Abstract
Precision cell signaling activities of reactive electrophilic species (RES) are arguably among the most poorly-understood means to transmit biological messages. Latest research implicates native RES to be a chemically-distinct subset of endogenous redox signals that influence cell decision making through non-enzyme-assisted modifications of specific proteins. Yet, fundamental questions remain regarding the role of RES as bona fide second messengers. Here, we lay out three sets of criteria we feel need to be met for RES to be considered as true cellular signals that directly mediate information transfer by modifying “first-responding” sensor proteins. We critically assess the available evidence and define the extent to which each criterion has been fulfilled. Finally, we offer some ideas on the future trajectories of the electrophile signaling field taking inspiration from work that has been done to understand canonical signaling mediators.
Keywords: cell signaling and response, electrophile signaling, reactive electrophilic species, reactive oxygen species, redox signaling
Graphical Abstract
A role for native reactive electrophilic species (RES) as true cellular messengers has yet to be unequivocally established. We define three thresholds RES must pass to be considered true signals. We describe how the first two thresholds have been traversed, and offer ideas on how the third can be crossed.
1. Introduction
Coordination of cellular responses is an inordinate task. The human genome contains over 20 000 protein-coding genes, and each gene product has, on average, 10 modified forms.[1] Although many of these genes are not strictly essential, it is likely that each gene contributes to a peak healthy state. For instance, the number of haploinsufficient genes appears to have been significantly underestimated.[2,3] Conversely, misregulation of any protein can, in principal, directly lead to diseased states. It is therefore unsurprising that a significant portion of our genome (at least 10%) regulates communication and interaction between different proteins and the pathways they control. These proteins/ enzymes coordinate, among other important responses, changes in cell cycle, upregulation of defense pathways, and promotion of cell death when a cell has outlived its usefulness or it has become a threat to the organism as a whole. The cell uses various different modifications to write specific codes that ultimately coordinate sophisticated responses. Common codes include small-molecule modifications, like phosphate, acetate, and methyl, and also small proteins such as ubiquitin and SUMO.
Canonical signals are “written” by specific enzymes that receive and relay intra- and/or extracellular cues to ultimately marshal specific responses. Signals can be passed from one protein to another while maintaining the type of modification (e.g., in MAP-kinase signaling cascades), or they can be exchanged (e.g., glycogen synthase kinase 3β-controlled β-TRCP- dependent Nrf2 ubiquitination).[4] Thus, these signaling activities are exquisitely regulated and intertwined. For instance, classical ubiquitination pathways are controlled by about 1000 proteins altogether: classes of these proteins include three groups of ubiquitinating enzymes (activating, conjugating, and ligases) that act sequentially to direct ubiquitin to a specific protein, and deubiquitinases (DUBs) that proofread these ubiquitination events and correct for errors. Ubiquitination can happen multiple times on the same protein, creating either polyubiquitin chains or a protein with multiple single ubiquitination sites. Once ubiquitinated, depending on the specific residue modified and the nature of the polyubiquitin linkages, proteins can change location, alter their activity/function, or be degraded, among various other possible outcomes. Biochemical, structural, and systems biology understanding has proven to be critical to unravel the “trade secrets” of these pathways: specific ubiquitination sites regulated by specific conjugating enzymes have been identified; functional ramifications have been assigned to linkages; recruitment factors are also known. A similar complexity is appreciated for kinase cascades and other forms of canonical signaling codes (Figure 1).[5]
Figure 1.
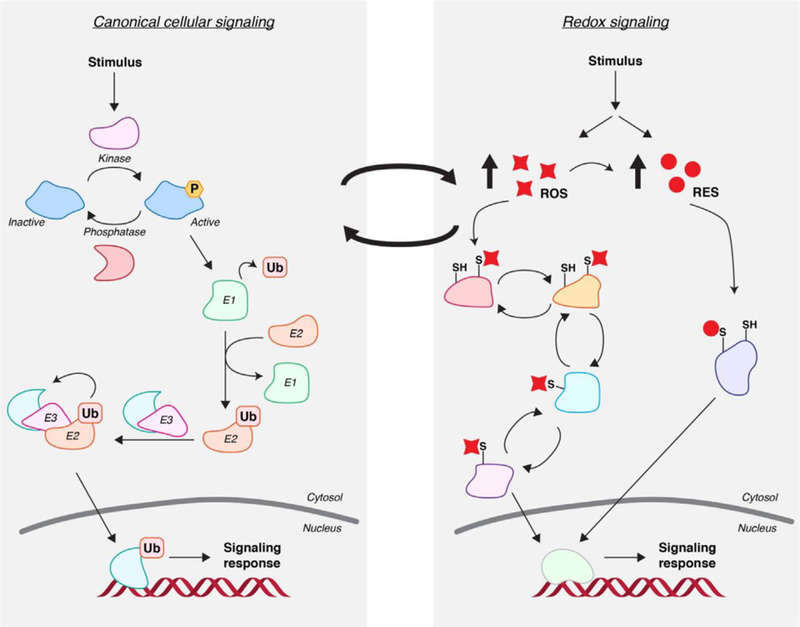
Intersecting canonical signaling with redox signaling. (Left). Canonical signaling paradigms are defined by enzyme-mediated installation/ removal of covalent modifications such as phosphate (P) or ubiquitin (Ub). (Right). Redox-linked signaling pathways—especially those regulated by reactive electrophilic species (RES)—defy these paradigms because no enzyme mediation occurs at the point of signal adduction to target proteins. ROS modifications are further differentiated by the established disulfide/thiol-sulfenic-acid-based signal relay mechanisms as well as enzyme-assisted reversal of certain ROS modifications. By contrast, RES modifications are largely irreversible. Importantly, redox-linked chemical messages often cross-talk with canonical messages in propagating biological signals.
2. Biological Processes Are Uniquely Complex
This complex level of orchestration at a molecular level is unique to life processes. But this level of organization is not unlike the way a multi-national company or a group of nations, such as the European Union, operates. In these organizations, multiple parallel, and intersecting units have to communicate and coordinate to ensure that the whole organization functions cohesively to achieve specific goals. In such an analogy, the different signaling codes in a cell would be represented by different languages used by member countries; yet, languages are not only spoken and written: subtle inflections/gesticulations/signals help us to convey our messages. To function in a society or multinational organization, one must understand these nuanced aspects. Are there parallels in cellular communication?
There are aspects of cellular signaling that may be akin to idioms. Redox signaling is an emerging field where it is proposed that ostensibly non-specifically reactive molecules are used by cells to propagate signals that appear to be essential for fitness. There is, for instance, significant evidence that reactive oxygen species (ROS; Figure 2) act as cellular-information brokers. These transient signals function in numerous pathways including neutrophil migration,[6] lifespan extension,[7] and cell differentiation.[8] However, elevation in ROS levels can contribute to several disease states, including driving proliferation in cancer.[9] This is at first glance surprising: ROS are typically considered bad for us, so why would any cell (healthy or otherwise) use ROS as a signal? This paradox is likely explained by the fact that ROS are unavoidable because of respiration, environmental oxygen, and the abundance of redox-active metals in cells, among other factors. Thus, cells need a way to sense and respond to ROS and one simple way to do this is through chemical reaction with specific proteins. The jury is still out as to whether ROS signaling pathways proceed via (1) direct modification of multiple “bystander” proteins; and/or (2) through modification of a few dedicated “professional sentinels” that then propagate the signal.[10,11] Proposed sentinels include peroxiredoxins, enzymes that have close to diffusion-controlled reaction kinetics with peroxide[12] (a 108-fold rate-enhancement relative to that of cysteine with peroxide). These sentinels then propagate the message by oxidizing downstream proteins (changing of hands of information[13]). In either case, ROS is the initial starting point of the message, even if “go-betweens” are required. In the “sentinel model,” most of the downstream signaling orchestration is enzymatic, i.e., the sentinels act as “translators” for ROS-based signals and these sentinels “spread the word” to other downstream proteins through their own intrinsic associations. As discussed in our other perspectives,[13–15] this model is possible for ROS-modification as disulfides and free thiols can be interchanged in a seemingly enthalpy-neutral process.[16,17] Ultimately, one of the proteins in this ROS-cascade must translate the ROS-modification to another line of canonical cellular communication such as kinase or ubiquitin signaling. For instance, oxidation of epidermal growth factor receptor (EGFR) by ROS enhances its kinase activity;[18] several ubiquitin specific proteases and ubiquitin carboxyl-terminal hydrolase enzymes are inactivated upon oxidation by ROS.[19] However, in general, there could be several exchange steps prior to this information transfer. Importantly, ROS-modification is generally reversible, so ROS-signaling proteins can be turned off efficiently by reduction. Several elaborate mechanisms have been identified to regulate reduction of oxidized signaling proteins.[20–22]
Figure 2.
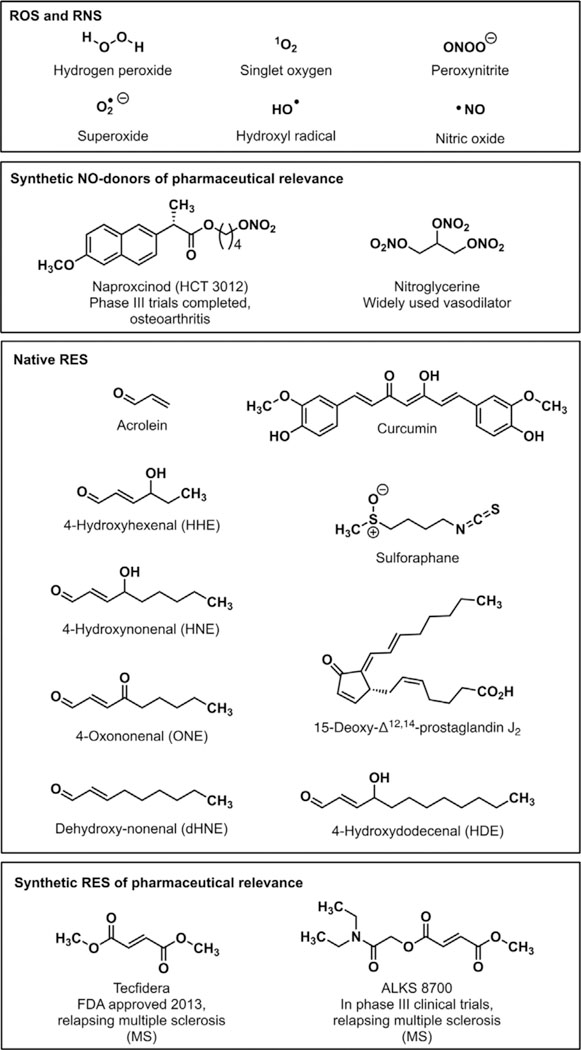
Representative ROS, RNS, and RES.
3. Cells Can Use Reactive Species for Communication
The fundamental importance of ROS-signaling sets a key precedent that the cell can harness reactive, seemingly-destructive molecules for signaling, turning a weakness into an advantage. There are several other classes of reactive redox-linked small signaling mediators that are generated endogenously. These include reactive nitrogen species (RNS) and reactive electrophilic species (RES) (Figure 2). RNS-protein adducts have been observed experimentally,[23] and these signaling events are involved in vasodilation,[24] cGMP production,[25] and apoptosis regulation,[26] among others.[27] RES are among the most prevalent native reactive signals. RES, such as the prototypical electrophile 4-hydroxynonenal (HNE), are derived from polyunsaturated fatty acids (PUFAs) either during non-enzymatic peroxidation, or under specifically-choreographed conditions catalyzed by enzymes, such as COX and LOX. COX enzymes are upregulated by several reactive species[28] including peroxide[29] and nitric oxide.[30] Thus, RES levels are intrinsically elevated upon ROS-upregulation.
However, RES have unique characteristics that distinguish them from ROS. Electrophilic enal/enone-derived RES are known to adduct proteins and endogenous nucleophilic species like glutathione (GSH) in cells and in purified systems, typically under conditions of excess RES exposure. However, unlike ROS, where specific proteins undergo a 108-fold rate-enhancement relative to the background conjugation rate (≈1M–1 s–1), RES metabolism is dominated by the various classes of glutathione-S-transferases (GSTs),[31] enzymes that accelerate HNE degradation by 103-105-fold above the uncatalyzed rate (≈1M–1s–1).[32] GSTs are abundant, but are unevenly distributed in cells. Assuming GSTs dominate RES-metabolism,[33] RES are degraded relatively slowly in cells. Thus, it is likely that to behave as effective signal transducers, kinetically-privileged RES-sensor proteins do not have to sense RES as effectively as privileged ROS-sensors must sense ROS. Obviously, distribution of different GSTs may also affect RES-signaling burdens in cells,[34] just like differential distribution of ROS-sensors can for ROS-signaling. However, this heterogeneity is not currently addressable.
Nevertheless, consistent with the above discussion, experimental evidence on the whole points to RES being more mobile and longer-lived in cells than ROS.[35–37] Indeed, protein-RES adducts have been isolated from blood of healthy subjects,[38,39] patient samples,[40,41] and model organisms,[42] indicating that RES- adducted proteins are physiologically accessible. However, precise quantitation of “target occupancy” under these “native” conditions remains almost entirely unknown. Fortunately, several endogenous RES function through mechanisms that require electrophilic adduction, such as 15-deoxy-Δ12,14-prostaglandin J2 (15-d-Δ12,14- PGJ2).[43] Furthermore, different RES are associated with different responses,[44–49] indicating that the RES chemotype itself influences cellular decision-making. However, this correlation certainly does not prove that RES make a necessary contribution to wellness or disease. It is our goal in this essay to critically evaluate the arguments for and against electrophile signaling as a bona fide mode of cellular communication. Importantly, progress has been hampered because traditional methods to assess targets (knockout/gene manipulation) are not readily applicable to study of RES/ ROS signaling because these are pleotropic signals and controlling their release/localization/duration is difficult. We will discuss how the field has dealt with these issues below. Although it is proper to group RES- and ROS-signaling together in some instances, there are several aspects of RES as signaling mediators that render them distinct from ROS signals:
RES signals typically cannot change hands/interconvert (unlike ROS-signals). This means that the protein modified by RES must be able to directly translate RES-signals to another form of cellular communication. Thus, RES-sensor proteins need to both sense (react rapidly with) RES and translate RES-signals into other canonical signaling codes (such as phosphate,[50] ubiquitin,[51] etc.).
Aside from a few exceptions (mainly involving RES-modified lysine residues),[47,52–55] RES-modification is largely irreversible (or at least relatively long-lived; half-life of HNEylated protein in cells >4h),[53] and signaling can likely only be turned off by degradation of the RES-modified protein.[56] Thus, unlike reversible ROS-modifications, RES-modifications are likely to have prolonged impacts on downstream pathways.
4. Thresholds That Must Be Passed to Establish Res as a Biological Signal
With this perspective, we propose that there are three thresholds that must be traversed for RES to be established as a bona fide means of cellular communication:
-
1)
Suitability and Feasibility: Can Res Modifications Constitute a Viable Post-Translational Regulatory Mechanism to Control Cell Response?
The notion that nature makes use of toxic and damaging signals as non-canonical post-translational modifications (PTMs) in signaling was difficult for the field to accept. Indeed, in marked contrast to how cells integrate and process enzyme-assisted PTMs, making convincing cases as to whether the cell can perceive and decode non-enzyme-mediated RES modifications and harness them as a means to transmit information was itself a fundamental challenge and paradigm shift in the principles of cell signaling. Nevertheless, as we detail below, careful pioneering experiments and recent efforts to profile targets of RES on a global scale have collectively made a clear case for RES as regulatory signals.
-
2)
Specificity and Sufficiency: Can Low-Occupancy Res Modifications of Specific Proteins in Otherwise Unperturbed Cells/Animals Engender Dominant Responses?
In contrast to ROS, measurement of endogenous RES levels in intact cells is hard to do accurately. Many methods do not function at equilibrium (like ROS-detection methods can[57]), and general irreversibility of RES modifications further hampers measurement efforts. Hence these methods are not dynamic (Cf. genetically-encoded ROS-sensors[57,58]) and can only give a cumulative measurement of RES present over a given period. Furthermore, studies of RES largely rely upon whole-cell/organism RES-treatment at RES-concentrations likely not readily attainable in cells. The issue with RES in particular is that once a RES has adducted a protein, another molecule of RES from the bulk media (under typical experimental conditions, an effectively inexhaustible supply of RES) can enter the cell. Thus, the effective concentration of RES in cells can be much higher than predicted based on what is present in the media. Similar effects have been reported in whole organisms.[59] Furthermore, it is important to try to link target-specific occupancy of RES-modification to target-specific phenotypic output while addressing RES signaling events. This is because the lower the occupancy required to sufficiently elicit phenotypically-relevant responses, the more likely it is that a signaling response could be elicited under “physiological” signaling conditions. It is also critical that experiments are performed under electrophile-limited conditions.
-
3)
Functionality and Physiology: Can Res Modification and Ensuing Downstream Signaling Operate under Accessible Signaling Conditions?
Obviously, satisfying both criteria 1 and 2 above will go a long way to proving that endogenous RES-sensing of a single protein is a novel and functional signaling pathway. However, this finding alone does not necessitate that RES-signaling occurs.
4.1. Passing Threshold 1: Suitability and Feasibility—can Res Act as a Viable Means of Cellular Communication?
4.1.1. Early Work with Endogenous Res in Cells Indicated Electrophiles May Function in Signaling Pathways, but the Data Were Confounding
A considerable body of work has been generated by treating cells with excess endogenous or synthetic electrophiles.[60] One interesting test case is 15-deoxy-Δ12,14-prostaglandin J2 (15-d- Δ12,14-PGJ2).[43] This endogenously-produced prostaglandin— present at nanomolar concentrations in 3T3-L1 cells[61]—can function as an agonist of PPARγ, a protein that regulates adipocyte differentiation and controls glucose levels.[62] However, 15-d-Δ12,14-PGJ2 also forms electrophilic adducts to other proteins, such as Keap1,[43] GSTs,[63] and thioredoxin.[64] It has been postulated that this covalent interactome may explain some properties unique to 15-d-Δ12,14-PGJ2, including antioxidant response (AR) upregulation.[43] However, micromolar concentrations of exogenous 15-d-Δ12,14-PGJ2 are required for these responses to be observed.[43] Obviously, there is an unsatisfying discrepancy between “accessible concentrations” in cells and what is required experimentally to observe a phenotype.
These issues have been addressed by some very careful experiments from the Yamamoto lab.[65] This group used shear stress as a means to induce AR (Figure 3). Under certain conditions, AR upregulation was shown to be both Nrf2 and COX2 dependent. As mentioned, 15-d-Δ12,14-PGJ2 modifies Keap1, the cytosolic gatekeeper of Nrf2/AR. Thus, a simple interpretation of these data is that COX2-mediated generation of 15-d-Δ12,14-PGJ2 upon shear stress modifies Keap1, thereby eliciting AR. However, this reasoning does not take into consideration the fact that NO and various ROS (Figure 3) can also be generated under shear stress[66] and these species can also modify Keap1. This model also neglects other “unknowns” caused by shear stress. Thus, despite the strong evidence for the dependence on specific genes, underlying reasons as to why a specific pathway is selectively activated are less clear-cut. Given the confounding data from these experiments, focus has turned to identifying proteins modified under RES-limited conditions.
Figure 3.
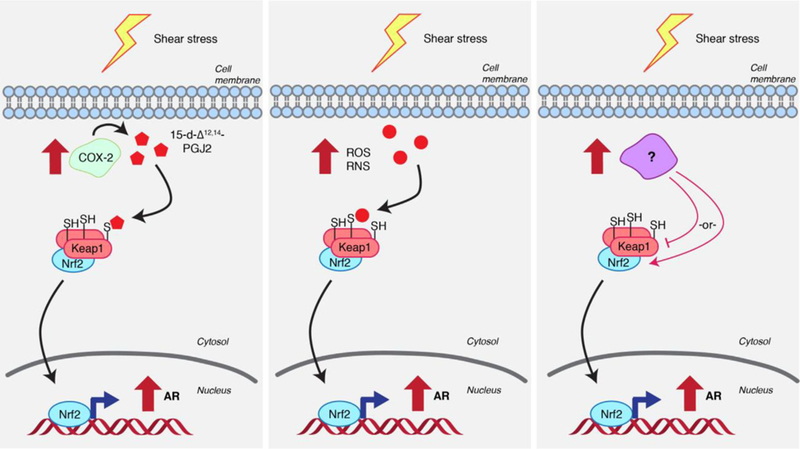
The challenge of deconvoluting redox signaling pathways: three possibilities to engender AR upregulation under shear stress. (Left) Shear stress upregulates COX-2 activity, leading to the generation of 15-d-Δ12,14-PGJ2 (Refer to Figure 2 for structure). 15-d-Δ12,14-PGJ2 modifies Keap1, leading to AR upregulation. (Center) Shear stress upregulates ROS/RNS, which directly modify Keap1 and cause AR upregulation. (Right) An unknown factor affected under shear stress regulates Keap1 or Nrf2 (or other interconnected players) to upregulate AR.
4.1.2. Proteomic Studies Identify Efficient Res Sensors; Res Modification of These Proteins Affects Activity In Vitro
Continued inspirational work from many pioneering contributors has established that some proteins are uniquely sensitive to RES modification. Intriguingly, these experiments were carried out in different ways. One simple proteomic approach involves treatment of cells or lysates with an alkynylated RES such as HNE or 4-oxononenal (ONE). The proteome is digested and the modified peptides are biotinylated using Click chemistry, enriched, and identified by mass spectrometry.[52–54,67] Isotopic labeling of the biotin tag allows for comparison between multiple treatment conditions. An alternative proteomic approach is the ground-breaking mass spectrometry-based “competitive activity-based protein profiling (ABPP)” method. This method uses a reporter electrophile such as iodoacetamide to assay the reactivity of 800–2000 specific cysteines. The method follows a simple regimen (Figure 4): pretreatment of cells or lysates/homogenates with a user-defined RES of interest (such as HNE) “caps” reactive cysteines; subsequent treatment with the reporter electrophile (such as iodoacetamide) then labels remaining (iodoacetamide-reactive) cysteines. When compared to a parallel experimental set treated with reporter electrophile, but not the RES, RES-modified proteins are identified by loss of reporter labeling. This powerful method thus allows high-throughput, albeit indirect, identification of proteins uniquely sensitive to a given RES. In vitro, these modifications were found to modulate activity of RES-sensitive proteins.[68]
Figure 4.
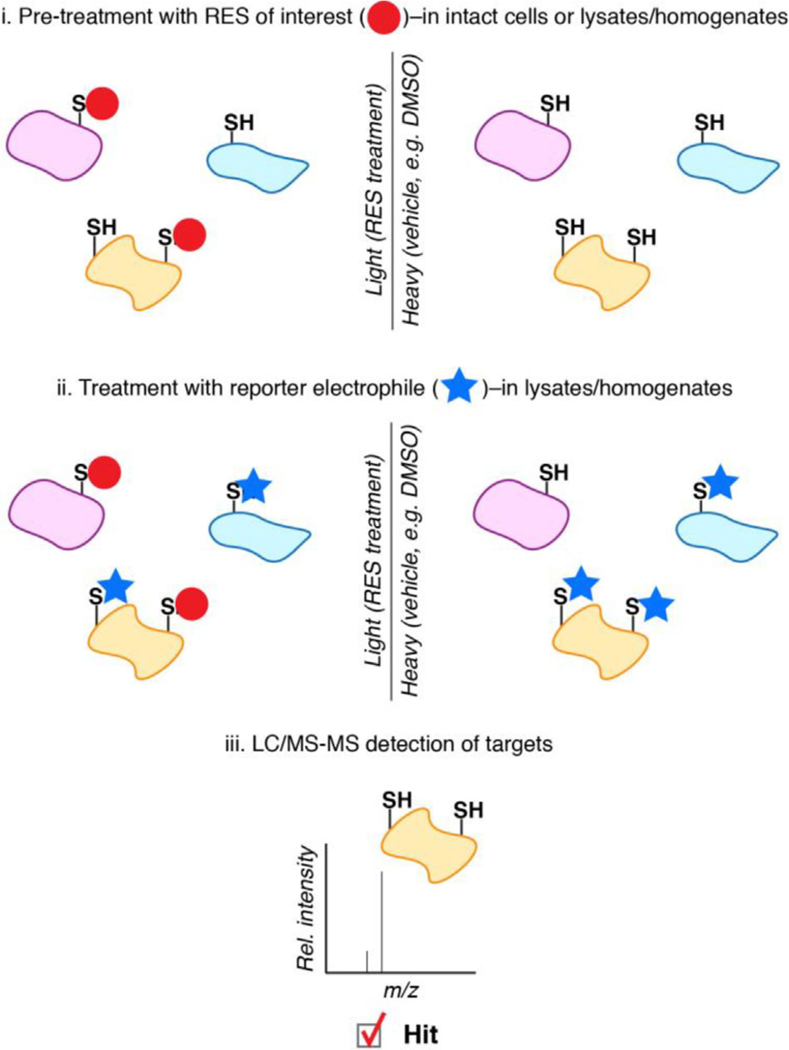
RES target identification by competitive ABPP. In competitive ABPP, parallel sets of intact cells or lysates/homogenates are treated with the RES of interest or vehicle (typically DMSO). Following lysis, the lysates are both treated with a reporter electrophile (e.g., iodoacetamide) which non-specifically labels cysteines. LC/MS-MS analysis allows for indirect detection of RES-modified cysteines by loss of reporter electrophile labeling.
This observation makes a strong case that these sensor proteins are tuned to react quickly with RES and engender a functional impact. But such data cannot rule out that RES modifications of other proteins change the functions of multiple proteins triggering an ensemble response. However, providing a specific protein is inherently sensitive over the global proteome, and assuming high-occupancy labeling can be obtained (as must occur to score as a hit in ABPP system), selective pathway modulation can nonetheless occur. If we put the in vitro and the bolus RES dosing data together, we get a strong hint that RES modifications could be modulating specific signaling pathways.
Despite the broad applicability of these approaches, limitations exist due to indirect nature of the set-up combined with the non-enzymatic nature of RES modifications (Figure 1). Significant challenges arise from inherent toxicity/reactivity/ metabolic activity of RES themselves along with the limited scope in addressing spatiotemporal nuances of RES signaling following bulk RES exposure of cells/animals or lysates. These can muddy functional validations of identified targets and could confound conclusions. ABPP assumes that there is a linear relationship between occupancy and phenotypic output. If this were the case, some proteins would have to have hugely elevated RES-conjugation rates relative to the bulk, to be able to selectively intercept RES and achieve high occupancy (assuming that the target is not a haploinsufficient gene). One challenge in iodoacetamide-based ABPP is associated with the general constraint of RES target identification to cysteine conjugation. Reactive RES like ONE may target residues other than cysteine. Native RES such as HNE for instance modifies protein lysine and histidine beyond cysteines.[14] Recently reported proxy probes that can profile lysine residues have begun to address this general challenge.[69]
However, in light of seminal studies by many laboratories on ROS-signaling[70–85] and our own work with on-target RES- signaling[13–15,50,51,86–93] discussed below, it is likely that phenotypically-relevant outputs may occur at low occupancy. Furthermore, the competitive ABPP-method intrinsically assumes that the targets of iodoacetamide (the reporter electrophile) and the RES are the same (although, chemically, iodoacetamide is an sp3-hybridized electrophile, whereas Michael-acceptor-based RES—the largest class of RES in mammalian systems—are sp2-hybridized; thus their reactivity and “nucleophile-electrophile matching”[15] likely differ). This assumption may result in lack of scoring for some sensorproteins that are reactive to the RES of interest but not reactive to the reporter-electrophile (Figure 4).
Recently, a variation on competitive ABPP has emerged that does not rely on a reporter electrophile. This method uses an aminooxy probe to label RES-modified proteins through the carbonyl moiety remaining post-RES adduction.[94] One attractive feature of this approach is its expanded scope of electrophile adduct detection: In contrast to “classic” ABPP where reactivity of the reporter electrophile limits detection of hits to cysteine adducts, this method can also detect lysine and histidine adducts. Overall, this method provides a more direct readout than the reporter-electrophile method. However, one limitation of this strategy is its reliance on the presence of a carbonyl post RES adduction: we and others have observed reduction of the carbonyl post-target-adduction in intact cells.[87,88,90,95]
4.2. Passing Threshold 2. Specificity and Sufficiency—Can Res-Modification of a Target Protein at Low-Occupancy Sufficiently Drive a Signaling Output Against a Largely Non-Modified Cellular Backdrop?
We were intrigued by these thought-provoking seminal experiments, and resolved to investigate RES signaling in living systems at a level of specificity, timing, dosage, and resolution previously inaccessible. To this end, we developed a method to shepherd a specific RES to a specific—typically-overexpressed—protein of interest (POI) in cells/whole-organisms, allowing RES-sensitivity of a specific POI to be individually assayed in vivo. The method works by a novel exploitation of Class II proximity-enhancement[89] that allows for “pseudo-intramolecular” delivery of a native diffusible and reactive RES in situ (Figure 5). Critically, the method is both self-limiting (i.e., RES is supplied maximally at concentrations equal to the POI) and is more or less independent of POI-expression (similar results are obtained in various models—fish,[50,51] worms,[92,96] and cultured cells[50,51,86–88,90,91,93]—wherein transgene expression ranges from close to endogenous levels to several fold in excess), allowing association kinetics to be ranked fairly across different proteins.
Figure 5.
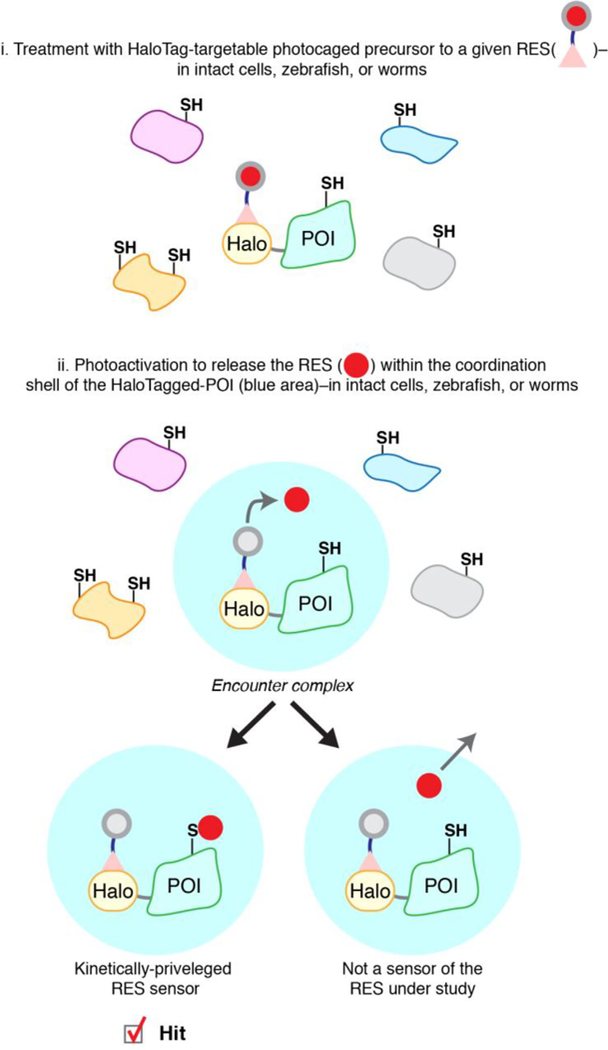
Interrogating precision RES signaling by T-REX. A protein of interest (POI) is expressed as a functional Halo-fusion POI in live cells, worms, or fish. Following non-invasive treatment of these living systems with a HaloTag-targetable bioinert cell/organism-permeable photocaged precursor to a given RES, a 1:1 covalent complex of Halo:photocaged-RES is achieved. Photoactivation following washing away of excess probe liberates the RES (in an amount stoichiometric to POI) in the immediate vicinity of the POI, resulting in the formation of an “encounter complex” (blue shell). The POI is given first refusal of the RES and competition between native reactivity of the POI to a specific RES and native diffusivity of the RES is allowed for. Provided the POI is a bona fide sensor able to react rapidly with the RES before its diffusion, fractional target occupancy is achieved between a specific RES-chemotype and a specific POI. If this occupancy fulfills biological sufficiency, T-REX enables on-target functional redox response to be measured in real time in living systems.
4.2.1. T-REX in Living Systems Directly Identifies Specific “First-Responding” Res-Sensors Whose Discrete Modification Sufficiently Drives Functional Redox Responsivity
Briefly, T-REX is based on expression of a functional Halo- tagged-POI and the design of a bioinert cell/fish/worm-permeable small-molecule photocaged precursor to a given RES. The Halo-protein-tag reacts irreversibly and highly specifically with an alkylchloride that is built into the photocaged RES, resulting in a Halo-POI:photocaged-RES covalent complex in a 1:1 ratio in vivo following probe wash out. Upon light exposure, the RES is rapidly liberated in the vicinity of the POI, giving the POI first refusal for covalent conjugation to the RES prior to its diffusion (Figure 5).
During conception of the project, we postulated that unless reaction occurred through the initial encounter complex formed post photouncaging, there would be no labeling of the POI as there are 200000 cysteines (a concentration of 1–10 mM protein cysteines in cells) within the proteome, and large amounts (around 1–10 mM at least) of free small- molecule thiols such as GSH. This postulate ultimately proved to be almost entirely correct. The use of Halo-tag is not always optimal: for each POI, it must be shown that Halo-tagging generally does not affect protein function. To further validate that the sensors identified by T-REX are genuine firstresponders, our most-recently-completed studies have recapitulated HNE-sensitivity of some targets using bulk RES- exposure of live specimens under low RES-doses and short treatment-periods.[51,93] Our published work has also demonstrated that the extent of covalent RES modification is not affected whether Halo resides on the C- or N-terminus of the POI,[90] consistent with the encounter complex adduction versus diffusion concept (Figure 5).
The first protein on which we road-tested the T-REX idea was Keap1.[86–88,90,92] Substiochiometric RES modification of Halo-Keap1 under T-REX conditions led to a Keap1-specific AR-activation.[87,88,90] Critically, if the Halo and Keap1 were expressed separately (i.e., non-fused), neither modification of Keap1 nor AR-activation was observed. This result proves that low-occupancy Keap1-specific modification through “pseudo-intramolecular” RES-delivery was the sole pathway responsible for AR activation. Of note, the overexpression system gives ≈5 μM Halo-Keap1 (or Halo); yielding the maximum theoretical amount of RES liberated around 5 μM.[51] Thus, RES not reacting with Keap1 is likely captured by GSH or other protein cysteines. The fact that no pathway activation was measurable in the nonfused system[87,88,90] demonstrates that occupancy of RES averaged over the background cysteome is not sufficient to drive AR activation. GSH:GSSG ratio, cell viability, and other stress-sensitive pathways were not affected by the T-REX system,[87,88,90] in stark contrast to bolus RES-exposure methods. We also demonstrated the tolerance of this platform to live C. elegans,[92,96] opening up exciting avenues to study lifespan regulation in this validated model organism with previously- inaccessible resolution.
To further document that on-target RES-modifications are sufficient drivers of functional signaling responses, we validated T-REX in another established redox-sensitive pathway involving the PTEN-redox-sensor.[86,90] T-REX coupled with established knockdown methods also uncovered redox-dependent pathway crosstalk hidden under conditions of bolus RES administration.[91]
More recently, we identified Akt3 as a novel electrophile sensor in a medium-throughput screen.[50] RES delivery to Akt3 led to downregulation of downstream signaling pathways. We also examined signaling in live zebrafish embryos, where the expression level of Halo-Akt3 is similar to that of the endogenous protein. RES labeling of Akt3 (delivery efficiency of about 20%; 12% occupancy) and downstream signaling responses were observed, further substantiating that T-REX is unaffected by POI-expression-levels. A cysteine to serine mutation, that rendered Akt3 RES-sensing-defective but otherwise functional, ablated both labeling and downstream phenotypes, underscoring the necessity of specific RES modification events to drive signaling. Of broader importance, in both fish and cells, a significantly greater degree of pathway modulation was observed than would have been expected based on RES-POI occupancy alone. We thus concluded that RES-signaling elicited dominant effects on signaling pathways. Indeed, Akt-enzymes manifest dominant-negative signaling functions.[97,98] Our latest work that combines studies using isolated proteins and in cells and fish has identified what transpired to be further examples of privileged—and non- catalytically-essential—protein-cysteines wherein low-occupancy modifications are functionally significant[51,93] and capable of accommodating signaling crosstalk.[51]
Although T-REX is not ideal, the extension to fish and worms, the careful “non-fused” protein controls assayed and sensing-defective functional mutants collectively make a strong argument that quasi-endogenous levels of RES can label POIs provided they are first responders to the RES in question. Thus, the 2nd Threshold has been passed. However, as RES concentrations in the cell are unknown, and second-order rates are proportional to both RES and target sensor-protein(s), it is presently unclear how T-REX data directly commute to endogenous sensing.
4.3. Is Native Res up to the Task of Labeling Endogenous Sensors?
It is becoming increasingly clear that context is important for effective ROS-sensing. The best ROS-scavengers in the cell are relatively abundant and have near diffusion-controlled reaction rates with ROS (108 M–1 s–1).[99] However, putative ROS- signaling sensors have second-order rate constants likely very much slower than this (often quoted around 102 M–1 s–1).[11,18] Unsurprisingly, the multi-target model of ROS-sensing[14] requires local depletion of cellular ROS-sensing sentinels to allow less-efficient sensors to catch up. The local sentinel depletion is typically suggested to occur through oxidation of sensing sentinels, but there are a few examples of inactivation of sentinels, such as peroxiredoxin I (by phosphorylation during redox signaling).[100] Of course, given the ability to change hands (Figure 1), ROS does not have to hit its intended target first, thus second-order reaction rates with ROS may not be wholly relevant for postulated ROS “sensors” like EGFR (second-order rate constant for oxidation by H2O2 > 102M–1 s–1).[18] But as outlined above, RES is different. Once a protein is modified, the RES is largely “stuck,” and regardless of reversibility, a changing-of-hands mechanism is much-less- readily envisioned because a free RES must be generated to allow re-conjugation. Thus, privileged RES-sensors are likely always the first point-of-contact with the RES, and enzymatic assistance is not required for sensing.
Nevertheless, ABPP and T-REX give some clues that endogenous sensing is likely possible. Both methods can function under RES-limited conditions, yet labeling of proteins occurs selectively and efficiently. T-REX forces an encounter complex (Figure 5) and asks whether a given protein will be modified by RES and at what frequency. For instance, a 60% delivery efficiency means that if the protein sees RES, there is a very high chance of labeling. Yet a very high chance does not mean that labeling can occur naturally and it does not mean that a threshold required to trigger signaling will be cleared endogenously, even if there are amplification effects at play. Furthermore, it is currently unknown how this T-REX delivery efficiency parameter correlates with second-order reaction kinetics between RES and the POI.
5. Could We Ever Pass Threshold 3: Functionality and Physiology?
Several syllogisms can lead to the conclusion that RES performs such a function. Arguably the most powerful is: the cell is exposed to “some” RES endogenously; RES can modify proteins; RES-modification leads to modulation of some significant pathways; and some of these modulations require only low occupancy. Thus, sensor proteins have likely evolved spontaneously to sense RES, and therefore, it is likely that such responses are occurring in the cell. It has certainly been postulated that ROS-sensing has had a hand in protein and organelle evolution.[101–103] Furthermore, we have traced the evolutionary tree of various Akts and shown that the RES-sensing cysteine in Akt3 is present in all Akt3s from fish to mammals.[15,50] The dominant-negative effects we have outlined above, in our opinion, serve to strengthen this line of reasoning, as many modifications of proteins (such as by conventional drugs) do not follow this sort of behavior. However, these observations are a far cry from clear experimental proof.
There are several methods that may be able to unequivocally prove redox signaling at endogenous proteins is a “real” phenomenon. Halo knock-in lines could prove to be very useful. Such lines could be created in fish or C. elegans, both of which are compatible with T-REX.[50,51,92,96] As we and many laboratories have independently shown, many RES sensors have specific RES-sensor cysteines that are themselves kinetically-privileged to react with RES. Thus, use of Halofusion POIs with privileged-cysteine to serine mutations—such as in Akt3 case wherein the mutant exhibits hypomorphism of RES-sensing—would be an ideal control, provided the sensing-defective mutation does not affect folding/activity/localization/ ground-state regulation.
5.1. Selectivity May Be Critical for Proof of Binding
One of the classic aspects of biological systems is their unique and often unexpected chemo- and regioselectivity. Early reports of the RES-sensor glutathione reductase (GSR) stated that this protein is HNE sensitive, as GSR activity was inhibited by HNE.[93,104] However, the same protein is not able to react with 2-oxoaldehydes, RES with higher electrophilicity than HNE. More recent reports reflect this finding on a global scale: in both THP-1 and RKO cells, the target spectra of HNE and 4-oxononenal (ONE; Figure 2) differ significantly, with 30% of targets on average modified by only one of the RES. Interestingly, the more electrophilic ONE modifies fewer targets overall in both cell types.[67] Making the sweeping assumption that modifications by HNE and ONE are both limited by protein targets, and not by permeability/stability/ subcellular distribution/adduct stabilities, these data indicate that many proteins may show preference for specific RES. Such chemoselectivity that bucks the chemically-expected trend is strongly indicative of a biological recognition process. Additionally, chirality (i.e., enantioselectivity) is the hallmark of biological materials.[105] Reports have surfaced that that different enantiomers of HNE elicit widely different phenotypes in mouse hepatocytes, depending on which pathway is investigated.[106–108] One simple interpretation of this result is that there is a significantly different protein target profile between the two different enantiomers of HNE. Indeed, GAPDH shows preference for inhibition by the (S)-enantiomer of HNE.[109] Understanding the specific proteins and sensing mechanisms involved in these putative enantioselective processes may help not only identify privileged sensors, but prove that proteins have evolved to sense specific RES.
5.2. Genetics May Ultimately Help to Provide a Framework to Understand Origins of Res-Signaling and Through Which to Explain/Predict Res Sensors
The kinase field has benefited from investigation of evolutionary history. We and others have started to look at sequence motifs, phylogeny, and structure of RES sensors.[15,51,93,95] Unfortunately, as of right now there are too few bona fide privileged- sensors to make any solid conclusions. What is clear though is that cysteines that sense RES are strongly conserved. However, cysteines are quite highly conserved in general, so this argument does not, on its own, sound particularly compelling. Interestingly, the RES-sensing cysteine of Akt3 is more strongly- conserved than the ROS-sensing cysteine of Akt2.[15] However, how this transmutes to other proteins is unknown. With the opening of the study of RES-sensing by T-REX in worms[92,96] and zebrafish[50,51] (the former being a quite distant ancestor of humans and the latter an organism that recently underwent a whole-genome duplication), it is possible that more phylogenetic information can be derived. Only time will tell how useful these model systems will be as mines for functional sensors and springboards to new information.
If we draw on analogy from the kinase playbook, we can postulate how such snapshots of evolution may help us understand RES-sensing. We can envision a hypothetical scenario wherein a bulky hydrophobic residue (e.g., tryptophan) on a genetically-duplicated POI mutates to a cysteine (a single base pair mutation), leaving the mutant POI of low or no activity (Figure 6). It is likely that due to change of the bulky hydrophobic residue to cysteine, there is already a vacant hydrophobic pocket rendering this mutant able to accommodate a hydrophobic- chain extension (e.g., from a RES such as HNE) on the new cysteine. The evolution of a protein containing a RES-binding site in proximity to a cysteine certainly could lead to a privileged sensing behavior, especially if subsequent mutations affected, for instance, pKa of the cysteine. Since RES-modified cysteine could potentially fill the void left by tryptophan, such a modification could chemically complement the loss of tryptophan and restore lost/diminished function. Thus, a RES-dependent function is created. One prediction of this model is that mutation of a RES-sensor cysteine to a bulky hydrophobic residue may recapitulate some of the phenotypes associated with RES modification, just like serine to aspartate mutations can turn on activity of proteins activated by phosphorylation. Conversely, identifying phylogenetic changes from large bulky residues to cysteine across evolution may be a good way to mine the genome for sensor cysteines. Success of these (or likely more refined) predictions would form a sound basis for establishing that RES, as bona fide signaling molecules, have shaped evolution of individual sensors.
Figure 6.
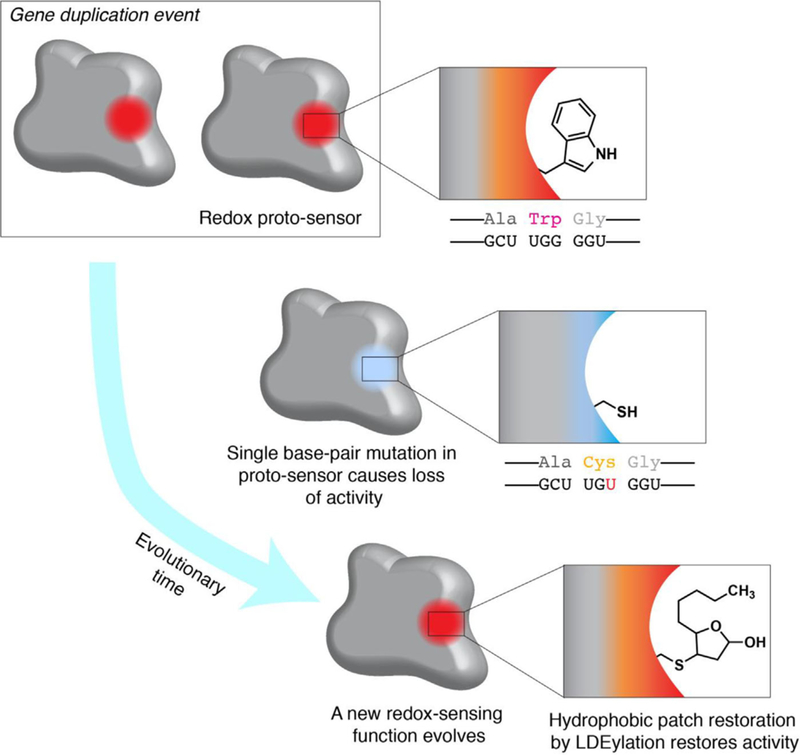
Evolution of an electrophile sensor. Following a gene duplication event, a single base-pair mutation causes a Trp to Cys mutation, ablating a hydrophobic patch and enzyme activity/function. Modification of the new Cys residue by RES (shown is a cyclized form of HNE-modification; see ref. [14] for details) restores enzyme activity. Thus, a RES-dependent function evolves.
5.3. Do We Need to Pass Through Threshold 3 for Res-Sensing to Be of Use?
The theoretical aspects of RES-sensing are intellectually stimulating, technically challenging, and of fundamental importance. However, RES have a very practical application that as academics we must not ignore. The drug community widely appreciates the importance of covalent inhibitors. Most of the recent rationally-designed covalent inhibitors target specific reactive cysteines.[15] Intriguingly, many of the reactive appendages on electrophilic drugs resemble natural RES-motifs (Figure 2). We have thus postulated that RES-sensitive proteins are natural resources for covalent drug development.[15] The Cravatt laboratory has pioneered elegant methods to identify what they term “ligandable” cysteines, or cysteines that lie proximal to ligand-binding pockets suitable for modification.[110] Regardless of the method of identification, a link to function and the ability to (semi)selectively target a POI by any given RES-based-molecule are sufficient to open new avenues for drug discovery. Thus, it is paramount that RES-sensors be identified and new ligands investigated.
6. Conclusion
For much of their history, RES have been an enigma to the scientific community due to their inherent reactivity and toxicity at high doses. However, it is now clear that these reactive signals are essential for cellular and organismal fitness, and mounting evidence collectively presents a strong case that the cell has harnessed RES as bona fide signaling messengers. Threshold 1 has been passed by pioneering experiments in the electrophile signaling field that have linked the generation of RES to signaling responses. Along with more recent powerful approaches to profile reactive cysteines on a global scale, these experiments have clearly shown the feasibility and suitability of RES as cellular signals. Our own work with the T-REX system has allowed us to more closely mimic endogenous signaling conditions by interrogating privileged RES-sensor proteins under electrophile-limited conditions. Because we have shown that low-occupancy RES modifications of specific sensor proteins are sufficient to drive downstream signaling responses, Threshold 2 has also been passed.
Passing Threshold 3—the “functionality and physiology” threshold—remains a formidable challenge that requires innovative new approaches to surmount. Genetic manipulation, such as knock-in of HaloTag-fused privileged RES sensors or mutation of privileged cysteines housed within endogenous RES-sensors (identified by T-REX or global profiling) may hold the key to proving that RES modifications are physiological cellular signals. A more thorough understanding of the evolution of RES-sensor cysteines formed by phylogenetic analysis of privileged RES-sensors as more are identified may also prove useful in cementing the functional significance of RES to cellular signaling. Ultimately, only time and more diligent work by RES-signaling trailblazers will tell if we will ever be able to pass Threshold 3. Nevertheless, we feel that RES signaling holds promise for understanding cellular fitness and also for developing new ways to treat disease.
Acknowledgments
J.R.P. and M.J.C.L. contributed equally to this work. The electrophile signaling research program in the Aye Lab is supported by a National Science Foundation CAREER grant (CHE-1351400), a Sloan Fellowship (FG-2016–6379), a Beckman Young Investigator Award (2014-BYI), an Office of Naval Research Young Investigator Award (N00014–17-1–2529), and a National Institutes of Health New Innovator Award (1DP2GM114850) (to Y.A.). J.R.P. acknowledges an American Heart Association Predoctoral Fellowship (17PRE33670395), and the Cornell CBI Training Grant (NIGMS T32GM008500).
Abbreviation
- 15-d-Δ12,14-PGJ2
15-deoxy-Δ12,14-prostaglandin J2
- ABPP
activity-based protein profiling
- AR
antioxidant response
- DUB
deubiquitinase
- EGFR
epidermal growth factor receptor
- GSH
glutathione
- GST
glutathione S-transferase
- HNE
4-hydroxynonenal
- ONE
4-oxononenal
- POI
protein of interest
- PTM
post-translational modification
- PUFA
polyunsaturated fatty acid
- RES
reactive electrophilic species
- ROS
reactive oxygen species
- SUMO
small ubiquitin-like modifier protein
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Contributor Information
Jesse R. Poganik, Department of Chemistry and Chemical Biology Cornell University Ithaca, NY 14853, USA ya222@cornell.edu
Dr. Marcus J. C. Long, Department of Chemistry and Chemical Biology Cornell University Ithaca, NY 14853, USA ya222@cornell.edu
Yimon Aye, Department of Chemistry and Chemical Biology Cornell University Ithaca, NY 14853, USA ya222@cornell.edu Department of Biochemistry Weill Cornell Medicine New York, NY 10065, USA.
References
- [1].Gunning P, Weinberger R, Jeffrey P, Hardeman E, Annu. Rev. Cell. Dev. Biol. 1998, 14, 339. [DOI] [PubMed] [Google Scholar]
- [2].Huang N, Lee I, Marcotte EM, Hurles ME, PLoS Genet. 2010, 6, e1001154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Santarosa M, Ashworth A, Biochim. Biophys. Acta 2004, 1654, 105. [DOI] [PubMed] [Google Scholar]
- [4].Hunter T, Mol. Cell 2007, 28, 730. [DOI] [PubMed] [Google Scholar]
- [5].Wang Z, Cole PA, Methods Enzymol. 2014, 548, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Yan B, Han P, Pan L, Lu W, Xiong J, Zhang M, Zhang W, Li L, Wen Z, Immunol J. 2014, 192, 5998. [DOI] [PubMed] [Google Scholar]
- [7].Sasakura H, Moribe H, Nakano M, Ikemoto K, Takeuchi K, Mori I, Cell Sci J. 2017, 130, 2631. [DOI] [PubMed] [Google Scholar]
- [8].Sato A, Okada M, Shibuya K, Watanabe E, Seino S, Narita Y, Shibui S, Kayama T, Kitanaka C, Stem Cell Res. 2014, 12, 119. [DOI] [PubMed] [Google Scholar]
- [9].Weinberg F, Hamanaka R, Wheaton WW, Weinberg S, Joseph J, Lopez M, Kalyanaraman B, Mutlu GM, Budinger GRS, Chandel NS, Proc. Nat. Acad. Sci. USA 2010, 107, 8788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Reczek CR, Chandel NS, Curr. Opin. Cell Biol. 2015, 33, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Sobotta MC, Liou W, Stocker S, Talwar D, Oehler M, Ruppert T, Scharf AN, Dick TP, Nat. Chem. Biol. 2015, 11, 64. [DOI] [PubMed] [Google Scholar]
- [12].Cox AG, Peskin AV, Paton LN, Winterbourn CC, Hampton MB, Biochemistry 2009, 48, 6495. [DOI] [PubMed] [Google Scholar]
- [13].Long MJ, Poganik JR, Ghosh S, Aye Y, ACS Chem. Biol. 2017, 12, 586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Long MJ, Aye Y, Chem. Res. Toxicol. 2016, 29, 1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Long MJC, Aye Y, Cell Chem. Biol. 2017, 24, 787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Singh R, Whitesides GM, in: Patai S, Rappoport Z (Ed.), Sulfur-Containing Functional Groups, 1993. [Google Scholar]
- [17].Nagy P, Antioxid. Redox Signal. 2013, 18, 1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Truong TH, Ung PM, Palde PB, Paulsen CE, Schlessinger A, Carroll KS, Cell Chem. Biol. 2016, 23, 837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lee JG, Baek K, Soetandyo N, Ye Y, Nat. Commun. 2013, 4, 1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Matsuzawa A, Arch. Biochem. Biophys. 2017, 617, 101. [DOI] [PubMed] [Google Scholar]
- [21].Schieber M, Chandel NS, Curr. Biol. 2014, 24, R453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Salmeen A, Andersen JN, Myers MP, Meng T-Z, Hinks JA, Tonks NK, Barford D, Nature 2003, 423, 769. [DOI] [PubMed] [Google Scholar]
- [23].Zhu JH, Zhang X, Roneker CA, McClung JP, Zhang S, Thannhauser TW, Ripoll DR, Sun Q, Lei XG, Free Radic. Biol. Med. 2008, 45, 611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zhao Y, Vanhoutte PM, Leung SW, J. Pharmacol. Sci. 2015, 129, 83. [DOI] [PubMed] [Google Scholar]
- [25].Francis SH, Busch JL, Corbin JD, Sibley D, Pharmacol. Rev. 2010, 62, 525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Franco MC, Estevez AG, in: Maurer M (Ed.), Amyotophic Lateral Sclerosis: InTech, 2012. [Google Scholar]
- [27].Martinez MC, Andriantsitohaina R, Antioxid. Redox Signal. 2009, 11, 669. [DOI] [PubMed] [Google Scholar]
- [28].Reddy KK, Vidya Rajan VK, Gupta A, Aparoy P, Reddanna P, BMC Res. Notes 2015, 8, 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Onodera Y, Teramura T, Takehara T, Shigi K, Fukuda K, FEBS Open Bio. 2015, 5, 492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Yang T, Zhang A, Pasumarthy A, Zhang L, Warnock Z, Schnermann JB, Am. J. Physiol. Renal. Physiol. 2006, 291, F891. [DOI] [PubMed] [Google Scholar]
- [31].Yang Y, Yang Y, Xu Y, Lick SD, Awasthi YC, Boor PJ, Toxicol. Appl. Pharmacol. 2008, 230, 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Fritz KS, Petersen DR, Free Radic. Biol. Med. 2013, 59, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Hayes JD, Pulford DJ, Crit. Rev. Biochem. Mol. Biol. 1995, 30, 445. [DOI] [PubMed] [Google Scholar]
- [34].Petersen DR, Doorn JA, Free Radic. Biol. Med. 2004, 37, 937. [DOI] [PubMed] [Google Scholar]
- [35].Warren EA, Netterfleld TS, Sarkar S, Kemp ML, Payne CK, Sci. Rep. 2015, 5, 16929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Aldini G, Facino RM, Beretta G, Carini M, BioFactors 2005, 24, 77. [DOI] [PubMed] [Google Scholar]
- [37].Doorn JA, Petersen DR, Chem. Res. Toxicol. 2002, 15, 1445. [DOI] [PubMed] [Google Scholar]
- [38].Méndez D, Hernáez ML, Kamali AN, Diez A, Puyet A, Bautista JM, Infect. Genet. Evol. 2012, 12, 1780. [DOI] [PubMed] [Google Scholar]
- [39].Chavez JD, Wu J, Bisson W, Maier CS, Proteom J. 2011, 74, 2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Castellani RJ, Perry G, Siedlak SL, Nunomura A, Shimohama S, Zhang J, Montine T, Sayre LM, Smith MA, Neurosci. Lett. 2002, 319, 25. [DOI] [PubMed] [Google Scholar]
- [41].Yoritaka A, Hattori N, Uchida K, Tanaka M, Stadtman ER, Mizuno Y, Proc. Nat. Acad. Sci. USA 1996, 93, 2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Andreoletti O, Levavasseur E, Uro-Coste E, Tabouret G, Sarradin P, Delisle M-B, Berthon P, Salvayre R, Schelcher F, Negre-Salvayre A, Neurobiol. Dis. 2002, 11, 386. [DOI] [PubMed] [Google Scholar]
- [43].Oh JY, Giles N, Landar A, Darley-Usmar V, Biochem. J. 2008, 411, 297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Long EK, Picklo MJ Sr., Free Radic. Biol. Med. 2010, 49, 1. [DOI] [PubMed] [Google Scholar]
- [45].Riahi Y, Cohen G, Shamni O, Sasson S, Am. J. Physiol. Endocrinol. Metab. 2010, 299, E879. [DOI] [PubMed] [Google Scholar]
- [46].Chipuk JE, McStay GP, Bharti A, Kuwana T, Clarke CJ, Siskind LJ, Obeid LM, Green DR, Cell 2012, 148, 988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Jin J, He B, Zhang X, Lin H, Wang Y, J. Am. Chem. Soc. 2016, 138, 12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Galligan JJ, Rose KL, Beavers WN, Hill S, Tallman KA, Tansey WP, Marnett LJ, J. Am. Chem. Soc. 2014, 136, 11864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Kastrati I, Siklos MI, Calderon-Gierszal EL, El-Shennawy L, Georgieva G, Thayer EN, Thatcher GR, Frasor J, J. Biol. Chem. 2016, 291, 3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Long MJC, Parvez S, Zhao Y, Surya SL, Wang Y, Zhang S, Aye Y, Nat. Chem. Biol. 2017, 13, 333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Zhao Y, Long MJC, Wang Y, Zhang S, Aye Y, ACS Cent. Sci. In press 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Sun R, Fu L, Liu K, Tian C, Yang Y, Tallman KA, Porter NA, Liebler DC, Yang J, Mol. Cell. Proteomics 2017, 16, 1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Yang J, Tallman KA, Porter NA, Liebler DC, Anal. Chem. 2015, 87, 2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Cui Y, Li X, Lin J, Hao Q, Li XD, ACS Chem. Biol. 2017, 12, 47. [DOI] [PubMed] [Google Scholar]
- [55].Bosch-Morell F, Flohé L, Marín N, Romero FJ, Free Radic. Biol. Med. 1999, 26, 1383. [DOI] [PubMed] [Google Scholar]
- [56].Copeland RA, Pompliano DL, Meek TD, Nat. Rev. Drug Discov. 2006, 5, 730. [DOI] [PubMed] [Google Scholar]
- [57].Schwarzlander M, Dick TP, Meyer AJ, Morgan B, Antioxid. Redox Signal. 2016, 24, 680. [DOI] [PubMed] [Google Scholar]
- [58].Mishina NM, Markvicheva KN, Bilan DS, Matlashov ME, Shirmanova MV, Liebl D, Schultz C, Lukyanov S, Belousov VV, Methods Enzymol. 2013, 526, 45. [DOI] [PubMed] [Google Scholar]
- [59].Gupta P, Kim B, Kim SH, Srivastava SK, Mol. Nutr. Food Res. 2014, 58, 1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Jacobs AT, Marnett LJ, Acc. Chem. Res. 2010, 43, 673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Bell-Parikh LC, Ide T, Lawson JA, McNamara P, Reilly M, FitzGerald GA, J. Clin. Invest. 2003, 112, 945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Murphy GJ, Holder JC, Trends Pharmacol. Sci. 2000, 21, 469. [DOI] [PubMed] [Google Scholar]
- [63].Paumi CM, Smitherman PK, Townsend AJ, Morrow CS, Biochemistry 2004, 43, 2345. [DOI] [PubMed] [Google Scholar]
- [64].Shibata T, Yamada T, Ishii T, Kumazawa S, Nakamura H, Masutani H, Yodoi J, Uchida K, J. Biol. Chem. 2003. , 278, 26046. [DOI] [PubMed] [Google Scholar]
- [65].Hosoya T, Maruyama A, Kang MI, Kawatani Y, Shibata T, Uchida K, Warabi E, Noguchi N, Itoh K, Yamamoto M, J. Biol. Chem. 2005, 280, 27244. [DOI] [PubMed] [Google Scholar]
- [66].McSweeney SR, Warabi E, Siow RC, Hypertension 2016, 67, 20. [DOI] [PubMed] [Google Scholar]
- [67].Codreanu SG, Ullery JC, Zhu J, Tallman KA, Beavers WN, Porter NA, Marnett LJ, Zhang B, Liebler DC, Mol. Cell. Proteomics 2014, 13, 849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Cravatt BF, Wright AT, Kozarich JW, Annu. Rev. Biochem. 2008, 77, 383. [DOI] [PubMed] [Google Scholar]
- [69].Hacker SM, Backus KM, Lazear MR, Forli S, Correia BE, Cravatt BF, Nat. Chem. 2017, 9, 1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Winterbourn CC, Nat. Chem. Biol. 2008, 4, 278. [DOI] [PubMed] [Google Scholar]
- [71].Dickinson BC, Chang CJ, Nat. Chem. Biol. 2011, 7, 504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Paulsen CE, Carroll KS, Chem. Rev. 2013, 113, 4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Meyer AJ, Dick TP, Antioxid. Redox Signal. 2010, 13, 621. [DOI] [PubMed] [Google Scholar]
- [74].Labunskyy VM, Gladyshev VN, Antioxid. Redox Signal. 2013, 19, 1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Murphy MP, Biochem. J. 2009, 417, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].B Poole L, Nelson KJ, Curr. Opin. Chem. Biol. 2008, 12, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Devarie-Baez NO, Silva Lopez EI, Furdui CM, Free Radic. Res. 2016, 50, 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Finkel T, FEBS Lett. 2000, 476, 52. [DOI] [PubMed] [Google Scholar]
- [79].Banerjee R, J. Biol. Chem. 2012, 287, 4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Fukuto JM, Carrington SJ, Tantillo DJ, Harrison JG, Ignarro LJ, Freeman BA, Chen A, Wink DA, Chem. Res. Toxicol. 2012, 25, 769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Abo M, Weerapana E, Antioxid. Redox Signal. In press. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Sies H, Redox Biol. 2015, 4, 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Lukyanov KA, Belousov VV, Biochim. Biophys. Acta. 2014, 1840, 745. [DOI] [PubMed] [Google Scholar]
- [84].Kloppel C, Michels C, Zimmer J, Herrmann JM, Riemer J, Biochem. Biophys. Res. Commun. 2010, 403, 114. [DOI] [PubMed] [Google Scholar]
- [85].Willems PH, Rossignol R, Dieteren CE, Murphy MP, Koopman WJ, Cell Metab. 2015, 22, 207. [DOI] [PubMed] [Google Scholar]
- [86].Fang X, Fu Y, Long MJ, Haegele JA, Ge EJ, Parvez S, Aye Y,J. Am. Chem. Soc. 2013, 135, 14496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Parvez S, Fu Y, Li J, Long MJ, Lin HY, Lee DK, Hu GS, Aye Y, J. Am. Chem. Soc. 2015, 137, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Lin HY, Haegele JA, Disare MT, Lin Q, Aye Y, J. Am. Chem. Soc. 2015, 137, 6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Long MJC, Poganik JR, Aye Y, J. Am. Chem. Soc. 2016, 138, 3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Parvez S, Long MJ, Lin HY, Zhao Y, Haegele JA, Pham VN, Lee DK, Aye Y, Nat. Protoc. 2016, 11, 2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Long MJ, Lin HY, Parvez S, Zhao Y, Poganik JR, Huang P, Aye Y, Cell Chem. Biol. 2017, 24, 944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Long MJC, Urul DA, Chawla S, Lin HY, Zhao Y, Haegele JA, Wang Y, Aye Y, Biochemistry 2017, 57, 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Surya SL, Long MJC, Urul DA, Zhao Y, Mercer EJ, IM EI, Evans T, Aye Y, ACS Chem. Biol. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Chen Y, Cong Y, Quan B, Lan T, Chu X, Ye Z, Hou X, Wang C, Redox Biol. 2017, 12, 712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].McMahon M, Lamont DJ, Beattie KA, Hayes JD, Proc. Natl. Acad. Sci. USA 2010, 107, 18838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Van Hall-Beauvais A, Zhao Y, Urul DA, Long MJC, Aye Y, Curr. Protoc. Chem. Biol. in press 2018, 10.1002/cpch.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Zhou BP, Hu MC-T, Miller SA, Yu Z, Xia W, Lin S-Y, Hung M-C, J. Biol. Chem. 2000, 275, 8027. [DOI] [PubMed] [Google Scholar]
- [98].Dudek H, Datta SR, Franke TF, Birnbaum MJ, Yao R, Cooper GM, Segal RA, Kaplan DR, Greenberg ME, Science 1997, 275, 661. [DOI] [PubMed] [Google Scholar]
- [99].Winterbourn CC, Hampton MB, Free Radic. Biol. Med. 2008, 45, 549. [DOI] [PubMed] [Google Scholar]
- [100].Woo HA, Yim SH, Shin DH, Kang D, Yu DY, Rhee SG, Cell 2010, 140, 517. [DOI] [PubMed] [Google Scholar]
- [101].Wood ZA, Poole LB, Karplus PA, Science 2003, 300, 650. [DOI] [PubMed] [Google Scholar]
- [102].Gacesa R, Dunlap WC, Barlow DJ, Laskowski RA, Long PF, Sci. Rep. 2016, 6, 27740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Speijer D, Biochem. J. 2016, 473, 4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Vander Jagt DL, Hunsaker LA, Vander Jagt TJ, Gomez MS, Gonzales DM, Deck LM, Royer RE, Biochem. Pharmacol. 1997, 53, 1133. [DOI] [PubMed] [Google Scholar]
- [105].Thiemann WH-P, Rosenbauer H, Meierhenrich UJ, Adv. Space Res. 2001, 27, 323. [DOI] [PubMed] [Google Scholar]
- [106].Dabrowski MJ, Zolnerciks JK, Balogh LM, Greene RJ, Kavanagh TJ, Atkins WM, Chem. Res. Toxicol. 2010, 23, 1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Wakita C, Maeshima T, Yamazaki A, Shibata T, Ito S, Akagawa M, Ojika M, Todoi J, Uchida K, J. Biol. Chem. 2009, 284, 28810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].West JD, Ji C, Duncan ST, Amarnath V, Schneider C, Rizzo CJ, Brash AR, Marnett LJ, Chem. Res. Toxicol. 2004, 17, 454. [DOI] [PubMed] [Google Scholar]
- [109].Hiratsuka A, Hirose K, Saito H, Watabe T, Biochem. J. 2000, 349, 729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Backus KM, Correia BE, Lum KM, Forli S, Horning BD, Gonzalez-Paez GE, Chatterjee S, Lanning BR, Teijaro JR, Olson AJ, Wolan DW, Cravatt BF, Nature 2016, 534, 570. [DOI] [PMC free article] [PubMed] [Google Scholar]


