpression modules were further used for functional annotation and preservation tests (Supplementary Materials). Module membership (MM) is a measurement used to assess the correlation between a gene and corresponding ME.
Key driver analysis
To identify key drivers in the CNV-lncRNA coexpression modules, we performed key driver analysis (KDA) using the KDA R package (version 0.1) (33). KDA takes an input gene list of interest (generally a disease-associated gene list) and a gene network as input files. It first generates a sub-network consisting of nodes that are no more than H-layers away from each node in the target gene list in the network. Then, for each node in the sub-network, it assesses the enrichment of its H-layer downstream neighbors for target gene list. In this study, a combined SCZ-related gene set (343 SCZ GWAS genes, 291 SCZ DNM genes, 747 BrainGVEX DEGs and 693 CommonMind DEGs) was used as the input gene list. To identify key drivers in the large coexpression module, a cut-off of weight value > 0.08 was used to filter out those gene pairs with low correlations in the network. H = 1 was found to be optimal for both networks of the BrainGVEX and Developmental Capstone data. The KDA results were visualized using Cytoscape (v3.5.1).
DGCR5 knockdown in hNPCs
We used a KD experiment in hNPCs to validate the role of DGCR5 in regulating other protein-coding genes, because DGCR5 is a shared hub CNV-lncRNA (MM ≥ 0.8, P < 0.05) in the neuronal module of both BrainGVEX and Developmental Capstone data, and is specifically and highly expressed in the brain (fig. S5). As DGCR5 was expressed in both the nucleus and cytoplasm (fig. S6), we used LncRNA smart silencer (Ribobio, China) for DGCR5 KD. The lncRNA smart silencer is a mixture of three anti-sense oligonucleotides (ASO) and three small interference RNAs (siRNAs), which could effectively knock down both nuclear and cytoplasmic lncRNAs. The smart silencer at the optimal final concentration (50nM) and Lipofectamine® RNAiMAX reagent (13778030, Invitrogen, USA) was used for DGCR5 KD. Total RNA was extracted at 24 hours after cell transfection and used for qPCR analysis, as DGCR5 was the most efficiently knocked down at this time point (fig. S7). To validate the specificity of DGCR5 in regulating gene expression, CNV-lncRNA LINC01637 from a non-neuronal module with a similar module size was used as a negative control. With the same experiment conditions, we knocked down LINC01637 and checked the effect on detected DGCR5 coexpressed genes. The sequences of lncRNA smart silencer are listed in table S2. More details about DGCR5 and LINC01637 can be found in Supplementary Materials.
DGCR5 overexpression in hNPCs
For DGCR5 OE assay, full length of the major transcript of DGCR5 (transcript ID: NR_002733) was synthesized and cloned into the transient OE vector GV144 (GeneChem, China), using the restriction enzymes XhoI and BamHI (NEB, USA). The constructed DGCR5-GV144 OE vector or empty GV144 vector was transfected into hNPCs using Lipofectamine® LTX and PLUS™ Reagents (A12621, Invitrogen, USA). Total RNA was extracted at 24 hours after transfection and used for qPCR analysis.
Quantitative PCR analysis
Total RNA was extracted with the miRNeasy mini kit (217004, Qiagen, Germany). cDNA was generated using HiScript II Q RT SuperMix plus gDNA wiper (R223–01, Vazyme, China). ChamQ SYBR qPCR Master Mix (Q311–01, Vazyme, China) was used for qPCR analysis on the CFX 96 qPCR instrument (Bio-Rad, USA). GAPDH and β-actin were both used as internal reference genes, the geometric mean of their expression was used for normalization. Triplicates per gene were used for quantitative analysis. The sequences of qPCR primers are listed in table S3.
Statistical analysis
A threshold FDR of < 0.05 and P < 0.01 were used for differential gene expression analysis in the BrainGVEX data. Hypergeometric test was used to assess the significance of enrichment for the coexpressed genes of DGCR5 with SCZ-related genes in both the BrainGVEX and Developmental Capstone data sets. The qPCR data shown in Fig. 5 and fig. S4 are means ± SEM. Statistical analysis was performed with GraphPad Prism (v6.0) and described in each figure legend. The two-tailed t test was used for comparison between two groups (α = 0.05). P values were adjusted for multiple testing using the Benjamini-Hochberg method.
Supplementary Material
Data file S1 (Excel file format). LncRNAs and protein-coding genes inside SCZ risk-associated CNV deletion regions.
Data file S2 (Excel file format). Summary information for CNV-lncRNA coexpression modules.
Data file S3 (Excel file format). Top 500 genes coexpressed with DGCR5 in the module.
Data file S4 (Excel file format). Differentially expressed genes between SCZ cases and controls in the BrainGVEX data set.
Data file S5 (Excel file format). Key driver genes in the DGCR5 coexpression module.
Data file S6 (Excel file format). Some of the DGCR5-regulated genes within CD1 module are predicted targets of hsa-miR-1227.
Data file S7 (Excel file format). Sample information of four transcriptome data sets (BrainGVEX, Developmental Capstone, GTEx, and BrainCloud).
Acknowledgements:
We thank Gina Giase for editing the manuscript. Funding: This work was supported by NIH grants 1 U01 MH103340-01, 1R01ES024988 (to C. Liu), and National Natural Science Foundation of China (NSFC) grants 81401114, 31571312, the National Key Plan for Scientific Research and Development of China (2016YFC1306000), Innovation-Driven Project of Central South University (# 2015CXS034, 2018CX033; to C. Chen). We sincerely thank Chicago Biomedical Consortium with support from the Searle Funds at The Chicago Community Trust. Data were generated as part of the PsychENCODE Consortium, supported by: U01MH103392, U01MH103365, U01MH103346, U01MH103340, U01MH103339, R21MH109956, R21MH105881, R21MH105853, R21MH103877, R21MH102791, R01MH111721, R01MH110928, R01MH110927, R01MH110926, R01MH110921, R01MH110920, R01MH110905, R01MH109715, R01MH109677, R01MH105898, R01MH105898, R01MH094714, P50MH106934 awarded to: Schahram Akbarian (Icahn School of Medicine at Mount Sinai), Gregory Crawford (Duke University), Stella Dracheva (Icahn School of Medicine at Mount Sinai), Peggy Farnham (University of Southern California), Mark Gerstein (Yale University), Daniel Geschwind (University of California, Los Angeles), Fernando Goes (Johns Hopkins University), Thomas M. Hyde (Lieber Institute for Brain Development), Andrew Jaffe (Lieber Institute for Brain Development), James A. Knowles (University of Southern California), Chunyu Liu (SUNY Upstate Medical University), Dalila Pinto (Icahn School of Medicine at Mount Sinai), Panos Roussos (Icahn School of Medicine at Mount Sinai), Stephan Sanders (University of California, San Francisco), Nenad Sestan (Yale University), Pamela Sklar (Icahn School of Medicine at Mount Sinai), Matthew State (University of California, San Francisco), Patrick Sullivan (University of North Carolina), Flora Vaccarino (Yale University), Daniel Weinberger (Lieber Institute for Brain Development), Sherman Weissman (Yale University), Kevin White (University of Chicago), Jeremy Willsey (University of California, San Francisco), and Peter Zandi (Johns Hopkins University).
Footnotes
Competing interests: K.P.W is President and a shareholder in Tempus Labs, Inc.
Data and materials availability: Detailed protocols for brain sample preparation and RNA-Seq are available on Synapse (Synapse number: syn4590909). Data can be requested on a specific page on Synapse (Synapse number: syn4921369) for The PsychENCODE Consortium.
References and Notes
- 1.Cardno AG, Gottesman II, Twin studies of schizophrenia: from bow-and-arrow concordances to star wars Mx and functional genomics. Am J Med Genet 97, 12–17 (2000). [PubMed] [Google Scholar]
- 2.Sullivan PF, Kendler KS, Neale MC, Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch Gen Psychiatry 60, 1187–1192 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Knapp M, Mangalore R, Simon J, The global costs of schizophrenia. Schizophr Bull 30, 279–293 (2004). [DOI] [PubMed] [Google Scholar]
- 4.C. International Schizophrenia, Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature 455, 237–241 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kirov G, Grozeva D, Norton N, Ivanov D, Mantripragada KK, Holmans P, C. International Schizophrenia, C. Wellcome Trust Case Control, Craddock N, Owen MJ, O’Donovan MC, Support for the involvement of large copy number variants in the pathogenesis of schizophrenia. Hum Mol Genet 18, 1497–1503 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cnv C Schizophrenia Working Groups of the Psychiatric Genomics, C. Psychosis Endophenotypes International, Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat Genet 49, 27–35 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, Nord AS, Kusenda M, Malhotra D, Bhandari A, Stray SM, Rippey CF, Roccanova P, Makarov V, Lakshmi B, Findling RL, Sikich L, Stromberg T, Merriman B, Gogtay N, Butler P, Eckstrand K, Noory L, Gochman P, Long R, Chen Z, Davis S, Baker C, Eichler EE, Meltzer PS, Nelson SF, Singleton AB, Lee MK, Rapoport JL, King MC, Sebat J, Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science 320, 539–543 (2008). [DOI] [PubMed] [Google Scholar]
- 8.Sudhof TC, Neuroligins and neurexins link synaptic function to cognitive disease. Nature 455, 903–911 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Egan MF, Goldberg TE, Kolachana BS, Callicott JH, Mazzanti CM, Straub RE, Goldman D, Weinberger DR, Effect of COMT Val108/158 Met genotype on frontal lobe function and risk for schizophrenia. Proc Natl Acad Sci U S A 98, 6917–6922 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farrell MS, Werge T, Sklar P, Owen MJ, Ophoff RA, O’Donovan MC, Corvin A, Cichon S, Sullivan PF, Evaluating historical candidate genes for schizophrenia. Mol Psychiatry 20, 555–562 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, Bell I, Cheung E, Drenkow J, Dumais E, Patel S, Helt G, Ganesh M, Ghosh S, Piccolboni A, Sementchenko V, Tammana H, Gingeras TR, RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316, 1484–1488 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Banfai B, Jia H, Khatun J, Wood E, Risk B, Gundling WE Jr., Kundaje A, Gunawardena HP, Yu Y, Xie L, Krajewski K, Strahl BD, Chen X, Bickel P, Giddings MC, Brown JB, Lipovich L, Long noncoding RNAs are rarely translated in two human cell lines. Genome Res 22, 1646–1657 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Briggs JA, Wolvetang EJ, Mattick JS, Rinn JL, Barry G, Mechanisms of Long Non-coding RNAs in Mammalian Nervous System Development, Plasticity, Disease, and Evolution. Neuron 88, 861–877 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Ward M, McEwan C, Mills JD, Janitz M, Conservation and tissue-specific transcription patterns of long noncoding RNAs. J Hum Transcr 1, 2–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vance KW, Ponting CP, Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends Genet 30, 348–355 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ng SY, Johnson R, Stanton LW, Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J 31, 522–533 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fatemi SH, Folsom TD, The neurodevelopmental hypothesis of schizophrenia, revisited. Schizophr Bull 35, 528–548 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li L, Chang HY, Physiological roles of long noncoding RNAs: insight from knockout mice. Trends Cell Biol 24, 594–602 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gudenas BL, Wang L, Gene Coexpression Networks in Human Brain Developmental Transcriptomes Implicate the Association of Long Noncoding RNAs with Intellectual Disability. Bioinform Biol Insights 9, 21–27 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liao Q, Liu C, Yuan X, Kang S, Miao R, Xiao H, Zhao G, Luo H, Bu D, Zhao H, Skogerbo G, Wu Z, Zhao Y, Large-scale prediction of long non-coding RNA functions in a coding-non-coding gene co-expression network. Nucleic Acids Res 39, 3864–3878 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quackenbush J, Genomics. Microarrays--guilt by association. Science 302, 240–241 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Lee HK, Hsu AK, Sajdak J, Qin J, Pavlidis P, Coexpression analysis of human genes across many microarray data sets. Genome Res 14, 1085–1094 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langfelder P, Horvath S, WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Psych EC, Akbarian S, Liu C, Knowles JA, Vaccarino FM, Farnham PJ, Crawford GE, Jaffe AE, Pinto D, Dracheva S, Geschwind DH, Mill J, Nairn AC, Abyzov A, Pochareddy S, Prabhakar S, Weissman S, Sullivan PF, State MW, Weng Z, Peters MA, White KP, Gerstein MB, Amiri A, Armoskus C, Ashley-Koch AE, Bae T, Beckel-Mitchener A, Berman BP, Coetzee GA, Coppola G, Francoeur N, Fromer M, Gao R, Grennan K, Herstein J, Kavanagh DH, Ivanov NA, Jiang Y, Kitchen RR, Kozlenkov A, Kundakovic M, Li M, Li Z, Liu S, Mangravite LM, Mattei E, Markenscoff-Papadimitriou E, Navarro FC, North N, Omberg L, Panchision D, Parikshak N, Poschmann J, Price AJ, Purcaro M, Reddy TE, Roussos P, Schreiner S, Scuderi S, Sebra R, Shibata M, Shieh AW, Skarica M, Sun W, Swarup V, Thomas A, Tsuji J, van Bakel H, Wang D, Wang Y, Wang K, Werling DM, Willsey AJ, Witt H, Won H, Wong CC, Wray GA, Wu EY, Xu X, Yao L, Senthil G, Lehner T, Sklar P, Sestan N, The PsychENCODE project. Nat Neurosci 18, 1707–1712 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li M, Santpere G, Kawasawa YI, Evgrafov OV, Integrative Functional Genomic Analysis of Human Brain Development and Neuropsychiatric Risk. Science, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.G. T. Consortium, The Genotype-Tissue Expression (GTEx) project. Nat Genet 45, 580–585 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaladjian A, Jeanningros R, Azorin JM, Grimault S, Anton JL, Mazzola-Pomietto P, Blunted activation in right ventrolateral prefrontal cortex during motor response inhibition in schizophrenia. Schizophr Res 97, 184–193 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Colantuoni C, Lipska BK, Ye T, Hyde TM, Tao R, Leek JT, Colantuoni EA, Elkahloun AG, Herman MM, Weinberger DR, Kleinman JE, Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature 478, 519–523 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.C. Schizophrenia Working Group of the Psychiatric Genomics, Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li J, Cai T, Jiang Y, Chen H, He X, Chen C, Li X, Shao Q, Ran X, Li Z, Xia K, Liu C, Sun ZS, Wu J, Genes with de novo mutations are shared by four neuropsychiatric disorders discovered from NPdenovo database. Mol Psychiatry 21, 298 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Fromer M, Roussos P, Sieberts SK, Johnson JS, Kavanagh DH, Perumal TM, Ruderfer DM, Oh EC, Topol A, Shah HR, Klei LL, Kramer R, Pinto D, Gumus ZH, Cicek AE, Dang KK, Browne A, Lu C, Xie L, Readhead B, Stahl EA, Xiao J, Parvizi M, Hamamsy T, Fullard JF, Wang YC, Mahajan MC, Derry JM, Dudley JT, Hemby SE, Logsdon BA, Talbot K, Raj T, Bennett DA, De Jager PL, Zhu J, Zhang B, Sullivan PF, Chess A, Purcell SM, Shinobu LA, Mangravite LM, Toyoshiba H, Gur RE, Hahn CG, Lewis DA, Haroutunian V, Peters MA, Lipska BK, Buxbaum JD, Schadt EE, Hirai K, Roeder K, Brennand KJ, Katsanis N, Domenici E, Devlin B, Sklar P, Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci 19, 1442–1453 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zylka MJ, Simon JM, Philpot BD, Gene length matters in neurons. Neuron 86, 353–355 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang B, Zhu J, Identification of key causal regulators in gene networks. Lecture Notes in Engineering & Computer Science 2205, (2013). [Google Scholar]
- 34.Tang H, Hammack C, Ogden SC, Wen Z, Qian X, Li Y, Yao B, Shin J, Zhang F, Lee EM, Christian KM, Didier RA, Jin P, Song H, Ming GL, Zika Virus Infects Human Cortical Neural Progenitors and Attenuates Their Growth. Cell stem cell 18, 587–590 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stachowiak EK, Benson CA, Narla ST, Dimitri A, Chuye LEB, Dhiman S, Harikrishnan K, Elahi S, Freedman D, Brennand KJ, Cerebral organoids reveal early cortical maldevelopment in schizophrenia—computational anatomy and genomics, role of FGFR1. Translational psychiatry 7, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tam GW, Redon R, Carter NP, Grant SG, The role of DNA copy number variation in schizophrenia. Biol Psychiatry 66, 1005–1012 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Malhotra D, Sebat J, CNVs: harbingers of a rare variant revolution in psychiatric genetics. Cell 148, 1223–1241 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Merico D, Costain G, Butcher NJ, Warnica W, Ogura L, Alfred SE, Brzustowicz LM, Bassett AS, MicroRNA Dysregulation, Gene Networks, and Risk for Schizophrenia in 22q11.2 Deletion Syndrome. Front Neurol 5, 238 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Warnica W, Merico D, Costain G, Alfred SE, Wei J, Marshall CR, Scherer SW, Bassett AS, Copy number variable microRNAs in schizophrenia and their neurodevelopmental gene targets. Biol Psychiatry 77, 158–166 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mishra A, Traut MH, Becker L, Klopstock T, Stein V, Klein R, Genetic evidence for the adhesion protein IgSF9/Dasm1 to regulate inhibitory synapse development independent of its intracellular domain. J Neurosci 34, 4187–4199 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pohlack ST, Nees F, Ruttorf M, Witt SH, Nieratschker V, Rietschel M, Flor H, Risk variant for schizophrenia in the neurogranin gene impacts on hippocampus activation during contextual fear conditioning. Mol Psychiatry 16, 1072–1073 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gandal MJ, Zhang P, Hadjimichael E, Walker R, Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gandal MJ, Haney JR, Parikshak NN, Leppa V, Ramaswami G, Hartl C, Schork AJ, Appadurai V, Buil A, Werge TM, Liu C, White KP, CommonMind C, Psych EC, P.-B. W. G. i, Horvath S, Geschwind DH, Shared molecular neuropathology across major psychiatric disorders parallels polygenic overlap. Science 359, 693–697 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I, A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147, 358–369 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen EG, Zhang JS, Xu S, Zhu XJ, Hu HH, Long non-coding RNA DGCR5 is involved in the regulation of proliferation, migration and invasion of lung cancer by targeting miR-1180. American journal of cancer research 7, 1463–1475 (2017). [PMC free article] [PubMed] [Google Scholar]
- 46.Yong S, Yabin Y, Bing Z, Chuanrong Z, Dianhua G, Jianhuai Z, Weidong Y, Shuming W, Ling L, Reciprocal regulation of DGCR5 and miR-320a affects the cellular malignant phenotype and 5-FU response in pancreatic ductal adenocarcinoma. Oncotarget 8, 90868–90878 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dong HX, Wang R, Jin XY, Zeng J, Pan J, LncRNA DGCR5 promotes lung adenocarcinoma (LUAD) progression via inhibiting hsa-mir-22–3p. Journal of cellular physiology 233, 4126–4136 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Wang R, Dong HX, Zeng J, Pan J, Jin XY, LncRNA DGCR5 contributes to CSC-like properties via modulating miR-330–5p/CD44 in NSCLC. Journal of cellular physiology, (2018). [DOI] [PubMed] [Google Scholar]
- 49.Luo J, Zhu H, Jiang H, Cui Y, Wang M, Ni X, Ma C, The effects of aberrant expression of LncRNA DGCR5/miR-873–5p/TUSC3 in lung cancer cell progression. Cancer medicine, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Casanova MF, Functional and anatomical aspects of prefrontal pathology in schizophrenia. Schizophr Bull 23, 517–519 (1997). [DOI] [PubMed] [Google Scholar]
- 51.Firth HV, Richards SM, Bevan AP, Clayton S, Corpas M, Rajan D, Van Vooren S, Moreau Y, Pettett RM, Carter NP, DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am J Hum Genet 84, 524–533 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data file S1 (Excel file format). LncRNAs and protein-coding genes inside SCZ risk-associated CNV deletion regions.
Data file S2 (Excel file format). Summary information for CNV-lncRNA coexpression modules.
Data file S3 (Excel file format). Top 500 genes coexpressed with DGCR5 in the module.
Data file S4 (Excel file format). Differentially expressed genes between SCZ cases and controls in the BrainGVEX data set.
Data file S5 (Excel file format). Key driver genes in the DGCR5 coexpression module.
Data file S6 (Excel file format). Some of the DGCR5-regulated genes within CD1 module are predicted targets of hsa-miR-1227.
Data file S7 (Excel file format). Sample information of four transcriptome data sets (BrainGVEX, Developmental Capstone, GTEx, and BrainCloud).