Summary
Methanogenic archaea in the bovine rumen are responsible for the reduction of carbon molecules to methane, using various electron donors and driving the electron flow across the microbial food webs. Thus, methanogens play a key role in sustaining rumen metabolism and function. Research of rumen methanogenic archaea typically focuses on their composition and function in mature animals, while studies of early colonization and functional establishment remain scarce. Here, we investigated the metabolic potential and taxonomic composition of the methanogenic communities across different rumen developmental stages. We discovered that the methanogenesis process changes with age and that the early methanogenic community is characterized by a high activity of methylotrophic methanogenesis, likely performed by members of the order Methanosarcinales, exclusively found in young rumen. In contrast, higher hydrogenotrophic activity was observed in the mature rumen, where a higher proportion of exclusively hydrogenotrophic taxa are found. These findings suggest that environmental filtering acts on the archaeal communities and select for different methanogenic lineages during different growth stages, affecting the functionality of this ecosystem. This study provides a better understanding of the compositional and metabolic changes that occur in the rumen microbiome from its initial stages of colonization and throughout the animals' life.
Introduction
The rumen microbiome residing in the upper digestive tract of ruminant animals is essential for the viability of the host animal, as it is responsible for the fermentation of the ingested plant material and its transformation into molecules that are readily usable for the animals' energy requirements. In this sense, this host–microbiome relationship is unique (Hungate, 2013). The anaerobic metabolism necessary for the degradation and fermentation of plant material carried out by this complex microbial community is driven by an electron flow across a redox potential gradient which mostly ends with the reduction of carbon to methane (Thauer et al., 2008). The methanogenic archaea responsible for this process occupy approximately 2–5% of the mature rumen microbiome (Brulc et al., 2009), and are a driving force for the complex rumen microbial communities' metabolism, serving as an electron sink for the entire rumen microbial ecosystem (Sharp et al., 1998; Hook et al., 2010). Methanogens can utilize different substrates obtained from the fermentation process for methanogenesis, each characterized by distinct mechanisms (Rouviere and Wolfe, 1988; Garcia et al., 2000; Borrel et al., 2013). The most studied mechanism for methanogenesis in the rumen uses the hydrogenotrophic pathway, the most common pathway in the mature rumen, in which CO2 is reduced to methane using hydrogen. Methane can also be produced in the rumen through the methylotrophic pathway, using other fermentation products such as methylamines and methanol, as well as acetate which is used by acetoclastic methanogens to produce methane (Rouviere and Wolfe, 1988).
Methanogenesis plays a crucial role in terms of the animal's energy‐harvesting capability from feed and its effect on the environment. Methane cannot be absorbed by the host and is therefore emitted to the atmosphere along with its retained energy, resulting in substantial energy loss for the animal. It is also a potent greenhouse gas that contributes significantly to the greenhouse effect (Hook et al., 2010). A large body of research exists on the composition and functionality of these methanogenic archaeal communities in the mature rumen, where the microbiome is considered to be more stable in composition (Janssen and Kirs, 2008; Jeyanathan et al., 2011; Jami and Mizrahi, 2012; Jami et al., 2013; Morgavi et al., 2013). However, less emphasis has been given to the initial development and functionality of these communities in early stages of rumen colonization.
During early developmental stages, the functionality of the rumen differs greatly from that found in mature animals. At the early stage, animals are still feeding mostly on suckled milk which is digested directly in the true stomach, bypassing the rumen via the oesophageal groove (Van Soest, 1994). Hence, prior to full development of its function in plant fibre degradation and digestion, the rumen is exposed only to fractions of the ingested feed. Rumen colonization is characterized by rapid and dynamic changes in composition, beginning with early colonization by diverse aerobic and facultative anaerobic microorganisms, which are then gradually replaced by the obligate anaerobes responsible for the fermentation of feed products and proper physiological development (Fonty et al., 1987; Stewart et al., 1988; Li et al., 2012; Jami et al., 2013). Given the importance of the rumen methanogenic communities and the enormous significance of early gut microbial colonization on mammalian physiology throughout the individuals' lives (Bäckhed et al., 2015; Malmuthuge et al., 2015), it is essential to understand when these archaeal communities become established and active and whether compositional and functional changes can be observed as a function of development.
Early acquisition and establishment of methanogenic communities in ruminants has been reported to occur a few days after birth (Fonty et al., 1987; Minato et al., 1992; Morvan et al., 1994; Skillman et al., 2004). More recent studies have revealed that methanogenic cells can be found in the calf rumen as early as 20 min after birth (Guzman et al., 2015). However, questions remain as to their actual functionality in the first days of life, their viability and substrate requirements for methanogenesis during the initial stages of rumen colonization, when environmental conditions are vastly different from those of the mature rumen ecosystem. In the mature rumen environment, hydrogenotrophic methanogenesis is thought to be the dominant pathway for the production of methane (Fonty and Morvan, 1996; Janssen and Kirs, 2008) mainly due to the availability of CO2 and hydrogen. However, whether this holds true during the early stages of the rumen microbiome development, in which considerable differences in environmental conditions exist, remains to be investigated. Furthermore, as already noted, most studies focus on the taxonomic composition of the methanogenic population in the rumen of mature animals (Jarvis et al., 2000; Janssen and Kirs, 2008; Jeyanathan et al., 2011) and research into the methanogenic composition, dynamic changes and metabolic function in young calves, remains scarce.
The current study focuses on the metabolic nature of the methanogenic community during the early stages of rumen colonization and attempts to link these parameters to compositional changes in this community across different stages of development.
Results
The rumen methanogenic communities of newborn calves carry a metabolic potential facilitated by environmental factors
A methanogenic community in the rumen at early stages of bovine development has been reported to appear as early as 20 min after birth (Guzman et al., 2015). Nevertheless, questions remain as to whether during the early developmental stages the population is alive and has the potential for methanogenic activity. To answer this question, we performed a long‐term incubation and quantification experiment by sampling the rumen fluids of newborn calves at 2 and 3 days after birth. The rumen fluids with their resident microbiome were directly incubated for a period of up to 1 week, without the addition of growth factors or any other additives targeted at enriching these microbial communities. Rumen fluids from each calf were incubated under anaerobic conditions in five tubes. At each time point, microbial cells and DNA were extracted from one tube and methane gas was monitored (see Experimental procedures). This setup allowed us to avoid contamination related to handling and therefore any observation of methane could confidently be associated with the microbiome found at the time of sampling.
There was no methane production in the first 24 h of incubation. However, longer incubation times revealed methane production from the microbiome of the 2‐day‐old calf after 4 days of incubation, and of the 3‐day‐old calf after 2 days of incubation (Fig. 1). The increase in methane production was associated with an increase in abundance of methanogenic archaea, suggesting that the methanogenic community arising from these newborns has the potential for methanogenesis. Further, it suggested that archaeal development and potential for methanogenesis are partly independent of colonization or host influence subsequent to the days of sampling. In contrast, we observed that the methanogenic community of the 3‐day‐old calf expands much faster in terms of methane production and cell numbers than that of the 2‐day‐old calf.
Figure 1.
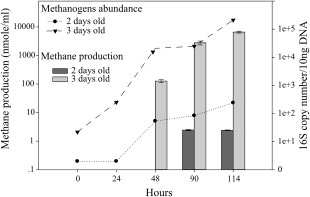
Methanogenic communities are metabolically active in the first days of life. Rumen fluids of 2‐day‐old and 3‐day‐old calves were sampled and monitored each day for methane production by GC. In addition, genomic DNA was extracted from these samples and archaeal 16S rRNA copy number was quantified. The dark grey bars represent the methane concentration measured for the 2‐day‐old calf and the light grey bars represent the methane concentration measured for the 3‐day‐old calf. The triangle and circle symbols represent the archaeal 16S rRNA copy number for the 2‐day‐old and 3‐day‐old calves, respectively. For each timepoint, at least 3 independent samples were analyzed for methane production and 3 technical repeats were subjected to qRT‐PCR analysis. Mean for each timepoint is plotted ± SEM.
Hydrogen concentration decreases rapidly with age
We further asked whether methanogenesis is driven by similar factors in newborn calves and mature animals. As methanogens in the rumen of mature animals are mostly considered to be hydrogenotrophic (Hungate, 1967), we assessed the presence and potential changes in hydrogen concentration during several stages of development. Rumen fluids sampled from newborn calves during the first 2 weeks of life and from mature animals were incubated in vitro and hydrogen concentration was quantified after 24 h (Supporting Information Table S1). We observed a high concentration of hydrogen in the ruminal fluids of calves 1 day after birth, that gradually decreased to levels similar to those of mature animals after 2 weeks (Fig. 2). This suggested that consumption of the produced hydrogen is already occurring during the primary stages of development. However, this consumption is not necessarily linked to methanogenesis, as a previous study revealed the existence of a large population of hydrogenotrophic bacteria such as acetogens and sulfate‐reducing bacteria, which may compete for the hydrogen present, during early colonization (Morvan et al., 1994; Fonty et al., 2007).
Figure 2.
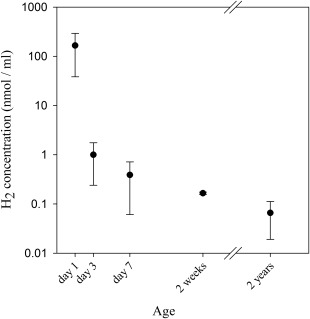
Hydrogen concentration decreases gradually with age. Rumen fluids of young calves and mature animals were sampled and anaerobically incubated for 24 h and hydrogen was measured by GC. At least 2 independent samples were analyzed for each timepoint. Mean for each timepoint is plotted ± SEM.
Different response to methanogenesis precursors between the young and mature rumen communities
Although the hydrogenotrophic methanogenesis is thought to be the most prominent pathway in mature animals, substrates other than hydrogen and CO2 are known to be used by various species of methanogens via the acetoclastic and methylotrophic pathways (Rouviere and Wolfe, 1988; Garcia et al., 2000). However, information regarding the type and prevalence of these pathways across different development stages of the rumen is still lacking.
We therefore assessed whether the methanogenic communities functionally differ between young and mature animals, in terms of the prevalence of the pathways used for methanogenesis. We subjected freshly extracted rumen fluids from 2‐year‐old heifers and 2‐week‐old calves to the addition of different substrates known to be essential precursors for the various methanogenesis pathways. Rumen fluids from 16 animals (eight per group) were incubated under a saturating amount of hydrogen and 8 of these samples (four per group) were also separately incubated with the addition of acetate, methanol and methylamines (composed of an equimolar combination of mono‐methylamine, di‐methylamine and tri‐methylamine). The effect of the different substrates on methane production was then measured after 24 h of incubation by gas chromatography (GC) and was compared with their respective control (incubation without the addition of substrate), for intrinsic methane production. For this analysis, 2‐week‐old calves were selected, following our observation that methane production after 24 h of incubation can only be detected in calves older than 1 week, making them an appropriate group for comparative analysis of the methanogenic activity between young and mature animals.
The addition of hydrogen to rumen samples taken from 2‐year‐old heifers, resulted in a significantly higher fold increase in methane production compared to those taken from young calves. A ∼6‐fold increase was observed in 2‐year‐old heifers compared to ∼1.5‐fold increase in 2‐week‐old calves (t‐test p < 0.01; Fig. 3). This suggested a higher prevalence of hydrogenotrophic pathways for methane production in this group. In contrast, incubation of samples taken from young calves with methylamines, exhibited a significantly higher fold increase in methane production compared to mature animals. A ∼6‐fold increase was observed for young calves, compared to a ∼2.7 fold for 2 years old heifers (t‐test p < 0.05; Fig. 3). Similar results were observed when methanol was added, with a 4.5‐fold increase in methane production in samples from young calves compared to the 2.6‐fold increase in mature animals (t‐test p <0.01). Addition of acetate did not result in a significant increase in methane production in either group. These differences in substrate utilization profiles suggest that the methanogenic community in the early developing stages of the rumen relies more on substrates, other than hydrogen for methanogenesis compared to the mature microbiome.
Figure 3.
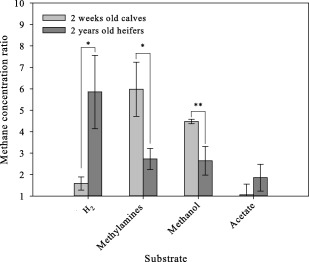
Methane production is affected by different methanogenesis precursors in young and mature animals. Rumen fluids from each animal were anaerobically incubated for 24 h with the addition of either 80%/20% H2/CO2 at 202 kPa pressure, acetate (20mM), methanol (30 mM), methylamines mix (30 mM; 10 mM for each) or with 80%/20% N2/CO2 at 202 kPa pressure as a control, and methane concentration was measured by GC. Each bar represents the average fold change in methane concentration observed when samples were incubated with the different substrates, compared to their respective control for intrinsic methane production, for each group (light grey bars represent 2‐weeks‐old calves and dark grey bars represent 2‐years‐old heifers). Significance was calculated using Student t‐test for the differences in response to substrates between 2‐weeks‐old calves and 2‐years‐old heifers (* denotes p < 0.05, ** denotes p < 0.01).
Dynamic changes in methanogenic population and abundance across different developmental stages
We subsequently assessed whether the different substrate utilization patterns observed can be linked to compositional changes in the rumen methanogenic population across different stages of development. Therefore, we quantified the abundance of the overall methanogenic community and the relative abundance of the four main archaeal orders known to reside in the rumen; Methanobacteriales, Methanosarcinales, Methanomicrobiales and Methanoplasmatales (Janssen and Kirs, 2008; Paul et al., 2012) (Fig. 4). Overall, these included samples from, 1‐day‐old calves (n = 5) and 3‐day‐old calves (n = 6) representing the first steps of colonization in the rumen, 2‐week‐old calves (n = 6), 2‐month‐old calves (n = 5), 6‐month‐old (n = 5) and 2‐year‐old (n = 8). For details of each group's diet and animals used, see Experimental procedures and Supporting Information (Supporting Information Tables S1 and S2). Quantification of the overall methanogenic communities showed the presence of methanogens one day after birth and a rapid increase in their abundance with age, reaching levels similar to those observed in mature animals after 2 weeks (Fig. 4A). All four orders tested could be observed in the samples from newborn calves, whereas only two orders could be detected in the mature rumen (Fig. 4B). The two orders that could only be detected in young calves were Methanosarcinales and Methanomicrobiales. The Methanosarcinales, considered to be the most metabolically diverse order in terms of its versatility of methanogenesis pathway and substrate utilization (Balch et al., 1979; Lambie et al., 2015), could only be detected in the rumen of young calves, and was proportionally the second most abundant order in the first two weeks. The Methanobacteriales and the recently‐characterized Methanoplasmatales were the only orders detected in older samples, increasing with age until, at the age of 2 months, reaching levels similar to those observed in 2‐year‐old heifers. The most dominant order found in all samples and across all age groups, was the Methanobacteriales accounting for more than 70% of the total archaeal population (Fig. 4B). Hence, we investigated whether the observed compositional changes across age also exist at the genus level within this order, and quantified its three commonly found genera in the rumen: Methanobrevibacter, Methanobacterium and Methanosphaera, using quantitative RT‐PCR analysis (Jarvis et al., 2000; Skillman et al., 2004). Methanosphaera, which was the least abundant genus of the three quantified, was detected only in trace amounts in samples from 1 to 3‐day‐old calves, peaked in abundance in samples from the 2‐month‐old animals with an average of 3.3% of the total Methanobacteriales, then steadily decreased in older animals (Fig. 5). Both the Methanobrevibacter and Methanobacterium could be found in all age groups. However, their ratio changed across age groups, with Methanobacterium being the dominant genus in 1–3‐day‐old and 2‐month‐old calves, whereas in the older animals, Methanobrevibacter became the most abundant of the tested genera (Fig. 5).
Figure 4.
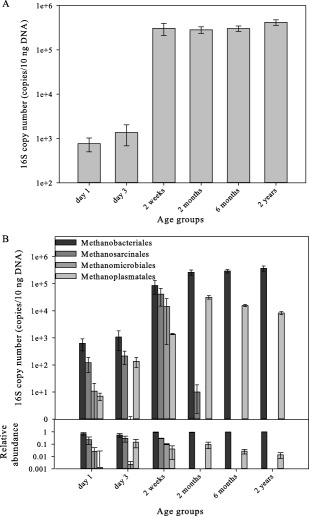
Methanogenic community abundance and composition changes with age. Quantification of (A) total methanogenic communities and (B) the four major methanogenic orders: Methanobacteriales, Methanomicrobiales, Methanosarcinales and Methanoplasmatales by qRT‐PCR. Each timepoint represents the average of at least four animals. Mean for each timepoint is plotted ± SEM.
Figure 5.
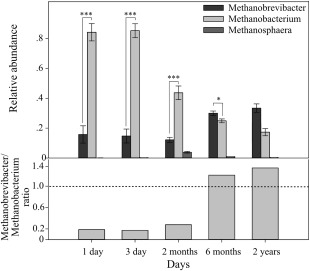
Quantification of Methanobrevibacter, Methanobacterium and Methanosphaera (order Methanobacteriales) and age‐dependent interplay between the two main genera. The relative abundance of 16S rRNA copy number of each genus out of the total copy number observed for the order Methanobacteriales, is shown in the upper panel. The ratio between the Methanobacteriales genera Methanobrevibacter and Methanobacterium is shown in the lower panel. Mean for each timepoint is plotted ± SEM. *p < 0.05, ***p < 0.001 (Student's t‐test).
Discussion
We explored the viability and metabolic activity of rumen methanogenic communities in early stages of the host animal's life compared to mature stages, as well as their compositional changes. We show that the methanogenic communities in newborn calves are capable of metabolic functions, as revealed by their ability to produce methane as early as 2 days after birth. Therefore, these communities are composed of live cells that can actively take part in the colonization and establishment of the developing rumen environment (Fig. 1). This observation is in accordance with our previous finding that microorganisms typically residing in the mature rumen are present at early stages of development (Jami et al., 2013). Similar conclusions were reached in previous studies in newborn lambs, where methane production could also be observed in the rumen sample of 2‐days‐old lamb (Fonty et al., 1987; Morvan et al., 1994). These processes of colonization and enrichment of microbial components may be exclusively dependent upon the environmental conditions created by the consecutively residing microbiome, in order to select toward the mature phenotype. Results obtained using the in vitro setup in the present study also suggested that at least some of these processes are independent of further colonization or communication with the host. It would be interesting to investigate the extent to which this is true by assessing the ability of the primary microbial community to be directed towards its mature phenotype both taxonomically and functionally, independent of further exposure to host‐mediated factors or further acquisition. The more extensive development of the methanogenic community and methane emission found on day 3, suggested that more favourable environmental conditions exist in those samples for the development of this community, whereas the environmental conditions on day 2 limit the methanogenic population's ability to thrive. However, we cannot completely rule out the existence of a more abundant – but still below the qPCR‐detection threshold – methanogenic population in the rumen of 3‐day‐old calves resulting in more rapid expansion of the community.
Quantification of hydrogen concentrations in young and mature animals revealed a higher concentration of hydrogen in newborn compared to mature animals, which decreased to a level similar to that of the mature animals over the course of the first 2 weeks. This kinetic could result from accumulation of hydrogen on the first day of life due to the lack of an established potent hydrogenotrophic microbial community which, when established, decreases its partial pressure in the rumen.
Our results showed that the hydrogen consumed during early stages of development may not be the most prevalent substrate for methanogenesis. This is represented by the different response in methane production between 2 week‐old calves and 2 year‐old heifers when their respective rumen fluids were incubated with substrates known to be the main precursors for different methanogenesis pathways. We observed that methanol and methylamine elicited a higher response in young calves compared to mature animals, while incubation with H2 resulted in a higher methane production response in mature animals. These different responses to methanogenesis precursors suggest that (i) during the early stages of life, hydrogen is not the main substrate relied upon for methanogenesis in young calves; other products, specifically methylamine and methanol may have a relatively more prominent role at this stage and might serve as alternative substrates for the early methanogenic population; (ii) hydrogen utilization at this stage may not be dominantly governed by methanogens but by other taxonomic groups for their metabolism, explaining the observed decrease in hydrogen. Regarding the latter, several studies have shown that the microbiome of newborn animals is populated by a vast community of hydrogenotrophic bacteria such as acetogens, sulfate‐reducing bacteria and others (Fonty et al., 1987; Morvan et al., 1994; Fonty et al., 2007). These bacteria may therefore make up a large proportion of the hydrogen consumers at this stage of life. Indeed, when we analyzed the bacterial 16S rRNA sequence composition in our previous study, characterizing the composition of the bacterial community across developmental stages, we could identify 26 different operational taxonomic units (OTUs) that were found exclusively on the first days of life, and were completely absent from other age groups. These included 16 OTUs taxonomically associated with species known to be hydrogen‐utilizers, such as acetogens and sulfate‐, nitrate‐ and fumarate‐reducing bacteria (Supporting Information Table S3).
The observation that the early methanogenic population relies on substrates other than hydrogen to support their growth is also reflected in the observed significant difference in the methanogenic community composition between young and mature animals (Fig. 4). Methanogenic taxa found exclusively in the rumen of newborn and developing calves such as the order Methanosarcinales (Fig. 4B), have been shown to grow on a broad range of substrates other than H2 and CO2, such as methanol, methylamines and acetate (Balch et al., 1979; Patterson and Hespell, 1979; Penger et al., 2012; Lambie et al., 2015), providing alternative strategies for methane production which allow them to thrive during these primary developmental stages (Jarvis et al., 2000). Conversely, in the mature animals, the overwhelming dominance of the order Methanobacteriales (>95% in 2‐years‐old heifers; Fig. 4B), which is known to predominantly produce methane via the hydrogenotrophic pathway, could explain the significantly higher increase in methane production when hydrogen was added. In addition to the Methanobacteriales, the recently defined order Methanoplasmatales (Paul et al., 2012), was also observed in the rumen of mature animals and is known to produce methane via the methylotrophic pathway and therefore may be responsible for the significant, albeit lower, response to methanol and methylamine in mature animals. However, the 2.6‐fold increase in methane observed when methanol or methylamine was added might indicate a significant contribution of this order to the production of methane, despite its low relative abundance in the samples. Our study strengthens the previously proposed notion, that Methanoplasmatales may play an important role in methanogenesis in the rumen and that its members are prime targets for potential methane mitigation (Poulsen et al., 2013). Our findings show that different development stages may be characterized by different substrate reliance and bottlenecks for methane production; while hydrogen may be the bottleneck substrate in mature animals, other substrates may have a greater influence on methanogenesis in newborn animals. Much like the Methanosarcinales, the high abundance of Methanobacterium and Methanosphaera from the order Methanobacteriales in the early stages of life (1 and 3 days and 2 months), may be linked to more favourable rumen conditions in young calves related to the availability of specific substrates. This may explain their dominance over Methanobrevibacter species during early colonization and the later switch in dominance at later growth stages. The assumption that compositional changes at the order and genus levels in the methanogenic population could be governed by different ecological niches, was also demonstrated in our previous work where environmental parameters such as redox potential were shown to be an important factor which affects various methanogenic groups (Friedman et al., 2016).
This study provides evidence of methanogenic activity in the early days of rumen development and reveals that different metabolic pathways for methanogenesis come into play in young vs. mature animals. This is further supported by the shift in composition across different developmental stages which may shed light on the forces driving these changes in the rumen. Our findings may prove to be seminal for the understanding of microbial colonization and in future potential applications for methane‐mitigation strategies toward improving many aspects of agriculture and its impact on the environment.
Experimental procedures
Animal handling and rumen sampling
All of the experimental procedures described in this study were approved by the Faculty Animal Policy and Welfare Committee of the Agricultural Research Organization (ARO), approval numbers IL‐326/11 and IL‐691/16, and were in accordance with the guidelines of the Israel Council for Animal Care. Holstein Friesian cows were housed at the ARO's experimental dairy farm in Beit Dagan, Israel. All animals used in this experiment were housed and fed according to conventional practice at the ARO. Newborns were separated from their mothers immediately after birth and raised in individual pens until weaning, at 2 months of age. During the first 3 days after birth, the calves were fed exclusively colostrum. From day 4 until 2 month of age (included), calves were fed a combination of milk and solid starter diet. All subsequent age groups used for this experiment were provided a diet ad libitum consisting of 70% concentrated food, mineral and vitamin mix and 30% roughage. The animals sampled, their age at the time of sampling and the experiments for which they were used is summarized in Supporting Information Table S1.
One hour after the morning feeding, rumen fluids were extracted via the mouth using a stomach tube attached to a vacuum pump. Samples were transferred to CO2‐containing bottles to maintain anaerobic conditions. An aliquot of the samples was stored after collection at −20°C for subsequent DNA extraction, and the rest was immediately transferred to an anaerobic chamber (Coy Laboratories Inc., Grace Lake, MI, USA) to maintain anaerobic conditions for further incubation assays.
In vitro incubation assays and GC measurements
Duplicates of 10‐ml aliquots from each animal were poured into sterile anaerobic‐designated tubes (Hungate Anaerobic Culture Tube, BeLLCo Glass, and VWR). The samples were incubated at 39°C without shaking in the dark for 24 h, with the addition of the following substrates; 80%/20% H2/CO2 at 202 kPa pressure, 20 mM of acetate, 30 mM of methanol, 30 mM of a combination of methylamines (equimolar solution of mono‐methylamine, di‐methylamine, tri‐methylamine; 10 mM each) and a control for intrinsic CH4 production with 80%/20% N2/CO2. The concentrations used for the incubations were taken from similar experiments performed for the assessment of methanogenic taxa substrate requirements in pure cultures (Tanner and Wolfe, 1988; Borrel et al., 2011). After 24 h, headspace gas was analyzed by sampling and injecting aliquots of 0.1 ml into a GC system (HP‐5890 series II) equipped with a flame ionization detector for methane measurements and a thermal conductivity detector for hydrogen measurements. Calibration for the methane measurements was performed using pure 100% methane gas and for the hydrogen measurements, 80%/20% H2/CO2 gas. Calibration samples were manually injected into the GC using a gas‐tight syringe and standard curves were obtained from five different volumes (50 µl, 100 µl, 200 µl, 300 µl and 400 µl) which were measured in triplicate. Each point on the standard curve was calculated as the average of those triplicates.
The aliquots were injected into a 45/60 Mol Sieve 5A column (Supelco Analytical), with helium carrier gas set to a flow rate of 10 ml/min and initial oven temperature of 200°C. The temperatures at the inlet and the detector were set at 200°C and 250°C, respectively.
Isolation of microbial fraction and DNA extraction from rumen samples
Treatment of rumen fluid was performed as described in (Stevenson and Weimer, 2007). Rumen fluids were centrifuged at 10 000g for 10 min, and the pellet was suspended in extraction buffer (100 mM Tris‐HCl, 10 mM EDTA, 150 mM NaCl pH 8.0, 0.15% v/v Tween‐80) and incubated for 1 h at 4°C, to detach particle‐associated microorganisms from the rumen content. Following slow centrifugation (500g) for 15 min at 4°C, the microbiota‐containing supernatant was filtered through eight layers of cheesecloth, centrifuged at 6000g for 10 min and resuspended in extraction buffer. The pellets were kept at −20°C until DNA extraction, which was performed as described by (Yu and Morrison, 2004) with a few modifications. Briefly, the microbial cells were lysed using bead disruption and lysis buffer (500 mM NaCl, 50 mM Tris‐HCl pH 8.0, 50 mM EDTA, 4% v/v sodium dodecyl sulfate). The final supernatant was precipitated using ammonium acetate and isopropanol. The precipitate was then dissolved in TE buffer (10 mM Tris‐HCl pH 7.5, 1 mM EDTA pH 8.0), checked for DNA concentration, diluted to 10 ng/µl and stored at −20°C.
Quantitative real‐time PCR (qRT‐PCR)
A qRT‐PCR analysis was performed to investigate the relative abundance of specific methanogen orders through amplification of their 16S rRNA gene. A standard curve was generated for the methanogen orders, genera and total archaea by amplifying serial 10‐fold dilutions of gel‐extracted PCR products obtained by the amplification of each amplicon. The standard curves consisted of four‐to‐six dilution points, and were calculated using Rotorgene 6000 series software (Qiagen). Subsequent quantifications were calculated with the same program using the standard curve generated in each run, and one known purified product diluted for the standard curves was added to each quantification reaction to assess its reproducibility. All obtained standard curves met the required standards of efficiency (R 2 > 0.99, E > 90%). Reactions were performed in a 10‐µl reaction mixture containing 5 µl of 2X Absolute Blue SYBR Green Master Mix (Thermo Scientific), 0.5 µl of each primer (500 nM final concentration), 2 µl double‐distilled water and 2 µl of 10 ng/µl DNA template. Amplification consisted of 1 cycle at 95°C for 5 min for initial denaturation and activation of the polymerase system, then 45 cycles at 95°C for 10 s followed by annealing for 20 s at 60°C for total Archaea, Methanobacteriales and Methanoplasmatales (RCC clade) primers, 62°C for Methanosarcinales primers, 63°C for Methanomicrobiales primers, 57°C for Methanobacterium, Methanobrevibacter and Methanosphaera primers; then extension was performed at 72°C for 20 s for Archaea, Methanobacteriales, Methanosarcinales, Methanomicrobiales and Methanoplasmatales primers and for 45 s for Methanobacterium, Methanobrevibacter and Methanosphaera primers. Primer sequences were obtained from previous studies (Wolin and Miller, 1997; Jarvis et al., 2000; Whitford et al., 2001; Skillman et al., 2004; Skillman et al., 2006; Zhou and Hernandez‐Sanabria, 2009).
Sequencing data and analyses
The sequencing data used for the analysis of the bacterial community across different age groups were taken from Jami et al. (2013). Sequencing data and information were deposited in the MG‐RAST server under IDs 4514864.3 to 4514868.3.
Statistical analysis
In general, significance was calculated with the Student's t‐test. In all analyses, significance was set at p < 0.05 unless otherwise stated.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's web‐site:
Table S1. List of the animals sampled, the age of each animal at the time of sampling and the experiment performed.
Table S2. Formulated ingredients (g/kg dry matter) of the respective diets for each age group.
Table S3. Hydrogen‐utilizing bacterial species present exclusively in newborn calves (1 and 3 days old) and absent in older animals according to 16S rRNA pyrosequencing analysis.
Acknowledgements
The research described here was supported by grants from the Israel Science Foundation (No. 1313/13), by the European Research Council under the European Union's Horizon 2020 research and innovation program (grant agreement No 640384), Israel Dairy Board Foundation Projects No. 362‐0300 and 362‐0524 and by ICA grant 713 02‐15‐08a.
The copyright line for this article was changed on 16 April 2019 after original online publication
References
- Bäckhed, F. , Roswall, J. , Peng, Y. , Feng, Q. , Jia, H. , Kovatcheva‐Datchary, P. , et al (2015) Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 17: 690–703. [DOI] [PubMed] [Google Scholar]
- Balch, W. , Fox, G. , Magrum, L. , Woese, C. , and Wolfe, R. (1979) Methanogens: reevaluation of a unique biological group. Microbiol Rev 43: 260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrel, G. , O'Toole, P.W. , Harris, H.M. , Peyret, P. , Brugère, J.‐F. , and Gribaldo, S. (2013) Phylogenomic data support a seventh order of methylotrophic methanogens and provide insights into the evolution of methanogenesis. Genome Biol Evol 5: 1769–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrel, G. , Joblin, K. , Guedon, A. , Colombet, J. , Tardy, V. , Lehours, A.‐C. , and Fonty, G. (2011) Methanobacterium lacus sp. nov., a novel hydrogenotrophic methanogen from the deep cold sediment of a meromictic lake. Int J System Evol Microbiol 62: 1625–1629. [DOI] [PubMed] [Google Scholar]
- Brulc, J.M. , Antonopoulos, D.A. , Miller, M.E. , Wilson, M.K. , Yannarell, A.C. , Dinsdale, E.A. , et al (2009) Gene‐centric metagenomics of the fiber‐adherent bovine rumen microbiome reveals forage specific glycoside hydrolases. Proc Natl Acad Sci U S A 106: 1948–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonty, G. , and Morvan, B. (1996) Ruminal methanogenesis and its alternatives. Ann Zootech 45: 313–318. [Google Scholar]
- Fonty, G. , Gouet, P. , Jouany, J.‐P. , and Senaud, J. (1987) Establishment of the microflora and anaerobic fungi in the rumen of lambs. J Gen Microbiol 133: 1835–1843. [Google Scholar]
- Fonty, G. , Joblin, K. , Chavarot, M. , Roux, R. , Naylor, G. , and Michallon, F. (2007) Establishment and development of ruminal hydrogenotrophs in methanogen‐free lambs. Appl Environ Microbiol 73: 6391–6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman, N. , Shriker, E. , Gold, B. , Durman, T. , Zarecki, R. , Ruppin, E. , and Mizrahi, I. (2016) Diet‐induced changes of redox potential underlie compositional shifts in the rumen archaeal community. Environ Microbiol 19: 174–184. [DOI] [PubMed] [Google Scholar]
- Garcia, J.‐L. , Patel, B.K. , and Ollivier, B. (2000) Taxonomic, phylogenetic, and ecological diversity of methanogenic Archaea. Anaerobe 6: 205–226. [DOI] [PubMed] [Google Scholar]
- Guzman, C.E. , Bereza‐Malcolm, L.T. , De Groef, B. , and Franks, A.E. (2015) Presence of selected methanogens, fibrolytic bacteria, and proteobacteria in the gastrointestinal tract of neonatal dairy calves from birth to 72 hours. PLoS One 10: e0133048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hook, S.E. , Wright, A.D. , and McBride, B.W. (2010) Methanogens: methane producers of the rumen and mitigation strategies. Archaea 2010: 945785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hungate, R. (1967) Hydrogen as an intermediate in the rumen fermentation. Arch Microbiol 59: 158–164. [DOI] [PubMed] [Google Scholar]
- Hungate, R.E. (2013) The Rumen and Its Microbes. New York, NY: Elsevier Academic Press. [Google Scholar]
- Jami, E. , and Mizrahi, I. (2012) Composition and similarity of bovine rumen microbiota across individual animals. PLoS One 7: e33306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jami, E. , Israel, A. , Kotser, A. , and Mizrahi, I. (2013) Exploring the bovine rumen bacterial community from birth to adulthood. ISME J 7: 1069–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen, P.H. , and Kirs, M. (2008) Structure of the archaeal community of the rumen. Appl Environ Microbiol 74: 3619–3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis, G.N. , Strömpl, C. , Burgess, D.M. , Skillman, L.C. , Moore, E.R. , and Joblin, K.N. (2000) Isolation and identification of ruminal methanogens from grazing cattle. Curr Microbiol 40: 327–332. [DOI] [PubMed] [Google Scholar]
- Jeyanathan, J. , Kirs, M. , Ronimus, R.S. , Hoskin, S.O. , and Janssen, P.H. (2011) Methanogen community structure in the rumens of farmed sheep, cattle and red deer fed different diets. FEMS Microbiol Ecol 76: 311–326. [DOI] [PubMed] [Google Scholar]
- Lambie, S.C. , Kelly, W.J. , Leahy, S.C. , Li, D. , Reilly, K. , McAllister, T.A. , et al (2015) The complete genome sequence of the rumen methanogen Methanosarcina barkeri CM1. Stand Genomic Sci 10: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, R.W. , Connor, E.E. , Li, C. , Baldwin, V. , R.L., and Sparks, M.E. (2012) Characterization of the rumen microbiota of pre‐ruminant calves using metagenomic tools. Environ Microbiol 14: 129–139. [DOI] [PubMed] [Google Scholar]
- Malmuthuge, N. , Griebel, P.J. , and Guan, L.L. (2015) The Gut Microbiome and its potential role in the development and function of newborn calf gastrointestinal tract. Front Vet Sci 2: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minato, H. , Otsuka, M. , Shirasaka, S. , Itabashi, H. , and Mitsumori, M. (1992) Colonization of microorganisms in the rumen of young calves. J Gen Appl Microbiol 38: 447–456. [Google Scholar]
- Morgavi, D.P. , Kelly, W. , Janssen, P. , and Attwood, G. (2013) Rumen microbial (meta) genomics and its application to ruminant production. Animal 7: 184–201. [DOI] [PubMed] [Google Scholar]
- Morvan, B. , Dore, J. , Rieu‐Lesme, F. , Foucat, L. , Fonty, G. , and Gouet, P. (1994) Establishment of hydrogen‐utilizing bacteria in the rumen of the newborn lamb. FEMS Microbiol Lett 117: 249–256. [DOI] [PubMed] [Google Scholar]
- Patterson, J.A. , and Hespell, R.B. (1979) Trimethylamine and methylamine as growth substrates for rumen bacteria and Methanosarcina barkeri . Curr Microbiol 3: 79–83. [Google Scholar]
- Paul, K. , Nonoh, J.O. , Mikulski, L. , and Brune, A. (2012) “Methanoplasmatales,” Thermoplasmatales‐related archaea in termite guts and other environments, are the seventh order of methanogens. Appl Environ Microbiol 78: 8245–8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penger, J. , Conrad, R. , and Blaser, M. (2012) Stable carbon isotope fractionation by methylotrophic methanogenic archaea. Appl Environ Microbiol 78: 7596–7602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen, M. , Schwab, C. , Jensen, B.B. , Engberg, R.M. , Spang, A. , Canibe, N. , et al (2013) Methylotrophic methanogenic Thermoplasmata implicated in reduced methane emissions from bovine rumen. Nat Commun 4: 1428. [DOI] [PubMed] [Google Scholar]
- Rouviere, P. , and Wolfe, R. (1988) Novel biochemistry of methanogenesis. J Biol Chem 263: 7913–7916. [PubMed] [Google Scholar]
- Sharp, R. , Ziemer, C.J. , Stern, M.D. , and Stahl, D.A. (1998) Taxon‐specific associations between protozoal and methanogen populations in the rumen and a model rumen system. FEMS Microbiol Ecol 26: 71–78. [Google Scholar]
- Skillman, L. , Evans, P. , Strömpl, C. , and Joblin, K. (2006) 16S rDNA directed PCR primers and detection of methanogens in the bovine rumen. Lett Appl Microbiol 42: 222–228. [DOI] [PubMed] [Google Scholar]
- Skillman, L.C. , Evans, P.N. , Naylor, G.E. , Morvan, B. , Jarvis, G.N. , and Joblin, K.N. (2004) 16S ribosomal DNA‐directed PCR primers for ruminal methanogens and identification of methanogens colonising young lambs. Anaerobe 10: 277–285. [DOI] [PubMed] [Google Scholar]
- Stevenson, D.M. , and Weimer, P.J. (2007) Dominance of Prevotella and low abundance of classical ruminal bacterial species in the bovine rumen revealed by relative quantification real‐time PCR. Appl Microbiol Biotechnol 75: 165–174. [DOI] [PubMed] [Google Scholar]
- Stewart, C. , Fonty, G. , and Gouet, P. (1988) The establishment of rumen microbial communities. Animal Feed Sci Technol 21: 69–97. [Google Scholar]
- Tanner, R.S. , and Wolfe, R.S. (1988) Nutritional requirements of Methanomicrobium mobile. Appl Environ Microbiol 54: 625–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thauer, R.K. , Kaster, A.‐K. , Seedorf, H. , Buckel, W. , and Hedderich, R. (2008) Methanogenic archaea: ecologically relevant differences in energy conservation. Nat Rev Microbiol 6: 579–591. [DOI] [PubMed] [Google Scholar]
- Van Soest, P.J. (1994) Nutritional Ecology of the Ruminant. New York, USA: Cornell University Press. [Google Scholar]
- Whitford, M.F. , Teather, R.M. , and Forster, R.J. (2001) Phylogenetic analysis of methanogens from the bovine rumen. BMC Microbiol 1: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolin, M. , and Miller, T. (1997) Microbe–microbe interactions In The Rumen Microbial Ecosystem. Hobson P.N., and Stewart C.S. (eds.). London: Chapman & Hall, pp. 467–491. [Google Scholar]
- Yu, Z. , and Morrison, M. (2004) Improved extraction of PCR‐quality community DNA from digesta and fecal samples. Biotechniques 36: 808–813. [DOI] [PubMed] [Google Scholar]
- Zhou, M. , and Hernandez‐Sanabria, E. (2009) Assessment of the microbial ecology of ruminal methanogens in cattle with different feed efficiencies. Appl Environ Microbiol 75: 6524–6533. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's web‐site:
Table S1. List of the animals sampled, the age of each animal at the time of sampling and the experiment performed.
Table S2. Formulated ingredients (g/kg dry matter) of the respective diets for each age group.
Table S3. Hydrogen‐utilizing bacterial species present exclusively in newborn calves (1 and 3 days old) and absent in older animals according to 16S rRNA pyrosequencing analysis.


