Abstract
Leishmaniasis is a zoonotic disease of worldwide relevance. Visceral leishmaniasis is endemic in Brazil, where it is caused by Leishmania infantum with Lutzomyia longipalpis being the most important invertebrate vector. Non-human primates are susceptible to L. infantum infection. However, little is known about the role of these species as reservoirs. The aim of this study was to evaluate the transmissibility potential of visceral leishmaniasis by non-human primates through xenodiagnosis using the phlebotomine Lu. longipalpis as well as to identify phlebotomine species prevalent in the area where the primates were kept in captivity, and assess infection by Leishmania in captured phlebotomine specimens. Fifty two non-human primates kept in captivity in an endemic area for leishmaniasis were subjected to xenodiagnosis. All primates were serologically tested for detection of anti-Leishmania antibodies. Additionally, an anti-Lu. longipalpis saliva ELISA was performed. Sand flies fed on all animals were tested by qPCR to identify and quantify L. infantum promastigotes. Eight of the 52 non-human primates were positive by xenodiagnosis, including three Pan troglodytes, three Leontopithecus rosalia, one Sapajus apella, and one Miopithecus talapoin, with estimated numbers of promastigotes ranging from 5.67 to 1,181.93 per μg of DNA. Positive animals had higher levels of IgG anti-Lu. longipalpis saliva when compared to negative animals, prior to xenodiagnosis. Captive non-human primates are capable of infecting Lu. longipalpis with L. infantum. Our findings also demonstrate the relevance of non-human primates as sentinels to zoonotic diseases. Several phlebotomine species, including Lu. longipalpis, have been identified in the area where the primates were maintained, but only one pool of Lutzomyia lenti was infected with L. infantum. This study has implications for public health strategies and conservation medicine.
Author summary
Visceral leishmaniasis is a zoonotic disease with worldwide distribution. The disease is endemic in several Brazilian regions, including the city of Belo Horizonte, where visceral leishmaniasis is caused by Leishmania infantum and transmitted by Lutzomyia longipalpis. This study evaluated the competence of non-human primates to infect Lutzomyia longipalpis with Leishmania infantum. Eight of 52 non-human primates were positive to leishmaniasis by xenodiagnosis, i.e. capable of infecting sand flies, with averages of 5.67 to 1,181.93 promastigotes/μg of DNA. Positive animals had higher levels of IgG anti-Lu. longipalpis saliva when compared to negative animals, prior to xenodiagnosis. This study highlights the importance of non-human primates in the leishmaniasis cycle, providing information that is relevant for development of better public health strategies, and to conservation medicine.
Introduction
Visceral leishmaniasis is a zoonotic disease caused by obligate intracellular protozoa of the genus Leishmania (order Kinetoplasta: family Trypanosomatidae), which infect macrophages of several mammalian species, including man [1]. There are different species of Leishmania that can cause visceral leishmaniasis in the world. L. infantum (synonym L. chagasi) has the broadest distribution [2]. The main form of transmission is through the bite of female flies of the subfamily Phlebotominae [1], and the most relevant biological vector in Brazil is Lutomyia longipalpis [3].
Although leishmaniasis has a high incidence, morbidity and lethality, it is one of the most neglected zoonotic diseases in the world, affecting mainly deprived human populations from developing countries in tropical areas of the Americas, Asia and Africa, extending to temperate regions of Latin America [1]. Despite its known geographic distribution, leishmaniasis, as others vector born diseases, is a dynamic disease, which transmission circumstances undergo continuous changes dependent on environmental, demographic, and human behavior factors [4].
According to the World Health Organization [1], potential wild reservoirs of visceral leishmaniasis in the New World are wild canids, especially the crab-eating fox (Cerdocyon thous), and opossums (Didelphis marsupialis and D. albiventris), although the domestic dog is recognized as the most important reservoir in urban areas [5]. Many other species of wild and synanthropic mammals have also been identified as potential reservoirs, including some species of bats, felines, and neotropical primates [6,7].
To classify an animal as a reservoir, some criteria must be met, among which the capacity of the reservoir host to maintain the availability of parasites in the skin in sufficient numbers to be transmitted to the vectors [1,8]. Xenodiagnosis is an efficient tool to evaluate the capacity of a mammalian host to transmit this pathogen, which characterizes the host as a potential reservoir [9,10]. Although the risk of transmission of leishmaniasis by potential wild reservoirs, such as crab-eating fox [11], wild rabbits [12], maned wolves [9], and bush dogs [9], has already been evaluated by xenodiagnosis, such risk is completely unknown in the case of non-human primates.
Non-human primates can be affected by leishmaniasis, with clinical and pathological manifestations that are similar to those reported in human patients, sometimes remaining asymptomatic [13–15]. However, there are only a few epidemiological studies on the occurrence of leishmaniasis in non-human primates [16], and a complete absence of scientific data on the role they play in transmission of leishmaniasis. This study aimed to evaluate the transmissibility potential of visceral leishmaniasis by non-human primates through xenodiagnoses using the phlebotomine Lu. longipalpis. In addition, we performed identification of the phlebotomine species prevalent in the area where the primates were kept in captivity and assessed infection by Leishmania in captured phlebotomine specimens.
Methods
Animals
The experimental protocol employed in this study has been approved by the Ethics Committee on the Use of Animals of the Universidade Federal de Minas Gerais (CEUA/UFMG), under protocol number 94/2013. CEUA/UFMG adheres to the Brazilian legislation (law 11794 –October 8, 2008) under supervision of the Conselho Nacional de Controle de Experimentação Animal—CONCEA.
Fifty two non-human primates kept in captivity at the zoological garden in Belo Horizonte (Brazil) were included in this study, totaling 13 species, 11 neotropical primates and two old world primate species (Table 1). Most of the primates (27/52) were born at the Belo Horizonte zoo, and all of them were housed at the zoo for at least one year prior to this study. A detailed description of the origin of each primate included in this study is provided in a Supplementary Table (S1 Table).
Table 1. Species of non-human primates kept at FZB-BH and included at present study.
| Neotropical Primates | |||
| Family | Scientific Name | N | Identification |
| Atelidae |
Alouatta caraya Alouatta guariba Lagothrix cana |
4 2 7 |
01–04 05–06 07–13 |
| Aotidae | Aotus nigriceps | 2 | 14–15 |
| Callitrichidae |
Leontopithecus chrysomelas Leontopithecus rosalia Leontopithecus chrysopygus Saguinus imperator |
1 15 2 4 |
16 17–31 32–33 34–37 |
| Cebidae | Sapajus paella | 6 | 38–43 |
| Pitheciidae |
Callicebus nigrifrons Pithecia irrorata |
1 3 |
44 45–47 |
| Old World Primates | |||
| Family | Scientific Name | N | Identification |
| Cercopithecidae | Miopithecus talapoin | 2 | 48–49 |
| Hominidae | Pan troglodytes | 3 | 50–52 |
Serological tests
Serum sampling
Blood samples of 52 non-human primates were obtained immediately before exposure to the sand flies for xenodiagnosis. The animals were anesthetized with variable protocols according to the species (S2 Table). Blood samples were collected in sterile tubes without anti-coagulant and centrifuged at 2,500 x g for 5 minutes at 4°C. Serum samples were separated and stored at -20°C until serological evaluation.
ELISA anti- Leishmania (rKDDR and rK39)
ELISA was performed using 96-well-plates (Costar, Cornig, USA) coated with rKDDR (Safetest Diagnósticos, Brazil) or rK39 antigen in carbonate buffer (15 mM sodium carbonate and 34 mM sodium bicarbonate adjusted at pH 9.6) for 24 hours at 4°C. After coated, wells were blocked with 2% PBS-BSA (pH 7.4) for two hours. Serum samples were diluted 1:100 in 0.05% PBS-Tween 20, added to the wells, and incubated for 12 hours at 4°C. Plates were washed five times with 0.05% PBS- Tween 20 solution. Human anti-IgG diluted 1:2500 in 0.05% PBS-Tween 20 solution was added to each well and incubated for 1.5 hours. Plates were then washed, and finally incubated in citrate buffer (0.1 M acid citric and 0.2 M bibasic sodium phosphatase) containing 0.05% o-phenylenediamine (OPD) and 0.1% hydrogen peroxide. After ten minutes reactions were stopped with sulfuric acid, and optical densities (O.D.) were measure in an ELISA reader (BioRad 550, Brazil) at 490 nm. Human sera known to be positive or negative were used as positive and negative controls, respectively. Cut off was established at two standard deviations above average O.D. of negative controls.
rKDDR Immunochromatographic assay (RAPID test)
The rKDDR Immunochromatographic assay (Safetest Diagnósticos, Brazil) was used according to the manufacturer’s instructions. Briefly, 20 μL of the serum sample was applied without any further processing to the sample well of the test cassette and one drop of PBS buffer (approximately 40 μL) was subsequently added. The result was read visually after 10 to 20 minutes of incubation at room temperature.
ELISA anti-Lu. longipalpis saliva
Salivary glands from Lu. longipalpis raised at the Hematophagous Insect Physiology Laboratory at the Institute of Biological Sciences at UFMG (LFIH/ICB–UFMG) were dissected and stored in 1% PBS at -80°C. Immediately before use, glands were disrupted by ultrasonication in 1.5 mL conical tubes, and centrifuged at 15,680 x g for 2 minutes. Supernatant was collected and diluted in carbonate buffer (15 mM sodium carbonate and 34 mM sodium bicarbonate, pH 9.6) at concentration of 2.5 salivary glands pairs/mL. ELISA was performed as previously described [17] with few modifications. Human anti-IgG was used as secondary antibody. Positive control was the serum from one chimpanzee (animal 50) of the present study sampled three months after been exposed to sand flies for xenodiagnosis. Wells containing positive serum and no antigen (saliva) were used as negative controls.
Xenodiagnosis
Exposure to sand flies
Xenodiagnosis was performed using 50 four-day-old female Leishmania-free Lu. longipalpis sand flies, raised under controlled conditions at the LFIH/ICB–UFMG. Sand flies were placed in a FleboContainer [9,18], and anesthetized animals were exposed to sand flies directly on the ear for 30 minutes as previously described [9]. Ingurgitation of female flies was assessed visually, and although our parameter of success is at least 70% of females becoming ingurgitated, all animals equal or very close to 100% of female sand flies became ingurgitated. Sand flies were then fed with 50% sucrose for five days at 28°C, frozen, separated in individuals microtubes and stored at -80°C until further analysis.
DNA extraction
Ten female sand flies from each animal were randomly selected and individually macerated, homogenized in a 1.5 mL microtube with 50 μL of lysis buffer (0.08 M sodium chloride, 0.16 M sucrose, 0.06 M EDTA, 0.5% SDS, 0.1 M Tris-Cl, pH 8.6) and incubated for 30 minutes at 65°C. Then, 7.1 μL of 8 M potassium acetate were added to the homogenate to a final concentration of 1 M. The material was vortex homogenized and incubated for 30 minutes at 4°C. After incubation, the homogenate was centrifuged at 26,500 x g for 10 minutes. The supernatant was transferred to another 1.5 mL microtube and 100 μL of 95% ethanol were added, then, the mixture was centrifuged at 26,500 x g for 10 minutes. The supernatant was removed and 100 μL of 70% ethanol were added to wash the DNA pellet. The mixture was centrifuged at 26,500 x g for 10 minutes. After complete drying of residual ethanol, the pellet was resuspended in 30 μL of ultrapure water.
Identification and quantification of L. infantum in sand flies
Identification of positive sand flies was performed using qPCR. Primers targeting Leishmania sp. minicircle kinetoplast DNA (kDNA) specific for the donovani complex were used for qPCR [19]: sense 5’-CTTTTCTGGTCCCGCGGGTAGG-3’, anti-sense 5’-CCACCTGGCCTATTTTACACCA-3’, with final product of 145 base pairs; and primers targeting GAPDH as housekeeping gene: sense 5’-TTCGCAGAAGACAGTGATGG-3’, anti-sense 5’- CCCTTCATCGGTCTGGACTA-3’, with final product of 132 base pairs. qPCR was performed in a final volume of 10 μL, with 0.2 μM of each primer, 5 μL of 1x SYBR Green PCR master mix (Applied Biosystems, USA) and 4 μL of DNA (5 ng/μL). Reaction was performed using an ABI Prism 7500 (Applied Biosystems, USA) and followed initial denaturation at 95°C for 10 minutes, 40 cycles of denaturation at 95°C for 15 seconds and annealing and extension at 60°C for 1 minute.
Ten sand flies from each animal, including both seropositive and seronegative animals, were tested by qPCR in two pools with five sand flies each. Animals that had positive sand fly pools were included in quantitative analysis, and DNA from each individual sand fly that was included in the pools was evaluated separately. Quantification of promastigotes in each sand fly was based on a standard curve established using serial dilutions of L. infantum pure culture at initial concentration of 108 promastigotes/μL. Calculation of promastigotes/μg of DNA was performed based on Ct of kDNA amplification of each positive sand fly and on slope and y-intersection of L. infantum standard curve, given the following equation:
Survey of phlebotomine species in the area where primates were kept in captivity
Phlebotomine capture was performed at 10 sites within the zoo area from February 2014 to February 2015 using traps as previously described [20]. Phlebotomines were identified by morphologic examination based on previously described criteria [21]. DNA samples were extracted from captured phlebotomines and used for nested PCR (LnPCR) amplification of the Leishmania SSUrRNA gene [22]. All reactions included positive (20 ng of L. infantum genomic DNA) and negative controls (target DNA replaced with water). PCR products were sequenced and blasted against the GenBank database for identification of the Leishmania species.
Statistical analysis
Frequencies of positivity of non-human primates by xenodiagnosis were compared using the chi-square test with confidence interval of 95% (p < 0.05). Kruskall-Wallis and Mann-Whitney tests were performed to compare all other data. Agreement between the three serologic tests was calculated by Kappa analysis. All statistical analyses were performed using the Prism software version 7.0 (GraphPad).
Results
Serology anti-Leishmania
Seven of the 52 non-human primates tested (13.46%) were serologically positive for Leishmania spp. using rKDDR as antigen (Table 2). The others two tests (ELISA with rK39 and RAPID test) were a little less sensitive, with 9.61% (5/52) of positivity. However, all serological tests had strong agreement, with kappa coefficient equal to 0.87, using ELISA rKDDR as the main test. One L. rosalia was positive only at qPCR pool of sand flies, but serologically negative. In total, positive animals included: three P. troglodytes (100% 3/3), three L. rosalia (20% 3/15), one S. apella (16.67% 1/6), and one M. talapoin (50% 1/2). P. troglodytes were more predisposed to be serologically positive than the other species included in this study (p = 0.015).
Table 2. Serologically positive non-human primates for leishmaniasis with three different serological assays.
Legend: ID: individual identification of each animal. NA: not applicable.
| Species | ID | rKDDR (O.D.) | rK39 (O.D.) | RAPID test |
|---|---|---|---|---|
| Pan troglodytes | 50 | + (0.3232) | – (0.2352) | – |
| 51 | + (1.2434) | + (1.5628) | + | |
| 52 | + (1.8469) | + (2.1753) | + | |
| Leontopithecus rosalia | 19 | + (0.5595) | + (0.4301) | + |
| 27 | + (0.3964) | + (0.4298) | + | |
| 30* | – (0.0443) | – (0.0605) | – | |
| Sapajus paella | 43 | + (0.7985) | + (0.3595) | + |
| Myopithecus talapoin | 48 | + (0.3510) | – (0.2157) | – |
| Cut off | 0.2326 | 0.2563 | NA |
* Animal positive only by the qPCR of sand flies (xenodiagnosis).
Xenodiagnosis
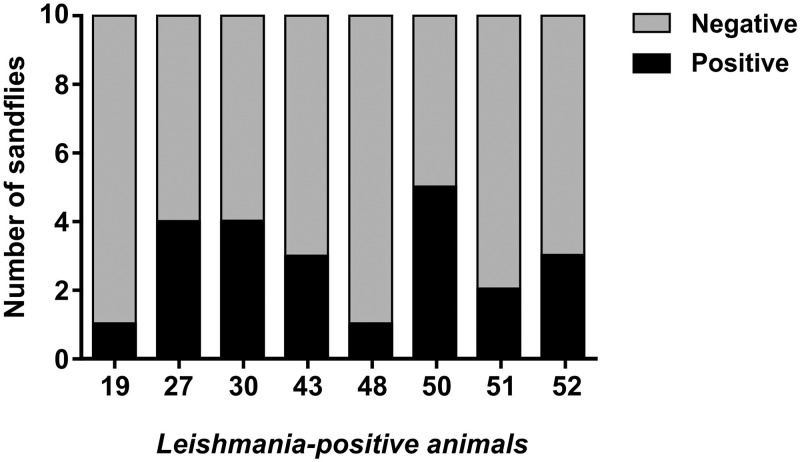
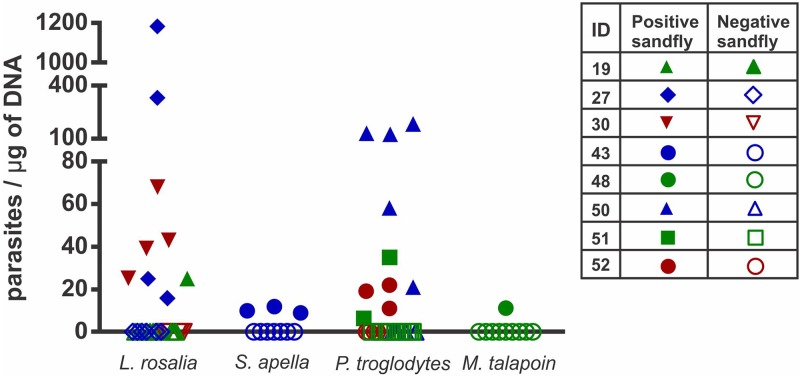
Both seropositive and seronegative animals were subjected to xenodiagnosis. All positive animals were capable to infect at least one sand fly, whereas one animal (Leontopithecus rosalia) that was serologically negative was also positive by xenodiagnosis (Fig 1). The number of promastigotes/μg of DNA varied from 5.67 to 1,181.93 in positive sand flies (Fig 2).
Fig 1. Transmission of Leishmania infantum from non-human primates to Lutzomyia longipalpis.
Ratio of positive and negative sand flies (n = 10) exposed to each of the positive animals, and tested for detection L. infantum DNA by qPCR.
Fig 2. Quantitative analysis of Leishmania infantum in Lutzomyia longipalpis exposed to non-human primates.
Relative quantification of L. infantum promastigotes in each individual positive sand fly according to the non-human primate species. Symbols refer to individual animals: open symbols indicate negative sand flies, and solid symbols indicate infected sand flies.
We observed that L. rosalia was more efficient than S. apella to infect sand flies (p = 0.0328), with average of 194 and 10 promastigotes/μg of sand fly DNA, respectively. Although M. talapoin had one infected sand-fly with also 10 promastigotes/μg of sand fly DNA we could not perform a statistical test because the low number of infected sand fly in this case. P. troglodytes had an average of 59 promastigotes/μg of sand fly DNA. Fig 2 shows the quantity of promastigotes/μg of DNA of each positive sand fly from each species of the study.
ELISA anti-Lutzomyia longipalpis saliva
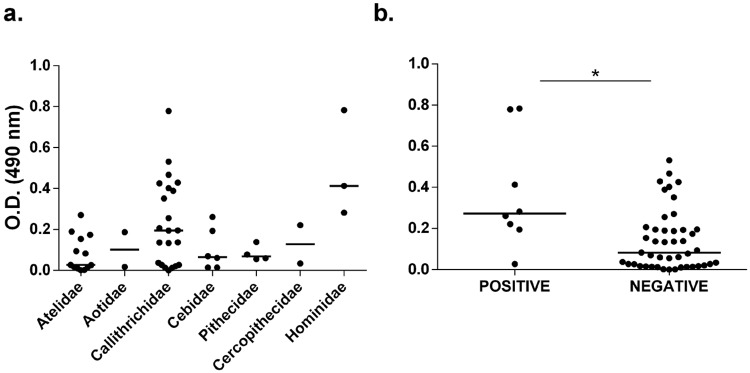
Although we did not observe significant differences between the O.D. of all families evaluated, animals positive to leishmaniasis had a significantly higher O.D. when compared to negative animals (p = 0.0049) (Fig 3).
Fig 3. ELISA anti- Lutzomyia longipalpis saliva.
(A) O.D. from each non-human primate evaluated distributed according their families. (B) O.D. from non-human primates grouped in positive and negative to Leishmania spp. by xenodiagnosis. Positive animals had statistically significant higher O.D. values when compared to negative animals (*p = 0.0049 –Mann-Whitney test). Line represents the median and dots indicate individual values.
Survey of the phebotomine population in the zoo area
A total of 1,392 phlebotomine specimens were captured, including the following species: Psathyromyia aragaoi (Costa Lima, 1912), Evandromyia bacula (Martins, Falcão and Silva, 1965), Evandromyia cortelezzii (Brèthes,1923), Lutzomyia ischnacantha (Martins, Souza and Falcão, 1962), Lutzomyia lenti (Mangabeira,1938), Pintomyia monticola (Costa Lima, 1932), Lutzomyia longipalpis (Lutz and Neiva, 1912), Pintomyia pessoai (Coutinho and Barretto, 1940), Microppygomyia quinquefer (Dyar, 1929), Sciopemyia sordellii (Shannon and Del Ponte, 1927) as detailed in Table 3. Seasonal distribution of captures is detailed in a Supplementary Table (S3 Table).
Table 3. Phlebotomines captured within the area of the Zoological Garden in Belo Horizonte (Brazil), from February 2014 to February 2015.
Legend: # Pools: indicate the number of pools from each phlebotomine species that were prepared for DNA extraction.
| Species | # Specimens | # Pools | ||
|---|---|---|---|---|
| Male | Female | Total | ||
| Psathyromyia aragoai | 1 | 0 | 1 | 0 |
| Evandromyia bacula | 0 | 1 | 1 | 1 |
| Evandromyia cortelezzii | 74 | 87 | 161 | 53 |
| Lutzomyia ischnacantha | 0 | 1 | 1 | 1 |
| Lutzomyia lenti | 92 | 64 | 156 | 35 |
| Lutzomyia longipalpis | 48 | 23 | 71 | 19 |
| Pintomyia monticola | 27 | 35 | 62 | 23 |
| Pintomyia pessoai | 485 | 379 | 864 | 82 |
| Micropygomyia quinquefer | 0 | 1 | 1 | 1 |
| Sciopemyia sordellii | 0 | 4 | 4 | 4 |
| Nyssomyia whitmani | 4 | 8 | 12 | 7 |
| Not identified | 23 | 35 | 58 | 0 |
| Total | 754 | 638 | 1392 | 226 |
After taxonomic identification of all captured females, 226 species-specific pools with up to 10 phlebotomines each (Table 3) were used for DNA extraction and PCR. These pools included all species with the exception of Psathyromyia aragaoi, which had only one male captured. Only one pool of Lutzomyia lenti was PCR positive for Leishmania spp. The amplified sequence had 97% identity with L. infantum.
Discussion
Natural infections and Leishmania-associated disease in non-human primates have been occasionally reported, and there is also evidences of infection based on serology and PCR affecting captive and free-living animals [7,13–15,23–26]. Non-human primates have been also extensively used for experimental infections with Leishmania spp., especially for vaccinology and clinical or immunopathological studies, with similar outcomes when compared to human patients [27–30]. Xenodiagnosis is the only tool capable of confirming the ability of a potential reservoir to infect the parasite vector. This study describes for the first time the competence of asymptomatic non-human primates to transmit L. infantum to Lu. longipalpis, the most important invertebrate vector of visceral leishmaniasis in the New World.
Parasite loads in infected sand flies observed in this study were considered low, although they were similar to parasite loads previously found in phlebotomines fed on asymptomatic or symptomatic Leishmania-infected dogs that had averages of 10 and 84 parasites, respectively [31]. However, in that particular study one symptomatic dog infected sand flies with 29,774 parasites. In our study there was also one L. rosalia that infected a sand fly resulting in a high parasite load (1,181.93 parasites/μg of DNA). Infected sand flies may be classified as “super-spreader” when they carry a high infectious dose (> 600 parasites), which is responsible for epidemic high-disease burden; and “endemic-spreader” with low parasite loads, and responsible for endemic low-disease burden [32]. Therefore, our results suggest that some non-human primate species may play a role in the endemic Leishmania cycle, transmitting lower infective doses of parasites to sand flies, favoring the circulation of “endemic-spreader” sand flies, which perpetuate a “mild/asymptomatic mode” of leishmaniasis. This situation is unlikely to favor emergence of clinical disease, but it may result in maintenance of Leishmania in a given population within an endemic area [32].
The infective dose of promastigotes in naturally infected sand flies is still unknown, with reports indicating hundreds to thousands promastigotes being required for establishment of infection in a mammalian host [31,33,34]. Some studies indicate that physical obstruction of the sand fly anterior midgut is required for actual transmission of the parasite to mammalian hosts [35,36]. Such obstruction is the result of an association of promastigotes and promastigote secretory gel (PSG) forming a sausage-like plug distending the sand fly anterior digestive tract [36,37]. Obstruction of the anterior gut leads to regurgitation of metacyclic promastigotes during blood feeding, resulting in infection of mammalian host [37]. A minimum number of promastigotes is needed to produce enough PSG to act as a blocking plug. However, the number of promastigotes increase about 16 times within the sand fly [36,37], and sand flies tend to become increasingly parasitized after a second blood meal even if this second meal takes place on an uninfected host [38]. Therefore, it is reasonable to consider that even low parasite loads, as observed in sand flies that fed on non-human primates in this study, could eventually lead to an infective parasite load in an endemic environment. Importantly, we performed qPCR using DNA extracted from sand flies at five days after the blood meal. At five days, ingested blood has been eliminated by the sand fly through defecation [39] so ingested Leishmania DNA fragments do not interfere with PCR amplification at that time point. Although later time points may result in higher parasite loads, the PCR technique employed in this study is highly sensitive so sampling of sand flies at 5 days post blood meal was considered appropriate for the goals of this study.
The competence of human individuals to infect sand flies is enhanced in symptomatic and immunocompromised patients [40,41]. Conversely, this tendency is still questionable in domestic dogs [31,42,43]. Previous reports demonstrated that two non-human primates housed at the same institution where this study was done have been diagnosed with symptomatic visceral leishmaniasis: one Callicebus nigrifrons in 2008 [13] and one Gorilla gorilla in 2016 [15], supporting the notion that captive non-human primates in endemic areas are susceptible to leishmaniasis.
Three serological diagnostic methods were performed in this study: two ELISAs with different antigens (rKDDR and rK39) and one Immunochromatographic (RAPID test) with rKDDR as antigen. Even though ELISA with rKDDR was more sensitive, all three tests had similar results. These results are in agreement with a recent study that demonstrated higher sensitivity and specificity of rKDDR for human or canine sera when compared to traditional serologic protocols [44]. Although the ELISA protocols employed in this study have anti-human secondary antibodies, their results were similar to those obtained with the RAPID test, which have direct interaction of primary antibody from the non-human primate serum with the specific antigen. Importantly, serological tests cannot discriminate infectious animals from those that were previously exposed to Leishmania, but are not infectious [9,45]. Therefore, we performed qPCR with a pool of sand flies from each animal, to ensure that positive sand flies could be detected even from serologically negative animals. Interestingly, we observed significantly higher levels of IgG anti-Lu. longipalpis saliva in positive animals, which corroborate previous studies in dogs and humans, which serology to sand fly saliva has been directly associated with host exposition to sand flies, and increased risk of infection [46–49].
Interestingly, one serologically negative animal (Leontopithecus rosalia) was capable of infecting female sand flies. Although in traditional experimental models the peak of parasite load coincides with higher serologic titers [50], non-human primates experimentally infected with L. infantum often have detectable parasites in the bone marrow or Leishmania-induced lesions prior to developing a humoral response [51,52]. Therefore, our hypothesis in this case is that the animal was infected and infectious, but had not yet seroconverted at that time.
Although naturally occurring symptomatic visceral leishmaniasis have been previously reported in non-human primates, it is considered uncommon. There is only one reported case of the disease in a neotropical primate [13], and three recently cases in old world primates, affecting two adults orangutans [14] and one infant gorilla [15]. All of these cases occurred in endemic regions for leishmaniasis. Carneiro and coworkers [53] suggest that neotropical primates have an innate immunological resistance to L. infantum infection, whereas our results suggest that chimpanzees were predisposed to the infection when compared to other non-human primate species under captivity in an endemic area. All chimpanzees included in this study were positive by xenodiagnosis, and their levels of -Lu. longipalpis saliva suggested high exposure to sand flies.
Among 1,392 phlebotomine specimens captured within the zoo area, and grouped into 226 pools, only one pool of Lutzomyia lenti was PCR positive for L. infantum. These results may suggest a possible role of Lu. lenti in the transmission of leishmaniasis. However, these data are insufficient to support this hypothesis since according Killick-Kendrick [54], in order to be considered a biological vector, a given invertebrate species must: (i) feed on humans and animal reservoir species; (ii) support the development of the parasite; (iii) carry parasites that are indistinguishable from the ones isolated from patients; and (iv) be capable of transmitting the parasite through bite.
In conclusion, this study demonstrated for the first time that captive non-human primates might be susceptible to Leishmania infection and capable of transmitting the pathogen to sand flies. This study raises awareness regarding the need for improved public health strategies focusing on controlling the vector and the disease. This study also emphasized the importance of non-human primates, wild or captive, as sentinels for zoonotic diseases [55,56], which is highly relevant under a conservation medicine point of view.
Supporting information
(PDF)
(PDF)
(PDF)
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
Work in RLS lab is supported by CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico, Brazil), FAPEMIG (Fundação de Amparo a Pesquisa do Estado de Minas Gerais, Brazil), and CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, Brazil). NFG, RTF, TAP, and RLS have fellowships from CNPq (Brazil). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.WHO Expert Committee on the control of leishmaniasis. 2010. http://apps.who.int/iris/bitstream/10665/44412/1/WHO_TRS_949_eng.pdf.
- 2.Lukes J, Mauricio IL, Schonian G. Evolutionary and geographical history of the Leishmania donovani complex with a revision of current taxonomy. Proc Natl Acad Sci U.S.A. 2007; 22: 9375–9380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lainson R, Rangel EF. Lutzomyia longipalpis and the eco-epidemiology of American visceral leishmaniasis, with particular reference to Brazil—A Review. Mem Inst Oswaldo Cruz. 2005; 100 (8): 811–827. [DOI] [PubMed] [Google Scholar]
- 4.Patz JA, Githeko AK, McCarty JP, Hussein S, Confaalonieri U, Wet N. Climate change and infectious disease In: McMichel AJ, Campbell-Lendrum DH, Corvalán CF, Ebi KL, Githeko AK, Scheraga JD, Woodwrd A, editors. Climate change and human health Risks and Responses. Genebra: WHO Library; 2003. pp 103–132. [Google Scholar]
- 5.Diniz SA, Silva FL, Carvalho NAV, Bueno R, Guerra RMSNC, Abreu-Silva AL, et al. Animal reservoirs for visceral leishmaniasis in densely populated urban areas. J Infect Dev Ctries. 2008; 2: 24–33. [DOI] [PubMed] [Google Scholar]
- 6.Lombardi MC, Turchetti AP, Tinoco HP, Pessanha AT, Soave SA, Malta MCC, et al. Diagnosis of Leishmania infantum infection by Polymerase Chain Reaction in wild mammals. Pesq Vet Bras. 2014; 34: 1243–1246. [Google Scholar]
- 7.Roque ALR, Jansen A. Wild and synanthropic reservoirs of Leishmania species in the Americas. Int J Parasitol Parasites Wildl. 2014; 3, 251–262. 10.1016/j.ijppaw.2014.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Millán J, Ferroglio E, Solano-Gallego L. Role of wildlife in the epidemiology of Leishmania infantum infection in Europe. Parasitol Res. 2014; 113: 2005–2014. 10.1007/s00436-014-3929-2 [DOI] [PubMed] [Google Scholar]
- 9.Mol JPS, Soave SA, Turchetti AP, Pinheiro GRG, Pessanha AT, Malta MCC, et al. Transmissibility of Leishmania infantum from maned wolves (Chrysocyon brachyurus) and bush dogs (Speothos venaticus) to Lutzomyia longipalpis. Vet Parasitol. 2015; 212: 86–91. 10.1016/j.vetpar.2015.08.024 [DOI] [PubMed] [Google Scholar]
- 10.Sadlova J, Seblova V, Votypka J, Warburg A, Volf P. Xenodiagnosis of Leishmania donovani in BALB/c mice using Phlebotomus orientalis: new laboratory model. Parasit Vectors. 2015; 8: 158 10.1186/s13071-015-0765-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Courtenay O, Quinnell RJ, Garcez LM, Dye C. Low infectiousness of a wildlife host of Leishmania infantum: the crab-eating fox is not important for transmission. Parasitol. 2002; 125: 407–414. [DOI] [PubMed] [Google Scholar]
- 12.Jiménez M, González E, Martín-Martín I, Hernández S, Molina R. Could wild rabbits (Oryctolagus cuniculus) be reservoirs for Leishmania infantum in the focus of Madrid, Spain? Vet Parasitol. 2014; 202: 296–300. 10.1016/j.vetpar.2014.03.027 [DOI] [PubMed] [Google Scholar]
- 13.Malta MCC, Tinoco HP, Xavier MN, Vieira AS, Costa EA, Santos RL. Naturally acquired visceral leishmaniasis in non-human primates in Brazil. Vet Parasitol. 2010; 169: 193–197. 10.1016/j.vetpar.2009.12.016 [DOI] [PubMed] [Google Scholar]
- 14.Miró G, Troyano A, Montoya A, Fariñas F, Fermín MA, Flores L, et al. First report of Leishmania infantum infection in the endangered orangutan (Pongo pygmaeus pygmaeus) in Madrid, Spain. Parasit Vectors. 2018; 11(185): 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tinoco HP, Costa MELT, Pessanha AT, Coelho CM, Carvalho TF, Mol JPS, Viana AG, Bueno LL, Fujiwara RT, Santos RL. Visceral leishmaniasis in na infant gorila (Gorilla gorila gorilla): clinical signs, diagnosis, and successful treatment with single-dose liposomal amphotericin B. J Med Primatol. 2018; 1–3. [DOI] [PubMed] [Google Scholar]
- 16.Silveira TF, Moraes MAP, Lainson R, Shaw JJ. Leishmaniose cutânea experimental III—aspectos histopatológicos do comportamento evolutivo da lesão cutânea produzida em Cebus apella (Primates: Cebidae) por Leishmania (Viannia) lainsoni, L. (V.) braziliensis e L. (Leishmania) amazonensis. Rev Inst Med Trop Sao Paulo. 1990; 32 (6): 387–394 [DOI] [PubMed] [Google Scholar]
- 17.Quinnell RJ, Soremekun S, Bates PA, Rogers ME, Garcez LM, Courtenay O. Antibody response to sand fly saliva is a marker of transmission intensity but not disease progression in dogs naturally infected with Leishmania infantum. Parasit Vectors. 2018; 11 (7): 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Costa-Val AP, Cavalcanti RR, Gontigo NF, Michalik MSM, Alexander B, Williams P, et al. Canine visceral leishmaniasis: relationships between clinical status, humoral immune response, haematology and Lutzomyia (Lutzomyia) longipalpis infectivity. Vet J. 2007; 174: 636–643. 10.1016/j.tvjl.2006.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lachaud L, Marchergui-Hammami S, Chabbert E, Dereure J, Dedet JP, Bastien P. Comparison of six PCR methods using peripheral blood for detection of canine visceral leishmaniasis. J Clin Microbiol. 2002; 40: 210–215. 10.1128/JCM.40.1.210-215.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lana RS, Michalsky ÉM, Lopes LO, Lara-Silva FO, Nascimento JL, Pinheiro LC, et al. Ecoepidemiological aspects of visceral leishmaniasis in an endemic area in the Steel Valley in Brazil: an ecological approach with spatial analysis. PLoS One. 2018;13(10):e0206452 10.1371/journal.pone.0206452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galati EAB. Morfologia e taxonomia. Classificação de Phlebotominae In Rangel EF, Lainson R, editors. Flebotomíneos do Brasil. Rio de Janeiro: Fiocruz; 2003. Pp. 23–206. [Google Scholar]
- 22.Cruz I, Cañavate C, Rubio JM, Morales MA, Chicharro C, Laguna F, et al. A nested polymerase chain reaction (Ln- PCR) for diagnosing and monitoring Leishmania infantum infection in coinfected patients with human immunodeficiency virus. Trans R Soc Trop Med Hyg. 2002; 96:185–189 [DOI] [PubMed] [Google Scholar]
- 23.Lima VMF, Santiago MEB, Sanches LC, Lima BD. Molecular diagnosis of Leishmania amazonensis in a captive spider monkey in Bauru, São Paulo, Brazil. J Zoo Wildl Med. 2012; 43(4): 943–945. 10.1638/2012-0059R1.1 [DOI] [PubMed] [Google Scholar]
- 24.Rovirosa-Hernández MJ, Cortes-Ortíz L, García-Orduña F, Guzmán-Gómez D, López-Monteon A, Gaba M, et al. Seroprevalence of Trypanosoma cruzi and Leishmania mexicana in free-ranging howler monkeys in Southeastern Mexico. Am J Primatol. 2013; 75: 161–169. 10.1002/ajp.22094 [DOI] [PubMed] [Google Scholar]
- 25.Souza TD, Turchetti AP, Fujiwara RT, Paixão TA, Santos RL. Visceral leishmaniasis in zoo and wildlife. Vet Parasitol. 2014; 200: 233–241. 10.1016/j.vetpar.2013.12.025 [DOI] [PubMed] [Google Scholar]
- 26.Bueno MG, Catão-Dias JL, Laroque PO, Vasconcelos SA, Ferreira Neto JS, Gennari SM, et al. Infectious diseases in free-ranging blonde capuchins, Sapajus flavus, in Brazil. Int J Primatol. 2017; 38(6): 1017–1031. [Google Scholar]
- 27.Broderson JR, Chapman Jr WL, Hanson WL. Experimental visceral leishmaniasis in the owl monkey. Vet Pathol. 1986; 23:293–302. 10.1177/030098588602300310 [DOI] [PubMed] [Google Scholar]
- 28.Carneiro LA, Silveira FT, Campos MB, Brígido COM, Gomes CMC, Corbett CEP, et al. Susceptibility of Cebus apella Monkey (Primates: Cebidae) to experimental Leishmania (L.) infantum chagasi-infection. Rev Inst Med Trop São Paulo. 2011; 53(1): 45–50. [DOI] [PubMed] [Google Scholar]
- 29.Porrozzi R, Pereira MS, Teva A, Volpini AC, Pinto MA, Marchevsky RS. Leishmania infantum-induced primary and challenge infections in rhesus monkeys (Macaca mulatta):a primate model for visceral leishmaniasis. Trans R Soc Trop Med Hyg. 2006; 100: 926–937. 10.1016/j.trstmh.2005.11.005 [DOI] [PubMed] [Google Scholar]
- 30.Grimaldi Jr G, Teva A, Porrozzi R, Pinto MA, Marchevsky RS, Rocha MGL, et al. Clinical and parasitological protection in a Leishmania infantum-macaque model vaccinated with adenovirus and the recombinant A2 antigen. Plos Negl Trop Dis. 2014; 8(6): e2853 10.1371/journal.pntd.0002853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borja LS, Sousa OMF, Solcà MDS, Bastos LA, Bordoni M, Magalhães JT, et al. Parasite load in the blood and skin of dogs naturally infected by Leishmania infantum is correlated with their capacity to infect sand fly vectors. Vet Parasitol. 2016; 229: 110–117. 10.1016/j.vetpar.2016.10.004 [DOI] [PubMed] [Google Scholar]
- 32.Courtenay O, Peters NC, Rogers ME, Bern C. Combining epidemiology with basic biology of sand flies, parasites, and host to inform leishmaniasis transmission dynamic and control. Plos Pathog. 2017; 13 (10): 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Warburg A, Schlein Y. The effect of post-bloodmeal nutrition of Phlebotomus papatasi on the transmission of Leishmania major. Am J Trop Med Hyg. 1986; 35:926–930. [DOI] [PubMed] [Google Scholar]
- 34.Rogers ME, Ilg T, Nikolaev AV, Ferguson MA, Bates PA. Transmission of cutaneous leishmaniasis by sand flies is enhanced by regurgitation of fPPG. Nature. 2004; 430 (6998): 463–467. 10.1038/nature02675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bates PA. Transmission of Leishmania metacyclic promastigotes by phlebotomine sand flies. Int J Parasitol. 2007; 37: 1097–1106. 10.1016/j.ijpara.2007.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rogers ME. The role of Leishmania proteophosphoglycans in sand fly transmission and infection of the mammalian host. Front Microbiol. 2012; 223 (3): 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rogers ME, Chance ML, Bates PA. The role of promastigote secretory gel in the origin and transmission of the infective stage of Leishmania mexicana by the sandfly Lutzomyia longipalpis. Parasitol. 2002; 124: 495–507. [DOI] [PubMed] [Google Scholar]
- 38.Serafim TD, Coutinho-Abreu IV, Oliveira F, Meneses C, Kamhawi S, Valenzuela JG. Sequential blood meals promote Leishmania replication and reverse metacyclogenesis augmenting vector infectivity. Nat Microbiol. 2018; 3(5): 548–555. 10.1038/s41564-018-0125-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Secundino NFC, Eger-Magrich I, Braga EM, Santoro MM, Pimenta PFP. Lutzomyia longipalpis peritrophic matrix: formation, structure, and chemical composition. J Med Entomol. 2005; 42(6):928–938. [DOI] [PubMed] [Google Scholar]
- 40.Costa CHN, Gomes RBB, Silva MRB, Garcez LM, Rampos PKS, Santos RS, et al. Competence of the human host as a reservoir of Leishmania chagasi. J Infect Dis. 2000; 182, 997–1000. 10.1086/315795 [DOI] [PubMed] [Google Scholar]
- 41.Ferreira GR, Ribeiro JCCB, Meneses Filho A, Pereira TJCF, Parente DM, Pereira HF, et al. Human competence to transmit Leishmania infantum to Lutzomyia longipalpis and the influence of human immunodeficiency virus infection. Am J Trop Med Hyg. 2018; 98 (1): 126–133. 10.4269/ajtmh.16-0883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soares MRA, Mendonça IL, Bonfim JM, Rodrigues JÁ, Werneck GL, Costa CHN. Canine visceral leishmaniasis in Teresina, Brazil: Relationship between clinical features and infectivity for sand flies. Acta Tropica. 2011; 117: 6–9. 10.1016/j.actatropica.2010.08.015 [DOI] [PubMed] [Google Scholar]
- 43.Laurenti MD, Rossi CN, da Matta VLR, Tomokane TY, Corbett CEP, Secundino NFC. Asymptomatic dogs are highly competent to transmit Leishmania (Leishmania) infantum chagasi to the natural vector. Vet Parasitol. 2013; 196: 296–300. 10.1016/j.vetpar.2013.03.017 [DOI] [PubMed] [Google Scholar]
- 44.Dhom-Lemos L, Viana AG, Cunha JLR, Cardoso MS, Mendes TAO, Pinheiro GRG. Leishmania infantum recombinant kinesin degenerated derived repeat (rKDDR): A novel potential antigen for serodiagnosis of visceral leishmaniasis. PLoS One. 2019; 14: e0211719 10.1371/journal.pone.0211719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mendonça IL, Batista JF, Werneck GL, Soares MRA, Costa DL, Costa CHN. Serological tests fail to discriminate dogs with visceral leishmaniasis that transmit Leishmania infantum to the vector Lutzomyia longipalpis. Rev Bras Med Trop. 2017; 50(4): 483–488. [DOI] [PubMed] [Google Scholar]
- 46.Barral A, Honda E, Caldas A, Costa J, Vinhas V, Rowton ED, et al. Human immune response to sand fly salivary gland antigens: a useful epidemiological marker? Am J Trop. Med Hyg. 2000; 62(6): 740–745. [DOI] [PubMed] [Google Scholar]
- 47.Batista LFS, Matta VLR, Tomokane TY, Pacheco AD, Silveira FT, Rossi CN, et al. Canine antibody response to Lutzomyia longipalpis saliva in endemic area of visceral leishmaniasis. Rev Soc Bras Med Trop. 2016; 49(3):361–364. 10.1590/0037-8682-0360-2015 [DOI] [PubMed] [Google Scholar]
- 48.Lestinova T, Rohousova I, Sima M, Oliveira CI, Volf P. Insights into the sand fly saliva: Blood-feeding and immune interactions between sand flies, hosts, and Leishmania. PLoS Negl Trop Dis. 2017; 11 (7): 1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quinnell RJ, Soremekun S, Bates PA, Rogers ME, Garcez LM, Courtney O. Antibody response to sand fly saliva is a marker of transmission intensity but not disease progression in dogs naturally infected with Leishmania infantum. Parasit Vectors. 2018; 11 (7): 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Riça-Capela MJ, Cortes S, Leandro C, Peleteiro MC, Santos-Gomes G, Campino L. Immunological and histopathological studies in a rodent model infected with Leishmania infantum promastigotes or amastigotes. Parasitol Res. 2003; 89: 163–169. [DOI] [PubMed] [Google Scholar]
- 51.Porrozzi R, Pereira MS, Teva A, Volpini AC, Pinto MA, Marchevsky RS. Leishmania infantum-induced primary and challenge infections in rhesus monkeys (Macaca mulatta): a primate model for visceral leishmaniasis. Trans R Soc Trop Med Hyg. 2006; 100: 926–937. 10.1016/j.trstmh.2005.11.005 [DOI] [PubMed] [Google Scholar]
- 52.Carneiro LA, Silveira FT, Campos MB, Brígido Mdo C, Gomes CM, Corbett CE, Susceptibility of Cebus apella monkey (Primates: Cebidae) to experimental Leishmania (L.) infantum chagasi-infection. Rev Inst Med Trop Sao Paulo. 2011; 53: 45–50. [DOI] [PubMed] [Google Scholar]
- 53.Carneiro LA, Laurenti MD, Campos MB, Gomes CMC, Corbett CEP, Silveira FT. Susceptibility of peritoneal macrophage from different species of neotropical primates to ex vivo Leishmania (L.) infantum chagasi-infection. Rev Inst Med Trop Sao Paulo. 2012; 54(2):95–101. [DOI] [PubMed] [Google Scholar]
- 54.Killick-Kendrick R. Phlebotomine vectors of the leishmaniases: a review. Med Vet Entomol. 1990; 4:1–24. [DOI] [PubMed] [Google Scholar]
- 55.Calvignac-Spencer S, Leendertz SAJ, Gillespie TR, Leendertz FH. Wild great apes as sentinels and sources of infectious disease. Clin Microbiol Infect. 2012; 18:521–527. 10.1111/j.1469-0691.2012.03816.x [DOI] [PubMed] [Google Scholar]
- 56.Moreno ES, Spinola R, Tengan CH, Brasil RA, Siciliano MM, Coimbra TL, et al. Yellow Fever epizootics in non-human primates, São Paulo State, Brazil, 2008–2009. Rev Inst Med Trop Sao Paulo. 2013; 55 (1): 45–50. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.