Abstract
SAMHD1 is a human restriction factor known to prevent infection of macrophages, resting CD4+ T cells, and dendritic cells by HIV-1. To test the contribution of MxB to the ability of SAMHD1 to block HIV-1 infection, we created human THP-1 cell lines that were knocked out for expression of MxB, SAMHD1, or both. Interestingly, MxB depletion renders SAMHD1 ineffective against HIV-1 but not SIVmac. We observed similar results in human primary macrophages that were knockdown for the expression of MxB. To understand how MxB assists SAMHD1 restriction of HIV-1, we examined direct interaction between SAMHD1 and MxB in pull-down experiments. In addition, we investigated several properties of SAMHD1 in the absence of MxB expression, including subcellular localization, phosphorylation of the SAMHD1 residue T592, and dNTPs levels. These experiments showed that SAMHD1 restriction of HIV-1 requires expression of MxB.
Keywords: SAMHD1, MxB, HIV-1, Restriction factors, SIVmac
1. Introduction
SAMHD1 is a host restriction factor that blocks HIV-1 infection prior to reverse transcription in primary macrophages, dendritic cells, and resting CD4+ T cells (Baldauf et al., 2012; Descours et al., 2012; Hrecka et al., 2011; Laguette et al., 2011). SAMHD1 is a deoxyguanosine triphosphate (dGTP)-regulated deoxynucleotide triphosphohydrolase (dNTPase) that helps to regulate cellular levels of dNTPs (Goldstone et al., 2011; Kim et al., 2012; Lahouassa et al., 2012; Powell et al., 2011). Phosphorylation of SAMHD1 at residue T592 by the CDK1/CDK2 and cyclin A2 complex prevents its anti-viral activity, linking SAMHD1 phosphorylation to the cell cycle (Cribier et al., 2013; Welbourn et al., 2013; White et al., 2013). Only in the unphosphorylated state, found in non-cycling cells, is SAMHD1 able to restrict HIV-1 (Cribier et al., 2013; Welbourn et al., 2013; White et al., 2013). We and others have shown that the dNTPase activity of SAMHD1 alone is not sufficient for retroviral restriction. Phosphomimetic SAMHD1 variants T592E and T592D lack HIV-1 restriction but still are enzymatically active and deplete dNTPs (Bhattacharya et al., 2016; Welbourn et al., 2013; White et al., 2013). These findings suggested that an overall decrease in level of dNTPs is not solely responsible for the block imposed by SAMHD1, and that another co-factor or entirely distinct mechanism could be involved.
Here we report a novel cofactor that is required for the ability of SAMHD1 to block HIV-1 infection. To test whether MxB is a co-factor for SAMHD1 restriction, we examined the ability of SAMHD1 to restrict HIV-1 in cells that are knockout (KO) for the expression of MxB. The restriction factor MxB binds to the incoming HIV-1 core and prevents the uncoating process (Busnadiego et al., 2014; Fribourgh et al., 2014a; Fricke et al., 2014; Goujon et al., 2013; Liu et al., 2013). SAMHD1 restriction of HIV-1 was found to be defective in non-dividing cells that did not express MxB. To elucidate the role of MxB in SAMHD1 restriction of HIV-1, we examined the known properties of SAMHD1 in the absence of MxB, focusing on protein-protein interactions, subcellular localization, phosphorylation state, and dNTPase activity. Interestingly, MxB was found to assist SAMHD1 restriction activity for HIV-1 but not SIVmac. This result sheds light on a novel instance of co-factor assistance of SAMHD1 restriction of HIV-1.
2. Results
2.1. MxB expression contributes to the ability of SAMHD1 to restrict HIV-1
To investigate whether MxB expression influences the restriction ability of SAMHD1, we tested the ability of SAMHD1 to block HIV-1 infection in human THP-1 cells that are knockout (KO) for the expression of MxB (THP-1-MxB-KO), which were generated using CRISPR/Cas9. Restriction was measured by treating the cells with phorbol 12-myristate 13-acetate (PMA), which triggers differentiation of monocytes into macrophage-like cells (Hrecka et al., 2011; Laguette et al., 2011). Because expression of MxB is low in PMA-treated THP-1 cells, we identified THP-1-MxB-KO clones by performing a Western blot against endogenous MxB in clones that were pretreated with 1000 U/mL of IFN-α for 24 h (Fig. 1A). To measure HIV-1 restriction by SAMHD1 in THP-1-MxB-KO cells, PMA-treated cells were challenged with increasing amounts of HIV-1-GFP. As shown in Fig. 1 B and C, SAMHD1 was unable to restrict HIV-1 in two independent THP-1-MxB-KO clones. Similar experiments were performed in SAMHD1 expression KO clones (THP-1-SAMHD1-KO). As expected, HIV-1 was not restricted in THP-1-SAMHD1-KO cells (Fig. 1B and C). As a control, we challenged THP-1 clones that were generated using the CRISPR/Cas9 technology but did not lose expression of MxB or SAMHD1 (THP-1-Control) with increasing concentrations of HIV-1-GFP (Fig. 1B and C). To further confirm these experiments, we performed similar infections with all KO clones simultaneously (Fig. S1). We also challenged PMA-treated THP-1 cells with wild-type HIV-1 expressing luciferase as a reporter of infection (HIV-1-luc) (Fig. 1D). Taken together, these results suggested that MxB is playing a role in the ability of SAMHD1 to block HIV-1 infection and that this phenotype is not due to induction of MxB expression.
Fig. 1. Expression of MxB is necessary for HIV-1 restriction of SAMHD1.

(A) THP-1-SAMHD1-KO and THP-1-MxB-KO cell lines were created using the CRISPR-Cas 9 system, as described in methods. Because expression of MxB is low in PMA-treated THP-1 cells, we identified THP-1-MxB-KO clones by performing a Western blot against endogenous MxB in clones that were pretreated with 1000 U/mL of IFN-α for 24 h. Samples were also blotted using anti-SAMHD1 antibodies. As a loading control, we analyzed samples for expression of GAPDH. THP-1 control cells undergo the CRISPR-Cas 9 protocol but resulted in cells that were not KO for SAMHD1 or MxB. Representative Western blots are shown for two different THP-1-control, THP-1-SAMHD1-KO, and THP-1-MxB-KO clones. (B) THP-1 KO cell lines were challenged with increasing amounts of HIV-1 expressing GFP (HIV-1-GFP) as a reporter of infection. Infection is shown as the percentage of GFP-positive cells 48 h post-challenge. Similar results were obtained in three independent experiments and a representative experiment is shown. 250 μL of HIV-1-GFP represents a MOI of 2. (C) Infection of HIV-1-GFP in all cells is shown. The result of four independent experiments with standard deviation is shown. * denotes P-value of < 0.05, ** denotes P-value < 0.005 as determined by One-way ANOVA Tukey’s multiple comparisons test. (D) THP-1 KO cell lines were infected with HIV-1-luc and analyzed for luciferase activity. The result of three independent experiments with standard deviation is shown. * denotes P-value of < 0.0005, ** denotes P-value of < 0.0001 as determined by One-way ANOVA Tukey’s multiple comparisons test.
MxB is an interferon-inducible protein (Liu et al., 2013; Goujon et al., 2013; Kane et al., 2013), which is not detected by Western blotting in THP-1-control cells that are not treated with IFNα (Fig. S2). However, wild type HIV-1 infection may or may not induce the type I IFN response (Decalf et al., 2017; Gao et al., 2013; Gendelman et al., 1990; Harman et al., 2011; Rasaiyaah et al., 2013; Saito et al., 2016; Szebeni et al., 1991; Yan et al., 2010), consequentially inducing MxB. To rule out the possibility that HIV-1 infection is inducing expression of endogenous MxB, we tested expression of MxB in HIV-1-infected wild type and KO clones. As shown in Fig. S2, HIV-1 infection of control and KO THP-1 clones did not induce expression of MxB measured 48 h post-infection.
Our experiments suggested MxB to be a possible cofactor required for the ability of SAMHD1 to restrict HIV-1 infection. Although the expression of MxB is very low in PMA-treated cells, one possibility is that MxB is contributing to the restriction imposed by PMA-treated THP-1 cells to HIV-1. To determine the contribution of MxB to the restriction imposed by PMA-treated THP-1 cells to HIV-1, we generated THP-1 cells that are KO for the expression of both MxB and SAMHD1 (THP-1-MxB/SAMHD1-KO) using CRISPR/Cas9 (Fig. 2A). MxB did not show an independent contribution to the restriction imposed by PMA-treated THP-1 cells to HIV-1 (Fig. 2B). As shown by two independent double KO clones, THP-1-MxB/SAMHD1-KO cells were as permissive as THP-1-MxB-KO or THP-1-SAMHD1-KO cells (Fig. 2B). To further confirm these experiments, we performed similar infections with all KO clones simultaneously (Fig. S1). These results suggested that MxB by itself does not have a contribution to the ability of PMA-treated THP-1 cells to block HIV-1 infection strengthening the conclusion that SAMHD1 requires MxB to restrict HIV-1 infection.
Fig. 2. MxB contribution to the ability of PMA-treated THP-1 cells to block HIV-1 infection.
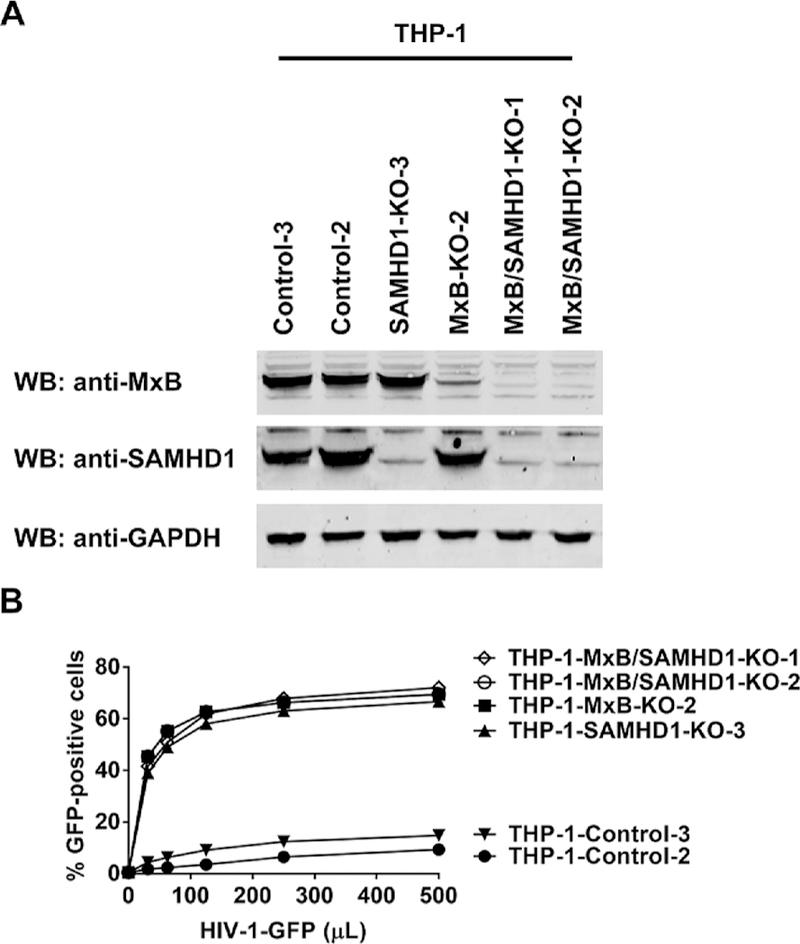
(A) THP-1 cells that are KO for the expression of both MxB and SAMHD1 (THP-1-MxB/SAMHD1-KO) were created using the CRISPR-Cas 9 system, as described in methods. Expression of endogenous SAMHD1 and MxB in THP-1 KO cells was analyzed by Western blot using anti-SAMHD1 and anti-MxB antibodies. Because expression of MxB is low in PMA-treated THP-1 cells, we identified THP-1-MxB-KO clones by performing a Western blot against endogenous MxB in clones that were pretreated with 1000 U/mL of IFN-α for 24 h. As a loading control, we analyzed samples for expression of GAPDH. THP-1 control cells undergo the CRISPR/Cas9 protocol but resulted in cells that were not KO for SAMHD1 or MxB (THP-1-Control). (B) THP-1 KO cell lines were challenged with increasing amounts of HIV-1 expressing GFP (HIV-1-GFP) as a reporter of infection. Infection is shown as the percentage of GFP-positive cells 48 h post-challenge. 125 μL of HIV-1-GFP represents an MOI of 2. Similar results were obtained in three independent experiments and a representative experiment is shown.
To efficiently infect human primary macrophages, resting CD4+ cells, and dendritic cells, SIVmac requires the virion-associated viral accessory protein Vpx (Arfi et al., 2008; Baldauf et al., 2012; Descours et al., 2012; Goujon et al., 2003, 2007, 2008). An important function of Vpx is its ability to recruit SAMHD1 for proteasomal degradation (Berger et al., 2011; Hrecka et al., 2011; Laguette et al., 2011). Next we tested whether MxB plays a role in the ability of SAMHD1 to block SIVmac. To this end, we challenged PMA-treated THP-1-MxB-KO cells with increasing amounts of SIVmac or SIVmacΔVpx. As shown in Fig. 3A, THP-1-MxB-KO cells were permissive to SIVmac infection. By contrast, THP-1-MxB-KO cells potently blocked SIVmacΔVpx when compared to wild type THP-1 cells (Fig. 3A). As a control, we performed similar experiments in THP-1-SAMHD1-KO cells. As shown in Fig. 3B, THP-1-SAMHD1-KO cells were permissive to infection by both SIVmac and SIVmacΔVpx. These experiments showed that SAMHD1 is blocking SIVmacΔVpx in the absence of MxB expression suggesting that MxB is not required for SAMHD1 to block SIVmacΔVpx. As shown in Fig. 3C, THP-MxB/SAMHD1-KO cells showed similar infectivity phenotypes when compared to THP-1-SAMHD1-KO cells suggesting that SAMHD1 does not require MxB to block SIVmacΔVpx. Three independent experiments revealed that MxB minimally contributes to the ability of SAMHD1 to block SIVmacΔVpx infection (Fig. 3D). To further confirm these experiments, we performed similar infections with all KO clones simultaneously (Fig. S3). Our results indicated that MxB is not required for the ability of SAMHD1 to block SIVmac.
Fig. 3. Expression of MxB is not necessary for the ability of SAMHD1 to restrict SIVmac.
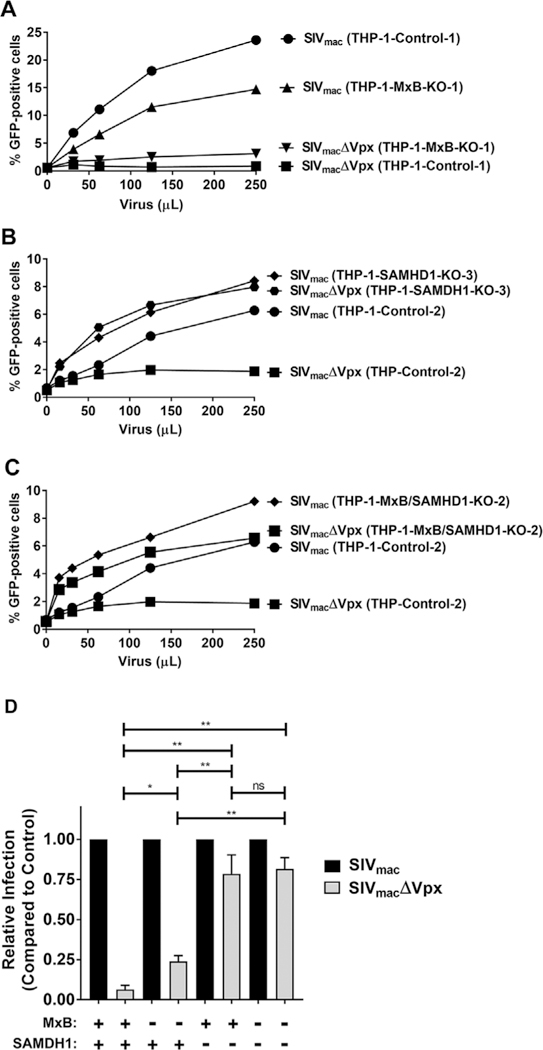
(A) Restriction of SIVmac in THP-1-MxB-KO cells. THP-1 and THP-1-MxB-KO cell lines were challenged with increasing amounts of SIVmac or SIVmacΔVPX expressing GFP as a reporter of infection. Infection is shown as the percentage of GFP-positive cells 72 h post-challenge as measured by flow cytometry. Experiments were performed in triplicate and a representative experiment is shown. 500 μL of SIVmac or SIVmacΔVPX represents a MOI of 2. (B) Restriction of SIVmac in THP-1-SAMHD1-KO cells. THP-1 and THP-1-SAMHD1-KO cell lines were challenged with increasing amounts of SIVmac or SIVmacΔVPX. Infection is shown as the percentage of GFP-positive cells 72 h post-challenge as measured by flow cytometry. Experiments were performed in triplicate and a representative experiment is shown. 500 μL of SIVmac or SIVmacΔVPX represents a MOI of 2. (C) Restriction of SIVmac in THP-1-MxB/SAMHD1-KO cells. THP-1 and THP-1-MxB/SAMHD1-KO cell lines were challenged with increasing amounts of SIVmac or SIVmacΔVPX. Infection is shown as the percentage of GFP-positive cells 72 h post-challenge as measured by flow cytometry. 500 μL of SIVmac or SIVmacΔVPX represents a MOI of 2. (D) Relative infection of SIVmac and SIVmacΔVPX in all cells. The results of three independent experiments with standard deviation are shown, * denotes P value of < 0.05, ** denotes P value of < 0.0001, ns denotes not significant as determined by One-Way ANOVA Tukey’s multiple comparisons test. Black bars represent SIVmac infection. Grey bars represent SIVmacΔVPX infection.
To examine the contribution of MxB to the restriction ability of SAMHD1, we knocked down (KD) expression of MxB in human primary macrophages, MxB-618-siRNA, from four different donors. Our control cells were primary macrophages treated with scrambled shRNA, Src-siRNA (Fig. 4A). We infected the primary macrophages with SIVmac, SIVmacΔVPX, or HIV-1. As expected, MxB KD macrophages released restriction of primary macrophages when infected with HIV-1 as previously shown in THP-1 cells. MxB-618-siRNA macrophages were more permissive to SIVmac compared to control macrophages, though infection of SIVmacΔVPX showed no significant difference between Scr-siRNA and MxB-618-siRNA macrophages (Fig. 4B). Overall these results suggested that SAMHD1 restriction of HIV-1 requires MxB. By contrast, MxB is less important for the ability of SAMDH1 to restrict infection of SIVmac.
Fig. 4. Ability of human primary macrophages that are knocked down for MxB to restrict HIV and SIVmac.
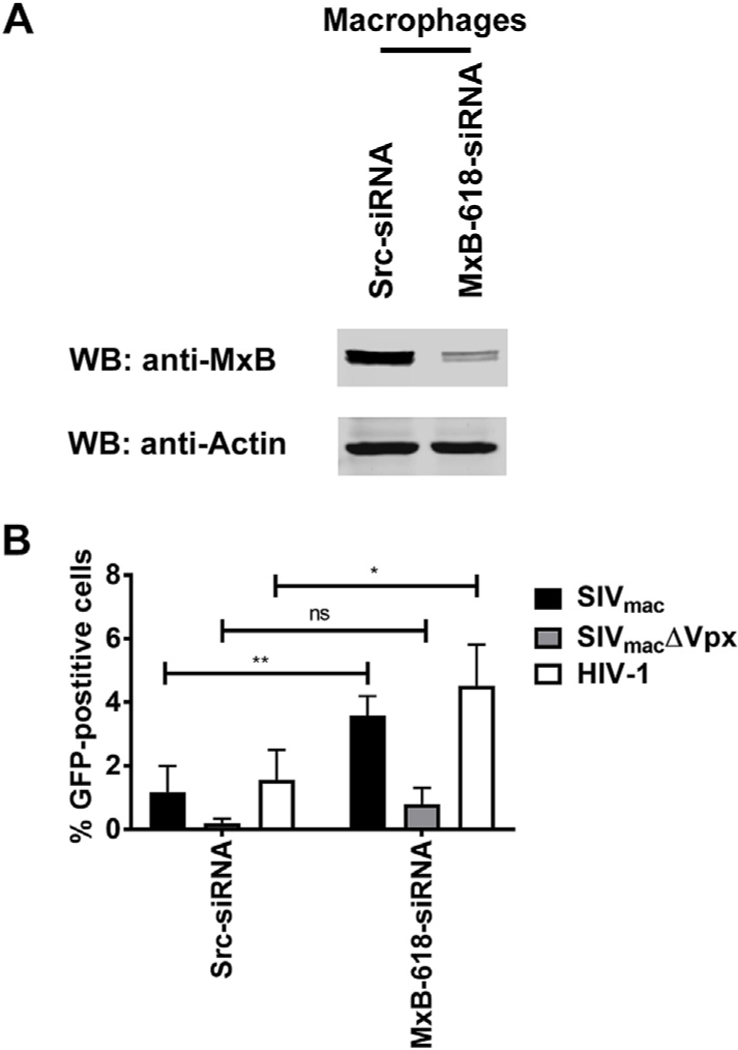
(A) Primary macrophages were knocked down for endogenous MxB expression using siRNA, as described in methods. Expression of endogenous MxB was analyzed by Western blot using anti-MxB antibodies. As a loading control, we analyzed samples for expression of actin. Primary macrophage control cells were treated with scrambled shRNA (Src-siRNA). The western blot shown is a representative blot derived from four different human donors. (B) Restriction of SIVmac and HIV-1 in primary macrophages. Src-siRNA and MxB-618-siRNA macrophages were infected with SIVmac, SIVmacΔVPX, or HIV-1 expressing GFP as a reporter of infection. Infection is shown as the percentage of GFP-positive cells 72 h post-challenge as measured by flow cytometry. Experiments were down with four different donor cells knocked down for endogenous MxB. The results of four independent experiments with standard deviation are shown, * denotes P value < 0.05, ** denotes P value < 0.005, ns denotes not significant as determined by Two-way ANOVA Sidak’s multiple comparisons test.
2.2. SAMHD1 does not directly interact with MxB
To test whether SAMHD1 interacts with MxB, we performed immunoprecipitation assays as previously described (White et al., 2013). For this purpose, human 293T cells were co-transfected with SAMHD1-FLAG and MxB-HA, and SAMHD1-FLAG was immunoprecipitated with anti-FLAG antibodies. Western blot analysis revealed that SAMHD1-FLAG was pulled down without MxB-HA being present (Fig. 5). Similarly, immunoprecipitation experiments were performed using extracts from HIV-1 infected cells in the presence of the chemical crosslinker ethylene-glycol-bis-sulfosuccinimidyl-succinate (sulfo-EGS) (Buffone et al., 2015). These experiments did not reveal an interaction between SAMHD1 and MxB (data not shown). Overall this suggests that SAMHD1 and MxB do not interact under the tested conditions.
Fig. 5. SAMHD1 does not interact with MxB.
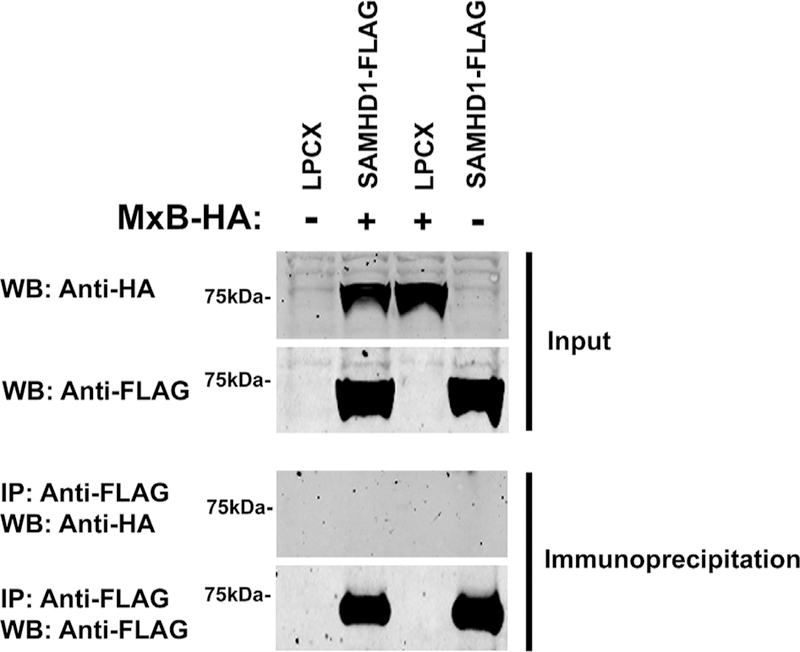
293T cells were co-transfected with SAMHD1-FLAG and either MxB-FLAG or empty vector LPCX. Cells were lysed 24 h post-transfection and analyzed by Western blot using anti-HA and anti-FLAG antibodies (Input). Subsequently, lysates were immunoprecipitated by using anti-FLAG agarose beads, as described in Methods. Anti-FLAG agarose beads were eluted using the 3X FLAG peptide. Elutions were analyzed by Western blot using anti-HA and anti-FLAG antibodies (Immunoprecipitation). WB, Western blot; IP, Immunoprecipitation.
2.3. MxB does not recruit SAMHD1 to the HIV-1 core, nor is MxB binding to the HIV-1 core required for SAMHD1 restriction
We and others have previously showed that the ability of MxB to restrict HIV-1 infection correlates with its ability to bind to the viral capsid (Fribourgh et al., 2014b; Fricke et al., 2014). One possibility is that MxB recruits SAMHD1 to the HIV-1 core by increasing the local concentration of SAMHD1 in the vicinity of the core, resulting in restriction. To test whether MxB recruits SAMHD1 to the surface of the HIV-1 core, we tested the ability of endogenously expressed SAMHD1 to bind to HIV-1 CA-NC complexes pre-assembled in vitro in the presence and absence of endogenously expressed MxB as described (Fricke et al., 2014). Endogenously expressed SAMHD1 was found to not bind to in vitro-assembled HIV-1 CA-NC complexes in either the presence or absence of MxB (Fig. 6), indicating that MxB does not recruit SAMHD1 to the HIV-1 core. As expected and in agreement with previous results, in vitro binding of endogenously expressed MxB to pre-assembled HIV-1 CA-NC complexes (Fricke et al., 2014) was observed.
Fig. 6. Contribution of MxB capsid-binding ability to SAMHD1 restriction of HIV-1.
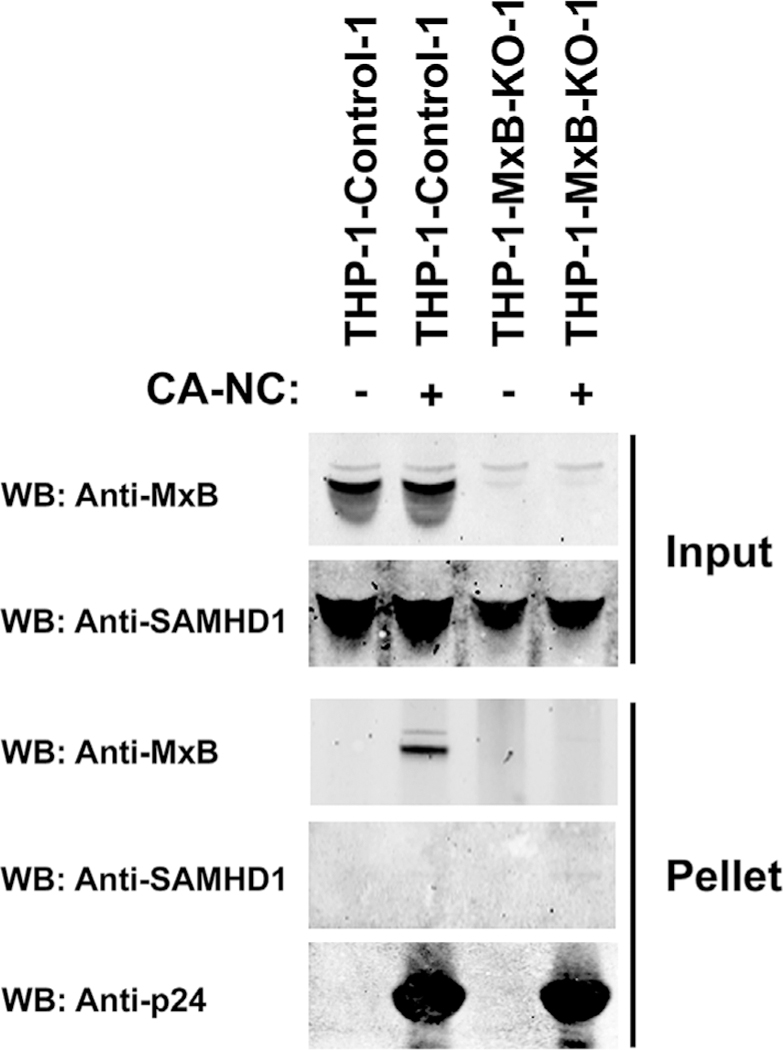
The ability of endogenous SAMHD1 to bind to in vitro assembled HIV-1 CA-NC complexes were measured in extracts from cells that do not express MxB (THP-1-MxB-KO) as described in Methods. As a control, similar experiments were performed using extracts from control THP-1 cells that do express MxB. Input and Pellets fractions were analyzed by Western blot using anti-MxB, anti-SAMHD1, and anti-p24 antibodies. Similar results were obtained in three independent experiments and a representative experiment is shown. * denotes P value < 0.05, ** denotes P value < 0.005, *** denotes P value < 0.0001 as determined by One-way ANOVA Tukey’s multiple comparisons test.
We decided to test whether the binding of MxB to the HIV-1 core is important for the ability of SAMHD1 to block HIV-1. To this end we made use of the previously described HIV-1 capsid mutant G208R, which is known to overcome MxB restriction without actually binding to MxB (Busnadiego et al., 2014; Opp et al., 2015). PMA-treated THP-1 cells were challenged using increasing concentrations of HIV-1-G208R and examined restriction in these cells. As shown in Fig. 7A, HIV-1-G208R was restricted when compared to wild-type HIV-1. By contrast, wild-type and mutant HIV-1 were both permissive in PMA-treated THP-1-MxB-KO cells. Statistically there was no significant difference between HIV-1 and HIV-1-G208R infection in THP-1-MxB-KO cells (Fig. S4 and Fig. 7B). These results suggested that MxB binding to capsid is not required for SAMHD1 to block HIV-1 infection.
Fig. 7. SAMHD1 blocks infection by HIV-1-G208R.
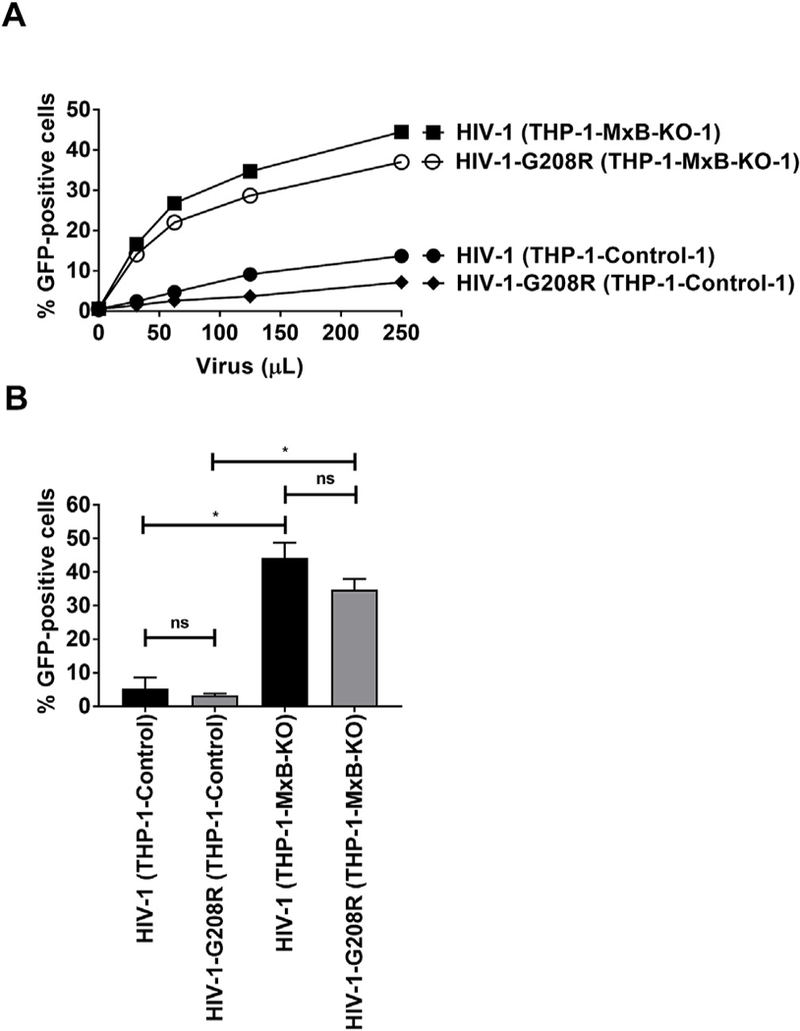
(A) THP-1 and THP-1-MxB-KO cells were challenged with increasing amounts of HIV-1 or HIV-1-G208R expressing GFP as a reporter of infection. G208R is a mutation in the capsid protein that prevents the interaction of HIV-1 with MxB during infection. Wild type and mutant viruses were normalized for p24. The percentage of GFP-positive cells was measured by flow cytometry at 48 h post-challenge. Similar results were obtained in three independent experiments and a representative experiment is shown. (B) Infection of HIV-1-G208R in THP-1-MxB-KO clones is shown. The results of three independent experiments with standard deviation are shown. * denotes P-value of < 0.0001, ns denotes not significant as determined by One-way ANOVA Tukey’s multiple comparisons test.
2.4. MxB does not affect cellular localization of SAMHD1
Both MxB and SAMHD1 have distinct cellular localizations. MxB co-localizes with nucleoporins on the cytoplasmic side of the nuclear envelope and is distributed in the cytoplasm as cytoplasmic punctae (Busnadiego et al., 2014; Fricke et al., 2014; Goujon et al., 2014; Kane et al., 2013; King et al., 2004; Matreyek et al., 2014; Melen et al., 1996). By contrast, SAMHD1 is exclusively localized within the nucleus (Brandariz-Nunez et al., 2012; Hofmann et al., 2012; Rice et al., 2009; Schaller et al., 2014). To test whether and how MxB affects cellular localization of SAMHD1, we examined endogenous localization of SAMHD1 in primary macrophages when MxB expression was knocked down. Depletion of MxB by siRNA was found not to affect cellular localization of SAMHD1 in primary macrophages (Fig. 8A and B). As a control, we studied cellular localization of SAMHD1 in non-treated and siRNA-Scr-treated macrophages. As shown in Fig. 8B, cellular localization of SAMHD1 did not change after any of the treatments. In addition, endogenous MxB and SAMHD1 localization in PMA-treated THP-1-WT and THP-1-MxB-KO cells was analyzed. Immunofluorescence showed that THP-1-KO-MxB cells retained nuclear localization of endogenous SAMHD1 (Fig. 8C). It can therefore be concluded no changes in SAMHD1 localization in macrophages or THP-1 occurs in cells that do not express MxB.
Fig. 8. Subcellular localization of endogenous SAMHD1 in the absence of MxB.
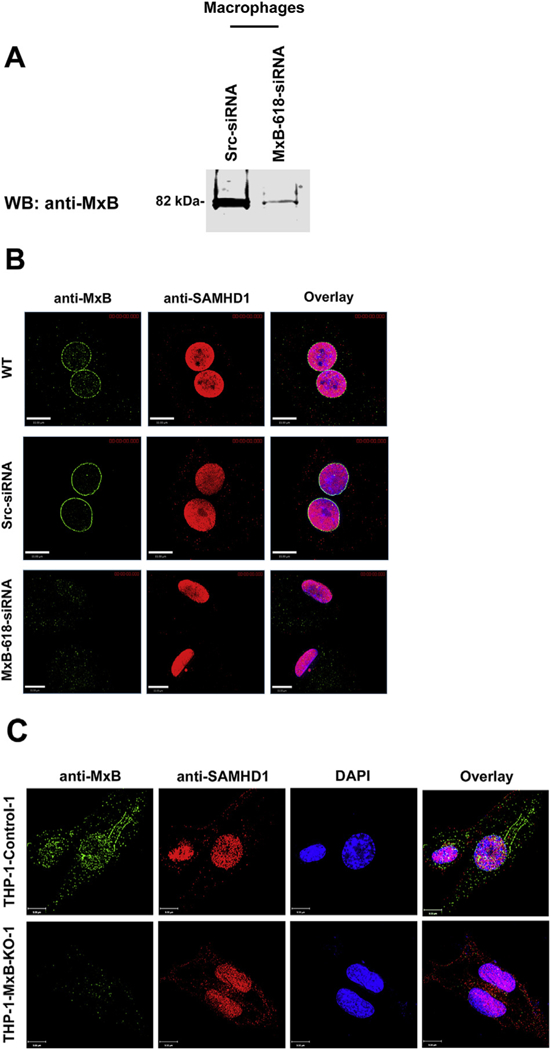
(A) Transient knockdown of MxB in monocyte-derived macrophages (MDM). Expression of MxB was analyzed in lysates from siRNA-Scramble (Src) and siRNA-MxB treated MDMs by Western blot using anti-MxB antibodies. (B) MDMs treated siRNA-Src, siRNA-MxB, or left untreated were fixed/permeabilized and analyzed by immunofluorescence for expression of endogenous MxB and SAMHD1. (C) Similarly, THP-1 control and THP-1-MxB-KO cells were fixed/permeabilized and analyzed by immunofluorescence for expression of endogenous MxB and SAMHD1.
2.5. MxB does not influence SAMHD1 phosphorylation
The ability of SAMHD1 to restrict HIV-1 has been shown to be linked to the dephosphorylation of SAMHD1 at residue T592 (Cribier et al., 2013; Welbourn et al., 2013; White et al., 2013). We therefore investigated whether MxB expression altered the phosphorylation state of SAMHD1 at T592 in PMA treated cells. Western blot analysis revealed that T592 remained unphosphorylated in differentiated THP-1-MxB-KO cells, just as it did in wild-type THP-1 cells (Fig. 9). As a positive control, we analyzed undifferentiated THP-1 cells (not treated with PMA), and showed phosphorylation of SAMHD1 at position T592. These results suggested that absence of MxB expression does not affect the phosphorylation state of SAMHD1 at T592.
Fig. 9. Phosphorylation state of SAMHD1 at position T592 in the absence of MxB.
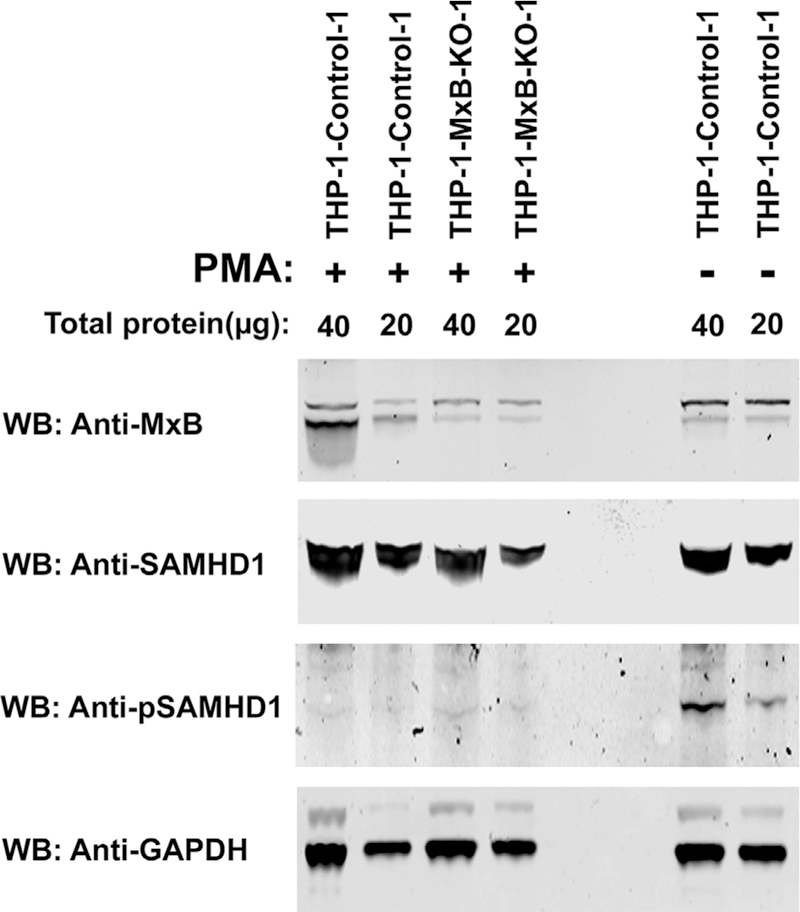
Lysates from PMA-treated THP-1 and THP-1-MxB-KO cells were analyzed by Western blot analysis using anti-MxB, anti-SAMHD1, and anti-phospho-T592-SAMHD1. As loading control, samples were analyzed for the expression of GAPDH. As a positive control for phosphorylation, THP-1 cells that were not treated with PMA were utilized. Experiments were done in triplicate and a representative experiment is shown.
2.6. The ability of SAMHD1 to decrease cellular levels of dNTPs is not affected in MxB-depleted cells
We and others have previously shown that the dNTPase activity of SAMHD1 is necessary but not sufficient to restrict HIV-1 (Cribier et al., 2013; Welbourn et al., 2013; White et al., 2013). To study in greater detail how MxB contributes to the dNTPase activity of SAMHD1, we measured cellular dNTP levels in differentiated THP-1, THP-1-MxB-KO, and THP-1-SAMHD1-KO cells. As expected, THP-1-SAMHD1-KO cells showed a ∼5-fold increase in dATP and dGTP levels when compared to THP-1 cells (Fig. 10). By contrast, THP-1-MxB-KO cells showed a ∼1.5 old and 2-fold increase respectively in dATP and dGTP levels (Fig. 10), suggesting that MxB has only a marginal effect on cellular dNTPs.
Fig. 10. Ability of SAMHD1 to decrease the cellular levels of dNTPs in the absence of MxB.
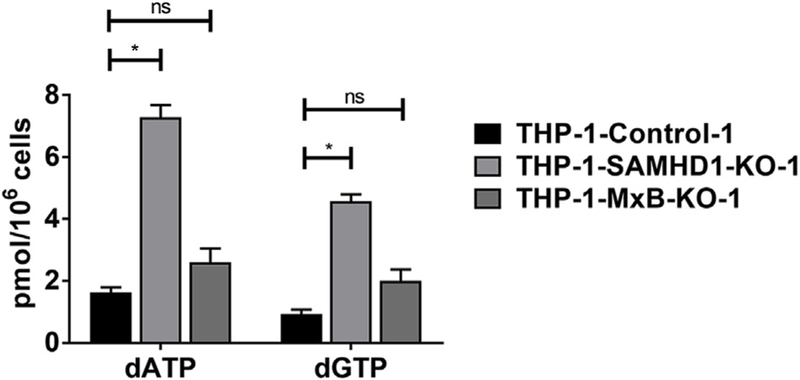
Intracellular dATP and dGTP concentrations from PMA treated THP-1, THP-1-MxB-KO, and THP-1-SAMHD1-KO cell cell were quantified as described in Methods. Experiments were performed in triplicates and standard deviation is shown. * denotes P value < 0.0001, ns denotes not significant as determined by One-way ANOVA Tukey’s multiple comparisons test.
3. Discussion
SAMHD1 restriction of HIV-1 is linked to the phosphorylation state of residue T592 (Cribier et al., 2013; Pauls et al., 2014; St Gelais et al., 2014; Welbourn et al., 2013; White et al., 2013), but the exact molecular details of how this phosphorylation contributes to restriction is less clear. Several studies have correlated HIV-1 restriction with decreased intracellular pools of dNTPs by SAMHD1 (Goldstone et al., 2011; Kim et al., 2012; Lahouassa et al., 2012; Powell et al., 2011; White et al., 2013). However, we and others have shown that dNTPase activity by SAMHD1 is necessary but not sufficient to block HIV-1 infection (Welbourn et al., 2013; White et al., 2013). It has also been proposed that SAMHD1 tetramerization is important for dNTPase activity and retroviral restriction (Arnold et al., 2015; Tang et al., 2015; Yan et al., 2015). In contrast, we have shown that the SAMHD1 tetramerization-defective mutant Y146S/Y154S can still restrict HIV-1 infection in addition to retaining dNTPase activity (Bhattacharya et al., 2016; Brandariz-Nuñez et al., 2013). Altogether this suggests that SAMHD1 restriction of HIV-1 might be regulated by a yet unidentified mechanism or co-factor. In this study we report MxB to be a novel co-factor governing the ability of SAMHD1 to block HIV-1 but not SIVmac restriction.
Our results demonstrated that knocking out MxB expression in THP-1 cells disrupted the ability of SAMHD1 to block HIV-1 infection, but did not affect expression of SAMHD1. By contrast, knocking out expression of MxB in THP-1 cells did not affect the ability of SAMHD1 to block SIVmac. A possible reason for the differences between HIV-1 and SIVmac could lie in the intrinsic properties of the two viruses. It is important to clarify that MxB is an interferon-inducible protein not able to restrict HIV-1 in PMA-treated THP-1 cells since PMA does not induce expression of MxB as seen with interferon treatment.
Previous investigations have revealed differences in the ability of SAMHD1 to restrict HIV-1 and SIVmac. Although use of Vpx or deoxynucleosides (dNs) increases infection in dendritic and THP-1 cells by HIV-1, only Vpx restores infectivity of SIVmac in dendritic and THP-1 cells (Barateau et al., 2015; Reinhard et al., 2014). Interestingly, the dichotomy seen with HIV-1 and SIVmac was found to map to the retroviral capsid. Infection of THP-1 cells by chimeric HIV-1 viruses containing the capsid of SIVmac was not increased by supplying exogenous dNs (Barateau et al., 2015). This suggests that SAMHD1 restriction of lentiviral infection is occurring by depletion of dNTPs and/or an unknown mechanism. In agreement with this evidence, we found that SAMHD1 restriction of HIV-1, but not SIVmac, requires MxB. Interestingly, MxB interacts with capsid (Buffone et al., 2015; Fribourgh et al., 2014b; Fricke et al., 2014; Schulte et al., 2015); however, we were unable to prove that the interaction of MxB with capsid is similarly important for the ability of SAMHD1 to block HIV-1.
To understand how MxB contributes to the ability of SAMHD1 to restrict HIV-1 infection, we first investigated a possible biochemical interaction between MxB and SAMHD1. We failed to observe an interaction between SAMHD1 and MxB using endogenous or tagged versions of the proteins. This might be due to not using the correct buffer for the interaction, or that the interaction involves a larger complex. Since we were unable to define a biochemical interaction between SAMHD1 and MxB, one possibility is that there is still yet an unidentified co-factor that directly interacts with both MxB and SAMHD1. We tested whether MxB recruits SAMHD1 to the viral core using endogenously expressed proteins from THP-1 cells; however, no evidence was found that MxB recruits SAMHD1 to the HIV-1 core. We also examined whether SAMHD1 blocks HIV-1 viruses with a capsid mutation that disrupts MxB binding to the HIV-1 core. The capsid mutation G208R is known to disrupt the ability of MxB to bind to capsid and restrict HIV-1 infectivity (Busnadiego et al., 2014; Opp et al., 2015). Accordingly, we tested whether HIV-1 viruses with the G208R capsid mutation are restricted by SAMHD1 in THP-1 cells that do not express MxB, and found that this does not affect the ability of SAMHD1 to block HIV-1. Because of the clear nuclear localization of SAMHD1 in the nucleus, we tested whether knocking out MxB in macrophages or THP-1 cells changes the subcellular localization. Our results showed that MxB depletion did not change SAMHD1 nuclear localization. We also demonstrated that MxB depletion did not change the phosphorylation state of SAMHD1, suggesting that phosphorylation is not the reason for the lack of restriction of HIV-1 infection in MxB knockouts. Lastly, we investigated the levels of dNTPs in THP-1 cells knocked out for MxB, and found MxB only minimally affected the ability of SAMHD1 to deplete cellular dNTPs. Overall, our work genetically links MxB to the ability of SAMHD1 to block HIV-1 infection.
Our work establishes a genetic link between MxB and the ability of SAMHD1 to block HIV-1 infection; however, it does not provide mechanistic information on how MxB is assisting the ability of SAMHD1 to block HIV-1 infection. One possibility is that MxB is in a large complex with SAMHD1 regulating the local levels of GTP in the complex (Melen et al., 1996). The levels of GTP in the complex may be affecting the oligomeric state of SAMHD1 (Bhattacharya et al., 2016; Brandariz-Nuñez et al., 2013; Hansen et al., 2014; Ji et al., 2014; Wang et al., 2016), which may lead to a change in HIV-1 restriction. Future experiments will further explore the link between SAMHD1 and MxB.
4. Methods
4.1. Cell lines
HEK293T cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% (vol/vol) fetal bovine serum and 1% (vol/vol) penicillin-streptomycin. Human THP-1 cells (ATCC TIB-202) were grown in RPMI medium supplemented with 10% (vol/vol) fetal bovine serum and 1% (vol/vol) penicillin-streptomycin. Human monocyte derived macrophages (MDMs) were prepared from peripheral blood mononuclear cells (PBMCs). PBMCs were isolated using Ficoll-Paque Plus (GE Health Care) separation from healthy donors. Monocytes were obtained via negative selection using a Monocyte Isolation Kit II human (MACS Miltenyi Biotec) and differentiated into MDMs by treatment with granulocyte-macrophages colony stimulating factor (GM-CSF) (10 ng/mL, Miltenyl Biotec) for 7 days.
4.2. HIV-1 CA-NC expression and purification
The CA-NC proteins of HIV-1 and HIV-1 bearing the G208R capsid mutant (HIV-1-G208R) were expressed, purified, and assembled as previously described (Lienlaf et al., 2011).
4.3. Generation of MxB and SAMDH1 knockout human cells
MxB and SAMHD1 knockout (KO) cell lines were generated by using the clustered regularly interspaced short palindromic repeat (CRISPR)-Cas9 gene system (Edit-R CRISPR-Cas9 gene engineered with SMARTCas9 nucleases; GE Healthcare) (Dominguez et al., 2016). The CRISPR genomic guide RNA sequences for human MxB in THP-1 were designed to target exon 2 near the start codon. The guide RNA used to target MxB in THP-1 cells were CCGCCATTCGGCACAGT GCC(complementary strand) and CACAAGCCTTGGCCCTACCGG (complementary strand). The CRISPR genomic guide RNA sequences for human SAMHD1 in THP-1 were designed to target exon 1 of the SAMHD1 gene. The guide RNA used to target SAMHD1 in THP-1 cells were CTCAAACACCCCTTCCGCAG and GGCAGACTGGTCCCCGGGCC as previously described (Martinez-Lopez et al., 2018). The guide RNA constructs, human cytomegalovirus hCMV-Casp9, and CMV-GFP were co-transfected into THP-1 cells. At 48 h of post-transfection, GFP-positive cells were sorted using the FACSAria system (BD Biosciences, Franklin Lakes, NJ, USA). THP-1 colonies were expanded individually and characterized for loss of MxB or SAMHD1 protein expression by Western blot analysis using anti-MxB or anti-SAMHD1 antibodies respectively. Exon 2 of MxB was amplified and sequenced from genomic DNA isolated from wild-type and knockout cells. KO generation was confirmed with 24 h 1000 U/mL IFN-α (IF007 human Interferon-α A protein, Millipore) treatment and analyzed by Western blot analysis.
4.4. Generation of MxB knockdown primary macrophages
MxB knockdown (KD) primary macrophages were generated using siRNA (MX2HSS106818, Invitrogen) in combination with stealth RNAi negative control duplexes (Invitrogen) following the RNAiMAX transfection protocol (Invitrogen). In a 12-well plate, 60 ρm of siRNA were added to macrophages with 3 μL of RNAiMAX (Lipofectamine RNAiMAX, Invitrogen). After 3–4 days, the cells were lysed and analyzed by Western Blot for KD of endogenous MxB.
4.5. Infection using GFP-reporter viruses
Recombinant HIV-1, HIV-1-G208R mutant, SIVmac, and SIVmacΔVPX expressing green fluorescent protein (GFP) were prepared as described (Diaz-Griffero et al., 2008). Recombinant viruses were pseudotyped with VSV-G glycoprotein. For infections, 24-well plates were seeded with 50,000 THP-1 cells and treated with PMA (40 ng/mL) for 24 h at 37 °C. The cells were then treated with virus and incubated for 24 h. A total of 500 μL of medium was added, and GFP-positive cells were analyzed using a flow cytometer (Becton Dickinson).
4.6. Co-immunoprecipitation assay
Approximately 107 human 293T embryonic kidney cells were co-transfected with plasmids expressing SAMHD1 tagged with FLAG and/or expressing MxB tagged with hemagglutinin (HA) using an LPCX vector. After 24 h, cells were lysed in 0.5 ml of whole-cell extract (WCE) buffer (50 mM Tris [pH 8.0], 280 mM NaCl, 0.5% Igepal, 10% glycerol, 5 mM MgCl2, 50 μg/ml ethidium bromide, 50 U/ml nuclease [Benzonase; Roche]). Lysates were centrifuged at 20,817×g for 1 h at 4 °C. Post-spin lysates were then pre-cleared with protein A-agarose (Sigma) for 1 h at 4 °C; a small aliquot of each lysate was stored for use as the input. Precleared lysates containing the tagged proteins were incubated with anti-FLAG-agarose beads (Sigma) at 4 °C. Anti-FLAG-agarose beads were washed three times in WCE buffer, and immune complexes were eluted using 200 μg of FLAG tripeptide/ml in WCE buffer. The eluted samples were fractionated by SDS-PAGE and analyzed by Western blotting with either anti-HA or anti-FLAG antibodies (Sigma).
4.7. EGS cross-linking with co-immunoprecipitation experiments of MxB variants
293T cells were transfected either with SAMHD1-FLAG, MxB-HA, or both. 24 h after transfection, cells were lysed in cross-link lysis buffer [5% IGEPAL in 1X PBS]. Lysates were centrifuged at 15,000 rpm for 1 hat 4 °C. 120 μL of supernatants were collected and incubated with 40 μL of sulfo-EGS (Thermo Scientific) for 30 min 10 μL of 1M Tris-HCl pH 7.5 were then added to the lysate EGS mixture. Samples were then co-immunoprecipitated as mentioned above. The eluted samples were fractionated by SDS-PAGE and analyzed by Western blotting with either anti-HA or anti-FLAG antibodies (Sigma).
4.8. Binding to HIV-1 CA-NC complexes pre-assembled in vitro
THP-1 control and KO MxB cells were seeded at 5 × 106 per plate (4 plates per sample) and treated with PMA for 24 h. Cell lysates were then prepared as follows: pre-washed cells were re-suspended in hypotonic lysis buffer (10 mM Tris, pH 7.4, 1.5 mM MgCl2, 10 m MKCl, 0.5 mM dithiothreitol [DTT]), and the cell suspension was frozen, thawed, and incubated on ice for 10 min. After this, the lysate was centrifuged at maximum speed in a refrigerated Eppendorf microcentrifuge (∼20,817×g) for 5 min. The supernatant was supplemented with 1/10 volume of 10X PBS for use in the binding assay. In some cases, samples containing the MxB variants were diluted with extracts prepared in parallel from non-transfected cells. To test binding, 5 μL of capsid-nucleocapsid (CA-NC) particles pre-assembled in vitro was incubated with 200 μL of cell lysate at room temperature for 1 h (Ganser et al., 1999; Yang et al., 2014). A fraction of this mixture was stored (input). The mixture was spun through a sucrose cushion (70% sucrose, 1X PBS, 0.5 mM DTT) at 100,000×g in an SW55 rotor (Beckman) for 1 h at 4 °C. After centrifugation, the supernatant was carefully removed and the pellet re-suspended in 1 X SDS-PAGE loading buffer (pellet). The level of MxB proteins was determined by Western blotting with anti-HA or anti-FLAG antibody as described above. The level of HIV-1 CA-NC protein in the pellet was assessed by Western blotting with anti-p24 antibodies.
4.9. Western blot analysis
Cells were lysed in lysis buffer (50 mM Tris [pH 8.0], 280 mM NaCl, 0.5% Igepal 40, 10% glycerol, 5 mM MgCl2). Detection of proteins by Western blotting was performed with anti-FLAG (1:1000 dilution; Sigma), anti-MxB (1:1000 dilution; Novus Biologicals), anti-SAMHD1 (1:1000 dilution), anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (1:5000; Ambion), or anti-p24 (1:1,000, catalog number 183-H12–5C; NIH) antibodies. Secondary antibodies against rabbit and mouse conjugated to IRDye 680LT or IRDye 800CW were obtained from Li-Cor (1:10,000 dilution). Bands were detected by scanning blots using the Li-Cor Odyssey imaging system in the 700-nm or 800-nm channel.
4.10. Quantification of cellular dNTPs by primer extension assay
2×106 cells THP-1 cells were induced to differentiate with PMA for 24 h, then washed twice with 1X PBS, pelleted and re-suspended in ice cold 65% methanol in Millipore-grade water. Cells were vortexed for 2 min, incubated at 95 °C for 3 min, and centrifuged at 20,817×g for 3 min. The supernatant was transferred to a new tube for complete removal of the methanol with a speed vac. The dried samples were resuspended in Millipore-grade water. An 18-nucleotide primer terminally 5′-labeled with [32P] (5′-GTCCCTGTTCGGGCGCCA-3′) was annealed at a 1:2 ratio to four different 19-nucleotide templates (5′-NTG GCGCCCGAACAGGGAC-3′), where ‘N’ represents nucleotide variation at the 5′ end. The assay mixture contains 200 fmoles of template primer, 2 μL of 0.5 mM dNTP mix for the positive control or the cell extract, 4 μL of excess HIV-1 RT, 25 mM Tris-HCl, pH 8.0, 2 mM dithiothreitol, 100 mM KCl, 5 mM MgCl2, and 10 μM oligo(dT) to a final volume of 20 μL. The reaction was incubated at 37 °C for 5 min before being quenched with 10 μL of 40 mM EDTA and 99% (vol/vol) formamide at 95 °C for 5 min. The extended-primer products were resolved on a 14% urea-PAGE gel and analyzed with a phosphoimager. The products were analyzed for percent volume of saturation using QuantityOne software. The quantified dNTP content of each sample was calculated from its dilution factor, so that each sample volume was adjusted to obtain a signal within the linear range of the assay (Kim et al., 2012; Lahouassa et al., 2012).
Supplementary Material
Acknowledgements
This work was funded by NIH R01 AI087390, R21 AI102824, and R56 AI108432 to F.D.-G and NIH R01 GM104198 and R01 AI049781–0 to B.K. We are grateful to the NIH/AIDS repository program for providing antibodies.
Footnotes
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.virol.2019.03.018.
References
- Arfi V, Riviere L, Jarrosson-Wuilleme L, Goujon C, Rigal D, Darlix JL, Cimarelli A, 2008. Characterization of the early steps of infection of primary blood monocytes by human immunodeficiency virus type 1. J. Virol 82, 6557–6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold LH, Groom HC, Kunzelmann S, Schwefel D, Caswell SJ, Ordonez P, Mann MC, Rueschenbaum S, Goldstone DC, Pennell S, Howell SA, Stoye JP, Webb M, Taylor IA, Bishop KN, 2015. Phospho-dependent regulation of SAMHD1 oligomerisation couples catalysis and restriction. PLoS Pathog 11, e1005194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldauf H-M, Pan X, Erikson E, Schmidt S, Daddacha W, Burggraf M, Schenkova K, Ambiel I, Wabnitz G, Gramberg T, Panitz S, Flory E, Landau NR, Sertel S, Rutsch F, Lasitschka F, Kim B, Konig R, Fackler OT, Keppler OT, 2012. SAMHD1 restricts HIV-1 infection in resting CD4+ T cells. Nat. Med 18, 1682–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barateau V, Nguyen XN, Bourguillault F, Berger G, Cordeil S, Cimarelli A, 2015. The susceptibility of primate lentiviruses to nucleosides and Vpx during infection of dendritic cells is regulated by CA. J. Virol 89, 4030–4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger A, Sommer AF, Zwarg J, Hamdorf M, Welzel K, Esly N, Panitz S, Reuter A, Ramos I, Jatiani A, 2011. SAMHD1-deficient CD14+ cells from individuals with Aicardi-Goutieres syndrome are highly susceptible to HIV-1 infection. PLoS Pathog 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A, Wang Z, White T, Buffone C, Nguyen LA, Shepard CN, Kim B, Demeler B, Diaz-Griffero F, Ivanov DN, 2016. Effects of T592 phosphomimetic mutations on tetramer stability and dNTPase activity of SAMHD1 can not explain the retroviral restriction defect. Sci. Rep 6, 31353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandariz-Nunez A, Valle-Casuso JC, White TE, Laguette N, Benkirane M, Brojatsch J, Diaz-Griffero F, 2012. Role of SAMHD1 nuclear localization in restriction of HIV-1 and SIVmac. Retrovirology 9, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandariz-Nuñez A, Valle-Casuso JC, White TE, Nguyen L, Bhattacharya A, Wang Z, Demeler B, Amie S, Knowlton C, Kim B, Ivanov DN, Diaz-Griffero F, 2013. Contribution of oligomerization to the anti-HIV-1 properties of SAMHD1. Retrovirology 10 131–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffone C, Schulte B, Opp S, Diaz-Griffero F, 2015. Contribution of MxB oligomerization to HIV-1 capsid binding and restriction. J. Virol 89, 3285–3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busnadiego I, Kane M, Rihn SJ, Preugschas HF, Hughes J, Blanco-Melo D, Strouvelle VP, Zang TM, Willett BJ, Boutell C, Bieniasz PD, Wilson SJ, 2014. Host and viral determinants of Mx2 antiretroviral activity. J. Virol 88, 7738–7752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cribier A, Descours B, Valadao AL, Laguette N, Benkirane M, 2013. Phosphorylation of SAMHD1 by cyclin A2/CDK1 regulates its restriction activity toward HIV-1. Cell Rep 3, 1036–1043. [DOI] [PubMed] [Google Scholar]
- Decalf J, Desdouits M, Rodrigues V, Gobert FX, Gentili M, Marques-Ladeira S, Chamontin C, Mougel M, Cunha de Alencar B, Benaroch P, 2017. Sensing of HIV-1 entry triggers a type I interferon response in human primary macrophages. J. Virol 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Descours B, Cribier A, Chable-Bessia C, Ayinde D, Rice G, Crow Y, Yatim A, Schwartz O, Laguette N, Benkirane M, 2012. SAMHD1 restricts HIV-1 reverse transcription in quiescent CD4+T-cells. Retrovirology 9, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Griffero F, Perron M, McGee-Estrada K, Hanna R, Maillard PV, Trono D, Sodroski J, 2008. A human TRIM5alpha B30.2/SPRY domain mutant gains the ability to restrict and prematurely uncoat B-tropic murine leukemia virus. Virology 378, 233–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez AA, Lim WA, Qi LS, 2016. Beyond editing: repurposing CRISPR-Cas9 for precision genome regulation and interrogation. Nat. Rev. Mol. Cell Biol 17, 5–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fribourgh JL, Nguyen HC, Matreyek KA, Alvarez FJ, Summers BJ, Dewdney TG, Aiken C, Zhang P, Engelman A, Xiong Y, 2014a. Structural insight into HIV-1 restriction by MxB. Cell Host Microbe 16, 627–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fribourgh JL, Nguyen HC, Matreyek KA, Alvarez FJD, Summers BJ, Dewdney TG, Aiken C, Zhang P, Engelman A, Xiong Y, 2014b. Structural insight into HIV-1 restriction by MxB. Cell Host Microbe 16, 627–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricke T, White TE, Schulte B, de Souza Aranha Vieira DA, Dharan A, Campbell EM, Brandariz-Nunez A, Diaz-Griffero F, 2014. MxB binds to the HIV-1 core and prevents the uncoating process of HIV-1. Retrovirology 11, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganser BK, Li S, Klishko VY, Finch JT, Sundquist WI, 1999. Assembly and analysis of conical models for the HIV-1 core. Science 283, 80–83. [DOI] [PubMed] [Google Scholar]
- Gao D, Wu J, Wu YT, Du F, Aroh C, Yan N, Sun L, Chen ZJ, 2013. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science 341, 903–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendelman HE, Baca LM, Turpin J, Kalter DC, Hansen B, Orenstein JM, Dieffenbach CW, Friedman RM, Meltzer MS, 1990. Regulation of HIV replication in infected monocytes by IFN-alpha. Mechanisms for viral restriction. J. Immunol 145, 2669–2676 (Baltimore, Md. : 1950). [PubMed] [Google Scholar]
- Goldstone DC, Ennis-Adeniran V, Hedden JJ, Groom HC, Rice GI, Christodoulou E, Walker PA, Kelly G, Haire LF, Yap MW, 2011. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature 480. [DOI] [PubMed] [Google Scholar]
- Goujon C, Arfi V, Pertel T, Luban J, Lienard J, Rigal D, Darlix JL, Cimarelli A, 2008. Characterization of simian immunodeficiency virus SIVSM/human immunodeficiency virus type 2 Vpx function in human myeloid cells. J. Virol 82, 12335–12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goujon C, Jarrosson-Wuilleme L, Bernaud J, Rigal D, Darlix JL, Cimarelli A, 2003. Heterologous human immunodeficiency virus type 1 lentiviral vectors packaging a simian immunodeficiency virus-derived genome display a specific postentry transduction defect in dendritic cells. J. Virol 77, 9295–9304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goujon C, Moncorge O, Bauby H, Doyle T, Barclay WS, Malim MH, 2014. Transfer of the amino-terminal nuclear envelope targeting domain of human MX2 converts MX1 into an HIV-1 resistance factor. J. Virol 88, 9017–9026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goujon C, Moncorge O, Bauby H, Doyle T, Ward CC, Schaller T, Hue S, Barclay WS, Schulz R, Malim MH, 2013. Human MX2 is an interferon-induced post-entry inhibitor of HIV-1 infection. Nature 502, 559–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goujon C, Riviere L, Jarrosson-Wuilleme L, Bernaud J, Rigal D, Darlix JL, Cimarelli A, 2007. SIVSM/HIV-2 Vpx proteins promote retroviral escape from a proteasome-dependent restriction pathway present in human dendritic cells. Retrovirology 4, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen EC, Seamon KJ, Cravens SL, Stivers JT, 2014. GTP activator and dNTP substrates of HIV-1 restriction factor SAMHD1 generate a long-lived activated state. Proc. Natl. Acad. Sci. U.S.A 111, E1843–E1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harman AN, Lai J, Turville S, Samarajiwa S, Gray L, Marsden V, Mercier SK, Jones K, Nasr N, Rustagi A, Cumming H, Donaghy H, Mak J, Gale M Jr., Churchill M, Hertzog P, Cunningham AL, 2011. HIV infection of dendritic cells subverts the IFN induction pathway via IRF-1 and inhibits type 1 IFN production. Blood 118, 298–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann H, Logue EC, Bloch N, Daddacha W, Polsky SB, Schultz ML, Kim B, Landau NR, 2012. The Vpx lentiviral accessory protein targets SAMHD1 for degradation in the nucleus. J. Virol 86, 12552–12560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrecka K, Hao C, Gierszewska M, Swanson SK, Kesik-Brodacka M, Srivastava S, Florens L, Washburn MP, Skowronski J, 2011. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X, Tang C, Zhao Q, Wang W, Xiong Y, 2014. Structural basis of cellular dNTP regulation by SAMHD1. Proc. Natl. Acad. Sci. U.S.A 111, E4305–E4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane M, Yadav SS, Bitzegeio J, Kutluay SB, Zang T, Wilson SJ, Schoggins JW, Rice CM, Yamashita M, Hatziioannou T, Bieniasz PD, 2013. MX2 is an interferon-induced inhibitor of HIV-1 infection. Nature 502, 563–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B, Nguyen LA, Daddacha W, Hollenbaugh JA, 2012. Tight interplay among SAMHD1 protein level, cellular dNTP levels, and HIV-1 proviral DNA synthesis kinetics in human primary monocyte-derived macrophages. J. Biol. Chem 287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MC, Raposo G, Lemmon MA, 2004. Inhibition of nuclear import and cell-cycle progression by mutated forms of the dynamin-like GTPase MxB. Proc. Natl. Acad. Sci. U.S.A 101, 8957–8962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laguette N, Sobhian B, Casartelli N, Ringeard M, Chable-Bessia C, Segeral E, Yatim A, Emiliani S, Schwartz O, Benkirane M, 2011. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahouassa H, Daddacha W, Hofmann H, Ayinde D, Logue EC, Dragin L, Bloch N, Maudet C, Bertrand M, Gramberg T, 2012. SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat. Immunol 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lienlaf M, Hayashi F, Di Nunzio F, Tochio N, Kigawa T, Yokoyama S, Diaz-Griffero F, 2011. Contribution of E3-ubiquitin ligase activity to HIV-1 restriction by TRIM5alpha(rh): structure of the RING domain of TRIM5alpha. J. Virol 85, 8725–8737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Pan Q, Ding S, Qian J, Xu F, Zhou J, Cen S, Guo F, Liang C, 2013. The interferon-inducible MxB protein inhibits HIV-1 infection. Cell Host Microbe 14, 398–410. [DOI] [PubMed] [Google Scholar]
- Martinez-Lopez A, Martin-Fernandez M, Buta S, Kim B, Bogunovic D, Diaz-Griffero F, 2018. SAMHD1 deficient human monocytes autonomously trigger type I interferon. Mol. Immunol 101, 450–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matreyek KA, Wang W, Serrao E, Singh P, Levin HL, Engelman A, 2014. Host and viral determinants for MxB restriction of HIV-1 infection. Retrovirology 11, 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melen K, Keskinen P, Ronni T, Sareneva T, Lounatmaa K, Julkunen I, 1996. Human MxB protein, an interferon-alpha-inducible GTPase, contains a nuclear targeting signal and is localized in the heterochromatin region beneath the nuclear envelope. J. Biol. Chem 271, 23478–23486. [DOI] [PubMed] [Google Scholar]
- Opp S, Vieira DA, Schulte B, Chanda SK, Diaz-Griffero F, 2015. MxB is not responsible for the blocking of HIV-1 infection observed in alpha interferon-treated cells. J. Virol 90, 3056–3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauls E, Ruiz A, Badia R, Permanyer M, Gubern A, Riveira-Muñoz E, Torres-Torronteras J, Álvarez M, Mothe B, Brander C, Crespo M, Menéndez-Arias L, Clotet B, Keppler OT, Martí R, Posas F, Ballana E, Esté JA, 2014. Cell cycle control and HIV-1 susceptibility are linked by CDK6-dependent CDK2 phosphorylation of SAMHD1 in myeloid and lymphoid cells. J. Immunol 193, 1988–1997. [DOI] [PubMed] [Google Scholar]
- Powell RD, Holland PJ, Hollis T, Perrino FW, 2011. Aicardi-Goutieres syndrome gene and HIV-1 restriction factor SAMHD1 is a dGTP-regulated deoxynucleotide triphosphohydrolase. J. Biol. Chem 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasaiyaah J, Tan CP, Fletcher AJ, Price AJ, Blondeau C, Hilditch L, Jacques DA, Selwood DL, James LC, Noursadeghi M, Towers GJ, 2013. HIV-1 evades innate immune recognition through specific cofactor recruitment. Nature 503, 402–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhard C, Bottinelli D, Kim B, Luban J, 2014. Vpx rescue of HIV-1 from the antiviral state in mature dendritic cells is independent of the intracellular deoxynucleotide concentration. Retrovirology 11, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice GI, Bond J, Asipu A, Brunette RL, Manfield IW, Carr IM, 2009. Mutations involved in Aicardi-Goutieres syndrome implicate SAMHD1 as regulator of the innate immune response. Nat. Genet 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito A, Henning MS, Serrao E, Dubose BN, Teng S, Huang J, Li X, Saito N, Roy SP, Siddiqui MA, Ahn J, Tsuji M, Hatziioannou T, Engelman AN, Yamashita M, 2016. Capsid-CPSF6 interaction is dispensable for HIV-1 replication in primary cells but is selected during virus passage in vivo. J. Virol 90, 6918–6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller T, Pollpeter D, Apolonia L, Goujon C, Malim MH, 2014. Nuclear import of SAMHD1 is mediated by a classical karyopherin alpha/beta1 dependent pathway and confers sensitivity to VpxMAC induced ubiquitination and proteasomal degradation. Retrovirology 11, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte B, Buffone C, Opp S, Di Nunzio F, De Souza Aranha Vieira DA, Brandariz-Nunez A, Diaz-Griffero F, 2015. Restriction of HIV-1 requires the N-terminal region of MxB as a capsid-binding Motif but not as a nuclear localization signal. J. Virol 89, 8599–8610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Gelais C, Silva S, Hach JC, White TE, Diaz-Griffero F, Yount JS, 2014. Identification of cellular proteins interacting with the retroviral restriction factor SAMHD1. J. Virol 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szebeni J, Dieffenbach C, Wahl SM, Venkateshan CN, Yeh A, Popovic M, Gartner S, Wahl LM, Peterfy M, Friedman RM, et al. , 1991. Induction of alpha interferon by human immunodeficiency virus type 1 in human monocyte-macrophage cultures. J. Virol 65, 6362–6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C, Ji X, Wu L, Xiong Y, 2015. Impaired dNTPase activity of SAMHD1 by phosphomimetic mutation of Thr-592. J. Biol. Chem 290, 26352–26359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Bhattacharya A, Villacorta J, Diaz-Griffero F, Ivanov DN, 2016. Allosteric activation of SAMHD1 protein by deoxynucleotide triphosphate (dNTP)-dependent tetramerization requires dNTP concentrations that are similar to dNTP concentrations observed in cycling T cells. J. Biol. Chem 291, 21407–21413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welbourn S, Dutta SM, Semmes OJ, Strebel K, 2013. Restriction of virus infection but not catalytic dNTPase activity is regulated by phosphorylation of SAMHD1. J. Virol 87, 11516–11524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TE, Brandariz-Nunez A, Valle-Casuso JC, Amie S, Nguyen LA, Kim B, Tuzova M, Diaz-Griffero F, 2013. The retroviral restriction ability of SAMHD1, but not its deoxynucleotide triphosphohydrolase activity, is regulated by phosphorylation. Cell Host Microbe 13, 441–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Hao C, DeLucia M, Swanson S, Florens L, Washburn MP, Ahn J, Skowronski J, 2015. CyclinA2-Cyclin-dependent kinase regulates SAMHD1 protein phosphohydrolase domain. J. Biol. Chem 290, 13279–13292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan N, Regalado-Magdos AD, Stiggelbout B, Lee-Kirsch MA, Lieberman J, 2010. The cytosolic exonuclease TREX1 inhibits the innate immune response to human immunodeficiency virus type 1. Nat. Immunol 11, 1005–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Luban J, Diaz-Griffero F, 2014. The fate of HIV-1 capsid: a biochemical assay for HIV-1 uncoating. Methods Mol. Biol 1087, 29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


