Abstract
Kidney organoids derived from human pluripotent stem cells exhibit glomerular- and tubular-like compartments that are largely avascular and immature in static culture. Here, we report an in vitro method for culturing kidney organoids under flow on millifluidic chips, which greatly expands their endogenous pool of endothelial progenitor cells (EPCs) and generates vascular networks with perfusable lumens surrounded by mural cells. Vascularized kidney organoids cultured under flow exhibit more mature podocyte and tubular compartments with enhanced cellular polarity and adult gene expression, compared to static controls. However, the association of vessels with these compartments is reduced upon disrupting the endogenous VEGF gradient. Glomerular vascular development progresses through intermediate stages akin to the embryonic mammalian kidney’s formation of capillary loops abutting foot processes. The ability to induce substantial vascularization and morphological maturation of kidney organoids in vitro under flow opens new avenues for studying kidney development, disease, and regeneration.
Introduction
The kidney continuously filters blood and maintains fluid homeostasis; functions that rely on specialized glomerular and tubular tissue compartments integrated with a complex vascular network. While kidney organoids exhibit such compartments 1–8, their vascular development, e.g. the formation of PECAM1+ networks with luminal architecture, is limited in static culture 3,9,10. Further, gene expression in podocytes and tubular epithelial cells in static organoids is reflective of less mature renal tissue compared to human adult kidneys in published work3,9. To date, researchers have depended upon animal transplantation to produce kidney organoids with a perfusable vasculature that facilitates nephron epithelial maturation10,11,12. However, the reliance on an animal host limits both the scalability and translation of organoid-based approaches, particularly for in vitro applications.
Given that multilineage communication with vasculature is implicated in epithelial maturation in vivo,13 we posit that enhanced vascularization and maturation may be promoted in hPSC-derived human kidney tissue in vitro when subject to environmental cues. To test our hypothesis, we developed a simple millifluidic culture system to probe the effects of extracellular matrix (ECM), media composition, fluidic shear stress (FSS), and co-culture with human endothelial cells on the in vitro development of kidney organoids.
Results
Developing kidney organoids exhibit enhanced vascularization under flow
Within our 3D printed millifluidic chips, organoids are subjected to superfusion (flow over their top surface) with a controlled wall shear, i.e., fluidic shear stress (FSS) (Fig. 1a top, Supplementary Fig. 1a-g). The developing kidney organoids adhere to and become partially embedded in a ~1 mm thick layer of gelatin-fibrin (gelbrin) ECM that coats the bottom of the printed chip14,15, permitting fluid to freely flow through the gap (2.6 mm in height) above the organoid/ECM surface (Supplementary Fig. 2a-f). Interestingly, the adherent gelbrin matrix leads to enhanced peripheral expression of vascular markers PECAM1 and its precursor, MCAM16 within 1 week in static conditions, compared to non-adherent matrices (e.g., glass, plastic, fibrin ± collagen type 1) (Fig. 1b). We tested several media compositions as well as co-culture with primary human endothelia ± fibroblasts; however, most inhibited nephron formation or failed to enhance vascularization under fluid flow (Supplementary Fig. 2g-i, 3a,b). We observed that a low FBS concentration of 1.5%, typically used in endothelial culture media, permits nephrogenesis and enhances vascular network formation in developing kidney organoids under static conditions (Supplementary Fig. 2h,i).
Figure 1. Developing kidney organoids cultured in vitro under high fluid flow exhibit enhanced vascularization during nephrogenesis.
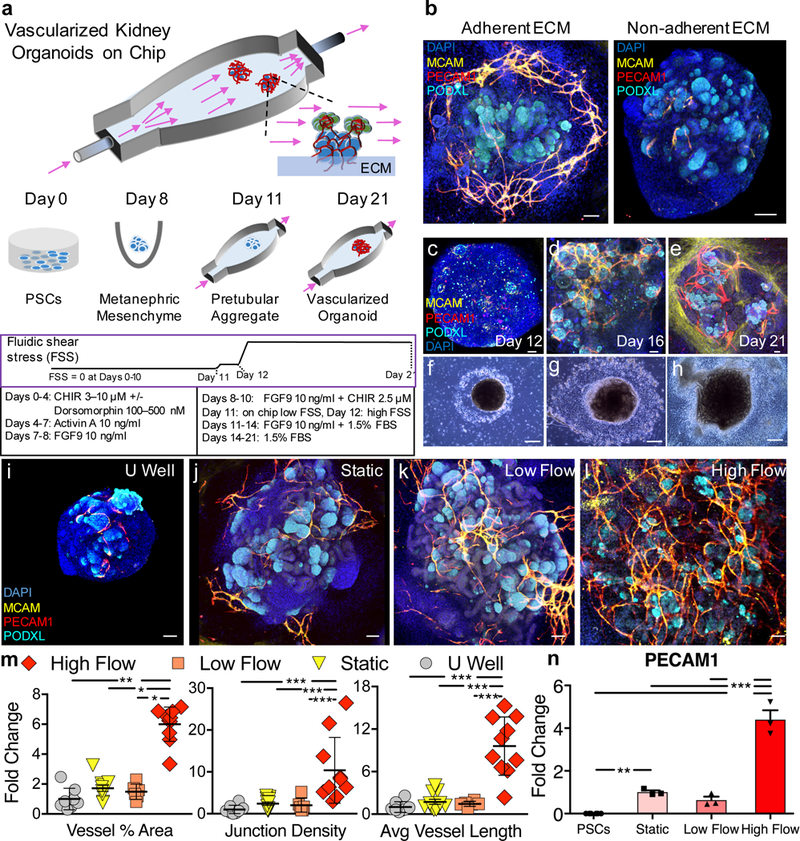
(a) Developing renal organoids are placed on an engineered extracellular matrix (ECM), housed within a perfusable millifluidic chip, and subjected to controlled fluidic shear stress (FSS), note organoids not drawn to scale. (b) Enhanced peripheral vascular network formation in adherent compared to non-adherent underlying ECMs, scale bars = 100 μm. (c-e) Immunostaining of whole mount organoids and (f-g) representative phase contrast images of entire organoids cultured under high FSS (days 12–21), scale bars = 50 μm and 300 μm, respectively, where perfusion direction is left to right. (i-l) Confocal 3D renderings for vascular markers in whole-mount organoids cultured under static U-well, static on engineered ECM, low FSS, and high FSS, scale bars = 100 μm. (m) Angiotool output, which quantifies the abundance and character of vasculature, reported as a fold change relative to the U-well condition. For (m), biological replicates of 8, 11, 6, and 10 were used per condition (U well, Static, Low Flow, and High Flow, respectively) in experiments using both iPSC- and hESC-derived organoids where the entire organoid represents one replicate (one dot) and mean +/− std is plotted. (n) qPCR depicting increased PECAM1 expression under high FSS. The graph is plotted with mean +/− std. Dots on the bar chart represent three technical replicates on RNA pooled from 6 organoids (biological replicates) per condition. DAPI: 4’,6-diamidino-2-phenylindole, PECAM1: CD31, MCAM: CD146, KDR: FLK1, PODXL: podocalyxin, CDH1: E-cadherin, CHIR: CHIR99021, FGF9: fibroblast growth factor 9, FBS: fetal bovine serum. Statistical analysis for (m) and (n) is performed using GraphPad Prism 7 and statistical significance is determined at a value of p < 0.05 as determined by a 1way and 2way ANOVA, respectively, using Tukey’s multiple comparisons test. Different significance levels (p values) are indicated with asterisks as such: *p<0.05, **p<0.01, ***p<0.001.
To determine the effects of fluid flow, developing kidney organoids are placed on the adherent gelbrin layer and superfused overnight with basal organoid media supplemented with 1.5% FBS in a closed-loop system over a range of flow rates from 0.04 mL/min (low FSS, ~0.0001 dyn/cm2) to 1 – 4.27 mL/min (high FSS, 0.008–0.035 dyn/cm2), while continuing the published differentiation protocol (Fig. 1a bottom)1,2. We observed enhanced formation of MCAM+PECAM1+ vascular networks in organoids cultured in vitro under high FSS after 10 days of perfusion (differentiation day 21), with nephrons forming over time (Fig. 1c-l, Supplementary Video 1). To quantify their vascularization, we evaluated confocal images of whole organoids using the Angiotool plugin to ImageJ17. We find that PECAM1+ vasculature of organoids cultured under high FSS exhibit a 5-fold increase in vessel % area compared to those cultured under low FSS (Fig. 1m, Supplementary Figs. 4a-l). Similarly, we find that PECAM1+ vasculature exhibits a 10-fold increase in junctional density (i.e., branch points per unit area) and average vessel length (i.e., inter-junctional distance) under high FSS compared to those cultured in static or low FSS (Fig. 1m). Concomitantly, PECAM1 transcripts are 5-fold upregulated under high FSS relative to controls (Fig. 1n). Note, such differences cannot be attributed solely to variations in media volume, since organoids cultured under high FSS with either 0.2 mL or 1 mL media per organoid in our closed-loop system lacked a statistically significant difference in their vasculature (Supplementary Fig. 5). Together, these observations suggest that FSS is a critical environmental cue that facilitates vascularization of kidney organoids in vitro.
Perfusable vessels form in developing kidney organoids under flow
During kidney development, vascular formation is believed to arise via a combination of vasculogenesis, de novo formation of blood vessels through the differentiation and coalescence of endothelial progenitor cells (EPCs), and angiogenesis, i.e., the formation of new blood vessels that sprout from pre-existing vessels18,19. In the developing mammalian kidney in vivo, fate mapping shows that KDR+ (FLK1) cells serve as EPCs that transform into intermediate MCAM+ cells and, ultimately, PECAM1+ mature endothelia (Fig. 2a)20,21. Akin to metanephric mesenchyme (MM) in vivo22, KDR+ EPCs are concurrently induced in vitro with SIX2+PAX2+SALL1+ nephron progenitor cells (NPCs) that differentiate into the epithelial cell types of the nephron (Supplementary Figs. 6–7). Under high FSS, KDR+ EPCs expand to 11.2% of the cell population in whole organoids compared to 4.25% in static culture on chip (Fig. 2b; Supplementary Fig. 8). Moreover, transcripts for KDR and MCAM are upregulated after 10 days of high flow (differentiation day 21) (Fig. 2c,d), further indicating that FSS is an important environmental cue to expand vascular potential. Meanwhile, expression of PDGFR-β is upregulated under high flow (Fig. 2e), where recruitment of PDGFR-β+ pericyte-like cells to PECAM1+ networks is consistent with vessel maturation (Fig. 2f). As vasculature within these kidney organoids evolves in vitro, we find heterogeneous expression of vascular precursor and mature markers, including areas that are MCAM+, PECAM1+, or MCAM+PECAM1+ after 10 days of perfusion (differentiation day 21) (Fig. 2g, Supplementary Fig. 9). Reflecting multi-scale vessel formation, we see open circular structures that lack nuclei (DAPI-) and are surrounded by MCAM and PECAM1 positive regions, which are indicative of lumens with varying diameter (Fig. 2h). Notably, analogous structures are observed in both hPSC-derived kidney organoids exposed to high FSS (Fig. 2i,j) and E14.5 embryonic mouse kidneys (Fig. 2k). To explore whether this embedded vasculature is perfusable, we carried out time-lapse, live cell monitoring of kidney organoids cultured in vitro under high FSS, and stained the vascular endothelium with Ulex europaeus I lectin. These experiments, carried out within minutes after introducing fluorescent beads (diameter = 100 nm) to the flowing media, confirm luminal perfusion in a subset of peripheral vessels (Supplementary Fig. 10a-g, Supplementary Video 2). To determine the extent of perfusable vasculature in whole kidney organoids, fluorescent beads are first superfused in media for several hours, then live confocal images (z-stack) are acquired at the base of the organoid embedded in ECM. The organoids are then fixed, immunostained for PECAM1, and co-registered with fiduciary markers. We observed the presence of beads in locations coincident with larger PECAM1+ vessels, several hundred micrometers from the superfused surface (Fig. 2l,m; Supplementary Fig. 11a-c). We also observed branched PECAM1+ networks that contain terminal sprout-like structures, which lack a PDGFR-β+ cell lining, suggestive of angiogenesis (Supplementary Fig. 12a,b). Interestingly, we find that kidney organoids fuse in as little as 24 h in culture under high FSS (Supplementary Video 3) and that robust vascular networks arise between adjacent organoids on chip (Fig. 2n). The association of PECAM1+ networks with ACTA2+ smooth muscle-like cells suggests maturation of arterial lineage cells23, while the venous marker, EMCN, stains presumptive venous lineage cells (Supplementary Fig. 12c,d). Hence, kidney organoids subjected to the appropriate combination of adherent ECM, culture media, and fluidic shear stress in vitro form increasingly mature, perfusable vasculature of varying size and lineage.
Figure 2. Intra and inter-organoid vascular networks with perfusable lumens supported by mural cells are observed for kidney organoids cultured under high flow in vitro.
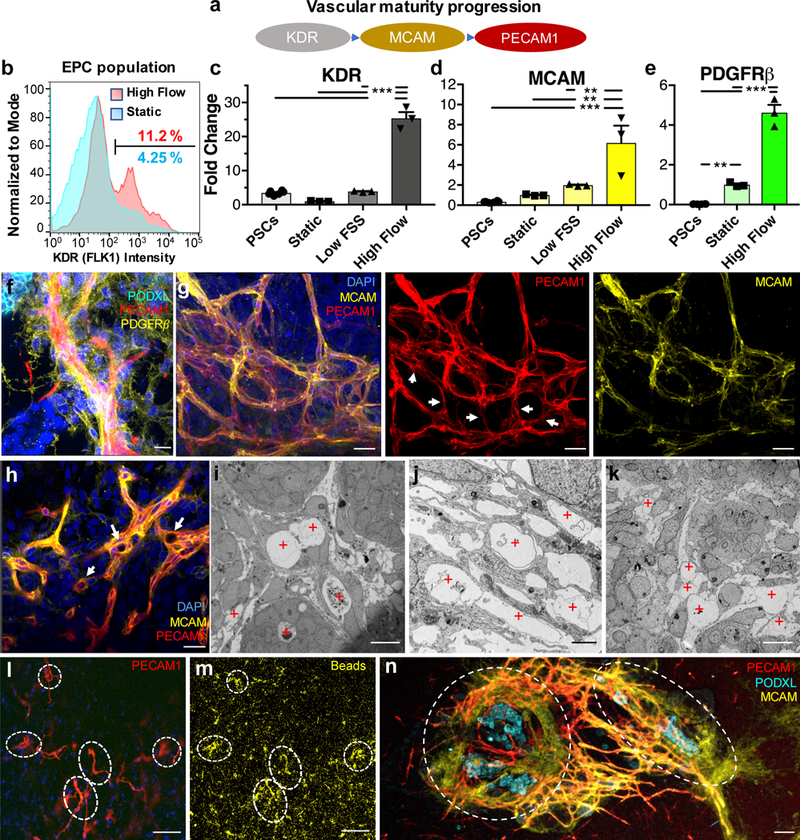
(a) Diagram of endothelial maturation in developing kidneys in vivo, from progenitor cells to sustained terminal marker expression. (b) Flow cytometry of dissociated whole organoids depicting ~3-fold expansion of the endothelial progenitor cell (EPC) population in response to high FSS, compared to static conditions on chip. (c,d) qPCR of endothelial cell markers in developing organoids showing their upregulation following high FSS (day 21). (e) A key stromal marker is upregulated under high FSS, possibly due to mural cells associating with enhanced vasculature as shown in (f), scale bar = 15 μm. (g) Confocal 3D renderings of vascular markers within whole-mount organoids reveal that some features are best visualized in co-stained samples (left), as opposed to only mature (middle) and intermediate (right) markers, scale bars = 30 μm. White arrows highlight areas that are PECAM1+MCAM-, showing the two markers are not always co-expressed. (h) A single z-slice from (g) in which white arrows highlight open lumens, scale bar = 30 μm. (i-k) TEM images showing circular openings encompassed by a thin membrane that reflect vascular lumens for (i,j) kidney organoids subjected to high FSS and (k) E14.5 mouse embryonic kidney in vivo [Note: Hierarchial luminal diameters vary from 2 to near 20 μm (red plus signs reflect vascular lumens), scale bars = 10 μm for (i,k) and 2 μm for (j)]. (l,m) Z-slice at the base of a kidney organoid cultured under high FSS, showing in (l) the vascular network and in (m) the accumulation of fluorescent beads within the vascular network, scale bars = 100 μm. (n) Confocal 3D rendering of bridging between two adjacent, whole organoids (outlined by dashed white lines, scale bar = 100 μm. DAPI: 4’,6-diamidino-2-phenylindole, PECAM1: CD31, MCAM: CD146, KDR: FLK1, PODXL: podocalyxin, PDGFRβ: platelet derived growth factor receptor beta. Dots on the bar charts (c-e) represent three technical replicates on RNA pooled from 6 organoids (biological replicates) per condition. All graphs are plotted with mean +/− std. Statistical analysis for (c-e) is performed using GraphPad Prism 7 and statistical significance is determined at a value of p < 0.05 as determined by a 1way ANOVA, using Tukey’s multiple comparisons test. Different significance levels (p values) are indicated with asterisks as such: *p<0.05, **p<0.01, ***p<0.001.
Tubular epithelia exhibit enhanced maturation under flow
Under static conditions, kidney organoids display more limited vasculature and tubular epithelia often exhibit immature gene expression profiles with morphology analogous to first trimester kidney3,9,10. Following subcapsular transplantation to the mouse kidney, progressive morphogenesis of tubular structures occurs, as evident by polarization, formation of a brush border, and ciliary assembly in vivo10. Native kidneys rely on extensive fluid flow both within and between tubular, interstitial, and vascular compartments to facilitate reabsorption of ~99% of the glomerular filtrate. We hypothesized that a similar progressive morphogenesis and maturation of gene expression profiles may occur in hPSC-derived tubular cells in vitro when subjected to high FSS. Indeed, we find that the polarity of tubules formed in vitro is enhanced, with apical enrichment of the brush border marker, Lotus tetragonolobus lectin (LTL) (Fig. 3a; Supplementary Fig. 13a-d) after 10 days (differentiation day 21) of culture under high FSS. Similarly, we find that primary cilia are apically enriched from 50% in static culture to 89% of LTL+ cells under high FSS (Figs. 3b-c; Supplementary Fig. 13e-j). Consistent with polarization and maturation-associated ciliary assembly, the expression of ciliary proteins (PKD1, PKD2, NPHP1, NPHP6, PKHD1) is upregulated (Fig. 3d). Concurrently, expression of tubular epithelial transporters, including AQP1, solute transporters (SLC34A1, ATP1A1, SLC6A19, SLC9A3, SLC2A2), and drug transporters (ABCB1, LRP2) are upregulated when compared to static controls and undifferentiated hPSCs (Fig. 3e; Supplementary Fig. 14a), indicative of enhanced functional potential. The maturation of tubular epithelial cells is evident by the upregulation of adult transcription factors (BNC2, NPAS2, TRPS1) (Fig. 3f), which are reported as mature proximal tubule markers by single cell RNA-seq in adult human kidneys9. We quantified the association between PECAM1+ networks and LTL+ tubules using confocal imaging rendered using Imaris surface functions, distance transformation, and masking tools (Fig. 3g), which revealed that, for organoids cultured under high FSS, the percent of vascular surface area in contact with tubules is increased by nearly 3-fold relative to organoids in static culture (Fig. 3h, Supplementary Fig. 15). Additionally, the mean distance between a given LTL+ tubule and the nearest blood vessel decreased ~70% under high FSS (Fig. 3i). This enhanced tubule-vascular association is observed in both transverse and longitudinal orientations (Fig. 3j,k; Supplementary Video 4). Importantly, we find that maintenance of endogenous VEGF gradients within organoids cultured in vitro is crucial for vascular-tubular interactions. For example, the close association of LTL+ tubules and blood vessels is disrupted when 100 ng/mL of VEGF or a VEGF inhibitor (bevacizumab24 at 250 μg/mL) is added to the media during the first 10 days of high FSS culture, reverting to that observed for organoids in static culture (Fig. 3h,i, Supplementary Fig. 15). In short, culturing organoids under fluid flow in vitro supports maturation and morphogenesis of tubular epithelia in kidney organoids likely due to interlineage endothelial-epithelial communication13.
Figure 3. Tubular epithelia mature and undergo morphogenesis to become a polarized, ciliated compartment in contact with vasculature in response to the high flow condition on chip.
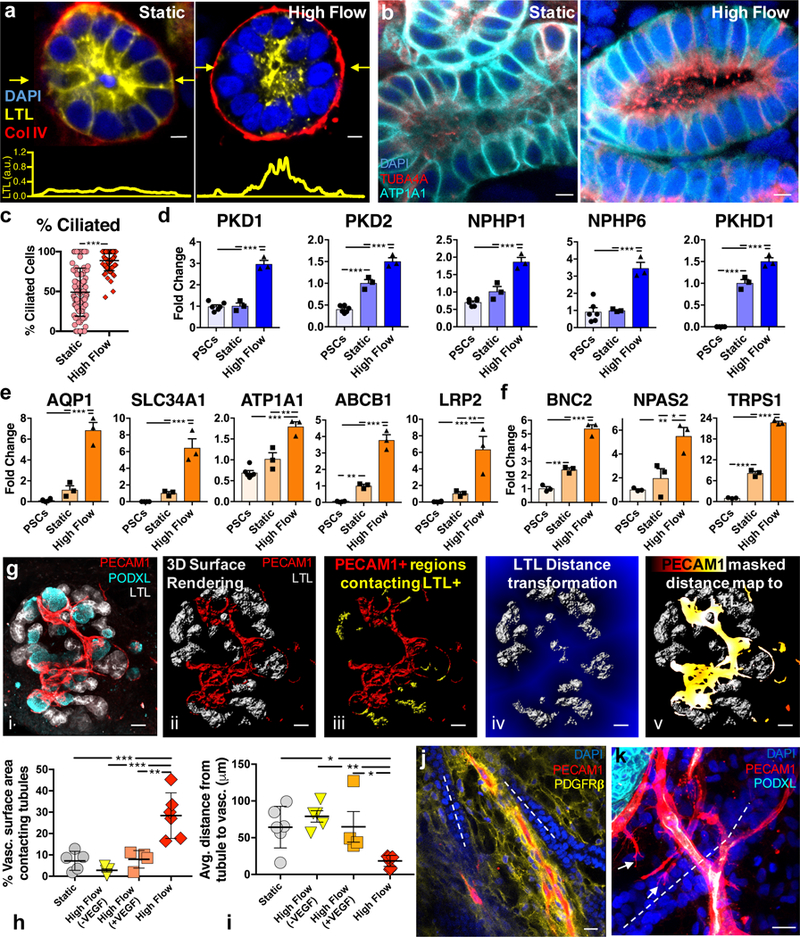
(a) Tubule cross-sections under static and high flow conditions showing proper basal expression of collagen IV in both cases, and proper apical expression of LTL under high flow at day 21, scale bars = 5 μm, the plots in yellow below show the intensity of LTL across a line scan denoted by yellow arrows in the images above. (b) Tubule cross-sections showing apical presence of cilia with higher prevalence (c) under high flow versus static conditions on chip and proper basolateral expression of ATP1A1 (Na/K ATPase) on day 21, scale bars = 5 μm. For (c), the static sample number is 101, and high flow is 64; samples are determined as a field of view in a LTL+ tubule where at least 8 cells could be viewed in cross section with cilia. (d-f) qPCR of ciliary markers, solute transporters, drug transporters, and adult transcription factors showing upregulation under high flow on day 21, compared to static conditions on chip and undifferentiated hPSCs. Whole organoid 3D confocal imaging stacks (gi, all scale bars = 50 μm) of a representative high flow sample are used to demonstrate the analysis method for the association of tubules with vasculature in Imaris 3D surface rendering (gii and giii) and distance transformation software (giv and gv) to find in (h) that the percent of vasculature surface area overlapping with LTL+ tubules within one voxel is significantly increased under high flow than in static conditions. Further, that tight vasculotubular association can be negated by dosing high amounts of VEGF or inhibiting VEGF in the media (h). Similarly, the average distance in 3D between the vasculature and the tubules decreases in the high flow condition (i) which does not significantly differ between static, high flow + VEGF, and high flow - VEGF (VEGF inhibition) conditions. Note the data in (h,i) represents biological replicates, or whole organoids, of 6, 5, 4, and 6 per condition (Static, High Flow (-VEGF), High Flow (+VEGF), and High Flow, respectively). (j,k) Immunostaining showing that PECAM1+ networks associate with tubular structures in both traverse and longitudinal planes in high flow at day 21, scale bars = 20 μm. DAPI: 4’,6-diamidino-2-phenylindole, LTL: lotus tetragonolobus lectin, PECAM1: CD31, PODXL: podocalyxin, TUBA4A: tubulin alpha 4a (also known as acetylated tubulin), AQP1: aquaporin 1, SLC34A1: Na/Phos cotransporter, ATP1A1: Na/K ATPase, ABCB1: MDR1, LRP2: Megalin, BNC2: basonuclin 2, NPAS2: neuronal PAS domain protein 2, TRPS1: transcription repressor GATA binding 1, PKD1: polycystin 1, PKD2: polycystin 2, NPHP1: nephrocystin 1, NPHP6: nephrocystin 6, PKHD1: fibrocystin. All statistical analysis is performed using GraphPad Prism 7 software. All graphs are plotted with mean +/− std. For (c) statistical significance is determined at a value of p < 0.05 as determined by an unpaired t test with Welch’s correction. Dots on the bar charts (d-f) represent three technical replicates on RNA pooled from 6 organoids (biological replicates) per condition. Statistical analysis for (d-f, h,i) is determined at a value of p < 0.05 as determined by a 1way ANOVA, using Tukey’s multiple comparisons test. Different significance levels (p values) are indicated with asterisks as such: *p<0.05, **p<0.01, ***p<0.001.
Glomerular vascularization and maturation in vitro mirrors early stages of development in vivo
The glomerular structures of kidney organoids in static culture are known to be largely avascular2,3,9,25. Upon animal transplantation, host-derived vascularization of kidney organoids promotes glomerular vascularization10. To determine whether FSS-induced vascularization of organoids in vitro extends to glomerular compartments, we quantified PODXL+ podocyte clusters invaded by MCAM1+PECAM1+ vascular structures in static and high FSS conditions using confocal imaging (Fig. 4a,b, Supplementary Fig. 16a,b). Under high FSS, MCAM+PECAM1+ vascular invasion and wrapping of PODXL+ clusters is significantly increased (Fig. 4b). Notably, invasion is greater than 60% in high FSS, as compared to 10 ~ 20% in static controls (Fig. 4b), consistent with the requirement of vascular flow for glomerular assembly in animal studies26. Given that there is significant upregulation of VEGF-A expression in organoids cultured under high FSS (Fig. 4c), we postulated that these samples could be used to elucidate the role of VEGF in glomerular vascularization in vitro. Interestingly, both VEGF inhibition (bevacizumab24 250 μg/mL for 10 days on chip) and VEGF addition (100 ng/mL for 10 days on chip) significantly reduced the incidence of invasion of PODXL+ glomeruli-like compartments by PECAM1+MCAM+ vascular networks under high FSS (Fig. 4b). As vessel % area under high flow conditions is unchanged between endogenous VEGF, VEGF addition, and VEGF inhibition, the difference in glomerular vascularization cannot be attributed to increased abundance of vasculature (Supplementary Fig. 17a-c, Supplementary Table 3). Unlike the random sporadic growth observed for organoids cultured in exogenously disrupted gradients, the reduced junctional density and increased average vessel length seen in unperturbed media conditions suggests that vessels grow towards an endogenous VEGF gradient (Supplementary Fig. 17b,c). The endogenous upregulation of VEGF under high FSS (Fig. 4c) appears to generate gradients that allow vessels to reach glomeruli-like compartments in time to invade rather than wrap Bowman’s capsule-like structures within these kidney organoids in vitro.
Figure 4. Flow-enhanced glomerular vascularization and morphogenesis of kidney organoids in vitro mirrors stages of glomerular development in vivo.
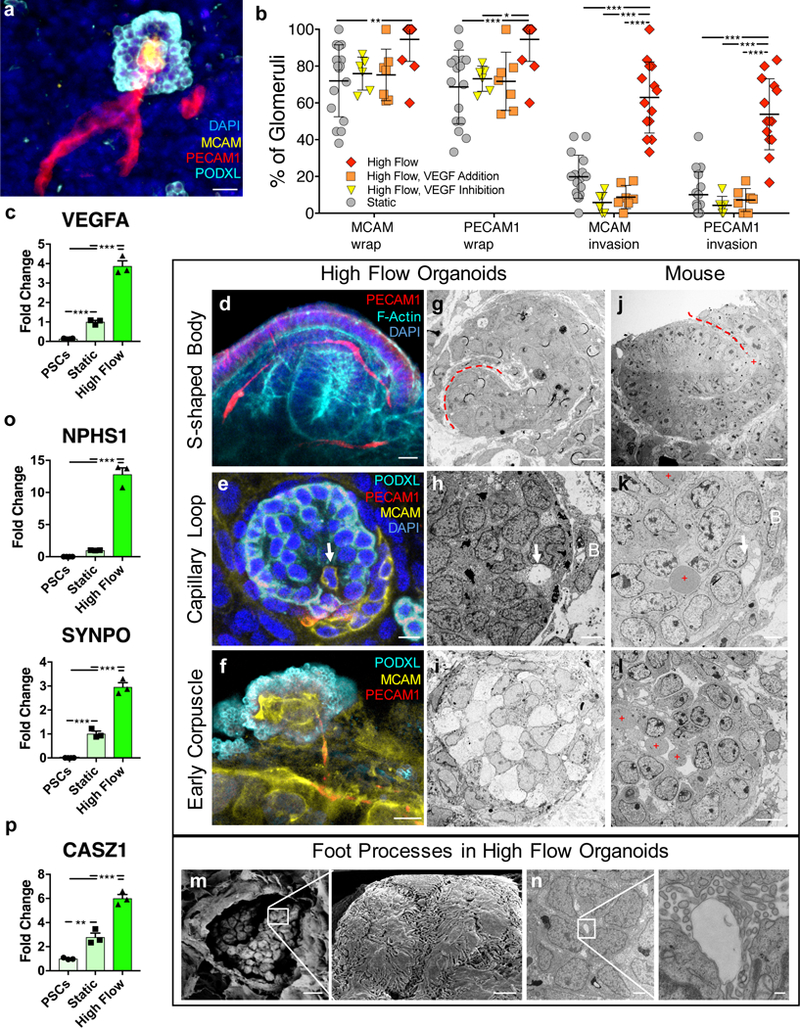
(a) A 3D rendered confocal image of vascular invasion in a PODXL+ cluster showing afferent and efferent vessels, scale bar = 40 μm. (b) Percent of PODXL+ clusters that exhibit vascular wrapping or invasion under static and high FSS ± VEGF (addition or inhibition) representing whole organoids of sample size 16, 6, 7, 14 for the conditions Static, High Flow VEGF Addition, High Flow VEGF Inhibition, and High Flow, respectively. (c) qPCR of VEGF showing significant upregulation in organoids cultured under high FSS in vitro (day 21). (d) A 3D rendered confocal image of capillary invasion in an S-shaped body in a vascularized organoid under high FSS in vitro (day 21), scale bar = 10 μm. (e) Single confocal z-slice showing capillary invasion with PECAM1+MCAM+ cell (white arrow) and MCAM+ vascular precursors (CD146+ cells), scale bar = 10 μm. (f) MCAM+PECAM1+ glomerular tuft-like formation shown as a single z-slice from confocal, scale bar = 10 μm. (g-i) TEM images of structures correlating with the IF images in kidney organoids (day 21), scale bars for (g,h) = 4 μm and (i) = 10 μm. Corresponding stages (j-l) in E14.5 mouse kidneys, where red dashed lines depict clefts, white arrows denote capillary invasion, B = Bowman’s capsule-like structure, and red plus signs denote RBCs, scale bars for (j,k) = 8 μm and (l) = 50 μm. (m) TEM images of a glomerular-like structure in organoids cultured under high FSS (day 21) showing a parietal membrane enclosing a visceral cluster of cells (left), which manifest interdigitating cytoplasmic projections extending across and into the plane of field on higher magnification (right), scale bars = 10 μm (left) and 1 μm (right). (n) TEM images of a glomerulus-like compartment in organoids cultured under high FSS (day 21) (left) in which higher magnification shows podocyte foot process abutting a glomerular tuft-like formation (right), scale bars = 2 μm (left) and 200 nm (left). (o,p) qPCR depicting significantly upregulated transcripts for podocyte foot process proteins (o) and an adult transcription factor (p). SSB: S-shaped body, CLS: capillary loop stage, DAPI: 4’,6-diamidino-2-phenylindole, MCAM: CD146, PECAM1: CD31, PODXL: podocalyxin, SYNPO: synaptopodin, NPHS1: nephrin, PDGFRβ: platelet derived growth factor receptor beta, VEGFA: vascular endothelial growth factor A, CASZ1: castor zinc finger 1. All statistical analysis is performed using GraphPad Prism 7 software. All graphs are plotted with mean +/− std. For (b) statistical significance is determined at a value of p < 0.05 as determined by a 2way ANOVA, using Tukey’s multiple comparisons test. Dots on the PCR plots in (c,o,p) represent three technical replicates on RNA pooled from 6 organoids (biological replicates) per condition. Statistical analysis for (c.o.p) is determined at a value of p < 0.05 as determined by a 1way ANOVA, using Tukey’s multiple comparisons test. Different significance levels (p values) are indicated with asterisks as such: *p<0.05, **p<0.01, ***p<0.001.
Vascularization of glomeruli in vivo commences with invasion of an S-Shaped body (SSB) by a single capillary loop around which podocytes coalesce with formation of a primitive Bowman’s capsule (capillary loop stage, CLS), followed by vascular expansion to form nascent glomerular tufts in early corpuscles27. Following 10 days of organoid culture under high FSS (differentiation day 21), glomeruli in vitro vary in a spectrum between SSB, CLS, and early corpuscle-like structures. Consistent with vascular invasion of an SSB, a PECAM1+ vessel invades a cleft in an SSB-like structure (Fig. 4d; Supplementary Video 5a). Meanwhile, the ‘luminal’ feature of an MCAM+PECAM1+ vessel invading a PODXL+ cellular cluster, surrounded by a putative Bowman’s capsule, suggests CLS-like developing glomeruli (Fig. 4e; Supplementary Video 5b,c). Renal corpuscle-like structures contain MCAM+PECAM1+ vasculature suggestive of capillary loops (Fig. 4f, Supplementary Fig. 16b, and Supplementary Video 6). TEM shows analogous structures between developing glomeruli in high FSS organoids and E14.5 mouse kidneys, revealing that these in vitro organoids may follow in vivo glomerular development through SSB, capillary loop, and early corpuscle stages (Fig. 4g-l). Importantly, capillary loop-like structures in organoid glomeruli in vitro exhibit open lumens without red blood cells, which are present in capillary loops in mice (Fig. 4j,k,l). Scanning electron microscopy (SEM) shows a thin-layered capsular structure suggestive of a parietal layer of epithelial cells (Bowman’s capsule) that contains round cellular bodies with elongated cytoplasmic projections indicative of a visceral layer of epithelium (podocytes) (Fig. 4m). In a manner similar to E14.5 embryonic mouse kidneys and human adult kidneys, grapelike clusters of visceral epithelial cells in organoids under high flow consist of round cellular bodies with interdigitating foot process-like structures (Supplementary Fig. 18a-f). The cytoplasmic projections, consisting of primary stalks and secondary side branches appear polarized and abut thin-layered membranes of capillary loop-like structures (Fig. 4n, Supplementary Fig. 19a-g), consistent with foot processes. Compared to static conditions, the foot process-like structures appear more prominent with significant upregulation of NPHS1 (nephrin) and SYNPO (synaptopodin), encoding foot process-associated proteins, under high FSS (Fig. 4o, Supplementary Fig. 20a-d). Further, these structures fuse in response to doxorubicin as observed for foot processes in mammalian kidneys (Supplementary Fig. 20e). Concurrently, transcriptional maturation is evident by enhanced expression of podocyte adult transcription factors (WT1, CASZ1, CUX1, TEAD1)9 (Fig. 4p; Supplementary Fig. S14b) with accompanying appropriate expression of the housekeeping gene, GAPDH, among all PCR samples (Supplementary Fig. 14c,d). These observations indicate that organoids cultured under high FSS exhibit enhanced glomerular vascularization and foot process maturation, which is required to facilitate functional morphogenesis of podocytes in vitro.
Discussion
In summary, kidney organoids subjected to high fluidic shear stress during development in vitro exhibit a significant enhancement in the abundance of vasculature and maturity in their tubular and glomerular compartments, along with a concomitant morphogenesis of tubular epithelial cells and podocytes. FSS is the key parameter driving our biological findings; however, other chemomechanical cues arising from the underlying ECM, flow profile or media composition do work collectively. While our method does not yet ensure that the microvascular networks present within these kidney organoids are readily perfusable, the ability to promote their flow-enhanced development in vitro opens new avenues for investigating organogenesis, nephrotoxicity, tubular and glomerular disease, and kidney regeneration using a simple millifluidic chip. Our findings could be applicable to other organoid types28–30 that may similarly benefit from controlled fluid flow during development from embryonic to more functional organ equivalents in vitro.
Online Methods:
Millifluidic chip fabrication.
A silicone-based ink is used to 3D print customized perfusion gaskets (Supplementary Fig. 1), in which developing kidney organoids are placed on an engineered ECM layer (1 mm thick) and subjected to a controlled fluidic shear stress environment. The ink is composed of a two-part silicone elastomer (SE 1700, DOW Chemical) with a 10:1 base to catalyst (by weight) that is homogenized using a centrifugal mixer for 2 min (2000 rpm, AE-310, Thinky Corp, Japan). The silicone ink is printed within 2 h of mixing with catalyst. This ink is loaded in a syringe (EFD Inc., East Providence, RI) and centrifuged to remove any air bubbles before printing at room temperature. The chips are fabricated using a custom-designed, multimaterial 3D bioprinter equipped with four independently addressable printheads mounted onto a 3-axis, motion-controlled gantry with a build volume of 725 mm x 650 mm x 125 mm (AGB 10000, Aerotech Inc., Pittsburgh, PA USA). The silicone (PDMS) ink is housed in a syringe barrel to which a 410 μm diameter nozzles is attached via a luer-lock (EFD Inc., East Providence, RI, USA). Ink is extruded through deposition nozzles by applying air pressure (800 Ultra dispensing system, EFD Inc., East Providence, RI, USA), ranging from 10–90 psi, corresponding to print speeds between 1 mm/s and 5 cm/s. The customized perfusion chip gasket is printed by depositing the silicone ink through a tapered 410 μm nozzle onto 50 mm x 75 mm glass slides. The gasket tool-path is created using a custom MATLAB script that generates G-code for a final gasket structure. After printing, the perfusion chip is cured at 80°C in an oven for > 1 h, stored at room temperature, and autoclaved prior to use. The organoid chamber is 15 mm wide by 3.6 mm high and 60 mm long; the ECM is placed on the base of the perfusion gasket and is 1 mm thick. The organoids, between 4 and 20 per chip, are placed centrally in an area of 8 mm wide by 3.6 mm high and 20 mm long as shown in Supplementary Fig. 1d.
Engineered extracellular matrix (ECM) preparation and rheology.
The extracellular matrix (ECM) is comprised of a network of gelatin and fibrin (gelbrin). To prepare the ECM components, a 15 wt/v% gelatin solution (Type A, 300 bloom from porcine skin, Sigma) is first produced by adding gelatin powder to a warm solution (70˚C) of DPBS (1X Dulbelcco’s phosphate buffered saline without Ca+2 and Mg+2). The gelatin is processed by stirring for 12 h at 70˚C, and the pH is then adjusted to 7.5 using 1 M NaOH. The solution is sterile filtered and stored at 4˚C in aliquots for later usage (< 3 months). A fibrinogen solution (50 mg/mL) is produced by dissolving lyophilized bovine blood plasma protein (Millipore) at 37˚C in sterile DPBS without Ca+2 and Mg+2. The solution is held at 37˚C without agitation for at least 45 min to allow complete dissolution. The transglutaminase (TG) solution (60 mg/mL) is prepared by dissolving lyophilized powder (Moo Gloo, TI) in DPBS without Ca+2 and Mg+2 and gently mixing for 20 sec. The solution is then held at 37˚C for 20 min and sterile filtered before using. A CaCl2 stock solution (250 mM) is prepared by dissolving CaCl2 pellets in sterile water. To prepare stock solutions of thrombin, lyophilized thrombin (Sigma Aldrich) is reconstituted at 500 U/mL using sterile water and stored at −20˚C. Thrombin aliquots are thawed immediately prior to use.
Prior to casting a layer of engineered ECM within the 3D printed chip, several components are mixed in advance at appropriate concentrations, including 10 mg/mL fibrinogen, 2 wt% gelatin, 2.5 mM CaCl2 and 0.2 wt% TG. This solution is then equilibrated at 37ºC for 15–20 min before use to improve optical clarity of the ECM.15 Next, the solution is rapidly mixed with stock thrombin solution at a ratio of 250:1, resulting in a final thrombin concentration of 2 U/mL. Within 2 min at 37ºC, soluble fibrinogen cures to a fibrin gel. For this reason, the ECM solution must be cast onto the base of the perfusion chip immediately after mixing with thrombin. The gasket with ECM is then placed in a sterile container and kept in the incubator for a minimum of 30 min prior to assembly with housing, media, and pretubular aggregate integration.
A controlled stress rheometer (DHR-3, TA Instruments, New Castle, DE) with a 40 mm diameter, 2˚ cone and plate geometry is used to measure the rheological properties of the ECM. The shear storage (G’) and loss (G’’) moduli are measured at a frequency of 1 Hz and an oscillatory strain (γ) of 0.01. Time sweeps are conducted by rapidly placing a premixed ECM solution that contains thrombin onto the Peltier plate held at 37˚C. The G’ of the final cured engineered ECM is approximately 800 Pa.
To prepare the ECM formulations shown in Supplementary Fig. 2, fibrin is used at either 10 mg/mL or 25 mg/mL with thrombin at 2 U/mL and 2.5 mM CaCl2. The fibrin/Col I ECM is prepared by mixing fibrinogen solution at a final concentration of 25 mg/mL with Collagen I (Rat Tail Collagen I from Corning, 1 mg/mL) at a pH = 7.5, a thrombin concentration of 2 U/mL, and 2.5 mM CaCl2. Matrigel (Corning) is diluted by 50% with sterile PBS and cured at 37˚C.
We also made and evaluated another ECM formulation, which included fibrin along with human umbilical vein endothelial cells (HUVEC) and human neonatal dermal fibroblast (HNDF) cells (Supplementary Fig. 3b). The pre-formed network of HUVECs and HNDFs are prepared by combining HUVECs:HNDFs at a 5:1 ratio at a concentration of 2M cells/mL in 10 mg/mL fibrin gel. The cells are cultured in 1:1 DMEM:EGM-2 (Dulbecco’s Modified Eagle Medium, Endothelial Growth Medium 2, Lonza) plus 5% FBS for 3 days to allow for spontaneous tubulogenesis to occur prior to loading pretubular aggregates on top of the fibrin gel supporting the HUVEC:HNDF network. At this point, the media is changed to 1:1 EGM2: ARPMI (Advanced Roswell Park Memorial Institute + 1x glutamax) and held in static conditions for 7 days.
Organoid assembly and perfusion on printed millifluidic chips.
To assemble the kidney organoids-on-chip, pretubular aggregates (with ages between Day 11 and Day 14) in media are pipetted onto the top of the ECM on gasket in the window / area shown in Supplementary Fig. 1d. A large number of organoids can fit on chip and we typically used between 4 and 25 per run, but upwards of 100 or more can fit if needed. The organoids are randomly spaced within the window of 8×20 mm; note the distance between the organoid surface on ECM and the upper acrylic lid is 2.6 mm. The gasket is then placed into a machined stainless steel base. Stainless steel tubes (Microgroup, Inc, grade 304, Gauge 18RW, ends are de-burred prior to use) are pushed through the PDMS at inlet and outlet and positioned such that they are above the ECM surface. Finally, a thick acrylic lid is placed on top (Supplementary Fig. 1a-e). The lid and base are clamped together by four screws, forming a seal around the printed silicone gasket. Next, sterile two-stop peristaltic tubing (PharMed BPT) is filled with media and connected to the outlet of a sterile filter that is attached to a 10 ml syringe barrel (EFD Nordson), which serves as a media reservoir. Organoid media (ARPMI + 1X glutamax + 1.5% FBS and 1% Antimycotic/Antibiotic solution) that is equilibrated for > 3 h in an incubator at 37°C, 5% CO2 is added to the media reservoir, and tubing from the reservoir is connected to the inlet of the chip (metal hollow perfusion pin). Tubing is also connected to the outlet of the chip through its respective stainless steel pin. A syringe is then used to exert slight pressure on the media in the barrel, forcing it to enter and completely fill the open gasket area, taking extra care not to disturb the pretubular aggregates. Hose pinch-off clamps are added at the inlet and outlet of the perfusion chip to prevent uncontrolled flow when disconnected from the peristaltic pump. To complete the closed perfusion circuit, tubing from the outlet is connected to the media reservoir. The media reservoir is equilibrated with atmospheric conditions in the incubator at all times by means of a sterile filter on top of the media reservoir. Media is changed every 2 to 3 days. The typical volume of media per organoid on chip and static on ECM is 0.5 to 0.8 mL. The typical volume of media per organoid in U well is 0.2 mL. We determined that volume of media per organoid in the range of 0.2 mL to 1 mL per organoid had no measurable effect on the resulting vasculature on chip in high flow conditions (Supplementary Fig. 5). Furthermore, we measured organoid height in order to understand the dynamic changes evolving on chip during organoid development. We find kidney organoids under low and high FSS initially flatten, then grow to heights greater than U-well controls. Thus, gross morphologic height changes are not a dominant variable controlling the enhancement in vascularization since the heights were similar in low and high FSS conditions by Day 21, while the vasculature is dramatically enhanced in high FSS only (Fig. 1 and Supplementary Fig. 22).
An Ismatec IPC-N low speed peristaltic pump is used to direct media into the gasket in a closed loop circuit at volumetric flow rates ranging from 40 μL/min to 4.27 mL/min. During the first 12 to 24 hours of culture on chip, pretubular aggregates are subjected to low flow rates of 40 μL/min or less. These extremely low flow rates provide nutrient supply without high shear stresses that could break the connection forming between the aggregates and the ECM below. After 24 h, the aggregates are strongly adherent to the ECM and the volumetric flow rate (Q) is raised to a value between 1.0 and 4.27 mL/min.
Flow Profile Analysis.
Flow modeling was performed using COMSOL Multiphysics simulation software. The fluid flow velocity profile was calculated by assuming a Stoke’s Flow, using a 1 mL/min volumetric flow rate. The channel comprised the curved surface of the gel, the silicone walls at the two sides, and the perfusion chip lid. For direct measurement of fluidic shear stress at the gel-channel interface, fluorescent beads were tracked within the organoid seeding region at various volumetric flow rates. To visualize the flow, millifluidic chips were mounted onto a confocal microscope stage, and were perfused with PBS containing 0.4% v/v of a 2% solids solution of 0.5 μm 488 nm fluorescent beads (Thermo Fisher). The pump was connected to the chip via 18.2 meters of loosely spooled silicone tubing (Cole-Parmer Peroxide-Cured Silicone Tubing, 1/32”ID x 3/32”OD) to dampen pulsatility to obtain a time-averaged shear-stress. Analysis was performed along an 8 mm long, transverse line centered on the mid-line of the channel and the organoid seeding region at two-thirds of the distance between the inlet and the outlet of the perfusion chip. Confocal videos of bead flow were captured using a window of 600 μm along the channel by 90 μm across. To estimate the velocity gradient for calculating shear stress at the gel-channel interface, four videos were captured at 40 μm-height intervals just above the gel surface. The mean bead velocity was extracted at each height by performing a cross-correlation of the video frames in a direction parallel to the bead flow. The peak location of the cross correlation represents the mean displacement of the beads over the timeframe of the two images used for cross-correlation. The cross-correlation timeframe was increased until the peak in the cross-correlation of the video dropped to below 6 standard deviations of the noise floor. The velocity was calculated as the ratio of the cross-correlation peak displacement and the time difference between the two frames used for cross-correlation. Velocities were calculated for each frame of the video, and averaged. The velocity gradient was measured using linear regression of the mean velocities at the four different heights. The fluidic shear stress was then calculated as the product of the gradient and a dynamic viscosity of 0.78 cP for DMEM at 37°C.31 To measure the flow pulsatility, the 18.2 meters of silicon tubing was removed and replaced with the standard 10 cm of tubing between the pump and chip. A 50 s video was captured at the midline of the channel, two-thirds of the distance between the inlet and the outlet, and the bead velocity was measured over time using the cross-correlation method described above (Supplementary Fig. 1e-g). The predicted flow profile using COMSOL and direct measurements using bead flow are in good agreement, if we assume a rectangular cross-section. Wall fluidic shear stress in flow through a rectangular cross-section (τ, denoted as FSS) is calculated using the equation τ = 6μQ/bh2, where μ is the medium viscosity, b is the channel width and h is the channel height (the empty channel through with fluid flows is approximated as a rectangular cross-section (b = 14 mm; h = 2.6 mm), where the organoids reside. In this study, we varied the volumetric flow rates to induce a low FSS that ranges from 1×10−7 to 1×10−4 dyn/cm2 and a high FSS that ranges from 8×10−3 to 3.5×10−2 dyn/cm2. Note the channel dimensions can be reduced simply by increasing the ECM height, which yields a higher FSS at a given volumetric flow rate. We have constructed channel heights as small as b = 0.5 mm, leading to FSS at ~1 dyn/cm2, and organoids cultured on this chip exhibit comparable enhancements in vascularity and tubular/glomerular maturation, as compared to those subjected to an FSS ranging from 8×10−3 to 3.5×10−2 dyn/cm2.
Cell Culture.
Human ESCs, H9 (WiCell) and human iPSCs, BJFF (provided by Prof. Sanjay Jain at Washington University) are maintained in feeder-free culture using StemFit ® Basic02 (Ajinomoto Co., Inc.) supplemented with 10 ng/ml FGF2 (Peprotech) as previously reported1. Human glomerular microvascular endothelial cells (GMECs), RFP expressing (Angio-Proteomie) are cultured using EGM2 media (Lonza) and used up to passage 9. Human umbilical vein endothelial cells (HUVECs), RFP expressing (Angio-Proteomie) are cultured using EGM-2 media (Lonza) and used up to passage 9. Human neonatal dermal fibroblasts (HNDF), GFP expressing (Angio-Proteomie) are cultured per supplier’s instructions and used up to passage 15.
Organoid preparation and culture.
Organoid preparation is covered in detail elsewhere1, but briefly hPSCs are differentiated into metanephric mesenchyme cells which included SIX2+ nephron progenitor cells with approximately 80–90% efficiency, by a 3-step directed differentiation protocol1 (Fig. 1a bottom). Metanephric mesenchyme cells are differentiated into pretubular aggregates in suspension culture, and then the aggregates are transferred onto the chip (Fig. 1a top), anytime between Days 11 and 14 work (Supplementary Fig. 2g). Further differentiation into kidney organoids are stimulated by the same differentiation protocol, reported previously except that 1.5% FBS (heat inactivated, Gibco) is added1. This same process can be used to massively scale up kidney organoid production using Elplasia™, a culture plate that has patterned microwells (Kuraray) (Supplementary Fig. 21). Using traditional methods, kidney organoids contain roughly 100,000 cells/aggregate. However, within the same footprint as a single well in a 6-well plate, approximately 1,000 mini-organoids can be produced, which contain approximately 5,000 cells/aggregate. Notably, when these mini-organoids are placed within our engineered microenvironment on chip, they behave similarly to larger organoids and exhibit enhanced vascularization under high FSS conditions.
We explored several experimental conditions that did not lead to enhanced vascularization. In particular, adding adult human primary GMECs, either by aggregating them with nephron progenitor cells at Day 8 or placing them in culture on ECM near renal aggregates or vesicles was not successful (Supplementary Fig. 3a). The developing kidney organoids either failed to form properly at Day 8 or HUVECS, HNDFs, and adult GMECs failed to integrate within the forming organoid, respectively (Supplementary Fig. 3b).
Doxorubicin (DOX) exposure.
The chemotherapeutic drug DOX (Sigma) is dosed at 10 μM for 24 h from Day 20 to Day 21 of culture in either static or high FSS conditions.
Bead Perfusion.
At Day 21 of differentiation in either static or perfused conditions, 100 nm fluorescent beads are added to the media (FluoSpheres from ThermoFisher, carboxylate terminated) at a dilution of 1:1000. For the static case, the organoids are gently shaken in the incubator for 3 h in the presence of bead-laden media. For the perfused conditions, the kidney organoids-on-chip are perfused with bead-laden media under high FSS conditions for 3 h in the incubator. Given the high degree of light scattering by the tissue, a two-step method employing fiduciary markers facilitated bead visualization within the whole organoids. The kidney organoids are then imaged using confocal microscopy to determine the distribution of the fluorescent beads within them. Fiduciary markers in the sample are used to ensure that after fixing, washing, and staining for PECAM1 (CD31), the same confocal Z-stack is collected with endothelial markers and can be properly correlated with bead location. Note, the beads are nearly completely flushed out during the washing and primary and secondary staining process. Z-stack images and reconstructions are rendered (Figs. 2l,m; Supplementary Fig. 11). While we find that the beads non-specifically bind to both static and perfused organoids, they concentrate in larger luminal CD31+ structures and are observed in those luminal spaces throughout the entire depth of those organoids under high FSS conditions.
To obtain a live perfusion bead movie, slightly different techniques were used. First, live imaging requires a very bright and lasting stain of the vasculature. We tried live tagging of CD31 and CD146 using fluorophore conjugated antibodies, but the signal was not strong. We switched to using a rhodamine-conjugated agglutinin (ULEX: Ulex Europaeus Agglutinin I (UEA I) from Vector Laboratories) as the signal overlaps with both CD31 and CD146 and was bright. As scattering from thick organoid tissue and non-specific deposition of beads from superfusion alone were known problems, we switched to imaging the vessels live within the first 15 minutes of starting bead perfusion. We imaged near the bottom of the organoid, close to the glass where perfusion is likely limited in comparison to the top of the organoid, but so is non-specific bead uptake. Before imaging we perfused in ULEX at a 1:200 dilution in media. ULEX was quickly rinsed away with fresh media at a 3x volume dilution and replaced with media + beads at a 1:1000 dilution. The imaging was taken for 900 frames at 400 ms between frames and half way through the ULEX laser was turned off in order to capture the bead flow alone on chip. The video was rendered at 20 frames per second (Supplementary Video 2, Supplementary Fig. 10).
Flow Cytometry.
To prepare live cells for flow cytometry, several digestion steps are required. First, the organoids (10 to 15 per condition) are cut away from the ECM and placed in a 15 mL Falcon tube. Excess media is removed and the sample is washed with PBS without Ca+2 and Mg+2. Next 100 uL of 2.5% trypsin (Corning) in 10 mM EDTA (Sigma-Aldrich) is added for 2 min at 37˚C. Then 1.9 mL of PBS without Ca+2 and Mg+2 is added and the tube is centrifuged at 300xg for 4 min. The supernatant is aspirated and 200 μL of collagenase IV (STEMCELL Technologies) is added and the suspension is pipetted to further break up the organoid. The samples are incubated for 10 min at 37˚C. After more pipetting, 5 mL of PBS is added and samples are centrifuged at 300xg for 4 min. The supernatants are aspirated and the cells are incubated on ice for 30 min with FLK1–555 (Bioss) at a 1:10 dilution in PBS without Ca+2 and Mg+2. The samples are washed 3X with PBS without Ca+2 and Mg+2 and then suspended in either DAPI (Sigma) or SYTOX Red (ThermoFisher 1:1000 dilution) in BD FACS Flow Buffer in a total of 300 μL of fluid. Samples are sent through a Falcon 70 μm filter prior to imaging in flow cytometry. Cells are analyzed by flow cytometry (BD LSR Fortessa) and data is collected from n = 100,000 cells per sample. All gates used to ensure live, homogenous cells are counted are shown in Supplementary Figs. 7 and 8. Note that in Supplementary Fig. 8d,f we observed a bimodal population of large and small cells sizes for Day 21 heterogenous kidney organoids. KDR+ EPCs were found predominantly in the small cellular fraction.
qRT-PCR.
Kidney organoids are manually extracted from perfusable chips by pipette. RNA is isolated from kidney organoid samples using TRIzol (Invitrogen) according to manufacturer’s protocol. A minimum of 6 organoids are used per sample. cDNA is synthesized using a High-capacity cDNA Reverse Transcription kit (Applied Biosystems). Quantitative Real-time PCR is performed using iTaq SYBR green supermix (Bio-Rad) and a Bio-Rad iQ5 Multicolor Real-time PCR Detection System. Primer sequences are designed using FASTA sequences (Pubmed) and verified using Primer3, and one of the primers from the pairs of primers is designed to include an exon-exon junction. Target genes are normalized to Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) expression. The mRNA expression is calculated using the 2-ΔΔCt method, expressed as an n-fold difference relative to the control group, and reported with standard error bars. Of note, cDNA quality was confirmed by DNA gel electrophoresis of the housekeeping gene, GAPDH, across samples (Supplementary Fig. 14c). A full primer list can be found in Supplementary Table 1.
Obtaining Mouse Embryonic Kidneys.
All procedures were in accordance with the NIH Guide for the Care and Use of Laboratory Animals and were approved by Institutional Animal Care and Use Committees at Brigham and Women’s Hospital. Embryonic kidneys at stage E14.5 (day of plug=E0.5) were isolated from timed pregnant females (Charles River).
Electron microscopy.
For transmission electron microscopy (TEM), kidney organoids or mouse embryonic kidneys are fixed in place using 2.5% glutaraldehyde, 1.25% paraformaldehyde, and 0.03% picric acid in 0.1 M sodium cacodylate buffer (pH 7.4) for a minimum of several hours. Small samples (1 mm x 1 mm) are excised and washed in 0.1 M cacodylate buffer and bathed in 1% osmiumtetroxide (OsO4) (EMS) and 1.5% potassium ferrocyanide (KFeCN6) (Sigma) for 1 h, washed in water 3x and incubated in 1% aqueous uranyl acetate (EMS) for 1 h followed by 2 washes in water and subsequent dehydration in varying grades of alcohol (10 min each; 50%, 70%, 90%, 2×10 min 100%). The organoids or mouse kidneys are then put in propyleneoxide (EMS) for 1 h and incubated overnight in a 1:1 mixture of propyleneoxide and TAAB Epon (Marivac Canada Inc. St. Laurent, Canada). The following day the samples are embedded in TAAB Epon and polymerized at 60ºC for 48 h. Ultrathin sections (about 60 nm) are cut on a Reichert Ultracut-S microtome, placed on copper grids, stained with lead citrate, and examined in a JEOL 1200EX Transmission electron microscope and images are recorded with an AMT 2k CCD camera.
For scanning electron microscopy (SEM), kidney organoids or mouse kidneys are again fixed in place using 2.5% glutaraldehyde, 1.25% paraformaldehyde, and 0.03% picric acid in 0.1 M sodium cacodylate buffer (pH 7.4) for a minimum of several hours. They are then washed 3x with PBS until picric acid (yellow color) is washed out. Organoids/mouse kidneys are placed in a 30% sucrose in PBS solution for 1 h. Then that solution is removed and replaced with a 1:1 mixture of 30% sucrose solution in PBS: optimal cutting temperature (OCT) freezing medium (Electron Microscopy Science) for 30 – 45 min at room temperature. Organoids are then set in a cryomold for freezing, excess fluid is removed and OCT is placed on top to fill in the mold. The organoids are frozen and placed in the −20°C freezer overnight. The samples are then cut in a cryotome in 5 μm sections (ThermoFisher), mounted on glass slides, and stained using hematoxylin and eosin. Once the opening to Bowman’s capsules is visible, sectioning is ceased. The organoids are then unembedded from OCT by heating the samples to 40°C, physically removing them from OCT, washing extensively with water and then dehydrating the tissue. Subsequent dehydration in varying grades of ethanol is required (20 min each; 30%, 50%, 70%, 90%, 3×20 min 100%). The samples are then placed in 50% ethanol and 50% hexamethyldisilazane (HMDS) for 30 min followed by 100% HMDS 3×30 min. All steps are performed in a closed glass container. After the final washing with HMDS, the samples are removed and placed in an open container in the fume hood to dry. Dried samples are mounted to aluminum pin mounts using conductive carbon tape, sputter coated with 5 nm of gold or platinum, and imaged with a UltraPlus Field Emission SEM (Zeiss) at 1 keV.
Immunostaining.
Immunostaining followed by confocal microscopy is used to assess the localization of cellular or extracellular proteins within or adjacent to organoids. Prior to immunostaining, each organoid sample is washed with PBS and then fixed for 1 h using 10% buffered formalin. The fixative is removed using several washes in PBS for several hours and then blocked overnight using 1 wt% donkey serum in PBS with 0.125 wt% TritonX-100. Primary antibodies to the protein or biomarker of interest are incubated with the constructs for 2 days at 4°C at the dilutions listed in Supplementary Table 2 in a solution of 0.5 wt% BSA and 0.125 wt% Triton X-100. Removal of unbound primary antibodies is accomplished using a wash step against a solution of PBS or 0.5 wt% BSA and 0.125 wt% Triton X-100 in PBS for 1 day. Secondary antibodies are incubated with the constructs for several hours at 1:500 dilution in a solution of 0.5 wt% BSA and 0.125 wt% Triton X-100 in PBS. Samples are counter-stained with DAPI and then washed for at least several hours in PBS prior to imaging. A full antibody list can be found in Supplementary Table 2.
Image rendering and analysis.
Phase contrast microscopy is performed using an inverted Leica DM IL scope with objectives ranging from 1.25X to 40X. Confocal microscopy is performed using an upright Zeiss LSM 710 with water immersion objectives ranging from 10X to 40X employing spectral lasers at 405, 488, 514, 561, and 633 nm wavelengths. Image reconstructions of z-stacks are performed in Imaris using the z-projection function with the maximum pixel intensity setting. Any increases in brightness are performed uniformly across an entire z-projected image. 3D image reconstructions and rotating movies are also performed using Imaris software. The CytoSMART (Lonza) in incubator system is used to capture time-lapse imaging (Supplementary Video 2). Confocal z-stacks are used to count the percent of ciliated cells (Fig. 3c), > 64 counts per condition including 4 biological replicates, and the amount of PODXL+ clusters that were invaded or wrapped by vascular cells (Fig. 4b, Supplementary Fig. 16), n > 14 biological replicates per condition, over 4 independent experiments for high flow and static conditions and n between 6 and 16 biological replicates (whole organoids) per conditions over 2 independent experiments for VEGF inhibition and addition.
Angiotool Analysis:
Confocal Z-stacks of PECAM1 are taken of fixed whole mount organoids, both iPSC and ESC derived, in the various culture conditions. The Z-stacks are taken at the limit of the confocal depth with each sample, nearly 250 μm per sample which represents approximately the same volume per organoid analyzed. Those z-stacks are then flattened using ImageJ to a 2D max intensity projection (as required by the Angiotool for input). The default settings were employed on the Angiotool for analysis and vessel diameters of 4, 7, 10, and 14 were analyzed for each organoid. In all cases, the whole organoid was used for analysis.
Imaris Analysis:
Confocal Z-stacks of PECAM1 and LTL are taken of fixed whole mount organoids, both iPSC and ESC derived, in the various culture conditions. The Z-stacks are taken at the limit of the confocal depth with each sample, roughly 250 μm per sample which represents approximately the same volume per organoid analyzed. Those z-stacks are then opened in Imaris imaging software. As shown in Supplementary Fig. 15, the confocal 3D rendering is turned into a vascular surface and a tubule surface. Then the Imaris surface surface contact area XTension is used to quantify the percent overlap between the vascular and tubular surfaces in 3D, values are reported in Fig. 3h. In order to assess the average distance the vasculature is away from a tubule, first the Imaris distance transformation XTension is run on the tubule surface. The resulting channel is then masked by the vascular surface to create a new masked distance transformation channel. That masked distance transformation channel is sent to Fiji (ImageJ) to retrieve a histogram of the masked voxels, from which we obtained a geometric mean for each sample which is reported in Fig. 3i.
Statistics and Reproducibility.
Data in all bar charts and dot plots are expressed as means ± standard deviation. Statistical analysis is performed using MATLAB and GraphPad Prism 7 and statistical significance is determined at a value of p < 0.05 as determined by ANOVA analysis as described in the figure legends. Different significance levels (p values) are indicated in each figure with asterisks as such: *p<0.05, **p<0.01, ***p<0.001. P values for Supplementary Figure 17c are summarized in Supplementary Table 3. For transparency, we state the number of times experiments were repeated independently with similar results to produce the data shown: Fig. 1b = 6, Fig. 1c-l = 15, Fig. 1m = 4, Fig. 1n = 1, Fig. 2b = 2, Fig. 2c-e = 1, Fig. 2f-h = 15, Fig. 2i-k = 5, Fig. 2l-m = 9, Fig. 2n = 10, Fig. 3a-b = 3, Fig. 3c = 3, Fig. 3d-f = 1, Fig. 3g = 15, Fig. 3h-i = 2, Fig. 3j-k = 5, Fig. 4a = 15, Fig. 4b = 3, Fig. 4c = 1, Fig. 4d-l = 15, Fig. 4m = 15, Fig. 4n = 15, Fig. 4o-p=1, Supplementary Fig. 1f-g = 2, Supplementary Fig. 2a-f = 6, Supplementary Fig. 2h-i = 15, Supplementary Fig. 3a-b = 2, Supplementary Fig. 4a-l = 4, Supplementary Fig. 5–8 = 2, Supplementary Fig. 9 = 15, Supplementary Fig. 10–11 = 9, Supplementary Fig. 12a-b = 15, Supplementary Fig. 12c-d = 2, Supplementary Fig. 13a-j = 3, Supplementary Fig. 14a-d = 1,Supplementary Fig. 16a-b = 15, Supplementary Fig. 17a = 3, Supplementary Fig. 17b-c = 4, Supplementary Fig. 18a-d,f = 15, Supplementary Fig. 19 = 15, Supplementary Fig. 20 = 3, Supplementary Fig. 21 = 10, Supplementary Fig. 22 = 3, Movie 2 = 9, Movie 3-6 = 15.
Supplementary Material
Acknowledgements
The authors thank Dr. Pierre Galichon for flow cytometry analyses, Ms. Yoko Yoda and Dr. Koichiro Susa for cell culture and immunocytochemistry, Dr. Sanjay Jain at The Washington University Kidney Translational Research Center (KTRC) for providing the BJFF hiPSC line, Annie Moisan, Christopher Chen, and Sebastian Uzel for insightful discussions, James Weaver, Benito Roman-Manso, Nanjia Zhou, and Maria Ericsson for imaging assistance, and Lori Sanders for videography.
This study was supported by a National Institutes of Health (NIH) T32 fellowship training grant (DK007527, to N.G.), a Harvard Stem Cell Institute interdisciplinary grant (to N.G.), an NIH Subaward (U01DK107350, to T.V.), an NIH R37 grant (DK039773, to J.V.B), , an NIH UG3 grant (TR002155, to J.V.B, T.V., J.A.L., and R.M.), a Brigham and Women’s Hospital Research Excellence Award (to N.G. and R.M.), a Brigham and Women’s Hospital Faculty Career Development Award (R.M.), a Harvard Stem Cell Institute Seed Grant (R.M. and J.A.L.), the NIDDK Diabetic Complications Consortium (DiaComp, www.diacomp.org) grant (DK076169, to R.M.), the NIH (Re)Building a Kidney Consortium (U01DK107350, K.A.H., J.A.L), the Office of Naval Research Vannevar Bush Faculty Fellowship program under award number N000141612823 (M.S.S. and J.A.L.), the Wyss Institute for Biologically Inspired Engineering (D.B.K., K.T.K, D.M., J.A.L.), and NIH grant supporting The Washington University KTRC (P30 DK079333, the BJFF line). J.A.L. thanks the GETTYLAB and Dr. Stan Lindenfeld for their generous donations in support of this research. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Conflict of Interest
J.V.B. and R.M. are co-inventors on patents (PCT/US16/52350) on organoid technologies that are assigned to Partners Healthcare. J.V.B. or his family has received income for consulting from companies interested in biomarkers: Sekisui, Millennium, Johnson & Johnson and Novartis. J.V.B. is a co-founder, consultant to, and owns equity in Goldfinch Bio. K.A.H. is a co-founder and chairwoman for NanoHybrids Inc. J.A.L. is a co-founder and owns equity in Voxel8 Inc.
Data Availability:
The data generated in this study are available from the corresponding authors upon request.
References:
- 1.Morizane R & Bonventre JV Generation of nephron progenitor cells and kidney organoids from human pluripotent stem cells. Nat Protoc 12, 195–207, doi: 10.1038/nprot.2016.170 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morizane R et al. Nephron organoids derived from human pluripotent stem cells model kidney development and injury. Nat. Biotechnol. 33, 1193–1200, doi: 10.1038/nbt.3392 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takasato M et al. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature 526, 564–568, doi: 10.1038/nature15695 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Lam AQ et al. Rapid and efficient differentiation of human pluripotent stem cells into intermediate mesoderm that forms tubules expressing kidney proximal tubular markers. J. Am. Soc. Nephrol. 25, 1211–1225, doi: 10.1681/ASN.2013080831 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freedman BS et al. Modelling kidney disease with CRISPR-mutant kidney organoids derived from human pluripotent epiblast spheroids. Nat Commun 6, 8715, doi: 10.1038/ncomms9715 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morizane R & Bonventre JV Kidney Organoids: A Translational Journey. Trends Mol Med 23, 246–263, doi: 10.1016/j.molmed.2017.01.001 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taguchi A et al. Redefining the in vivo origin of metanephric nephron progenitors enables generation of complex kidney structures from pluripotent stem cells. Cell Stem Cell 14, 53–67, doi: 10.1016/j.stem.2013.11.010 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Takasato M & Little MH A strategy for generating kidney organoids: Recapitulating the development in human pluripotent stem cells. Dev Biol, doi: 10.1016/j.ydbio.2016.08.024 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu H et al. Comparative analysis of kidney organoid and adult human kidney single cell and single nucleus transcriptomes. bioRxiv, doi: 10.1101/232561 (2017). [DOI] [Google Scholar]
- 10.van den Berg CW et al. Renal Subcapsular Transplantation of PSC-Derived Kidney Organoids Induces Neo-vasculogenesis and Significant Glomerular and Tubular Maturation In Vivo. Stem Cell Reports 10, 751–765, doi: 10.1016/j.stemcr.2018.01.041 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takebe T et al. Vascularized and Complex Organ Buds from Diverse Tissues via Mesenchymal Cell-Driven Condensation. Cell Stem Cell 16, 556–565, doi: 10.1016/j.stem.2015.03.004 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Bantounas I et al. Generation of Functioning Nephrons by Implanting Human Pluripotent Stem Cell-Derived Kidney Progenitors. Stem Cell Reports 10, 766–779, doi: 10.1016/j.stemcr.2018.01.008 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Camp JG et al. Multilineage communication regulates human liver bud development from pluripotency. Nature 546, 533–538, doi: 10.1038/nature22796 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Kolesky DB et al. 3D bioprinting of vascularized, heterogeneous cell-laden tissue constructs. Adv Mater 26, 3124–3130, doi: 10.1002/adma.201305506 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Homan KA et al. Bioprinting of 3D Convoluted Renal Proximal Tubules on Perfusable Chips. Sci Rep 6, 34845, doi: 10.1038/srep34845 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Halt KJ et al. CD146(+) cells are essential for kidney vasculature development. Kidney Int 90, 311–324, doi: 10.1016/j.kint.2016.02.021 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Zudaire E, Gambardella L, Kurcz C & Vermeren S A Computational Tool for Quantitative Analysis of Vascular Networks. PLOS ONE 6, e27385, doi: 10.1371/journal.pone.0027385 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Munro DAD, Hohenstein P & Davies JA Cycles of vascular plexus formation within the nephrogenic zone of the developing mouse kidney. Scientific reports 7, 3273, doi: 10.1038/s41598-017-03808-4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daniel E et al. Spatiotemporal heterogeneity and patterning of developing renal blood vessels. Angiogenesis, 1–18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robert B, St. John PL & Abrahamson DR Direct visualization of renal vascular morphogenesis in Flk1 heterozygous mutant mice. American Journal of Physiology - Renal Physiology 275, F164–F172 (1998). [DOI] [PubMed] [Google Scholar]
- 21.McMahon AP Development of the Mammalian Kidney. Curr Top Dev Biol 117, 31–64, doi: 10.1016/bs.ctdb.2015.10.010 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abrahamson DR Development of kidney glomerular endothelial cells and their role in basement membrane assembly. Organogenesis 5, 275–287 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scheppke L et al. Notch promotes vascular maturation by inducing integrin-mediated smooth muscle cell adhesion to the endothelial basement membrane. Blood 119, 2149–2158, doi: 10.1182/blood-2011-04-348706 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu S, Kim C, Baer L & Zhu X Bevacizumab increases risk for severe proteinuria in cancer patients. J. Am. Soc. Nephrol. 21, 1381–1389, doi: 10.1681/ASN.2010020167 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharmin S et al. Human Induced Pluripotent Stem Cell-Derived Podocytes Mature into Vascularized Glomeruli upon Experimental Transplantation. J. Am. Soc. Nephrol. 27, 1778–1791, doi: 10.1681/ASN.2015010096 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Serluca FC, Drummond IA & Fishman MC Endothelial signaling in kidney morphogenesis: a role for hemodynamic forces. Curr Biol 12, 492–497 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Ichimura K et al. Morphological process of podocyte development revealed by block-face scanning electron microscopy. J. Cell Sci. 130, 132–142, doi: 10.1242/jcs.187815 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Huch M, Knoblich JA, Lutolf MP & Martinez-Arias A The hope and the hype of organoid research. Development 144, 938–941, doi: 10.1242/dev.150201 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Oxburgh L & Carroll TJ The bioengineered kidney: science or science fiction? Current Opinion in Nephrology and Hypertension 25, 343–347, doi: 10.1097/mnh.0000000000000235 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Little MH Growing Kidney Tissue from Stem Cells: How Far from “Party Trick” to Medical Application? Cell Stem Cell 18, 695–698, doi: 10.1016/j.stem.2016.05.015 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only references:
- 31.Ainslie KM, Garanich JS, Dull RO & Tarbell JM Vascular smooth muscle cell glycocalyx influences shear stress-mediated contractile response. J Appl Physiol (1985) 98, 242–249, doi: 10.1152/japplphysiol.01006.2003 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


