Abstract
Adherent-invasive Escherichia coli (AIEC) has been reported as associated with the pathogenesis of inflammatory bowel disease (IBD). We aimed to investigate the characteristics of mucosa-associated E. coli and the clinical significance of AIEC in Korean IBD patients. E. coli strains were isolated from the mucosal tissues of 18 Crohn’s disease (CD) patients, 24 ulcerative colitis (UC) patients, and 9 healthy controls (HC). Adhesion, invasion, and survival assays were performed to evaluate phenotypic features of E. coli isolates and to identify AIEC. The presence of virulence genes and cytokine expression were examined using PCR. In addition, data on IBD-related hospitalization were collected. A total of 59 E. coli strains were isolated (25 from CD, 27 from UC, and 7 from HC). The average levels of adhesion, invasion, and survival were higher in E. coli strains from IBD patients than those from HC (adhesion: 1.65 vs. 0.71, p = 0.046; invasion: 1.68 vs. 0.52, p = 0.039; survival: 519.55 vs. 47.55, p = 0.363). Prevalence of AIEC in HC, CD and UC patients was 22.2%, 38.9% and 37.5%, respectively. E. coli isolates from IBD patients had various virulence genes and were associated with increased expression of TNF-α and IL-17. IBD-related hospitalization within 3 years was 18.8% in patients with AIEC and 11.5% in patients without AIEC. E. coli strains from IBD patients showed high levels of adhesion, invasion, and survival. AIEC strains were identified in both CD and UC patients at a similar rate. AIEC may be associated with sustaining inflammation in the pre-existing inflammatory mucosa.
Introduction
Inflammatory bowel disease (IBD), including Crohn’s disease (CD) and ulcerative colitis (UC), is characterized by chronic inflammation of the gastrointestinal tract. Although the etiology of IBD is not fully understood, it is generally accepted that genetic susceptibility, environmental factors, and altered intestinal microbiota are involved [1]. A dysbiosis, characterized by an increase in the number of mucosa-associated colitogenic bacteria and a reduction in overall biodiversity, has been suggested as an important contributor to the pathogenesis of IBD [2]. Previous studies have reported that increased numbers of mucosa-associated Escherichia coli strains were observed in patients with IBD [3, 4]. Adherent-invasive E. coli (AIEC), in particular, has been proposed as a possible pathogen that may potentially induce intestinal inflammation [5].
Several factors have been identified for the interaction between AIEC and the intestinal mucosa in IBD. AIEC strains are able to adhere to intestinal epithelial cells (IECs) using type 1 pili that bind to carcinoembryonic antigen-related cell-adhesion molecule 6 (CEACAM6) receptors on enterocytes, which are overexpressed on the surface of IECs in patients with CD [6]. In addition, AIEC is able to invade into the lamina propria and Peyer’s patches through M cells via long polar fimbriae [7]. AIEC not only can be internalized into macrophages, but also can survive and replicate within macrophages due to host autophagy defect. Then, AIEC induces the release of tumor necrosis factor alpha (TNF-α) by the activation of infected macrophages, and also induces Th17 and CD8+ cytotoxic responses [5, 8].
Thus, AIEC strains are considered to play an important role in the intestinal inflammatory responses. To date, most studies on AIEC have been conducted in Western countries, and inconsistent data exist regarding the prevalence and pathogenic role of AIEC in patients with IBD [1]. Therefore, we aimed to investigate the characteristics of mucosa-associated E. coli, identify AIEC, and evaluate the clinical significance of AIEC in Korean IBD patients.
Materials and methods
Study subjects
9 healthy controls (HCs), 18 CD patients, and 24 UC patients underwent colonoscopy and biopsy. Subjects signed an informed consent prior to undergoing endoscopy. HCs underwent colonoscopy for other reasons such as cancer screening, and tissues were taken from the ileocecal valve or rectum. In patients with IBD, tissues were taken from the inflamed areas for patients with active disease and from the ileocecal valve or rectum for patients with inactive disease. Disease activity was assessed endoscopically, using the simple endoscopic score for Crohn’s disease (SES-CD) [9] and ulcerative colitis endoscopic index of severity (UCEIS) [10]. SES-CD <3 and UCEIS ≤1 were considered endoscopically inactive disease. We reviewed medical records to obtain demographic characteristics, disease location and behavior, prior therapy, and IBD-related hospitalization rates. All samples were stored at -80°C until further analysis. The study protocol was approved by the Institutional Review Board of Hanyang University Guri Hospital (IRB No. 2014-10-011-001).
E. coli strains
Each biopsy sample was washed with phosphate-buffered saline (PBS) and homogenized using a pestle. Next, tissue lysates were diluted and plated onto MacConkey agar plates to obtain colonies. E. coli strains were identified using the matrix assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry system from Bruker Biotyper (Bruker Daltonics, Bremen, Germany). All E. coli strains were phylotyped into A, B1, B2, and D groups using a multiplex polymerase chain reaction (PCR) method. Reference E. coli strains K-12 and LF82 were used as a negative and positive control, respectively. When ≥2 strains were isolated from one subject, Enterobacterial Repetitive Intergenic Consensus (ERIC)-PCR was performed to evaluate clonality as previously described by Ardakani et al. [11].
Adhesion assays
HEp-2 cell monolayers were cultured in 12-well plates at a density of 1 x 105 cells/well at 37°C for 24 h, infected with 1 ml bacterial suspension (107 bacteria/ml; multiplicity of infection [MOI] = 100), and incubated at 37°C for 3 h [12]. After incubation, infected cell monolayers were washed with PBS, fixed in methanol and Giemsa-stained. Then cell layers were examined using a light microscope. Adhesion index, defined as the mean number of bacteria per cell after examination of 20 visual fields in three independent experiments, was measured for each isolate. Positive adhesion was defined when the adhesion index was ≥1. Grade of adhesion was defined as follows: grade 1, adhesion index ≥1; grade 2, adhesion index ≥10; grade 3, adhesion index ≥100 [13].
Invasion assays
The ability of all E. coli isolates to invade host cells was examined using the gentamicin protection assay, described by Darfeuille-Michaud et al. [14]. HEp-2 cells were seeded in 24-well tissue culture plates at a density of 1 x 105 cells/well, infected with 0.5 ml bacterial suspension (5 x 106 bacteria/ml; MOI = 50), and incubated at 37°C for 3 h. After incubation, infected cell monolayers were washed four times with PBS, supplemented with fresh medium containing 100 μg/ml gentamicin (Sigma) to kill extracellular bacteria, and further incubated for 1 h. Then cells were washed and lysed with a 1% Triton X-100 (Sigma) solution, and cell lysates dilutions were plated on MacConkey agar. Invasion level, defined as the percentage of intracellular bacteria compared with the initial inoculum, was measured. An isolate was considered invasive when the invasion level was ≥0.1%.
Survival assays
THP-1 cells were seeded in 24-well tissue culture plates at a density of 1 x 105 cells/well and infected with 0.1 ml bacterial suspension (1 x 106 bacteria/ml; MOI = 10). Bacteria were centrifuged onto cell monolayers at 500 x g for 10 min and incubated at 37°C for 10 min. After incubation, infected macrophages were washed twice with PBS and supplemented with fresh medium containing 100 μg/ml gentamicin to kill extracellular bacteria. After a 1 h of additional incubation period, the medium was removed and fresh medium with 50 μg/ml gentamicin was added for either 1 or 24 h. Then, cells were washed and lysed with a 1% Triton X-100 solution, and cell lysates dilutions were plated on MacConkey agar. Survival level, defined as the ratio between the number of intracellular bacteria recovered after 24 h of incubation and those recovered after 1 h, was measured. An isolate was considered to have survival capability when the survival level was ≥100% [14]. All assays were performed three times in separate experiments, and details of the used cell lines are shown in S1 Table.
Determination of AIEC
AIEC strains were determined when all of the following three conditions were met: (1) adhesiveness, adhesion index ≥1; (2) invasiveness, invasion level ≥0.1%; and (3) survival and replication, survival level ≥100% [1].
Virulence genotyping
The presence of 21 virulence factors (fimH, afa-dra, sfa-foc, hra, eaeA, ibeA, tia, ipaH, fyuA, chuA, kpsMT II, kps MT I(K1), hlyA, estA, estB, ompA, pic, aggR, and yjaA) was examined by PCR using appropriate primers. A detailed procedure for the identification of each virulence factor was performed with reference to a previous report [15].
In addition, to evaluate the genetic polymorphism of E. coli isolates, fimH gene was amplified by PCR and sequenced for identification of hotspot mutations reported in a previous study [16]. We searched for three points of mutation (G73A/E/R/W, T158A/P, R166C/H/S) in the amino acid sequence ranging from 15AA to 289AA.
Reverse transcription PCR (RT-PCR)
Total RNA was extracted from cell lines infected with E. coli isolates and controls using a Hybrid-R Total RNA Isolation Kit (GeneAll Biotechnology, Seoul, Korea), and quantified using a Biospec-nano spectrophotometer (Life Science, Columbia, MD, USA). The PCR products were resolved by electrophoresis on 1.5% agarose gels containing ethidium bromide, and bands were visualized using a ChemiDoc XRS+ System (Bio-Rad, CA, USA). The primer sequences used for PCR are shown in S2 Table.
Statistical analysis
Continuous and categorical variables among groups were compared using the Mann-Whitney test and the chi-square or Fisher’s exact test, respectively. A p value of <0.05 was considered statistically significant. All statistical procedures were conducted using IBM SPSS Statistics 20.0 (IBM Corp., Armonk, NY, USA).
Results
Characteristics of the study subjects
Table 1 shows the baseline characteristics of the study subjects. E. coli strains were isolated in 6 of 9 HC subjects, 16 of 18 CD patients, and 19 of 24 UC patients. A total of 83.3% of CD patients had ileocolonic disease, and 61.1% had non-stricturing and non-penetrating disease, while 54.2% of UC patients had pancolitis. 72.2% of CD patients and 29.2% of UC patients had been exposed to thiopurines, and 33.3% of CD patients had been exposed to anti-TNF agent.
Table 1. Characteristics of the study subjects.
| Variables | HC (n = 9) | CD (n = 18) | UC (n = 24) | |
|---|---|---|---|---|
| Age | 55.0 (37.0–57.0) | 25.0 (22.0–27.0) | 36.0 (26.0–46.0) | |
| Sex | Male | 4 (44.4) | 11 (61.1) | 10 (41.7) |
| Female | 5 (55.6) | 7 (38.9) | 14 (58.3) | |
| Biopsy site | Ileum | 0 (0) | 5 (27.8) | 0 (0) |
| Ileocecal valve | 0 (0) | 7 (38.9) | 3 (12.5) | |
| Colon | 9 (100) | 6 (33.3) | 21 (87.5) | |
| Number of subjects positive for E. coli | 6 (66.7) | 16 (88.9) | 19 (79.2) | |
| CD location* | Ileal | - | 1 (5.6) | - |
| Colonic | - | 2 (11.1) | - | |
| Ileocolonic | - | 15 (83.3) | - | |
| Isolated upper disease | - | 0 (0) | - | |
| CD behavior | Non-stricturing and non-penetrating | - | 11 (61.1) | - |
| Stricturing | - | 3 (16.7) | - | |
| Penetrating | - | 4 (22.2) | - | |
| UC extent* | Proctitis | - | - | 5 (20.8) |
| Left-sided colitis | - | - | 6 (25.0) | |
| Pancolitis | - | - | 13 (54.2) | |
| Disease activity† | Active | - | 14 (77.8) | 15 (62.5) |
| Inactive | - | 4 (22.2) | 9 (37.5) | |
| Treatment exposure | Aminosalicylates | - | 12 (66.7) | 15 (62.5) |
| Steroids | - | 10 (55.6) | 3 (12.5) | |
| Thiopurine | - | 13 (72.2) | 7 (29.2) | |
| Anti-TNF agent | - | 6 (33.3) | 0 (0) | |
| Methotrexate | - | 1 (5.6) | 0 (0) | |
HC, healthy control; CD, Crohn’s disease; UC, ulcerative colitis. Values are given as median (interquartile range) or numbers (%).
*Present location/extent when active disease, previous location/extent when inactive disease.
†Simple endoscopic score for Crohn’s disease (SES-CD) and ulcerative colitis endoscopic index of severity (UCEIS) were used to assess the endoscopic activity. SES-CD <3 and UCEIS ≤1 were considered endoscopically inactive diseases.
Characteristics of the isolated E. coli strains
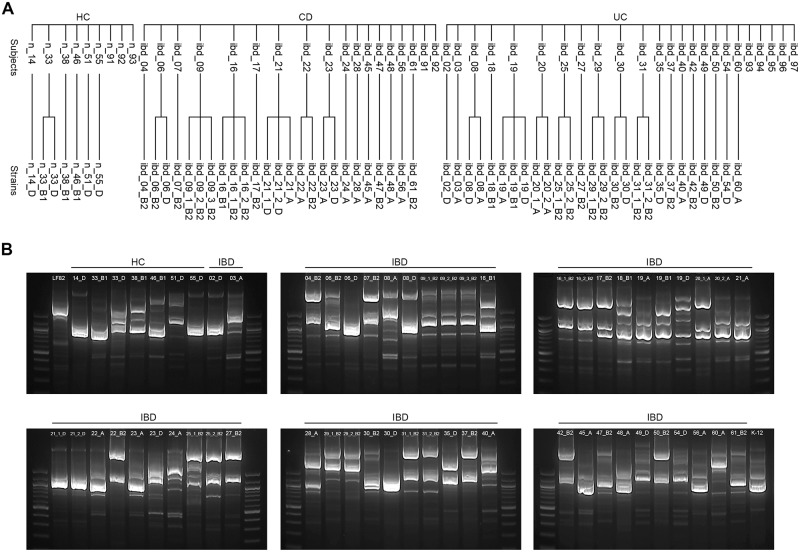
A total of 59 E. coli strains were isolated from 41 subjects: 7 strains from 6 HC subjects, 25 from 16 CD patients, and 27 from 19 UC patients. There were cases in which 2–3 strains were isolated from one subject. ERIC-PCR results demonstrated that the strains belonging to the same phylogroup showed similar electrophoresis patterns, suggesting that these strains are clonal. In contrast, the strains belonging to the different phylogroups showed distinct electrophoresis patterns, suggesting that these strains are not clonal (Fig 1).
Fig 1. Classification of E. coli isolates.
(A) Isolated E. coli strains from individual study subjects. (B) ERIC-PCR results. Prefex "n_" and "ibd_" represent healthy control and inflammatory bowel disease patients, respectively. Suffix "_A", "_B1", "_B2", and "_D" represent phylotypes. HC, healthy control; CD, Crohn’s disease; UC, ulcerative colitis.
Table 2 shows phylogenetic groups of E. coli isolates. Phylotype B2 made up 44.2% of E. coli strains in IBD patients, as compared with 0% in HC.
Table 2. E. coli isolate phylogenetic groups.
| Phylotype | HC (n = 7) | CD (n = 25) | UC (n = 27) | Total (n = 59) |
|---|---|---|---|---|
| A | 0 (0) | 8 (32.0) | 7 (25.9) | 15 (25.4) |
| B1 | 3 (42.9) | 1 (4.0) | 2 (7.4) | 6 (10.2) |
| B2 | 0 (0) | 12 (48.0) | 11 (40.7) | 23 (39.0) |
| D | 4 (57.1) | 4 (16.0) | 7 (25.9) | 15 (25.4) |
HC, healthy control; CD, Crohn’s disease; UC, ulcerative colitis. Values are given as the number (%).
E. coli strains from IBD patients showed significantly higher adhesion grade than those from HC subjects (1.65 ± 0.99 vs. 0.71 ± 0.49, p = 0.046). There were no significant differences in grade of adhesion according to phylotype and the presence of inflammation.
Although it was not statistically significant, the invasiveness of E. coli strains isolated from IBD patients was higher than those isolated from HC subjects (1.68 ± 0.27 vs. 0.52 ± 0.15, p = 0.055). Among them, E. coli strains from UC patients showed significantly higher invasion level than those from HC subjects (2.03 ± 0.34 vs. 0.52 ± 0.15, p = 0.032), and those from CD patients (2.03 ± 0.34 vs. 1.33 ± 0.42, p = 0.047). Phylotype A and B1 showed significantly higher invasion level than other phylotypes (p<0.001). There was no significant difference in invasion level according to the presence of inflammation.
E. coli strains from IBD patients showed higher average survival level than those from HC subjects, but it was not statistically significant (519.55 ± 130.82 vs. 47.55 ± 15.74, p = 0.363). Phylotype A showed significantly higher survival level than other phylotypes (p = 0.002), and E. coli strains isolated from inflamed tissues showed significantly higher survival level than those from non-inflamed tissues (628.73 ± 177.16 vs. 289.99 ± 157.98, p = 0.001).
E. coli strains from CD patients with stricturing or penetrating behavior showed increased grade of adhesion and survival level than those from CD patients without stricturing or penetrating behavior, but it was not statistically significant (grade of adhesion: 2.0 ± 0.29 vs. 1.31 ± 0.20, p = 0.074; survival level: 969.20 ± 545.20 vs. 338.0 ± 158.11, p = 0.061) (Table 3).
Table 3. Characteristics of the isolated E. coli strains.
| Characteristics | Grade of adhesiona | p value | Invasion levelb | p value | Survival levelc | p value | |
|---|---|---|---|---|---|---|---|
| Reference strains | K-12 | 0.05 ± 0.04 | - | 0.05 ± 0.02 | - | 2.61 ± 1.83 | - |
| LF82 | 1.05 ± 0.58 | 1.05 ± 0.16 | 116.07 ± 22.07 | ||||
| Disease group | HC (n = 7) | 0.71 ± 0.49 | 0.046 | 0.52 ± 0.15 | 0.039 | 47.55 ± 15.74 | 0.363 |
| CD (n = 25) | 1.56 ± 0.87 | 1.33 ± 0.42 | 634.10 ± 244.00 | ||||
| UC (n = 27) | 1.74 ± 1.10 | 2.03 ± 0.34 | 420.86 ± 123.84 | ||||
| IBD (n = 52) | 1.65 ± 0.99 | 1.68 ± 0.27 | 519.55 ± 130.82 | ||||
| Phylotype | A (n = 15) | 2.07 ± 0.03 | 0.064 | 1.88 ± 0.37 | <0.001 | 964.73 ± 374.76 | 0.002 |
| B1 (n = 6) | 1.67 ± 0.42 | 4.16 ± 1.40 | 543.84 ± 294.28 | ||||
| B2 (n = 23) | 1.39 ± 0.19 | 0.93 ± 0.26 | 229.69 ± 125.62 | ||||
| D (n = 15) | 1.20 ± 0.28 | 0.70 ± 0.21 | 290.99 ± 108.00 | ||||
| Inflammation | Absent (n = 18) | 1.72 ± 0.27 | 0.412 | 2.63 ± 0.66 | 0.105 | 289.99 ± 157.98 | 0.001 |
| Present (n = 34) | 1.62 ± 0.16 | 1.18 ± 0.21 | 628.73 ± 177.16 | ||||
| CD behavior | B1 (n = 16) | 1.31 ± 0.20 | 0.074 | 1.62 ± 0.67 | 0.450 | 338.00 ± 158.11 | 0.061 |
| B2/B3 (n = 9) | 2.00 ± 0.29 | 0.89 ± 0.22 | 969.20 ± 545.20 | ||||
HC, healthy control; CD, Crohn’s disease; UC, ulcerative colitis; IBD, inflammatory bowel disease; B1, nonstricturing and nonpenetrating; B2, stricturing; B3, penetrating. Values are given as the mean ± standard error of three independent experiments.
a1, adhesion index ≥1; 2, adhesion index ≥10; 3, adhesion index ≥100
bPercentage of inoculum surviving after 1 h of gentamicin treatment (number of intracellular bacteria / initial inoculum x 100)
cNumber of intracellular bacteria at 24 h post-infection / number of bacteria at 1h post-infection x 100 (%)
Identification of AIEC
AIEC strains were identified in 2 of 7 strains isolated from HC subjects (28.6%), 7 of 25 from CD patients (28.0%), and 12 of 27 from UC patients (44.4%) based on the criteria used to define AIEC strains as described in the methods. Among the 21 AIEC strains, 9 (42.9%), 2 (9.5%), 5 (23.8%), and 5 (23.8%) were identified as phylotype A, B1, B2, and D, respectively. Prevalence of AIEC was 22.2% (2 of 9) in HC subjects, 38.9% (7 of 18) in CD patients, and 37.5% (9 of 24) in UC patients. There were no significant differences in the prevalence of AIEC according to disease activity and treatment exposure (Table 4).
Table 4. Prevalence of adherent-invasive E. coli.
| Characteristics | Number of subjects positive for AIEC (%) | p value | |
|---|---|---|---|
| Disease group | HC (n = 9) | 2 (22.2) | 0.407 |
| CD (n = 18) | 7 (38.9) | ||
| UC (n = 24) | 9 (37.5) | ||
| Disease activity | Active IBD (n = 29) | 11 (37.9) | 0.618 |
| Inactive IBD (n = 13) | 5 (38.5) | ||
| Treatment exposure | Anti-TNF naïve (n = 36) | 14 (38.9) | 0.587 |
| Anti-TNF experienced (n = 6) | 2 (33.3) | ||
| Thiopurine naïve (n = 22) | 9 (40.9) | 0.470 | |
| Thiopurine experienced (n = 20) | 7 (35.0) | ||
AIEC, adherent-invasive E. coli; HC, healthy control; CD, Crohn’s disease; UC, ulcerative colitis; IBD, inflammatory bowel disease
Virulence genotyping
Various genes associated with virulence factors were identified. The fimH gene was identified in 100% of both control and IBD patients. Prevalence of yjaA gene was significantly higher in E.coli isolates from CD patients than those from UC patients and HC. Although it was not statistically significant, several virulence genes such as ibeA, ipaH, fyuA, kpsMT II, and K1 were more common in strains from IBD patients than those from HC (Table 5).
Table 5. Virulence genotyping.
| Function | Genes | Number of isolates with virulence genes (%) | p value | ||
|---|---|---|---|---|---|
| HC (n = 7) | CD (n = 25) | UC (n = 27) | |||
| Adhesin | fimH | 7 (100) | 25 (100) | 27 (100) | - |
| afa-dra | 6 (85.7) | 21 (84.0) | 22 (81.5) | 0.952 | |
| sfa-foc | 4 (57.1) | 11 (44.0) | 16 (59.3) | 0.527 | |
| hra | 1 (14.3) | 3 (12.0) | 5 (18.5) | 0.806 | |
| eaeA | 3 (42.9) | 11 (44.0) | 13 (48.1) | 0.943 | |
| Invasin | ibeA | 0 (0.0) | 4 (16.0) | 4 (14.8) | 0.532 |
| tia | 6 (86.0) | 19 (76.0) | 21 (77.8) | 0.860 | |
| ipaH | 2 (28.6) | 15 (60.0) | 15 (55.6) | 0.331 | |
| Siderophore | fyuA | 4 (57.1) | 19 (76.0) | 17 (63.0) | 0.491 |
| chuA | 4 (57.1) | 16 (64.0) | 18 (66.7) | 0.894 | |
| Capsule | kpsMT II | 2 (28.6) | 16 (64.0) | 14 (51.9) | 0.237 |
| kpsMT I (K1) | 1 (14.3) | 9 (36.0) | 6 (22.2) | 0.385 | |
| Toxin | hlyA | 7 (100) | 23 (92.0) | 26 (96.3) | 0.631 |
| estA | 0 (0.0) | 0 (0.0) | 0 (0.0) | - | |
| estB | 2 (28.6) | 5 (20.0) | 3 (11.1) | 0.475 | |
| Protectin | ompA | 7 (100) | 25 (100) | 27 (100) | - |
| Miscellaneous | pic | 7 (100) | 23 (92) | 23 (85.2) | 0.459 |
| aggR | 5 (71.4) | 22 (88.0) | 25 (92.6) | 0.304 | |
| yjaA | 0 (0.0) | 19 (76.0) | 13 (48.1) | 0.001 | |
| tsp | 5 (71.4) | 14 (56.0) | 17 (63.0) | 0.731 | |
HC, healthy control; CD, Crohn’s disease; UC, ulcerative colitis
In addition, mutation of the fimH gene was analyzed. Sequencing results of the fimH gene were obtained from 52 out of 59 isolated strains. As a result, R166H mutation was identified in three AIEC strains (ibd_07_B2 strain and ibd_28_A strain from CD patients, ibd_25_2_B2 strain from UC patient). All three patients had active disease. In contrast, no hotspot mutations were identified in non-AIEC strains (Table 6).
Table 6. Mutations of the fimH gene in E. coli isolates.
| Strain | Group | Phenotype | Amino acid | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 25 | 27 | 70 | 78 | 104 | 119 | 150 | 162 | 163 | 166 | 227 | |||
| K-12 | Reference | non-AIEC | A | V | N | S | P | A | A | D | V | R | R |
| LF82 | Reference | AIEC | A | S | N | ||||||||
| n_38_B1 | HC | AIEC | A | ||||||||||
| n_51_D | HC | AIEC | A | S | N | ||||||||
| ibd_03_A | UC | AIEC | A | ||||||||||
| ibd_06_B2 | CD | AIEC | L | ||||||||||
| ibd_06_D | CD | AIEC | A | V | S | ||||||||
| ibd_07_B2 | CD | AIEC | A | H | |||||||||
| ibd_19_B1 | UC | AIEC | A | ||||||||||
| ibd_22_B2 | CD | AIEC | A | S | |||||||||
| ibd_25_1_B2 | UC | AIEC | A | S | N | A | |||||||
| ibd_25_2_B2 | UC | AIEC | A | H | |||||||||
| ibd_28_A | CD | AIEC | A | H | |||||||||
| ibd_30_D | UC | AIEC | |||||||||||
| ibd_31_1_B2 | UC | AIEC | A | ||||||||||
| ibd_31_2_B2 | UC | AIEC | A | ||||||||||
| ibd_40_A | UC | AIEC | A | ||||||||||
| ibd_49_D | UC | AIEC | A | S | N | ||||||||
| ibd_54_D | UC | AIEC | A | S | N | ||||||||
| ibd_60_A | UC | AIEC | A | ||||||||||
| n_14_D | HC | non-AIEC | |||||||||||
| n_33_B1 | HC | non-AIEC | A | V | |||||||||
| n_33_D | HC | non-AIEC | A | V | |||||||||
| n_46_B1 | HC | non-AIEC | A | ||||||||||
| n_55_D | HC | non-AIEC | |||||||||||
| ibd_02_D | UC | non-AIEC | |||||||||||
| ibd_04_B2 | CD | non-AIEC | S | N | |||||||||
| ibd_08_A | UC | non-AIEC | T | A | N | ||||||||
| ibd_08_D | UC | non-AIEC | |||||||||||
| ibd_09_1_B2 | CD | non-AIEC | L | ||||||||||
| ibd_09_2_B2 | CD | non-AIEC | L | ||||||||||
| ibd_09_3_B2 | CD | non-AIEC | L | ||||||||||
| ibd_16_B1 | CD | non-AIEC | A | ||||||||||
| ibd_16_1_B2 | CD | non-AIEC | A | ||||||||||
| ibd_16_2_B2 | CD | non-AIEC | A | ||||||||||
| ibd_17_B2 | CD | non-AIEC | A | N | |||||||||
| ibd_18_B1 | UC | non-AIEC | A | ||||||||||
| ibd_19_D | UC | non-AIEC | A | S | |||||||||
| ibd_20_1_A | UC | non-AIEC | A | N | |||||||||
| ibd_21_1_D | CD | non-AIEC | |||||||||||
| ibd_21_2_D | CD | non-AIEC | |||||||||||
| ibd_23_D | CD | non-AIEC | A | S | N | ||||||||
| ibd_27_B2 | UC | non-AIEC | A | ||||||||||
| ibd_29_1_B2 | UC | non-AIEC | A | S | |||||||||
| ibd_29_2_B2 | UC | non-AIEC | A | S | |||||||||
| ibd_30_B2 | UC | non-AIEC | L | ||||||||||
| ibd_35_D | UC | non-AIEC | A | N | V | ||||||||
| ibd_37_B2 | UC | non-AIEC | S | N | |||||||||
| ibd_42_B2 | UC | non-AIEC | A | ||||||||||
| ibd_47_B2 | CD | non-AIEC | L | ||||||||||
| ibd_48_A | CD | non-AIEC | A | ||||||||||
| ibd_50_B2 | UC | non-AIEC | |||||||||||
| ibd_56_A | CD | non-AIEC | |||||||||||
| ibd_61_B2 | CD | non-AIEC | L | ||||||||||
Blanks indicate identical to wild type, and only the mutations are shown. Prefex "n_" and "ibd_" represent healthy control and inflammatory bowel disease patients, respectively. Suffix "_A", "_B1", "_B2", and "_D" represent phylotypes. HC, healthy control; CD, Crohn’s disease; UC, ulcerative colitis; AIEC, adherent-invasive E. coli
Expression of cytokines
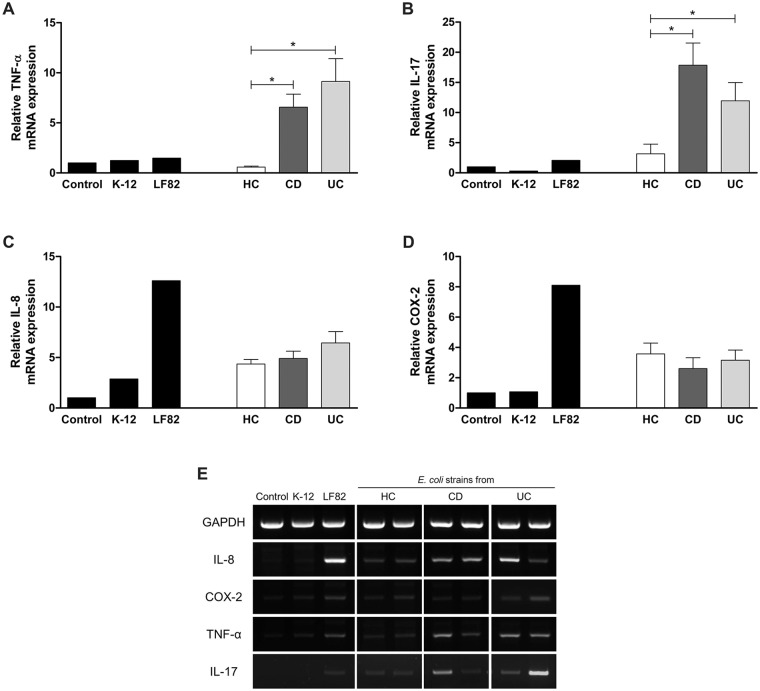
The expression of TNF-α and interleukin 17 (IL-17) was significantly increased in HEp-2 cells infected with E. coli isolates from IBD patients than those from HC subjects, whereas the expression of cyclooxygenase 2 (COX-2) and IL-8 was not significantly different between groups (Fig 2). In Caco-2 cells infected with E. coli isolates with survival capability, the relative mRNA expression level of CEACAM6 was increased by two-fold over baseline.
Fig 2. RT-PCR analyses for the expression of cytokines in E. coli-infected cell lines.
The expression of TNF-α (A) and IL-17 (B) was significantly increased by E. coli strains from IBD patients, while the expression of IL-8 (C) and COX-2 (D) was not increased. HC, healthy control; CD, Crohn’s disease; UC, ulcerative colitis; TNF-α, tumor necrosis factor alpha; IL, interleukin; COX-2, cyclooxygenase 2 *Statistically significant (p<0.05).
Clinical impact of AIEC
To determine the potential impact of AIEC on clinical outcomes, we assessed IBD-related hospitalization rates within 3 years according to the presence of AIEC at baseline. A total of 42 IBD patients (18 patients with CD, 24 with UC) were followed up for a median of 22.3 months (interquartile range, 14.3 months), and the causes of hospitalization included disease flares, IBD complications and surgery. IBD patients with AIEC showed a higher rate of IBD-related hospitalization than those without AIEC, but it was not statistically significant (3 of 16 patients, 18.8% vs. 3 of 26 patients, 11.5%, p = 0.658).
Discussion
In the present work, we evaluated characteristics of mucosa-associated E. coli and identified AIEC in Asian IBD patients. E. coli isolates from IBD patients showed higher adhesiveness, invasiveness, and survival capability than those from HC subjects. Prevalence of AIEC strains was relatively higher in IBD patients than HC subjects, and similar in CD and UC patients. AIEC strains from IBD patients were associated with increased expression of proinflammatory cytokines and increased rates of IBD-related hospitalization.
Phylotype B2 made up 44.2% of E. coli strains from IBD patients and together with phylotype A, made up the majority of strains. Previous studies reported that phylotype B2 was the most common among mucosa-associated E. coli isolates, and it was associated with inflammation [17, 18]. We observed phylotype B2 was the most common, but there were no significant increases in adhesion, invasion, and survival than other phylotypes.
Prevalence of AIEC in patients with IBD varies widely across studies in the West, ranging from 21.7% to 62.5% in CD patients [14, 19, 20] and from 0% to 10% in UC patients [14, 21, 22]. Little is known about the prevalence and pathogenic role of AIEC in UC patients. Although a few studies have reported a higher prevalence of adherent E. coli strains in UC patients, these studies did not investigate the presence of AIEC [23, 24]. In some previous studies, E. coli strains from UC patients were reported to be associated with severe disease activity, increased fecal calprotectin, and increased expression of pro-inflammatory cytokines [25, 26]. Our results showed that the prevalence of AIEC in UC patients was comparable to the prevalence in CD patients (37.5% vs. 38.9%). We also observed increased expression of TNF-α and IL-17. Interestingly, the average invasion level was higher in E. coli strains from UC patients than those from CD patients. In patients with CD, Darfeuille-Michaud et al. [14] reported that AIEC strains were found more frequently in the ileum than in the colon, whereas our results showed that AIEC strains were present not only in the ileum but also in the colon.
To our knowledge, this is the first study to identify AIEC in Asian IBD patients. As mentioned above, our results on the prevalence and distribution of AIEC differed from those have been reported in Western countries. These differences may be explained by the presence of various factors that determine AIEC colonization in the intestinal mucosa of IBD patients. The composition and distribution of the intestinal microbiota may vary according to disease phenotype and genotype in patients with IBD [27], and other host and/or environmental factors might be involved.
We found some virulence genes, including yjaA, ibeA, ipaH, fyuA, kpsMT II, and K1, were more common in E. coli strains from IBD patients than those from HC subjects. However, this does not mean that E. coli strains having these virulence factors are more pathogenic because E. coli can evolve according to the intestinal environment, and the expression of virulence genes can be increased by the intestinal inflammation [28]. It may be difficult to identify the cause-effect relationship between bacterial virulence genes and the pathogenesis of IBD.
The adherence of AIEC is dependent on type 1 pili expression on the bacterial surface and on CEACAM6 expression on the apical surface of IECs, which is increased in enterocytes of patients with CD [29]. In addition, AIEC isolates were reported to be able to induce intestinal inflammation by expressing CEACAM6 [30]. Our results showed the increased expression of CEACAM6 in cell lines infected with E. coli isolates with survival capability. In other words, the increased expression of CEACAM6 on enterocytes can increase the invasion of E. coli, while the intracellular infection of E. coli can increase the expression of CEACAM6. This implies that the interaction between enterocytes and microbiota plays an important role in inducing intestinal inflammation.
It is unclear whether AIEC strains trigger intestinal inflammation or whether they present as a consequence of inflammation in patients with IBD. AIEC strains are considered pathobionts because they are able to evolve in specific and susceptible hosts and promote intestinal inflammation by inducing the expression of proinflammatory cytokines [1, 5]. Our results showed that AIEC was present in the uninflamed mucosa as well as in the inflamed mucosa of IBD patients, which are consistent with previous findings [21]. There were no significant differences in adhesiveness and invasiveness of E. coli isolates according to the presence of inflammation, but the survival capability was significantly higher in E. coli isolates from the inflamed mucosa than those from the uninflamed mucosa. In addition, we found that E. coli strains were present in the normal mucosa of HC subjects. However, E. coli isolates from HC subjects did not increase the expression of TNF-α and IL-17, whereas those from IBD patients increased the expression of TNF-α and IL-17. Furthermore, although it was not statistically significant because of the small sample size, IBD patients with AIEC tended to have a higher rate of IBD-related hospitalization than those without AIEC. Taken together, these results suggest that AIEC is only pathogenic in a specific intestinal environment; AIEC may survive and induce further inflammation in the pre-existing inflammatory mucosa, whereas it may simply colonize without inducing inflammation in the uninflamed mucosa. AIEC may perpetuate chronic inflammation after inflammation has been evoked, leading to IBD-related complications and hospitalizations.
There are some limitations in this study. Because the number of HC subjects was relatively small, it was difficult to obtain statistical differences between IBD patients and HC subjects. Second, the presence of tissue inflammation was determined endoscopically, but not histologically. Thus, our results regarding characteristics of E. coli isolates according to the presence of inflammation may be inconclusive. Third, because we performed in vitro experiments with E. coli isolates only, it is not possible to understand the interactions between E. coli isolates and other abundant bacterial community, and the role of AIEC in the actual intestinal microenvironment. In addition, the abundance of E. coli as a whole was unable to confirm, because we did not count colonies after the tissue culture. Fourth, we could not assess the difference in E. coli phenotype according to patients’ genotype, because genomic data were not collected. Finally, MOI was adjusted for appropriate experimental results in our study, which may be an arbitrary measure. However, considering the previous study that there was no significant difference in the invasion assays tested with MOI = 10 and MOI = 100 [31], adjustment of MOI does not seem to be a significant drawback in interpreting the result.
Despite these limitations, this work will provide a better understanding of the possible role of mucosa-associated E. coli in the pathogenesis of IBD. E. coli isolates from IBD patients showed higher levels of adhesion, invasion, and survival than those from HC subjects. AIEC strains were identified from intestinal mucosal tissues of both CD and UC patients at a similar rate. AIEC may be associated with sustaining inflammation in the pre-existing inflammatory mucosa, leading to disease progression and related complications.
Supporting information
(PDF)
(PDF)
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (2018R1A2B6004475 to DSH). The funding source had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Palmela C, Chevarin C, Xu Z, Torres J, Sevrin G, Hirten R, et al. Adherent-invasive Escherichia coli in inflammatory bowel disease. Gut. 2018;67(3):574–87. Epub 2017/11/17. 10.1136/gutjnl-2017-314903 . [DOI] [PubMed] [Google Scholar]
- 2.Kaur N, Chen CC, Luther J, Kao JY. Intestinal dysbiosis in inflammatory bowel disease. Gut microbes. 2011;2(4):211–6. Epub 2011/10/11. 10.4161/gmic.2.4.17863 . [DOI] [PubMed] [Google Scholar]
- 3.Conte MP, Schippa S, Zamboni I, Penta M, Chiarini F, Seganti L, et al. Gut-associated bacterial microbiota in paediatric patients with inflammatory bowel disease. Gut. 2006;55(12):1760–7. Epub 2006/05/02. 10.1136/gut.2005.078824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kotlowski R, Bernstein CN, Sepehri S, Krause DO. High prevalence of Escherichia coli belonging to the B2+D phylogenetic group in inflammatory bowel disease. Gut. 2007;56(5):669–75. Epub 2006/10/10. 10.1136/gut.2006.099796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Darfeuille-Michaud A. Adherent-invasive Escherichia coli: a putative new E. coli pathotype associated with Crohn’s disease. International journal of medical microbiology: IJMM. 2002;292(3–4):185–93. Epub 2002/10/26. 10.1078/1438-4221-00201 . [DOI] [PubMed] [Google Scholar]
- 6.Barnich N, Darfeuille-Michaud A. Abnormal CEACAM6 expression in Crohn disease patients favors gut colonization and inflammation by adherent-invasive E. coli. Virulence. 2010;1(4):281–2. Epub 2010/12/24. 10.4161/viru.1.4.11510 . [DOI] [PubMed] [Google Scholar]
- 7.Chassaing B, Rolhion N, de Vallee A, Salim SY, Prorok-Hamon M, Neut C, et al. Crohn disease—associated adherent-invasive E. coli bacteria target mouse and human Peyer’s patches via long polar fimbriae. The Journal of clinical investigation. 2011;121(3):966–75. Epub 2011/02/23. 10.1172/JCI44632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Agus A, Massier S, Darfeuille-Michaud A, Billard E, Barnich N. Understanding host-adherent-invasive Escherichia coli interaction in Crohn’s disease: opening up new therapeutic strategies. BioMed research international. 2014;2014:567929 Epub 2015/01/13. 10.1155/2014/567929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daperno M, D'Haens G, Van Assche G, Baert F, Bulois P, Maunoury V, et al. Development and validation of a new, simplified endoscopic activity score for Crohn’s disease: the SES-CD. Gastrointest Endosc. 2004;60(4):505–12. Epub 2004/10/09. . [DOI] [PubMed] [Google Scholar]
- 10.Travis SP, Schnell D, Krzeski P, Abreu MT, Altman DG, Colombel JF, et al. Reliability and initial validation of the ulcerative colitis endoscopic index of severity. Gastroenterology. 2013;145(5):987–95. Epub 2013/07/31. 10.1053/j.gastro.2013.07.024 . [DOI] [PubMed] [Google Scholar]
- 11.Ardakani MA, Ranjbar R. Molecular typing of uropathogenic E. coli strains by the ERIC-PCR method. Electron Physician. 2016;8(4):2291–6. Epub 2016/06/10. 10.19082/2291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conte MP, Longhi C, Marazzato M, Conte AL, Aleandri M, Lepanto MS, et al. Adherent-invasive Escherichia coli (AIEC) in pediatric Crohn’s disease patients: phenotypic and genetic pathogenic features. BMC research notes. 2014;7:748 Epub 2014/10/24. 10.1186/1756-0500-7-748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mange JP, Stephan R, Borel N, Wild P, Kim KS, Pospischil A, et al. Adhesive properties of Enterobacter sakazakii to human epithelial and brain microvascular endothelial cells. BMC microbiology. 2006;6:58 Epub 2006/06/28. 10.1186/1471-2180-6-58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Darfeuille-Michaud A, Boudeau J, Bulois P, Neut C, Glasser AL, Barnich N, et al. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology. 2004;127(2):412–21. Epub 2004/08/10. . [DOI] [PubMed] [Google Scholar]
- 15.Frommel U, Lehmann W, Rodiger S, Bohm A, Nitschke J, Weinreich J, et al. Adhesion of human and animal Escherichia coli strains in association with their virulence-associated genes and phylogenetic origins. Applied and environmental microbiology. 2013;79(19):5814–29. Epub 2013/07/23. 10.1128/AEM.01384-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dreux N, Denizot J, Martinez-Medina M, Mellmann A, Billig M, Kisiela D, et al. Point mutations in FimH adhesin of Crohn’s disease-associated adherent-invasive Escherichia coli enhance intestinal inflammatory response. PLoS Pathog. 2013;9(1):e1003141 Epub 2013/01/30. 10.1371/journal.ppat.1003141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petersen AM, Nielsen EM, Litrup E, Brynskov J, Mirsepasi H, Krogfelt KA. A phylogenetic group of Escherichia coli associated with active left-sided inflammatory bowel disease. BMC microbiology. 2009;9:171 Epub 2009/08/22. 10.1186/1471-2180-9-171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sepehri S, Khafipour E, Bernstein CN, Coombes BK, Pilar AV, Karmali M, et al. Characterization of Escherichia coli isolated from gut biopsies of newly diagnosed patients with inflammatory bowel disease. Inflammatory bowel diseases. 2011;17(7):1451–63. Epub 2011/06/16. 10.1002/ibd.21509 . [DOI] [PubMed] [Google Scholar]
- 19.Martinez-Medina M, Aldeguer X, Lopez-Siles M, Gonzalez-Huix F, Lopez-Oliu C, Dahbi G, et al. Molecular diversity of Escherichia coli in the human gut: new ecological evidence supporting the role of adherent-invasive E. coli (AIEC) in Crohn’s disease. Inflammatory bowel diseases. 2009;15(6):872–82. Epub 2009/02/25. 10.1002/ibd.20860 . [DOI] [PubMed] [Google Scholar]
- 20.Raso T, Crivellaro S, Chirillo MG, Pais P, Gaia E, Savoia D. Analysis of Escherichia coli isolated from patients affected by Crohn’s disease. Current microbiology. 2011;63(2):131–7. Epub 2011/06/01. 10.1007/s00284-011-9947-8 . [DOI] [PubMed] [Google Scholar]
- 21.Elliott TR, Hudspith BN, Wu G, Cooley M, Parkes G, Quinones B, et al. Quantification and characterization of mucosa-associated and intracellular Escherichia coli in inflammatory bowel disease. Inflammatory bowel diseases. 2013;19(11):2326–38. Epub 2013/08/31. 10.1097/MIB.0b013e3182a38a92 . [DOI] [PubMed] [Google Scholar]
- 22.Negroni A, Costanzo M, Vitali R, Superti F, Bertuccini L, Tinari A, et al. Characterization of adherent-invasive Escherichia coli isolated from pediatric patients with inflammatory bowel disease. Inflammatory bowel diseases. 2012;18(5):913–24. Epub 2011/10/14. 10.1002/ibd.21899 . [DOI] [PubMed] [Google Scholar]
- 23.Schippa S, Conte MP, Borrelli O, Iebba V, Aleandri M, Seganti L, et al. Dominant genotypes in mucosa-associated Escherichia coli strains from pediatric patients with inflammatory bowel disease. Inflammatory bowel diseases. 2009;15(5):661–72. Epub 2008/12/11. 10.1002/ibd.20818 . [DOI] [PubMed] [Google Scholar]
- 24.Thomazini CM, Samegima DA, Rodrigues MA, Victoria CR, Rodrigues J. High prevalence of aggregative adherent Escherichia coli strains in the mucosa-associated microbiota of patients with inflammatory bowel diseases. International journal of medical microbiology: IJMM. 2011;301(6):475–9. Epub 2011/05/28. 10.1016/j.ijmm.2011.04.015 . [DOI] [PubMed] [Google Scholar]
- 25.Mirsepasi-Lauridsen HC, Halkjaer SI, Mortensen EM, Lydolph MC, Nordgaard-Lassen I, Krogfelt KA, et al. Extraintestinal pathogenic Escherichia coli are associated with intestinal inflammation in patients with ulcerative colitis. Scientific reports. 2016;6:31152 Epub 2016/10/01. 10.1038/srep31152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sasaki M, Sitaraman SV, Babbin BA, Gerner-Smidt P, Ribot EM, Garrett N, et al. Invasive Escherichia coli are a feature of Crohn’s disease. Laboratory investigation; a journal of technical methods and pathology. 2007;87(10):1042–54. Epub 2007/07/31. 10.1038/labinvest.3700661 . [DOI] [PubMed] [Google Scholar]
- 27.Frank DN, Robertson CE, Hamm CM, Kpadeh Z, Zhang T, Chen H, et al. Disease phenotype and genotype are associated with shifts in intestinal-associated microbiota in inflammatory bowel diseases. Inflammatory bowel diseases. 2011;17(1):179–84. Epub 2010/09/15. 10.1002/ibd.21339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patwa LG, Fan TJ, Tchaptchet S, Liu Y, Lussier YA, Sartor RB, et al. Chronic intestinal inflammation induces stress-response genes in commensal Escherichia coli. Gastroenterology. 2011;141(5):1842–51 e1-10. Epub 2011/07/06. 10.1053/j.gastro.2011.06.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barnich N, Carvalho FA, Glasser AL, Darcha C, Jantscheff P, Allez M, et al. CEACAM6 acts as a receptor for adherent-invasive E. coli, supporting ileal mucosa colonization in Crohn disease. The Journal of clinical investigation. 2007;117(6):1566–74. Epub 2007/05/26. 10.1172/JCI30504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carvalho FA, Barnich N, Sivignon A, Darcha C, Chan CH, Stanners CP, et al. Crohn’s disease adherent-invasive Escherichia coli colonize and induce strong gut inflammation in transgenic mice expressing human CEACAM. The Journal of experimental medicine. 2009;206(10):2179–89. Epub 2009/09/10. 10.1084/jem.20090741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boudeau J, Glasser AL, Masseret E, Joly B, Darfeuille-Michaud A. Invasive ability of an Escherichia coli strain isolated from the ileal mucosa of a patient with Crohn’s disease. Infection and immunity. 1999;67(9):4499–509. Epub 1999/08/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.