Summary
Biosynthesis of the nucleotide sugar precursor dTDP‐L‐rhamnose is critical for the viability and virulence of many human pathogenic bacteria, including Streptococcus pyogenes (Group A Streptococcus; GAS), Streptococcus mutans and Mycobacterium tuberculosis. Streptococcal pathogens require dTDP‐L‐rhamnose for the production of structurally similar rhamnose polysaccharides in their cell wall. Via heterologous expression in S. mutans, we confirmed that GAS RmlB and RmlC are critical for dTDP‐L‐rhamnose biosynthesis through their action as dTDP‐glucose‐4,6‐dehydratase and dTDP‐4‐keto‐6‐deoxyglucose‐3,5‐epimerase enzymes respectively. Complementation with GAS RmlB and RmlC containing specific point mutations corroborated the conservation of previous identified catalytic residues. Bio‐layer interferometry was used to identify and confirm inhibitory lead compounds that bind to GAS dTDP‐rhamnose biosynthesis enzymes RmlB, RmlC and GacA. One of the identified compounds, Ri03, inhibited growth of GAS, other rhamnose‐dependent streptococcal pathogens as well as M. tuberculosis with an IC50 of 120–410 µM. Importantly, we confirmed that Ri03 inhibited dTDP‐L‐rhamnose formation in a concentration‐dependent manner through a biochemical assay with recombinant rhamnose biosynthesis enzymes. We therefore conclude that inhibitors of dTDP‐L‐rhamnose biosynthesis, such as Ri03, affect streptococcal and mycobacterial viability and can serve as lead compounds for the development of a new class of antibiotics that targets dTDP‐rhamnose biosynthesis in pathogenic bacteria.
Introduction
The typical cell wall architecture of a Gram‐positive bacterium consists of a thick peptidoglycan layer that is decorated with wall teichoic acids, proteins and capsular polysaccharides. However, certain lactic acid bacteria, particularly streptococcal species, lack the expression of wall teichoic acids and instead express rhamnose cell wall polysaccharides, which are covalently anchored to peptidoglycan (Mistou et al., 2016). Rhamnose cell wall polysaccharides encompass about 40–60% of the cell wall mass and are considered to be functional homologs of wall teichoic acids (Sutcliffe et al., 2008; Caliot et al., 2012). Examples of rhamnose cell wall polysaccharides are the group A carbohydrate (GAC) of Group A Streptococcus (GAS; Streptococcus pyogenes) and the serotype‐determining polysaccharides, referred to as rhamnose‐glucose polysaccharides (RGP), of Streptococcus mutans. The GAC and RGP share structural similarities; both consist of an α‐1,2‐/α‐1,3‐linked polyrhamnose backbone with alternating N‐acetylglucosamine side chains for GAS and glucose or galactose side chains for S. mutans (Heymann et al., 1964; Linzer et al., 1985; Huang et al., 1986; Pritchard et al., 1986; St Michael et al., 2017) (Fig. 1). For S. mutans, the type of side chain as well as their linkage to the rhamnan backbone determines the S. mutans serotype (Nakano and Ooshima, 2009; St Michael et al., 2017) (Fig. 1). In contrast, all GAS serotypes express a structurally invariant GAC (Lancefield, 1933). Similar to classical wall teichoic acids, rhamnose cell wall polysaccharides are critical for maintaining cell shape, bacterial physiology and virulence, but in‐depth knowledge of their biosynthesis or host interactions at a molecular level is limited (van Sorge et al., 2014; van der Beek et al., 2015; Kovacs et al., 2017; Zhu et al., 2017). A better understanding of these mechanisms could aid the development of new classes of antibiotics, antibiotic adjuvants or vaccines (Sabharwal et al., 2006; van Sorge et al., 2014; St Michael et al., 2017).
Figure 1.

Schematic overview of streptococcal rhamnose polysaccharide structures. Schematic representation of the chemical composition of the GAC from GAS and the serotype‐specific (c, e, f and k) carbohydrates from S. mutans (SMU). All carbohydrates share an α‐1,2/α‐1,3 linked polyrhamnose backbone. Sugar residues in dashed boxes were recently identified by St Michael et al. (2017).
L‐Rhamnose is the main building block for both the GAC and RGP. The biosynthesis pathway of the nucleotide precursor, dTDP‐L‐rhamnose, is highly conserved among both Gram‐positive and Gram‐negative bacteria (Giraud and Naismith, 2000; Mistou et al., 2016) and critical or even essential for viability or virulence of a wide range of human pathogens including GAS (Le Breton et al., 2013; 2015), Group B Streptococcus (GBS) (Caliot et al., 2012; Hooven et al., 2016), some serotypes of Streptococcus pneumoniae (Kim et al., 1999; Magee and Yother, 2001), S. mutans (Tsuda et al., 2000; Engels‐Deutsch et al., 2003; Kovacs et al., 2017; Shields et al., 2018), Enterococcus faecalis (Xu et al., 2000; Rigottier‐Gois et al., 2015), Mycobacterium spp. (Ma et al., 2002; Li et al., 2006), Pseudomonas spp. (Engels et al., 1985) and Salmonella enterica serovar Typhimurium (Joiner, 1988). Consequently, this pathway is considered to be an interesting drug target, especially since dTDP‐L‐rhamnose is not produced or used by humans (Adibekian et al., 2011).
dTDP‐L‐rhamnose is produced through a four‐step enzymatic pathway catalyzed by the enzymes RmlABCD. In the first step of the pathway, RmlA, a glucose‐1‐phosphate thymidyltransferase, converts glucose‐1‐phosphate into dTDP‐glucose (Blankenfeldt et al., 2000), which is subsequently oxidized and dehydrated to form dTDP‐4‐keto‐6‐deoxy‐D‐glucose by the dTDP‐D‐glucose 4,6‐dehydratase RmlB (Beis et al., 2003). RmlC catalyzes an unusual double epimerization reaction (Giraud et al., 2000; Dong et al., 2003b; 2007), the product of which is finally reduced by RmlD, a dTDP‐4‐dehydrorhamnosereductase, to form dTDP‐L‐rhamnose (Blankenfeldt et al., 2002; van der Beek et al., 2015). The mechanisms of action as well as structural characteristics of the RmlABCD enzymes have been studied extensively (Blankenfeldt et al., 2000; Giraud and Naismith, 2000; Allard et al., 2001; Beis et al., 2003; Dong et al., 2003a; 2003b; Kantardjieff et al., 2004; van der Beek et al., 2015). Indeed, crystal structures are available for all four Rml enzymes from different Gram‐positive and Gram‐negative bacterial species and show high structural conservation. This structural and functional information has enabled the development of different screening methods to discover inhibitors against RmlBCD, yielding compounds that can inhibit dTDP‐L‐rhamnose biosynthesis in the low micromolar range (1–100 µM) in biochemical assay and inhibit growth of Mycobacterium in vitro (Ma et al., 2001; Babaoglu et al., 2003; Sivendran et al., 2010; Wang et al., 2011; Ren et al., 2015). Many of the identified compounds have limited solubility in aqueous solutions. Therefore, further biochemical and structural information on this pathway in additional species is important to optimize lead compounds for improved affinity and specificity through rational design.
Recently, we characterized GAS GacA as a RmlD homolog catalyzing the last step in dTDP‐L‐rhamnose biosynthesis (van der Beek et al., 2015). Surprisingly, this GAS RmlD enzyme was confirmed to be a monomer as opposed to previously characterized RmlD dimers. Additional bioinformatics analysis of 213 RmlD homologs revealed that the majority of RmlD enzymes are predicted to be monomeric, while only a subclass of RmlD enzymes in Gram‐negatives form metal‐dependent homodimers (Blankenfeldt et al., 2002). In this study, we extended our work to study GAS RmlB and RmlC on a functional level through heterologous expression in S. mutans and subsequent analysis of growth, morphology and cell wall composition. In addition, we report the identification of small chemical fragments that bind these enzymes and inhibit GAS growth with IC50s ranging from 100 to 300 µM (Ri01, Ri02, Ri03 and Ri06) to 2.7 mM (Ri08). For one compound, Ri03, we confirmed inhibition of dTDP‐L‐rhamnose in a biochemical assay. Furthermore, Ri03 could inhibit growth of S. mutans, S. equi subsp. zooepidemicus (Group C Streptococcus, GCS) and Mycobacterium tuberculosis with similar efficacy. These results demonstrate that rhamnose biosynthesis inhibitors can directly interfere with bacterial viability and could form a new class of antibiotics targeting nucleotide sugar production.
Results
GAS RmlB and RmlC functionally replace S. mutans homologs
As an extension of our previous work on GAS GacA (van der Beek et al., 2015), we sought to characterize GAS RmlB (GAS5448_RS05645) and RmlC (GAS5448_RS05650), the putative dTDP‐glucose‐4,6‐dehydratase and dTDP‐4‐keto‐6‐deoxyglucose‐3,5‐epimerase respectively. GAS rmlB and rmlC are clustered in an operon together with rmlA in the order rmlACB. Structural and biochemical analysis of RmlB and RmlC from Streptococcus suis, S. enterica and M. tuberculosis revealed that both enzymes are functional homodimers in these organisms (Giraud et al., 2000; Allard et al., 2001; Beis et al., 2003; Dong et al., 2003b; Kantardjieff et al., 2004). Protein sequence alignment of RmlB and RmlC homologs from several streptococcal species and S. enterica displayed high homology (Fig. 2, Table S2). Importantly, all catalytic residues in RmlB and RmlC are conserved (Fig. 2, Table S2).
Figure 2.
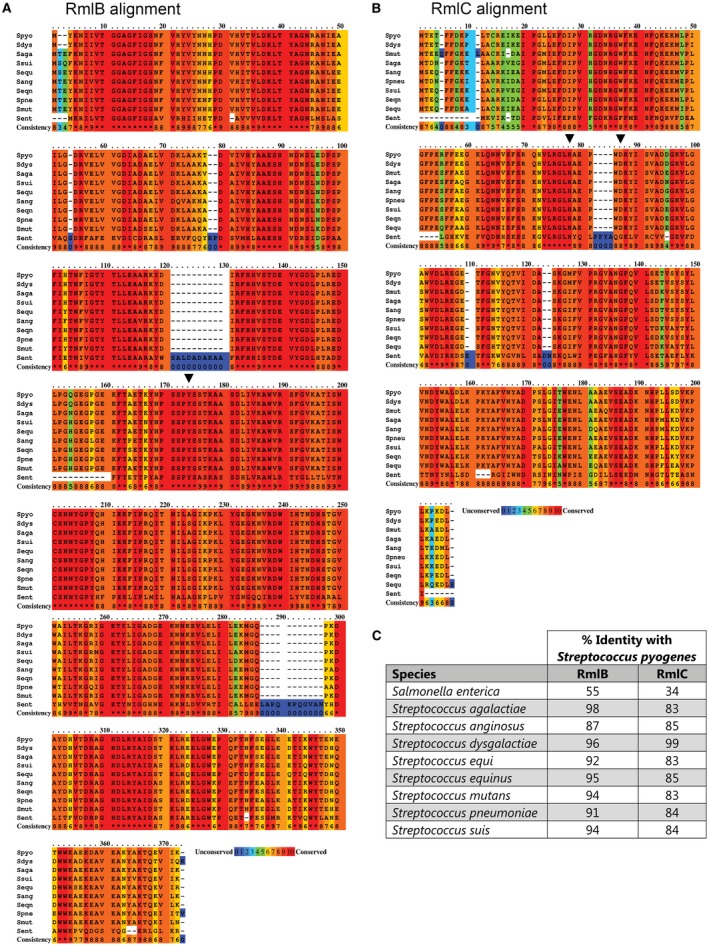
Protein sequence alignment and identity matrix of RmlB and RmlC homologs. Color‐coded representation of amino acid conservation for (A) GAS RmlB and (B) GAS RmlC to S. enterica and different streptococcal species. The amino acid conservation is scored from 0 to 10, with 0 (color blue) assigned to the least conserved residue and 10 (color red) to the most conserved residue. Critical enzymatic residues for RmlB (Y159) and RmlC (H76 and K82) are indicated with an inverted triangle. S. pyogenes (Spyo); Salmonella enterica; (Sent); S. agalactiae (Saga); Streptococcus anginosus (Sang); Streptococcus dysgalactiae (Sdys); Streptococcus equi (Sequ); Streptococcus equinus (Seqn); S. mutans (Smut); S. pneumoniae (Spneu); Streptococcus suis (Ssui). Protein accession numbers are described in the supplementary data (Table S2). C. Percentage identity matrix of RmlB and RmlC homologs.
Most genes directly or indirectly involved in the GAC biosynthesis pathway are essential for GAS viability, including all four dTDP‐L‐rhamnose biosynthesis genes rmlABC and gacA (Le Breton et al., 2013; 2015; van der Beek et al., 2015; Zhu et al., 2017). In contrast and for unknown reasons, the dTDP‐L‐rhamnose biosynthesis genes are not essential in S. mutans under noncompetitive conditions. However, gene deletions result in strongly attenuated growth and severe morphological defects (Tsukioka et al., 1997; van der Beek et al., 2015). GAS RmlB and RmlC proteins share 94% and 83% protein sequence identity with their S. mutans homologs respectively. In addition, the organization of these genes is identical in both species with the exception of a hypothetical protein (174 nucleotides, 57 amino acids) encoded between RmlC and RmlB.
We took advantage of the nonessential nature of the rhamnose biosynthesis genes in S. mutans to confirm the function of GAS RmlB and RmlC in dTDP‐L‐rhamnose biosynthesis. To this end, GAS RmlB and GAS RmlC‐encoding genes were heterologously expressed in S. mutans strains lacking rmlB or rmlC respectively. Deletion of S. mutans rmlB (SMU ΔrmlB) or rmlC (SMU ΔrmlC) by replacement with an erythromycin resistance cassette severely attenuated bacterial growth compared to the wild‐type strain (WT) (Fig. 3). This phenotype is characteristic for a rhamnose‐deficient S. mutans strain and in line with our previously constructed S. mutans rmlD deletion strain (van der Beek et al., 2015). Morphological analysis of rmlB and rmlC mutant bacteria by scanning electron microscopy revealed swelling and clumping of bacteria as a result of misplaced septa resulting in division errors and multidirectional growth (Fig. 4A). Subsequent analysis of the cell wall carbohydrate composition by HPLC/MS confirmed that SMUΔrmlB and SMUΔrmlC lacked rhamnose in their cell walls, which concordantly resulted in the loss of the glucose side chains (Fig. 4B). Introduction of either homologous S. mutans rmlB or heterologous GAS rmlB on an expression plasmid in the corresponding SMU ΔrmlB mutant restored rhamnose incorporation in the cell wall (Fig. 4B) as well as the defective morphological phenotype and growth (Figs 3 and 4A). Initially, we were unable to complement SMUΔrmlC with rmlC from S. mutans, whereas heterologous GAS rmlC could restore growth, morphology and rhamnose production of the SMUΔrmlC mutant (Figs 3B and 4). Upon reexamination of the UA159 genome, a S. mutans serotype c strain that we used as a reference genome for S. mutans Xc, we located an alternative ATG‐start site 135 bp upstream of the annotated rmlC gene, which is in line with the annotation of rmlC in GAS. Importantly, available structural information indicates that the first 45 amino acids are part of the RmlC dimerization interface, forming the extension of the beta‐sheet with two additional beta‐strands, which are required for nucleotide binding (Christendat et al., 2000; Giraud and Naismith, 2000). In agreement with structural and genetic predictions, complementation of SMU ΔrmlC with the extended PCR product complemented all observed defects (Figs 3B and 4), indicating that the extended genomic PCR product encodes a functional RmlC enzyme (198 amino acids), which is similar to the size of GAS and S. suis RmlC (197 amino acids).
Figure 3.
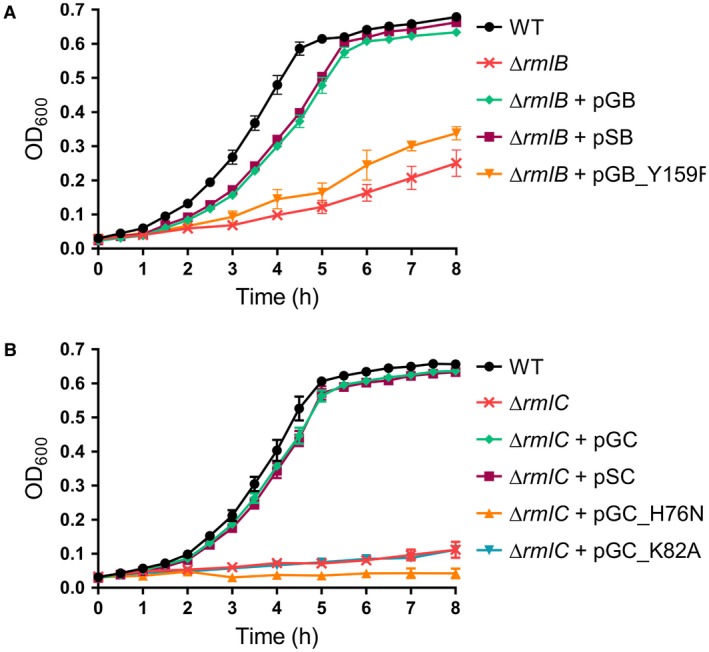
Heterologous expression of GAS RmlB and RmlC and catalytically inactive enzymes in S. mutans: growth curves. Growth curves of S. mutans (A) rmlB and (B) rmlC mutant sets: wild type (WT), ΔrmlB, ΔrmlC, ΔrmlB + pSB, ΔrmlB + pGB(_Y159F), ΔrmlC + pSC and ΔrmlC + pGC(_H76N/K82A). Growth curves represent mean ± standard error of mean (SEM) of at least three biological repeats.
Figure 4.
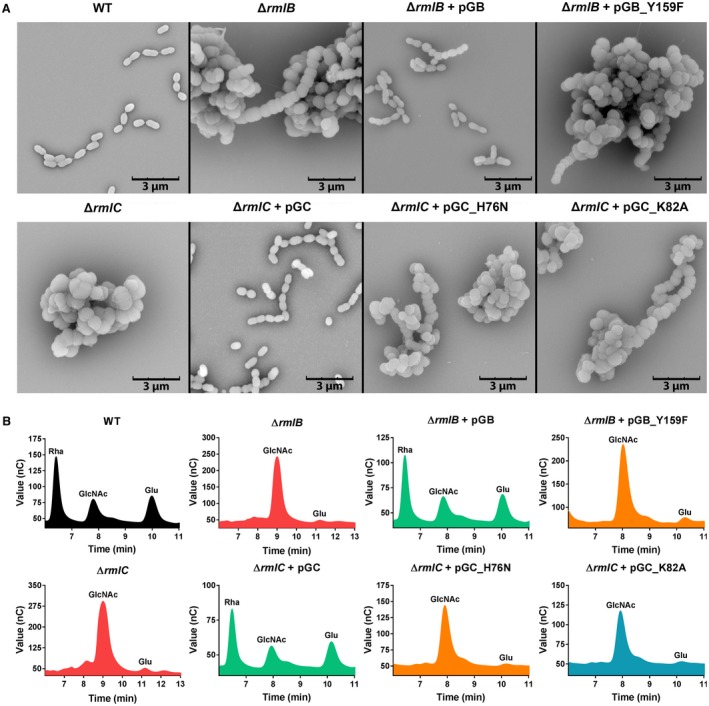
Heterologous expression of GAS RmlB and RmlC and catalytically inactive enzymes in S. mutans: morphology and cell wall content. A. Representative scanning electron microscopy images and (B) cell wall carbohydrate composition analysis of S. mutans wild type (WT), ΔrmlB, ΔrmlC, ΔrmlB + pGB(_Y159F), and ΔrmlC + pGC(_H76N/K82A). Rha, rhamnose; GlcNAc, N‐acetylglucosamine; Glu, glucose.
Catalytic residues of GAS RmlB and RmlC are conserved
Based on the conformation of catalytic residues in S. suis RmlB and RmlC (Allard et al., 2001; Dong et al., 2003b) and protein sequence alignment with the GAS RmlB and RmlC homologs (Fig. 2), we set out to functionally validate predicted catalytic residues of GAS RmlB (Y159) and RmlC (H76 and K82). Point mutations were individually introduced in GAS RmlB (Y159F) and RmlC (H76N and K82A) and overexpression vectors carrying these mutant genes were expressed in SMU ΔrmlB and SMUΔrmlC respectively. The residues that were mutated are all important for enzymatic activity, since the mutated genes were unable to complement the characteristic rhamnose‐depleted phenotype consisting of growth retardation and an aberrant morphology (Figs 3 and 4A). Correspondingly, our cell wall composition analysis revealed that the S. mutans strains carrying these three mutated constructs completely lacked incorporation of rhamnose in their cell wall (Fig. 4B). The functional characterization and confirmation of catalytic residues of streptococcal RmlB and RmlC is important for identification, characterization and optimization of potential dTDP‐L‐rhamnose biosynthesis inhibitors through a rational design approach.
Inhibitor screen against GAS Rml proteins and hit confirmation
Given the importance of the dTDP‐L‐rhamnose biosynthesis pathway for viability or virulence of many human pathogens, we aimed to identify chemical scaffolds that could act as starting points for future optimization and drug development. In addition, such compounds could also find applications in chemical biological studies aimed at understanding the biosynthesis of dTDP‐L‐rhamnose, particularly when a genetic approach is not possible or desired. Therefore, we conducted a bio‐layer interferometry (BLI) inhibitor screen against the commercially available Maybridge Library using the three recombinant GAS dTDP‐rhamnose biosynthesis enzymes RmlB, RmlC and GacA (Fig. S1A). The advantage of this approach is that it precludes the use of expensive or commercially unavailable enzyme substrates. Using a commercially available library of ~1,000 chemical fragments, our initial screen identified 12 hits (Figs 5, S1B‐D) of which seven fragments were found to specifically interact with GAS RmlB/RmlC/GacA as confirmed by BLI (Figs 5, S2). In addition, we confirmed the identified hits in dilution series for their binding affinity to these three enzymes (Fig. S2). Importantly, this method allowed us to estimate enzyme specificity by comparing the data obtained from one enzyme to the other two. Using this strategy, we identified chemical scaffolds with specificity for one enzyme, but also several fragments that bound to more than one enzyme of the dTDP‐rhamnose biosynthesis pathway (Figs 5, S2, and Table 1).
Figure 5.
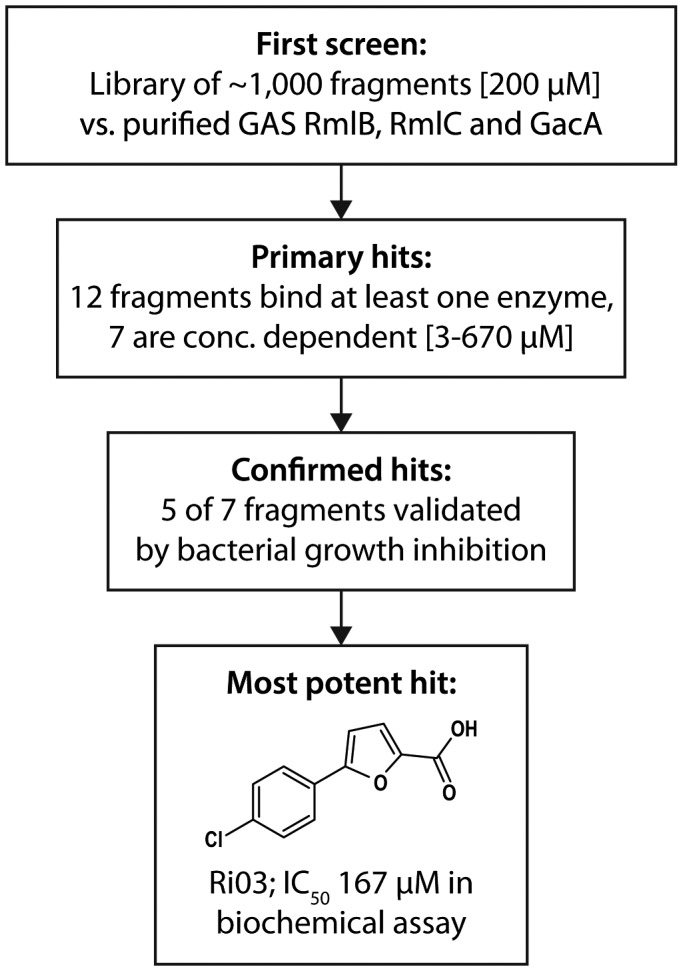
Screening and fragment validation flowchart.
Table 1.
Overview of identified compounds and IC50 for GAS growth inhibition
| Inhibitor | Compound | Binding target | IC50 (mM) GAS |
|---|---|---|---|
| Ri01 | N‐(1‐Naphthyl)ethylenediamine dihydrochloride | RmlB, RmlC, GacA | 0.27 |
| Ri02 | 2,5‐Difluorophenylhydrazine | RmlB, GacA | 0.27 |
| Ri03 | 5‐(4‐chlorophenyl)‐2‐furoic acid | RmlB, RmlC, GacA | 0.12 |
| Ri06 | 4,4'‐Thiodiphenol | RmlB, GacA | 0.11 |
| Ri08 | 2‐(2,5‐Dimethyl‐1H‐pyrrol‐1‐yl) benzoic acid | RmlB, RmlC | 2.67 |
Bactericidal activity of identified fragments
Since dTDP‐rhamnose is an essential nucleotide sugar in GAS (Le Breton et al., 2013; 2015), we assessed whether these fragments could affect GAS growth. All fragments identified in this study were water insoluble and were therefore dissolved in DMSO, except Ri07, which was already liquid at room temperature. GAS is tolerant to DMSO concentrations of 2%, therefore this was the maximum concentration used in bacterial assays. Even in DMSO, Ri04 and Ri07 were highly insoluble in bacterial culture medium and could therefore not be tested for inhibition of bacterial growth. The remaining five fragments were soluble at the highest concentration tested and were able to inhibit growth of GAS with IC50 values ranging from 110 µM to 2.67 mM (Fig. 6A, Table 1). Since Ri03 and Ri06 were most active against GAS, we continued with these two fragments. In accordance with growth inhibition, GAS morphology was also severely affected upon exposure to either 200 µM Ri03 or 100 µM Ri06 (Fig. 6C). Especially for Ri03, streptococci were swollen and chains were longer, a phenotype reminiscent of an inducible gacA GAS knockout strain (van der Beek et al., 2015).
Figure 6.
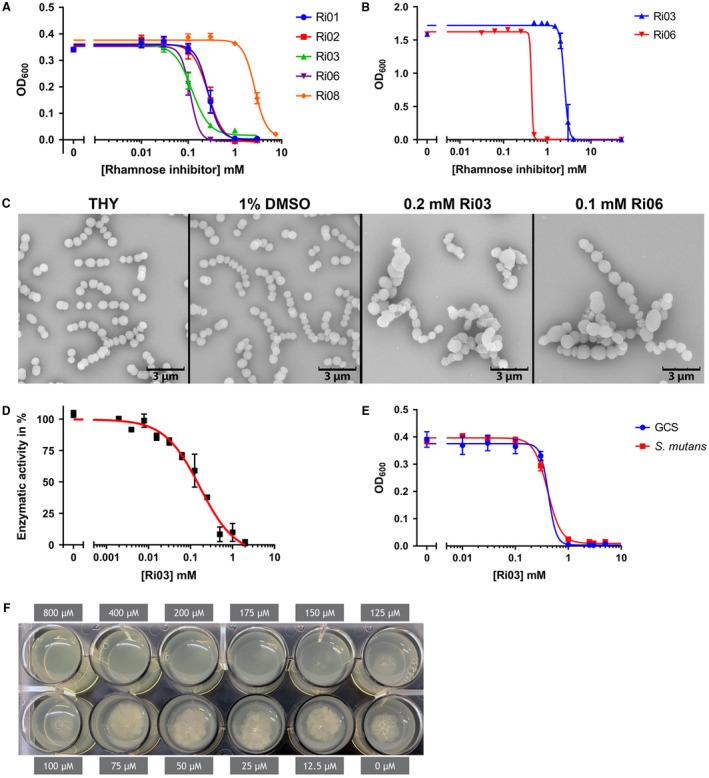
Fragment Ri03 inhibits dTDP‐L‐rhamnose biosynthesis in vitro and growth of streptococci and M. tuberculosis in vivo. A. Dose–response curves for growth inhibition of GAS with various concentrations of Ri01, Ri02, Ri03, Ri06 or Ri08. IC50 values range from 110 µM to 2.67 mM. B. Dose–response curves for growth inhibition of rhamnose‐deficient S. aureus with various concentrations of Ri03 and Ri06. IC50 values range are 2.48 mM and 440 µM respectively. C. Representative scanning electron microscopy images of GAS after 16 h incubation in growth medium (THY), or THY with the addition of 1% DMSO, 0.2 mM Ri03 or 0.1 mM Ri06. D. Dose‐dependent inhibition of dTDP‐L‐rhamnose biosynthesis using recombinant RmlB, C and GacA in a biochemical assay. The assay was initiated by the addition of 400 µM dTDP‐D‐glucose and conversion was measured through oxidation of NADPH to NADP+ at 340 nm. Background absorbance was subtracted and IC50 value was determined using the nonlinear fit model E. Dose–response curves for growth inhibition of S. equi subsp. zooepidemicus (Group C Streptococcus; GCS) and S. mutans with various concentrations of Ri03. IC50 values are 420 µM and 410 µM respectively. F. Ri03 prevents growth of M. tuberculosis with a minimal inhibitory concentration of 175 µM using an agar‐based spot culture growth inhibition assay. Dose–response curves represent mean ± SEM of at least three biological experiments performed in duplicate, except for M. tuberculosis MIC, which was performed twice.
The fragments that were identified here may not be completely specific for inhibition of dTDP‐L‐rhamnose since they generally have lower affinity. To assess potential off‐target effects, we incubated Ri03 and Ri06 with Staphylococcus aureus USA300 NRS384, a pathogenic methicillin‐resistant bacterium that lacks expression of rhamnose. Ri03 and Ri06 killed S. aureus with an IC50 ~ 2.48 mM and 440 µM, respectively (Fig. 6B), suggesting that the observed growth inhibition is due to off‐target effects. However, the approximately 20‐fold window in IC50 for Ri03 against GAS compared to S. aureus suggested that growth inhibition could be at least partially dependent on inhibition of dTDP‐L‐rhamnose biosynthesis pathway. To confirm this, we used a biochemical assay in which recombinant RmlB, RmlC and GacA form dTDP‐L‐rhamnose out of the substrate dTDP‐L‐glucose (van der Beek et al., 2015). Ri03 showed a concentration‐dependent inhibition of dTDP‐L‐rhamnose formation with an IC50 ~ 166 µM (Fig. 6D). These data suggest that Ri03 directly targets the dTDP‐L‐rhamnose biosynthesis pathway despite having additional off‐target effects in vivo.
Activity of Ri03 against other bacterial species
We extended our experiments to include other human and animal streptococcal species for which dTDP‐L‐rhamnose is an essential nucleotide sugar. Group C Streptococcus contains a related surface carbohydrate, the characteristic group C carbohydrate, composed of a polyrhamnose backbone decorated with a di‐GalNAc side chain (Mistou et al., 2016). Ri03 inhibited growth of GCS with an IC50 of 0.42 mM (Fig. 6E) and of S. mutans with an IC50 of 0.41 mM (Fig. 6E). In addition, we also assessed the effect of Ri03 against M. tuberculosis, the causative agent of tuberculosis, for which dTDP‐L‐rhamnose biosynthesis is also essential (Ma et al., 2002; Li et al., 2006). Again, Ri03 prevented growth of M. tuberculosis at a concentration of 175 µM (Fig. 6F).
Discussion
In this study, we show that GAS RmlB and RmlC are dTDP‐glucose‐4,6‐dehydratase and dTDP‐4‐keto‐6‐deoxyglucose‐3,5‐epimerase enzymes respectively. Both GAS RmlB and RmlC functionally replaced S. mutans homologs in a heterologous expression system. The rescue of growth and morphological phenotypes of the rmlB and rmlC mutants by plasmid overexpression excludes the occurrence of polar effects within the rmlACB operon. In addition to proving the function of both GAS RmlB and RmlC as rhamnose biosynthesis enzymes, we also identified Y159 in RmlB and H76 and K82 in RmlC as critical catalytic residues. Our complementation studies indicate that the mechanism of catalysis is conserved among streptococcal species.
Deletion of rmlB and rmlC in S. mutans resulted in a phenotype similar as previously observed for an rmlD deletion mutant (van der Beek et al., 2015), underscoring that inhibition of the dTDP‐rhamnose biosynthesis pathway severely impacts bacterial viability. Indeed, the rmlABCD genes are essential for S. mutans in the competitive environment of a mutant transposon library (Shields et al., 2018), similar to GAS and GCS (Le Breton et al., 2013; 2015; Charbonneau et al., 2017). We are however able to construct rml deletion mutants in S. mutans in isolation in contrast to similar attempts in GAS (van Sorge et al., 2014). The reason for this discrepancy is currently unclear but may be caused by differential compositions of the cell walls or possibly these deletions are partially rescued through unidentified interacting pathways.
Screening a chemical compound library to identify inhibitors of the dTDP‐L‐rhamnose biosynthesis pathway is methodically challenging. Previously, indirect methods were used to determine the production of the end product dTDP‐L‐rhamnose by monitoring the production of co‐factor NAD(P)H (Graninger et al., 1999; Sivendran et al., 2010; van der Beek et al., 2015). A superior assay would involve a HPLC or mass spectrometry approach where every single step of product formation and substrate consumption could be monitored. However, it is challenging to develop this method for medium or high throughput screening. We, therefore, investigated an approach using recombinant enzymes in their natural state, i.e. with bound co‐factors in their active site. RmlB enzymes require the co‐factors NAD(P), which appears to be tightly bound in the active site as it was co‐purified during RmlB crystallizations (Allard et al., 2002; Beis et al., 2003). This is in agreement with the regeneration of NAD during dTDP‐4‐keto‐6‐deoxy‐D‐glucose synthesis (Allard et al., 2002). GacA/RmlD enzymes also require NADPH as a co‐factor, which appears to be less tightly bound as it was not observed in the GAS GacA and S. suis RmlD crystal structures and therefore should be regenerated by a different enzyme (Blankenfeldt et al., 2002; van der Beek et al., 2015). Using the binding approach, we identified seven potential inhibitors that interacted with at least one enzyme of the dTDP‐L‐rhamnose biosynthesis pathway with affinities ranging from 0.1 mM to 80 mM. One of the most potent chemical fragments Ri03, 5‐(4‐chlorophenyl)‐2‐furoic acid, is a small molecule, which binds RmlB and GacA with an affinity of 0.2 and 0.3 mM, respectively, and RmlC with approximately 3 mM. The 2‐furoic acid scaffold of Ri03 is of interest since this organic compound is used as a preservative and flavoring agent in the food industry. The bactericidal and fungicidal activity of 2‐furoic acid is presumably unrelated to rhamnose biosynthesis inhibition, but may be an explanation for the observed rhamnose‐independent inhibition of S. aureus. Previous studies have also identified small molecules that inhibit Rml enzymes (Ma et al., 2001; Babaoglu et al., 2003; Sivendran et al., 2010; Wang et al., 2011; Ren et al., 2015). However, none of the predicted or confirmed inhibitors show a structural resemblance to Ri03.
Of particular interest is the observation that Ri03 also inhibits growth of M. tuberculosis. As drug‐resistant tuberculosis continues to be a public health crisis and specifically multi‐ and extensively drug‐resistant tuberculosis cure rates are below 60 and 30%, new chemical entities with activity against M. tuberculosis are urgently required. Interestingly, Ri03 has a similar chemical scaffold as another anti‐tuberculosis compound that was recently reported, which has an MIC50 of 135 µM (Negatu et al., 2018; 5‐(4‐Chlorophenyl)‐N,N,2‐trimethyl‐3‐furamide, compound F8). It is unknown whether F8 targets the dTDP‐L‐rhamnose biosynthesis pathway, but this is certainly something that should be investigated. If F8 also was inhibiting the dTDP‐rhamnose biosynthesis pathway, these lead structures together with available structural and biochemical information on enzymes of the rhamnose biosynthesis pathway, should guide future attempts for rational drug design to optimize specificity as well as solubility of Ri03 and F8.
Fragments such as the ones that we identified here, are generally known to have low affinity and limited specificity. Indeed, using a bacterial species which does not synthesize rhamnose (S. aureus), we observed that Ri03 has off‐target effects that attenuated bacterial growth. Nonetheless, Ri03 is approximately 10‐ to 20‐fold more effective at inhibiting growth of species for which rhamnose is essential, i.e. three streptococcal species as well as M. tuberculosis. The high sequence identity of RmlB, RmlC and GacA/RmlD among GAS, GCS and S. mutans (92–94%, 83% and 82–87% protein sequence identity respectively; Fig. 2C) supports the observation that Ri03 shows similar IC50 values against all three strains. For M. tuberculosis, protein sequence identity is less apparent and highest for RmlB (61%, 30% and 35% for RmlB, RmlC and GacA respectively). This may indicate that the observed mycobacterial growth inhibition of Ri03 predominantly occurs through interference with RmlB function, which coincides with the fact that Ri03 has the highest affinity for this rhamnose biosynthesis enzyme. Complementary, we have proven that Ri03 acts specifically on the dTDP‐L‐rhamnose biosynthesis pathway using a biochemical assay.
Future experiments aim to investigate the binding mode of Ri03 on a structural level to the proteins RmlB, RmlC and GacA to elucidate whether Ri03 inhibits RmlB, RmlC and GacA by competing with the nucleotide‐sugar and if a common binding mode is observed among the different enzymes. With more functional and structural knowledge, Ri03 can be further optimized to identify more potent and specific derivatives with greater potency targeting the GAS and related homologous Rml enzymes in other pathogenic species.
Experimental procedures
Bacterial strains and growth conditions
Bacterial strains used in this study are described in Table 2. S. mutans Xc (Koga et al., 1989), a serotype c wild‐type (WT) strain, was a kind gift of Dr. Y. Yamashita (Kyushu University, Japan) and was routinely grown in Todd‐Hewitt Broth (THB, Oxoid) or on THB agar at 37°C with 5% CO2. When appropriate, S. mutans was cultured with 10 µg ml−1 erythromycin (ERY) or 3 µg/ml chloramphenicol (CHL). GAS 5448, a representative of the epidemic M1T1 clone, was cultured in Todd‐Hewitt Broth (Becton Dickinson) supplemented with 1% yeast extract (THY; Oxoid) or on THY agar at 37°C. E. coli MC1061 was used for cloning purposes and was grown in lysogeny broth (LB, Oxoid) or on LB agar containing 10 µg/ml CHL at 37°C. Mycobacterium tuberculosis laboratory strain H37Rv was routinely cultured in Middlebrook 7H9 broth (Difco) supplemented with 10% (v/v) Albumin Dextrose Catalase (ADC) enrichment (Difco), 0.05% (v/v) tyloxapol and 0.02% (v/v) glycerol. Liquid cultures were grown at 37°C in 50 mL centrifugation tubes with rotation of 40 rpm until mid‐exponential phase (OD600 ~ 1). For solid growth drug susceptibility experiments, M. tuberculosis H37Rv was grown in Middlebrook 7H10 (Difco) agar medium supplemented with 10% (v/v) of Oleic acid ADC enrichment (OADC) (Difco) and 0.5% (v/v) glycerol (c7H10) at 37°C incubator.
Table 2.
Bacterial strains used in this study
| Bacterial strains | Abbreviation | Resistance |
|---|---|---|
| S. mutans Xc wild type (serotype c) | SMU WT | – |
| S. mutans Xc ΔrmlB | SMU ΔrmlB | ERY |
| S. mutans Xc ΔrmlB + pDC123_SMU_rmlB | SMU ΔrmlB + pSB | ERY + CHL |
| S. mutans Xc ΔrmlB + pDC123_GAS_rmlB | SMU ΔrmlB + pGB | ERY + CHL |
| S. mutans Xc ΔrmlB + pDC123_GAS_rmlB_Y159F | SMU ΔrmlB + pGB_Y159F | ERY + CHL |
| S. mutans Xc ΔrmlC | SMU ΔrmlC | ERY |
| S. mutans Xc ΔrmlC + pDC123_SMU_rmlC | SMU ΔrmlC + pSC | ERY + CHL |
| S. mutans Xc ΔrmlC + pDC123_GAS_rmlC | SMU ΔrmlC + pGC | ERY + CHL |
| S. mutans Xc ΔrmlC + pDC123_GAS_rmlC_H76N | SMU ΔrmlC + pGC_H76N | ERY + CHL |
| S. mutans Xc ΔrmlC + pDC123_GAS_rmlC_K82A | SMU ΔrmlC + pGC_K82A | ERY + CHL |
| S. pyogenes 5448 wild type (serotype M1T1) | GAS | – |
| S. equi subsp. zooepidemicus MGCS10565 | GCS | – |
| E. coli MC1061 | – | – |
| E. coli BL21 (DE3) | – | – |
| E. coli DH5alpha | – | – |
| Staphylococcus aureus NRS384 | – | MRSA |
| M. tuberculosis H37Rv | – | – |
Cloning, expression and purification of GAS enzymes and biochemical assay
GAS RmlB, RmlC and GacA proteins were produced and purified as described previously (van der Beek et al., 2015). The IC50 of Ri03 against the dTDP‐rhamnose biosynthesis pathway enzymes RmlB, RmlC and GacA was determined using the published procedure (van der Beek et al., 2015). In short, the assay buffer system contained 25 mM Tris‐base (pH 7.5), 150 mM NaCl, 0.2 mM NADPH and 5 pM of recombinant GAS RmlB, RmlC and GacA. The assay was started with the addition of 400 µM dTDP‐D‐glucose. The oxidation of NADPH to NADP+ was measured by the change in intrinsic absorbance at 340 nm using a SpectraMax M2 plate reader. Background absorbance was subtracted and IC50 value was determined using the nonlinear fit model in GraphPad Prism.
Genetic manipulation of S. mutans
Streptococcus mutans is naturally competent and was transformed as described previously. Shortly, bacteria were grown in THB containing 5% heat‐inactivated horse serum (BioRad) and supplied with 500 ng knockout construct or complementation plasmid (van der Beek et al., 2015). Cultures were plated on THB agar plates containing the appropriate antibiotics. Single colonies were selected and verified for the deletion of rmlB/rmlC and/or the presence of complementation plasmid using colony PCR and sequencing.
Deletion mutants of rmlB (SMU ΔrmlB) and rmlC (SMU ΔrmlC) were obtained via the addition of a knockout construct to competent S. mutans WT. This construct consisted of an ERY resistance cassette with ~700 bp flanking regions homologous to the up‐ and downstream regions of rmlB or rmlC. A detailed cloning strategy is available in the supplemental material and Table S1.
S. mutans complementation plasmids
S. mutans rmlB/rmlC and GAS rmlB/rmlC were amplified from gDNA using primers containing XbaI, XhoI or BamHI restriction site (Table S1). Digested PCR products were subsequently ligated into expression plasmid pDC123 and propagated in E. coli MC1061 to obtain large quantities of complementation plasmid. Point mutations were introduced by mutagenesis PCR, using overlapping primers with the corresponding mutation site integrated. The constructed plasmids with the single point mutations were all confirmed by DNA sequencing.
SMU ΔrmlB and SMU ΔrmlC knockout mutants were not transformable likely resulting from the severe growth defects. Therefore, SMU ΔrmlB and SMU ΔrmlC strains complemented with S. mutans rmlB/rmlC or GAS rmlB/rmlC were constructed using a two‐step approach. First, S. mutans WT was transformed with the complementation plasmid as described above. Next, these complemented strains were transformed with the respective rmlB or rmlC knockout constructs and selected for double antibiotic resistance.
Scanning Electron Microscopy
Scanning electron microscopy was performed as described previously by van der Beek et al. (2015). In short, bacteria were grown to mid‐exponential phase, except when incubated with rhamnose inhibitor. In this case, bacteria were cultured as described below in bacterial growth inhibition assays and grown overnight. Bacteria were subsequently washed, fixed, dehydrated, mounted onto 12.5 mm specimen stubs (Agar scientific, Stansted, Essex, UK) and coated with gold to 1 nm using a Quorum Q150R S sputter coater at 20 mA. Samples were visualized with a Phenom PRO desktop scanning electron microscope (Phenom‐World BV) at an acceleration voltage of 10 kV.
S. mutans growth curves
Overnight cultures of S. mutans strains with an optical density at 600 nm (OD600) higher than 0.35 were diluted 10 times and grown for 1.5 h to early exponential phase. Cultures were then diluted to OD600 0.03 and OD600 was measured manually every 30 min. For cultures with an overnight OD600 below 0.35, the initial 10‐fold dilution and growth step was omitted. Instead, such overnight cultures were directly diluted to OD600 0.03 and OD600 was measured every hour. All cultures were incubated at 37°C with 5% CO2 in between measurements.
Cell wall carbohydrate composition analysis
Cell wall polysaccharides were isolated from S. mutans, hydrolyzed and analyzed by chromatography as described previously by van der Beek et al. (2015). In short, bacterial cells were harvested from 2 to 5 L cultures and disrupted in 0.1 M citrate buffer (pH 4.6) using a bead beater (Biospec). Cell walls were collected by centrifugation and boiled for 1 h at 100°C in 0.1 M sodium acetate (pH 4.6) containing 4% sodium dodecyl sulfate. Samples were subsequently treated with RNase, DNase, pronase E and trypsin. Cell walls containing peptidoglycan and the serotype C carbohydrate were lyophilized before hydrolysis with TFA. Finally, carbohydrate analysis of monosaccharides was performed on a Dionex ICS‐3000 Ion Chromatography System (Dionex/Thermo Scientific) using a CarboPac PA20 (Dionex/Thermo Scientific) 3 × 150 mm column, equipped with a ICS‐3000 Electrochemical Detector (Dionex/Thermo Scientific). Monosaccharides from all samples were eluted from the column with 12% 0.2 M NaOH, except for monosaccharides from SMU ΔrmlB and ΔrmlC, which were eluted with 8% 0.2 M NaOH. The ‘Carbohydrates (Standard Quad)’ waveform was used for detection.
BLI screen using recombinant RmlB, RmlC and GacA
The Maybridge library of fragment‐like molecules (Ro3) was purchased from Maybridge (USA). All three GAS proteins, RmlB, RmlC and GacA, were screened against the library using identical protocols. Proteins were biotinylated on primary amine amino acid side chains using the Thermo ScientificTM EZ‐LinkTM NHS‐PEG4‐Biotin reagent. The enzymes were incubated with NHS‐PEG4‐Biotin in a 1:1 molar ratio at 150 µM for 45 min at room temperature. The reaction buffer contained 25 mM HEPES, pH 7.5, 150 mM NaCl and 0.02 mM TCEP. Excess NHS‐PEG4‐biotin reagent was removed using 2 mL Zeba desalting spin columns (Thermo Scientific). Successful biotinylation on primary amines was investigated via Western blotting and probing with ExtrAvidin®−Peroxidase antibody, 1:10,000 (Sigma E2886, Fig. S1A). Before compound screening, the proteins were loaded onto a parallel set of superstreptavidin biosensors (SSA) by incubation in buffer for 900 s. Protein concentrations suitable for the experiments were determined in a series of dilutions. Final screening was conducted at concentrations of: 50 µg/mL RmlB, 25 µg/mL RmlC and 12.5 µg/mL GacA. All compounds were at 2% DMSO concentration. The sensor was blocked by immersion in biocytin (10 µg/mL) for 30 s.
Initial library screening was performed at a compound concentration of 200 µM. Hits were identified by plotting the response rate of every single compound after background subtraction. In total, 12 compounds were validated in 6‐point concentration series using threefold dilutions, ranging from 670 µM to 3 µM. Data were processed and kinetic parameters were calculate using the ForteBio software. Binding curves were manually inspected and approximate binding constants KD are listed in Table S3. Fragments that did not induce concentration‐dependent signal changes, i.e. did not bind to the recombinant enzymes, are not displayed in Fig. S2.
Bactericidal activity
GAS 5448, GCS MGCS10565 and S. mutans Xc strains (Table 2) were grown in appropriate bacterial culture broth overnight at 37°C. Bacterial cultures were diluted 10 times and grown to mid‐log phase (OD600 0.4). Cultures were diluted 200 times in culture medium followed by a twofold dilution with various concentrations of rhamnose inhibitor or DMSO. OD600 was recorded every 15 min at 37°C with 5 sec medium shaking before measurement for GAS, GCS and S. mutans using either a Bioscreen C MBR machine (Growth Curves AB Ltd, Oy, Finland) or a Synergy 2 plate reader (Biotek). OD600 values were plotted against the concentration of compound after 9 h of growth using GraphPad Prism 7. IC50 values were calculated using a nonlinear four‐parameter dose–response curve with variable slope. Because high concentrations were not always feasible due to solubility issues, a high concentration of compound (50 mM) was artificially introduced where appropriate and set to zero to obtain more reliable dose–response curves.
Minimum inhibitory concentration assay in M. tuberculosis
To determine the effect of inhibitors of rhamnose biosynthesis against M. tuberculosis, we performed an agar‐based spot culture growth inhibition assay (Evangelopoulos and Bhakta, 2010). Briefly, compound Ri03 was dissolved in DMSO at 20 mM concentration and serial dilutions (800 µM to 12.5 µM) was added in each well of a 24‐well plate, followed by the addition of 2 mL of warm (55°C) c7H10 agar media. Once solid, the plates were inoculated with a diluted culture of M. tuberculosis H37Rv so that a spot (2 µL) of around 1000 CFUs was placed in the middle of each well. The plates were incubated at 37°C incubator for 14 days and the MIC99 was determined as the well with the lowest concentration of Ri03 where there was no bacteria growth. Rifampicin (a first line drugs against tuberculosis) was also used in parallel as control. The concentration of DMSO (0.1% v/v) remained constant in all wells, including the no drug control wells. Two independent experiments were performed for MIC determination.
Author contributions
Conception and design of the study: SvdB, AZ, HCD, NvS; Acquisition, analysis and interpretation of the data: SvdB, AZ, EC, RNC, BHM, DE, LPSC, GJB, HCD, NvS; Writing of the manuscript: SvdB, AZ, HCD, NvS.
Supporting information
Acknowledgements
The authors would like to acknowledge Dr Olawale Raimi (University of Dundee) for assistance with BLI and Dr Dejere Abate Negatu and Dr Thomas Dick for sharing the unpublished results concerning the compound F8. We are grateful for funding to HCD and his laboratory from Tenovus Scotland, Wellcome Trust and Royal Society (grant number 109357/Z/15/Z.), FEBS and the University of Dundee. This work was supported by a VIDI grant (91713303) from the Netherlands Organization for Scientific Research (NWO) to N.M.v.S and S.v.d.B. The authors declare no conflict of interest. Work in L.P.S.C.’s lab is funded by the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001060), the UK Medical Research Council (FC001060) and the Wellcome Trust (FC001060).
Contributor Information
Helge C. Dorfmueller, Email: h.c.z.dorfmueller@dundee.ac.uk
Nina M. van Sorge, Email: nsorge3@umcutrecht.nl.
References
- Adibekian, A. , Stallforth, P. , Hecht, M.‐L. , Werz, D.B. , Gagneux, P. and Seeberger, P.H. (2011) Comparative bioinformatics analysis of the mammalian and bacterial glycomes. Chemical Science, 2, 337–344. [Google Scholar]
- Allard, S.T. , Giraud, M.F. , Whitfield, C. , Graninger, M. , Messner, P. and Naismith, J.H. (2001) The crystal structure of dTDP‐D‐glucose 4,6‐dehydratase (RmlB) from Salmonella enterica serovar Typhimurium, the second enzyme in the dTDP‐l‐rhamnose pathway. Journal of Molecular Biology, 307, 283–295. [DOI] [PubMed] [Google Scholar]
- Allard, S.T. , Beis, K. , Giraud, M.F. , Hegeman, A.D. , Gross, J.W. , Wilmouth, R.C. , et al (2002) Toward a structural understanding of the dehydratase mechanism. Structure, 10, 81–92. [DOI] [PubMed] [Google Scholar]
- Babaoglu, K. , Page, M.A. , Jones, V.C. , McNeil, M.R. , Dong, C. , Naismith, J.H. and Lee, R.E. (2003) Novel inhibitors of an emerging target in Mycobacterium tuberculosis; substituted thiazolidinones as inhibitors of dTDP‐rhamnose synthesis. Bioorganic & Medicinal Chemistry Letters, 13, 3227–3230. [DOI] [PubMed] [Google Scholar]
- van der Beek, S.L. , Le Breton, Y. , Ferenbach, A.T. , Chapman, R.N. , van Aalten, D.M. , Navratilova, I. , et al (2015) GacA is essential for Group A Streptococcus and defines a new class of monomeric dTDP‐4‐dehydrorhamnose reductases (RmlD). Molecular Microbiology, 98, 946–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beis, K. , Allard, S.T. , Hegeman, A.D. , Murshudov, G. , Philp, D. and Naismith, J.H. (2003) The structure of NADH in the enzyme dTDP‐d‐glucose dehydratase (RmlB). Journal of the American Chemical Society, 125, 11872–11878. [DOI] [PubMed] [Google Scholar]
- Blankenfeldt, W. , Asuncion, M. , Lam, J.S. and Naismith, J.H. (2000) The structural basis of the catalytic mechanism and regulation of glucose‐1‐phosphate thymidylyltransferase (RmlA). EMBO Journal, 19, 6652–6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenfeldt, W. , Kerr, I.D. , Giraud, M.F. , McMiken, H.J. , Leonard, G. , Whitfield, C. , et al (2002) Variation on a theme of SDR. dTDP‐6‐deoxy‐L‐ lyxo‐4‐hexulose reductase (RmlD) shows a new Mg2+‐dependent dimerization mode. Structure, 10, 773–786. [DOI] [PubMed] [Google Scholar]
- Le Breton, Y. , Mistry, P. , Valdes, K.M. , Quigley, J. , Kumar, N. , Tettelin, H. and McIver, K.S. (2013) Genome‐wide identification of genes required for fitness of group A Streptococcus in human blood. Infection and Immunity, 81, 862–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Breton, Y. , Belew, A.T. , Valdes, K.M. , Islam, E. , Curry, P. , Tettelin, H. , et al, (2015) Essential genes in the core genome of the human pathogen Streptococcus pyogenes . Scientific Reports, 5, 9838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caliot, E. , Dramsi, S. , Chapot‐Chartier, M.P. , Courtin, P. , Kulakauskas, S. , Pechoux, C. , et al, (2012) Role of the Group B antigen of Streptococcus agalactiae: a peptidoglycan‐anchored polysaccharide involved in cell wall biogenesis. PLoS Pathogens, 8, e1002756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charbonneau, A.R.L. , Forman, O.P. , Cain, A.K. , Newland, G. , Robinson, C. , Boursnell, M. , et al, (2017) Defining the ABC of gene essentiality in streptococci. BMC Genomics, 18, 426–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christendat, D. , Saridakis, V. , Dharamsi, A. , Bochkarev, A. , Pai, E.F. , Arrowsmith, C.H. and Edwards, A.M. (2000) Crystal structure of dTDP‐4‐keto‐6‐deoxy‐D‐hexulose 3,5‐epimerase from Methanobacterium thermoautotrophicum complexed with dTDP. Journal of Biological Chemistry, 275, 24608–24612. [DOI] [PubMed] [Google Scholar]
- Dong, C. , Beis, K. , Giraud, M.F. , Blankenfeldt, W. , Allard, S. , Major, L.L. , et al (2003a) A structural perspective on the enzymes that convert dTDP‐d‐glucose into dTDP‐l‐rhamnose. Biochemical Society Transactions, 31, 532–536. [DOI] [PubMed] [Google Scholar]
- Dong, C. , Major, L.L. , Allen, A. , Blankenfeldt, W. , Maskell, D. and Naismith, J.H. (2003b) High‐resolution structures of RmlC from Streptococcus suis in complex with substrate analogs locate the active site of this class of enzyme. Structure, 11, 715–723. [DOI] [PubMed] [Google Scholar]
- Dong, C. , Major, L.L. , Srikannathasan, V. , Errey, J.C. , Giraud, M.F. , Lam, J.S. , et al (2007) RmlC, a C3' and C5' carbohydrate epimerase, appears to operate via an intermediate with an unusual twist boat conformation. Journal of Molecular Biology, 365, 146–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels, W. , Endert, J. , Kamps, M.A. and van Boven, C.P. (1985) Role of lipopolysaccharide in opsonization and phagocytosis of Pseudomonas aeruginosa . Infection and Immunity, 49, 182–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels‐Deutsch, M. , Pini, A. , Yamashita, Y. , Shibata, Y. , Haikel, Y. , Scholler‐Guinard, M. and Klein, J.P. (2003) Insertional inactivation of pac and rmlB genes reduces the release of tumor necrosis factor alpha, interleukin‐6, and interleukin‐8 induced by Streptococcus mutans in monocytic, dental pulp, and periodontal ligament cells. Infection and Immunity, 71, 5169–5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelopoulos, D. and Bhakta, S. (2010) Rapid methods for testing inhibitors of mycobacterial growth. Methods in Molecular Biology, 642, 193–201. [DOI] [PubMed] [Google Scholar]
- Giraud, M.F. and Naismith, J.H. (2000) The rhamnose pathway. Current Opinion in Structural Biology, 10, 687–696. [DOI] [PubMed] [Google Scholar]
- Giraud, M.F. , Leonard, G.A. , Field, R.A. , Berlind, C. and Naismith, J.H. (2000) RmlC, the third enzyme of dTDP‐L‐rhamnose pathway, is a new class of epimerase. Nature Structural Biology, 7, 398–402. [DOI] [PubMed] [Google Scholar]
- Graninger, M. , Nidetzky, B. , Heinrichs, D.E. , Whitfield, C. and Messner, P. (1999) Characterization of dTDP‐4‐dehydrorhamnose 3,5‐epimerase and dTDP‐4‐dehydrorhamnose reductase, required for dTDP‐L‐rhamnose biosynthesis in salmonella enterica serovar typhimurium LT2. Journal of Biological Chemistry, 274, 25069–25077. [DOI] [PubMed] [Google Scholar]
- Heymann, H. , Manniello, J.M. , Zeleznick, L.D. and Barkulis, S.S. (1964) Structure of streptococcal cell walls. Ii. Group a biose and group a triose from C‐polysaccharide. Journal of Biological Chemistry, 239, 1656–1663. [PubMed] [Google Scholar]
- Hooven, T.A. , Catomeris, A.J. , Akabas, L.H. , Randis, T.M. , Maskell, D.J. , Peters, S.E. , et al (2016) The essential genome of Streptococcus agalactiae . BMC Genomics, 17, 406–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, D.H. , Rama Krishna, N. and Pritchard, D.G. (1986) Characterization of the group a streptococcal polysaccharide by two‐dimensional 1H‐nuclear‐magnetic‐resonance spectroscopy. Carbohydrate Research, 155, 193–199. [DOI] [PubMed] [Google Scholar]
- Joiner, K.A. (1988) Complement evasion by bacteria and parasites. Annual Review of Microbiology, 42, 201–230. [DOI] [PubMed] [Google Scholar]
- Kantardjieff, K.A. , Kim, C.Y. , Naranjo, C. , Waldo, G.S. , Lekin, T. , Segelke, B.W. , et al (2004) Mycobacterium tuberculosis RmlC epimerase (Rv3465): a promising drug‐target structure in the rhamnose pathway. Acta Crystallographica. Section D, Biological Crystallography, 60, 895–902. [DOI] [PubMed] [Google Scholar]
- Kim, J.O. , Romero‐Steiner, S. , Sorensen, U.B. , Blom, J. , Carvalho, M. , Barnard, S. , et al (1999) Relationship between cell surface carbohydrates and intrastrain variation on opsonophagocytosis of Streptococcus pneumoniae . Infection and Immunity, 67, 2327–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga, T. , Asakawa, H. , Okahashi, N. and Takahashi, I. (1989) Effect of subculturing on expression of a cell‐surface protein antigen by Streptococcus mutans . Journal of General Microbiology, 135, 3199–3207. [DOI] [PubMed] [Google Scholar]
- Kovacs, C.J. , Faustoferri, R.C. and Quivey, R.G. Jr (2017) RgpF is required for maintenance of stress tolerance and virulence in Streptococcus mutans . Journal of Bacteriology, 199(24). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancefield, R.C. (1933) A serological differentiation of human and other groups of hemolytic streptococci. Journal of Experimental Medicine, 57, 571–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W. , Xin, Y. , McNeil, M.R. and Ma, Y. (2006) RmlB and rmlC genes are essential for growth of mycobacteria. Biochemical and Biophysical Research Communications, 342, 170–178. [DOI] [PubMed] [Google Scholar]
- Linzer, R. , Reddy, M.S. and Levine, M.J. (1985) Structural studies of the rhamnose‐glucose polysaccharide antigen from Streptococcus sobrinus B13 and 6715–T2. Infection and Immunity, 50, 583–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, Y. , Stern, R.J. , Scherman, M.S. , Vissa, V.D. , Yan, W. , Jones, V.C. , et al (2001) Drug targeting Mycobacterium tuberculosis cell wall synthesis: genetics of dTDP‐rhamnose synthetic enzymes and development of a microtiter plate‐based screen for inhibitors of conversion of dTDP‐glucose to dTDP‐rhamnose. Antimicrobial Agents and Chemotherapy, 45, 1407–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, Y. , Pan, F. and McNeil, M. (2002) Formation of dTDP‐rhamnose is essential for growth of mycobacteria. Journal of Bacteriology, 184, 3392–3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee, A.D. and Yother, J. (2001) Requirement for capsule in colonization by Streptococcus pneumoniae . Infection and Immunity, 69, 3755–3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistou, M.Y. , Sutcliffe, I.C. and van Sorge, N.M. (2016) Bacterial glycobiology: rhamnose‐containing cell wall polysaccharides in Gram‐positive bacteria. FEMS Microbiology Reviews, 40, 464–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano, K. and Ooshima, T. (2009) Serotype classification of Streptococcus mutans and its detection outside the oral cavity. Future Microbiology, 4, 891–902. [DOI] [PubMed] [Google Scholar]
- Negatu, D.A. , Liu, J.J.J. , Zimmerman, M. , Kaya, F. , Dartois, V. , Aldrich, C.C. , et al (2018) Whole‐cell screen of fragment library identifies gut microbiota metabolite indole propionic acid as antitubercular. Antimicrobial Agents and Chemotherapy, 62, e01571‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard, D.G. , Gregory, R.L. , Michalek, S.M. and McGhee, J.R. (1986) Characterization of the serotype e polysaccharide antigen of Streptococcus mutans . Molecular Immunology, 23, 141–145. [DOI] [PubMed] [Google Scholar]
- Ren, J.X. , Qian, H.L. , Huang, Y.X. , Zhu, N.Y. , Si, S.Y. and Xie, Y. (2015) Virtual screening for the identification of novel inhibitors of Mycobacterium tuberculosis cell wall synthesis: inhibitors targeting RmlB and RmlC. Computers in Biology and Medicine, 58, 110–117. [DOI] [PubMed] [Google Scholar]
- Rigottier‐Gois, L. , Madec, C. , Navickas, A. , Matos, R.C. , Akary‐Lepage, E. , Mistou, M.Y. and Serror, P. (2015) The surface rhamnopolysaccharide epa of Enterococcus faecalis is a key determinant of intestinal colonization. Journal of Infectious Diseases, 211, 62–71. [DOI] [PubMed] [Google Scholar]
- Sabharwal, H. , Michon, F. , Nelson, D. , Dong, W. , Fuchs, K. , Manjarrez, R.C. , et al (2006) Group A Streptococcus (GAS) carbohydrate as an immunogen for protection against GAS infection. Journal of Infectious Diseases, 193, 129–135. [DOI] [PubMed] [Google Scholar]
- Shields, R.C. , Zeng, L. , Culp, D.J. , and Burne, R.A. (2018) Genomewide identification of essential genes and fitness determinants of Streptococcus mutans UA159. mSphere, 3(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivendran, S. , Jones, V. , Sun, D. , Wang, Y. , Grzegorzewicz, A.E. , Scherman, M.S. , et al (2010) Identification of triazinoindol‐benzimidazolones as nanomolar inhibitors of the Mycobacterium tuberculosis enzyme TDP‐6‐deoxy‐d‐xylo‐4‐hexopyranosid‐4‐ulose 3,5‐epimerase (RmlC). Bioorganic & Medicinal Chemistry, 18, 896–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Sorge, N.M. , Cole, J.N. , Kuipers, K. , Henningham, A. , Aziz, R.K. , Kasirer‐Friede, A. , et al (2014) The classical lancefield antigen of group a Streptococcus is a virulence determinant with implications for vaccine design. Cell Host & Microbe, 15, 729–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Michael, F. , Yang, Q. , Cairns, C. , Vinogradov, E. , Fleming, P. , Hayes, A.C. , et al. (2017) Investigating the candidacy of the serotype specific rhamnan polysaccharide based glycoconjugates to prevent disease caused by the dental pathogen Streptococcus mutans . Glycoconjugate Journal, 35, 53–64. [DOI] [PubMed] [Google Scholar]
- Sutcliffe, I.C. , Black, G.W. and Harrington, D.J. (2008) Bioinformatic insights into the biosynthesis of the Group B carbohydrate in Streptococcus agalactiae . Microbiology, 154, 1354–1363. [DOI] [PubMed] [Google Scholar]
- Tsuda, H. , Yamashita, Y. , Toyoshima, K. , Yamaguchi, N. , Oho, T. , Nakano, Y. , et al (2000) Role of serotype‐specific polysaccharide in the resistance of Streptococcus mutans to phagocytosis by human polymorphonuclear leukocytes. Infection and Immunity, 68, 644–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukioka, Y. , Yamashita, Y. , Oho, T. , Nakano, Y. and Koga, T. (1997) Biological function of the dTDP‐rhamnose synthesis pathway in Streptococcus mutans . Journal of Bacteriology, 179, 1126–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Hess, T.N. , Jones, V. , Zhou, J.Z. , McNeil, M.R. and Andrew McCammon, J. (2011) Novel inhibitors of Mycobacterium tuberculosis dTDP‐6‐deoxy‐L‐lyxo‐4‐hexulose reductase (RmlD) identified by virtual screening. Bioorganic & Medicinal Chemistry Letters, 21, 7064–7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y. , Singh, K.V. , Qin, X. , Murray, B.E. and Weinstock, G.M. (2000) Analysis of a gene cluster of Enterococcus faecalis involved in polysaccharide biosynthesis. Infection and Immunity, 68, 815–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, L. , Charbonneau, A.R.L. , Waller, A.S. , Olsen, R.J. , Beres, S.B. , and Musser, J.M. (2017) Novel genes required for the fitness of Streptococcus pyogenes in human saliva. mSphere, 6(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


