Abstract
The most common and aggressive type of brain cancer in adults is glioblastoma multiforme (GBM), and hypoxia is a common feature of glioblastoma. As the histological features of glioma include capillary endothelial cell proliferation, they are highly prone to invading the surrounding normal brain tissue, which is often one of the reasons for the failure of treatment. However, the mechanisms involved in this process are not fully understood. MicroRNAs (miRs) are a class of non-coding RNA that are able to inhibit the malignant progression of tumor cells through the regulation of downstream genes. In the present study, the low expression of miR-576-3p was detected in glioma samples and hypoxia-treated glioma cells using a reverse transcription-quantitative polymerase chain reaction. The present study focused on the effects of miR-576-3p on hypoxia-induced glioma. The results of the functional experiments revealed that the overexpression of miR-576-3p significantly inhibited the migration and pro-angiogenic abilities of the glioma cells under hypoxic conditions (P<0.05) compared with in the lentivirus-miR-negative control group. Furthermore, luciferase reporter gene assays were used to validate the hypothesis that miR-576-3p interacts with the 3′-untranslated region of hypoxia-inducible factor-1α (HIF-1α) and induces a reduction in the protein levels of matrix metalloproteinase-2 and vascular endothelial growth factor. Rescue experiments demonstrated that the restoration of HIF-1α expression attenuated the effect of miR-576-3p on the migration and proangiogenic abilities of glioma cells. In conclusion, the present study confirms that miR-576-3p is a novel GBM inhibitor and its inhibition of the migration and proangiogenic capacity of hypoxia-induced glioma cells is mediated by HIF-1α.
Keywords: glioma, microRNA, hypoxia, hypoxia-inducible factor-1α, migration, angiogenesis
Introduction
The most common and fatal type of central nervous system tumor in adults is glioblastoma multiforme (GBM) (1). Although the surgical resection of gliomas has proved a useful treatment method, the overall survival of patients with glioma remains poor and the median survival time following diagnosis is ~1 year (2,3). Furthermore, <2% of patients survive for 3 years subsequent to the development of glioma (4). The ability of GBM to promote angiogenesis enables the development of novel blood vessels in pre-existing blood vessels, which is essential for its invasion and colonization in the brain and is the main reason for the poor prognosis and the high mortality rate for GBM (5). The high invasiveness of GBM contributes to its penetration into the normal tissues and results in the failure of conventional treatment, resulting in a poor prognosis (6). In addition, a previous study confirmed that glioblastoma is one of the most vascularized types of tumor, indicating the potential value of targeting angiogenesis in order to inhibit the malignant progression of gliomas (7).
Previous studies have demonstrated that in the malignant phenotype of brain tumor types, invasiveness is closely associated with the overexpression of hypoxia-inducible factor-1α (HIF-1α) (8), and the angiogenesis, growth, metastasis and apoptosis of tumor cells are substantially associated with HIF-1α (9-12). However, the detailed mechanisms involved require further investigation. As a class of non-coding RNAs, microRNAs (miRNAs/miRs) possess 19-25 nucleotides and induce mRNA degradation or inhibit protein translation and consequently exert biological functions in physiological and pathological processes through complete or incomplete binding to the 3′-untranslated region (3′-UTR) of its target mRNA (13,14). Aberrant miRNA expression profiles (15) are regarded as a common hallmark of human cancer and are used for tumor diagnosis and prognosis due to the multiple steps of miRNA biosynthesis disorder in human cancer, including GBM (16,17). Previous studies indicate that miRNAs regulate various vital biological functions, including cell invasion, proliferation, apoptosis and angiogenesis (18-20). To the best of our knowledge, there are no previous studies on the association between miR-576-3p and glioma.
The present study aimed to investigate the effects of miR-576-3p on the migration and proangiogenic abilities of hypoxia-treated glioma cells by targeting HIF-1α. These results may provide novel insights into clinically relevant treatments for glioma.
Patients and methods
Patient samples
A total of 158 glioma samples which included 48 astrocytomas [World Health Organization (WHO) grade II] (21), 13 oligodendrogliomas (WHO grade II), eight anaplastic astrocytomas (WHO grade III), 10 anaplastic oligodendrogliomas (WHO grade III), 15 anaplastic oligoastrocytomas (WHO grade III) and 64 GBMs (WHO grade IV) from the Chinese Glioma Genome Atlas (CGGA; www.cgga.org.cn; accessed December 18, 2016) database were used for the preliminary analysis. At the Department of Neurosurgery, The Second Affiliated Hospital of Nanchang University (Nanchang, China), 26 brain samples including 8 high-grade glioma (HGG) samples (WHO grade III and IV), 11 low-grade glioma (LGG) samples (WHO class II) and 7 non-neoplastic brain tissues (NBTs) used as a control collected from traumatic brain injury, were randomly screened following conventional treatment between February 2015 and November 2016. The Pathology Department of the hospital performed histological examination of the glioma tissues according to the WHO classification criteria (21). Each participant provided and signed written informed consent for the samples to be analyzed and the study was ethically approved by the Medical Ethics Committee of The Second Affiliated Hospital of Nanchang University.
Cell culture
Normal human astrocytes (NHAs), as a reference against the GBM cell lines, were provided by ScienCell Research Laboratories, Inc. (San Diego, CA, USA) and cultured in the provided astrocyte growth media supplemented with recombinant human epidermal growth factor, insulin, ascorbic acid, GA-1000, L-glutamine and 5% fetal bovine serum (FBS; SciencCell Research Laboratories, Inc.). U87, U251, T98, LN229 and U118 cells purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA) and were cultivated in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing 10% FBS. The U87 cell line is a glioblastoma of unknown origin and the catalogue number of the cell line is ATCC HTB-14. In addition, the short tandem repeat profiling method was used to authenticate the U87 cells. ScienCell Research Laboratories, Inc. provided the human umbilical vein endothelial cells (HUVECs) and they were raised in endothelial cell growth medium (ECM) (ScienCell Research Laboratories, Inc.) supplemented with 1X endothelial cell growth supplement (ScienCell Research Laboratories, Inc.), 100 U/ml penicillin, 100 µg streptomycin/ml and 5% FBS. All cells were cultured at 37°C in 5% CO2. Prior to each experiment, the cells were incubated for 24 or 48 h at 37°C in 1% O2, 5% CO2 and 94% N2.
Transfection and establishment of stable cell lines
Lentiviral plasmids (pGLVU6/GFP+Puro) carrying hsa-miR-576-3p (LV-miR-576-3p) and hsa-miR-negative control (NC) (LV-miR-NC) expression were purchased from Shanghai GenePharma Co., Ltd. (Shanghai, China). Lentiviruses carrying HIF-1α or HIF-1α-NC and short hairpin RNA (shRNA)-HIF-1α (shHIF-1α-1-1, 5′-GGAAGAACTATGAACATAA-3′; shHIF-1α-1-2, 5′-CTAACTGGACACAGTGTGT-3′; shHIF-1α-1-3, 5′-GCTGACCAGTTATGATTGT-3′) or shRNA-Control (shControl, 5′-TTCTCCGAACGTGTCACGT-3′) were provided by Shanghai Genechem Co., Ltd. (Shanghai, China). The lentiviral packing kit was obtained from Open Biosystems, Inc. (Huntsville, AL, USA). 293T cells were purchased from the Cell Bank of Chinese Academy of Sciences (Shanghai, China) and were cultured in DMEM containing 10% FBS. Lentiviruses were packaged in 293T cells and were collected from the supernatant, according to the manufacturer's protocol. U87 and U251 cells (5×104) were infected with 1×108 lentivirus-transducing units in the presence of 5 µg/ml polybrene in 6-well plates (MOI, 5). After 48 h at 37°C, stable cell lines were established followed by 5 µg/ml puromycin selection.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
The glioma tissues and cells were lysed with TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) for 30 min at 4°C, and the RNA was purified with a RNeasy Mini kit (Qiagen GmbH, Hilden, Germany). Complementary DNA was converted using 1 µg RNA template and RT-qPCR analysis was performed on an ABI-7500 Fast Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.). A miScript SYBR-Green PCR kit (Qiagen GmbH) was used for miRNA detection, and a SYBR-Green PCR Mixture (Invitrogen; Thermo Fisher Scientific, Inc.) was used for mRNA detection. The primers (Shanghai GenePharma Co., Ltd.) were as follows: hsa-HIF1α forward, 5′-GGACACAGATTTAGACTTGGAGATG-3′ and reverse, 5′-CTGCTTTCTAATGGTGACAACTGA-3′; hsa-β-actin mRNA forward, 5′-TCAAGATCATTGCTCCTCCTGAG-3′ and reverse, 5′-ACATCTGCTGGAAGGTGGACA-3′; U6 RT-primer, 5′-CGCTTCACGAATTTGCGTGTCAT-3′, forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′. The reaction conditions were as follows: 95°C for 10 min, followed by 40 cycles at 95°C for 5 sec, 60°C for 30 sec and extension at 72°C 30 sec. Bulge-loop™ miRNA RT-qPCR Primer sets (one RT primer and a pair of qPCR primers for each set) specific for miR-576-3p were designed by Guangzhou RiboBio Co., Ltd. β-actin or U6 were used as an internal reference. The results were analyzed using relative quantification (2−ΔΔCq) (22).
Enzyme-linked immunosorbent assay (ELISA)
U87 and U251 cells were cultured in DMEM containing 10% FBS, and vascular endothelial growth factor (VEGF) expression was detected in the supernatants of cultured U87 and U251 cells that were or were not transfected with miR-576-3p/miR-NC and shHIF-1α-1-3/shCtrl separately. Further analysis was performed post-transfection of glioma cells with miR-576-3p/miR-NC and hsa-HIF-1α/Ctrl separately or together. The levels of VEGF were evaluated using a VEGF human ELISA kit (cat. no. CSB-E11718h; Cusabio Technology LLC, Wuhan, China) according to the manufacturer's protocol. The absorbance was measured at 450 nm
Western blot analysis
The harvested cells were lysed using radioimmunoprecipitation assay buffer (Nanjing KeyGen Biotech Co., Ltd., Nanjing, China) for 30 min at 4°C. The extracted protein in the supernatant was detected using a bicinchonic acid Protein Assay kit (Pierce; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocols. Equal amounts of protein (40 µg) were separated using SDS-PAGE (10% gel) and transferred to polyvinylidene difluoride (PVDF) membranes (Corning Incorporated, Corning, NY, USA). Subsequent to blocking in 5% nonfat milk at room temperature for 2 h, the membranes were incubated with the following primary antibodies overnight at 4°C: Mouse anti-HIF-1α (1:1,000; cat no. sc-13515; Santa Cruz Biotechnology, Inc., Dallas, TX, USA), rabbit anti-matrix metalloproteinase (MMP)-2 (1:1,000; cat no. D8N9Y; Cell Signaling Technology, Inc., Danvers, MA, USA), mouse anti-VEGF (1:1,000; cat no. ab69479; Abcam, Cambridge, UK) and mouse anti-β-actin (1:2,000; cat no. ab8226; Abcam). Following washing three times with Tris-buffered saline Tween-20 (0.05%) at room temperature, the PVDF membranes were incubated with horseradish peroxidase (HRP)-labeled goat anti-rabbit immunoglobulin G (IgG) secondary antibody (1:5,000; cat no. A0208;) and HRP-labeled goat anti-mouse IgG secondary antibody (1:5,000; cat no. A0216; both Beyotime Institute of Biotechnology, Haimen, China) or 1 h at 37°C and then analyzed using an enhanced chemiluminescence detection system (GE Healthcare, Chicago, IL, USA).
Wound healing assay
U87 and U251 cells were seeded in 6-well plates at a density of 1×104 cells/well and grown until complete fusion. The cell layers were scratched using a 200-µl pipette tip to generate a linear wound. Phosphate buffered saline was used to remove the previous DMEM and then serum-free DMEM was added. Images of the migrated cells were captured at 0 and 24 h through an inverted bright-field microscope (Olympus Corporation, Tokyo, Japan). Each experiment was performed three times.
Transwell migration assay
U87 and U251 cells (2×104 cells/well) were added to the upper chamber of the Transwell insert (Corning Incorporated), which contained serum-free DMEM. Medium containing 10% FBS was added to lower chamber as an attractant. Subsequent to incubation for 24 h in 1% O2, images of the migrated cells were captured by an inverted bright-field microscope (Olympus Corporation) and the results were quantified by counting the number of cells.
Tube formation assay
Each well of 15-well angiogenesis slides (ibidi GmbH, Martinsried, Germany) was supplemented with 10 µl Matrigel (BD Biosciences, San Jose, CA, USA) and incubated at 37°C in 1% O2 for 30 min. DMEM containing 10% FBS was used to culture hypoxia-induced U87 and U251 cells (24 h in 1% O2, 5% CO2 and 94% N2 at 37°C) that were or were not transfected with miR-576-3p/miR-NC, hsa-HIF-1α/Ctrl and shHIF-1α/shCtrl, separately or together. Subsequently, the supernatants of cultured U87 and U251 cells were collected and used to suspend HUVECs (1×104 cells/well), which were incubated for 9 h at 37°C in an atmosphere containing 1% O2. Tube formation was analyzed under an inverted bright-field microscope (Olympus Corporation).
Luciferase reporter assay
Wild-type and mutated HIF-1α 3′-UTR reporter plasmids were constructed based on the predicted targets of miR-576-3p, which were detected using miRanda (http://www.microrna.org/microrna/home.do), TargetScan (http://www.targetscan.org/vert_72/) and miRWalk (http://mirwalk.umm.uni-heidelberg.de/). The plasmids were cloned into the pmiRNA-Report vector (Shanghai Genechem Co., Ltd.). U87 and U251 cells (5×104) were seeded in a 24-well plate; after being cultured overnight, the cells were cotransfected with 1 µg wild-type of mutated plasmids, 1 µg Renilla luciferase plasmid and equal amounts of miR-576-3p or miR-NC using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. After 48 h at 37°C, the luciferase activity was measured and compared with Renilla luciferase activity using a Dual Luciferase Reporter Assay kit (Promega Corporation, Madison, WI, USA) according to the manufacturer's protocol.
Statistical analysis
Data are presented as the mean ± standard deviation of three separate experiments. Comparisons were performed using a Student's t-test and one-way analysis of variance. Tukey's post hoc test was performed to compare the data of multiple groups with a normal distribution. The correlation between miR-576-3p expression and the mRNA levels of HIF-1α in glioma tissues were analyzed using Pearson's correlation analysis. GraphPad Prism 7 (GraphPad Software, Inc., La Jolla, CA, USA) was used to conduct the biostatistical analysis. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-576-3p is downregulated in glioma samples and hypoxia-treated glioma cells
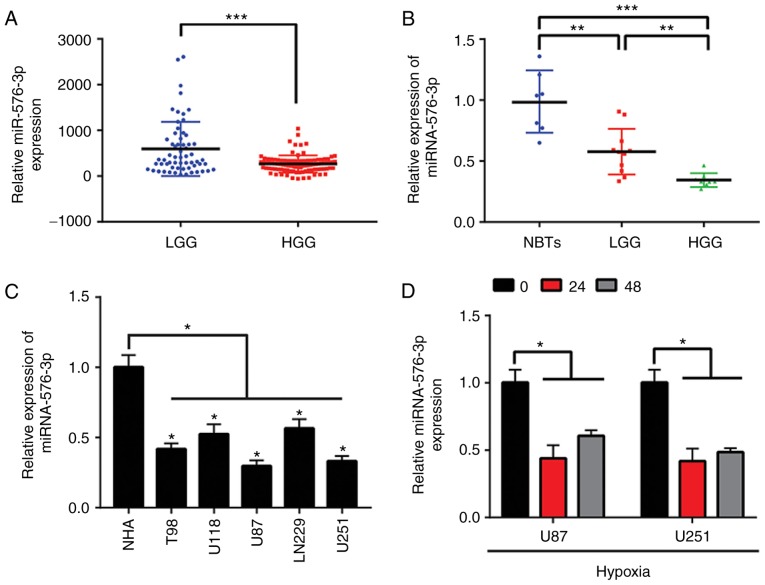
The data of 158 glioma samples were obtained from the CGGA in order to analyze the expression levels of miR-576-3p. The levels of miR-576-3p in the HGG samples (WHO grade III and IV) were significantly lower compared with the LGG samples (WHO class II), as presented in Fig. 1A (P<0.001). Furthermore, the levels of miR-576-3p in 19 clinical glioma tissues and 7 NBTs were evaluated. The experimental data demonstrated that miR-576-3p was significantly downregulated in glioma tissues compared with the NBTs (P<0.01), which was consistent with the CGGA data (Fig. 1B). Subsequently, using NHAs as a negative control, the expression levels of miR-576-3p in U87, U251, T98, LN229 and U118 cell lines were analyzed. The data revealed that miR-576-3p was significantly downregulated in these cell lines compared with the NHA cells (P<0.05) and this effect was most notable in U251 and U87 cells (Fig. 1C). In addition, the miR-576-3p levels in the glioma cells significantly decreased subsequent to hypoxic exposure compared with unexposed cells, as determined by RT-qPCR (P<0.05; Fig. 1D). Collectively, these results indicate that the glioma grade is negatively associated with the expression levels of miR-576-3p. Furthermore, there is a significant attenuation of miR-576-3p in hypoxia-treated glioma cells.
Figure 1.
miR-576-3p is downregulated in glioma samples and hypoxia-treated glioma cells. (A) A total of 158 glioma samples from the Chinese Glioma Genome Atlas database were used to determine the expression of miR-576-3p. (B) A total of 19 glioma specimens and 7 NBTs were used to determine the expression levels of miR-576-3p. (C) Expression levels of miR-576-3p in the indicated cell lines (NHA, T98, U118, U87, LN229 and U251). (D) Alteration in the levels of miR-576-3p in hypoxia-treated glioblastoma multiforme cells. *P<0.05, **P<0.01 and ***P<0.001 with comparisons shown by lines. miR/miRNA, microRNA; NBTs, non-neoplastic brain tissues; LGG, low-grade glioma; HGG, high-grade glioma; NHAs, normal human astrocytes.
Impact of miR-576-3p on hypoxia-treated glioma cells
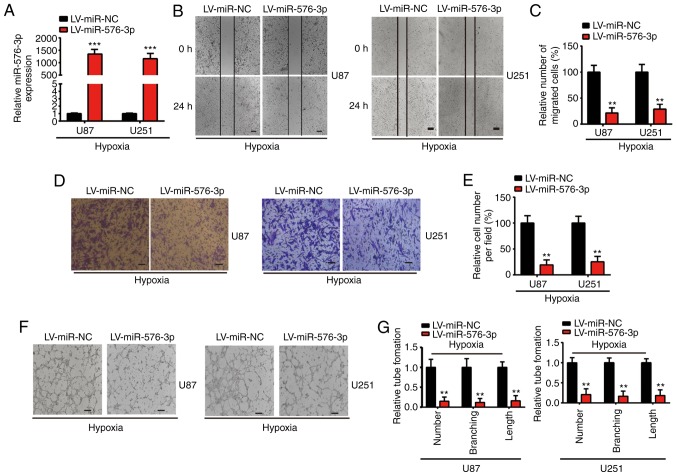
Glioma is a vascular-rich solid tumor, with necrotic and histological properties that include capillary endothelial cell proliferation (23). Therefore, the function of miR-576-3p in regulating the migration and proangiogenic capacity of glioma cells in hypoxic conditions was investigated. Subsequent to hypoxia treatment for 24 h, U87 and U251 cells with stable expression of miR-576-3p or miR-NC were established. Compared with those transfected with LV-miR-NC, the RT-qPCR results confirmed a significant increase in the levels of miR-576-3p in the cells transfected with LV-miR-576-3p (P<0.001; Fig. 2A). The wound healing and Transwell migration analyses revealed that the overexpression of miR-576-3p significantly attenuated the migration of hypoxia-treated U87 and U251 cells compared with cells transfected with LV-miR-NC (P<0.01; Fig. 2B-E). In addition, the impact of miR-576-3p on angiogenesis in glioma cells under hypoxic conditions was monitored using tube formation assays. The results demonstrated that the ability of U87 and U251 cells to promote angiogenesis was significantly suppressed by LV-miR-576-3p compared with cells transfected with LV-miR-NC (P<0.01; Fig. 2F and G). Overall, these results suggest that the dysregulation of miR-576-3p has a negative impact on the migration and angiogenesis of glioma cells under hypoxia in vitro.
Figure 2.
Effect of miR-576-3p on hypoxic-treated glioma cells in vitro. (A) Reverse transcription-quantitative polymerase chain reaction was used to detect the relative expression levels of the transfected LV-miR-576-3p in the indicated cells (U87 and U251). Effect of LV-miR-576-3p or LV-miR-NC on the (B) migration of U87 and U251 cells subsequent to hypoxic incubation for 24 h were determined using a wound healing assay (scale bar, 200 µm), which was then (C) quantified. Effect of LV-miR-576-3p or LV-miR-NC on the proangiogenic abilities of glioma cells subsequent to hypoxic incubation for 24 h were determined using (D) a Transwell migration assay (scale bar, 100 µm) and (E) quantified, and (F) tube formation assays (scale bar, 300 µm) which were then (G) quantified. All experiments were repeated three times. **P<0.01 and ***P<0.001 with comparisons shown by lines. miR, microRNA; LV, lentivirus; NC, negative control.
miR-576-3p targets HIF1-α
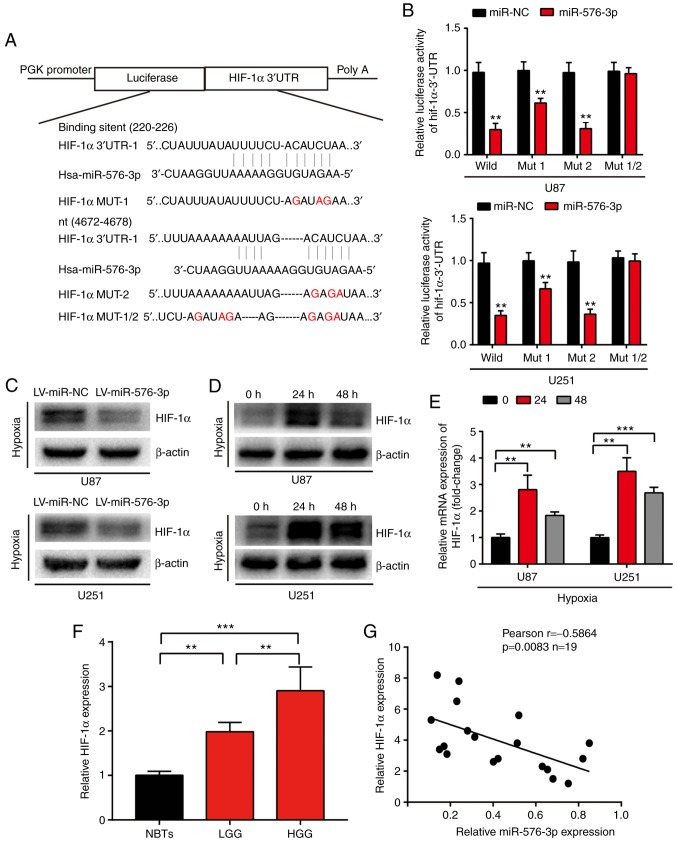
The targets of miR-576-3p were predicted using a number of algorithms, including miRanda, TargetScan and miRWalk. Among the candidates, HIF-1α was selected as a potential target gene and the corresponding plasmid was constructed (Fig. 3A). The results revealed that there was a significant reduction in luciferase activity in the cells transfected with miR-576-3p and wild-type HIF-1α compared with those transfected with the mutated reporter gene of the HIF-1α 3′-UTR together with miR-NC or miR-576-3p (P<0.01; Fig. 3B). Furthermore, the expression levels of HIF-1α protein in the glioma cells overexpressing miR-576-3p following 24 h of hypoxic exposure were evaluated. As presented in Fig. 3C, the expression of HIF-1α protein was substantially reduced in the miR-576-3p-transfected cells compared with the miR-NC transfected cells. In addition, western blot analysis and RT-qPCR were performed in order to determine the expression of HIF-1α without miR-576-3p intervention in glioma cells under a hypoxic environment. The results demonstrated that the expression levels were significantly elevated in glioma cells subsequent to hypoxic exposure compared with non-exposed cells (P<0.01; Fig. 3D and E). HIF-1α expression levels at different clinical stages of glioma and NBTs were investigated using RT-qPCR. The results indicated that HIF-1α expression was significantly positively associated with glioma malignancy and there were higher levels of HIF-1α expression in the glioma samples compared with the NBTs (P<0.01; Fig. 3F). In order to investigate the mechanism, the correlation between the expression levels of HIF-1α and miR-576-3p were determined using a Pearson's correlation test. The data revealed that there was a significant negative correlation between the expression levels of HIF-1α and miR-576-3p (P<0.01; Fig. 3G). These results strongly indicate that HIF-1α is a direct target of miR-576-3p, which may be a suppressor of glioma under hypoxia.
Figure 3.
HIF-1α is a direct target of miR-576-3p in glioma cells. (A) Predicted miR-576-3p binding sites in the 3′-UTR of HIF-1α mRNA. (B) Luciferase activities in HIF-1α-wild-type or HIF-1α-mut-type U251 and U87 cells transfected with miR-576-3p or miR-NC. **P<0.01 and ***P<0.001 vs. miR-NC. (C) miR-576-3p overexpression attenuated the protein expression of HIF-1α in the glioma cells following hypoxic exposure for 24 h. (D) Protein expression of HIF-1α was elevated in the glioma cells under hypoxic conditions. (E) Reverse transcription-quantitative polymerase chain reaction was performed to determine the expression of HIF-1α in the indicated cells. (F) HIF-1α was expressed in the indicated clinical brain tissue samples. (G) Pearson's correlation analysis revealed the correlation between the expression of miR-576-3p and mRNA levels of HIF-1α in 19 clinical glioma tissues. Data are presented as the mean ± standard deviation of three independent experiments. **P<0.01 and ***P<0.001 with comparisons shown by lines. HIF-1α, hypoxia-inducible factor-1α; 3′-UTR, 3′-untranslated region; miR, microRNA; NC, negative control; LV, lentivirus; NBTs, non-neoplastic brain tissues; LGG, low-grade glioma; HGG, high-grade glioma.
Downregulation of HIF-1α exerts similar effects to miR-576-3p in hypoxia-treated glioma cells
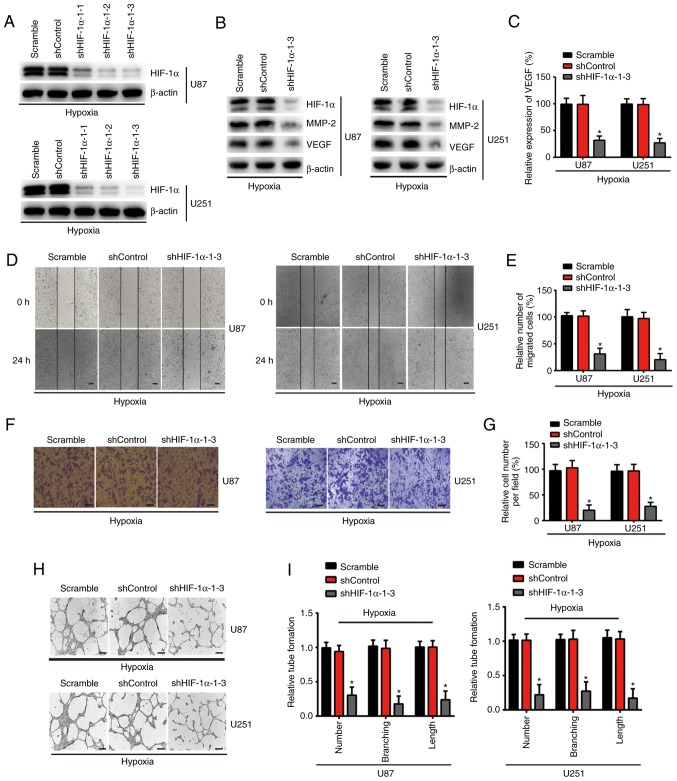
The present study aimed to validate that the malignant progression of glioma subsequent to the knockdown of HIF-1α alone is consistent with the intervention of miR-576-3p. In the present study, U87 and U251 cells were infected with three separate luciferase-encoding HIF-1α shRNAs or control shRNA. Detected using western blotting, the expression of HIF-1α was substantially decreased in the U87 and U251 cells compared with that in the scramble (non-targeting) group and the shControl groups (Fig. 4A). shHIF-1α-1-3 was selected for subsequent experiments based on its knockdown efficiency in the glioma cells. As expected, shHIF-1α-1-3 inhibited the expression of HIF-1α protein in the U87 and U251 cells following 24 h of hypoxia treatment compared with the scramble and shControl groups (Fig. 4B). No significant difference in the expression of HIF-1α protein was observed between the scramble group and the shControl group (P>0.05). In addition, compared with the scramble group and the shControl group, the expression of MMP-2, which is associated with tumor migration, and the expression of angiogenesis-associated VEGF were substantially reduced following HIF-1α silencing compared with the scramble and shControl groups (Fig. 4B). In order to evaluate the VEGF expression levels in the culture medium of these cells, an ELISA was performed and the results revealed that the expression levels of VEGF were significantly downregulated in the culture medium of the glioma cells following HIF-1α silencing compared with the scramble and shControl groups, whereas there were no significant difference between the scramble group and shControl group (P<0.05; Fig. 4C). The impact of HIF-1α knockdown on the migration and pro-angiogenesis of hypoxia-treated glioma cells was analyzed using wound healing, Transwell invasion and tube formation assays (Fig. 4D-I). As expected, the results demonstrated that HIF-1α knockdown exerted the same effect as miR-576-3p overexpression. This suggests that inhibition of HIF-1α may be a key mechanism by which miR-576-3p attenuates glioma cell migration and pro-angiogenic capacity.
Figure 4.
Knockdown of HIF-1α inhibits the migration and pro-angiogenesis of hypoxia-treated glioma cells in vitro. (A) Western blotting of the HIF-1α protein expression in glioblastoma multiforme cells. (B) Western blotting determined the expression of HIF-1α, MMP-2 and VEGF in each group following hypoxia exposure for 24 h. (C) Expression levels of VEGF in the conditioned medium of the indicated cells (U87 and U251) treated with hypoxia for 24 h, as determined by an ELISA. Effect of shHIF-1α-1-3 and shControl on the migration and pro-angiogenesis of glioma cells following hypoxic incubation for 24 h were determined using (D) a wound healing assay (scale bar, 200 µm) and (E) quantified, (F) a Transwell invasion assay (scale bar, 100 µm) and (G) quantified and (H) a tube formation assay (scale bar, 300 µm) and (I) quantified. Data are presented as the mean ± standard deviation of three independent experiments. *P<0.05 vs. scramble group and shControl group. HIF-1α, hypoxia-inducible factor-1α; MMP, matrix metalloproteinase; VEGF, vascular endothelial growth factor; shRNA, short hairpin RNA.
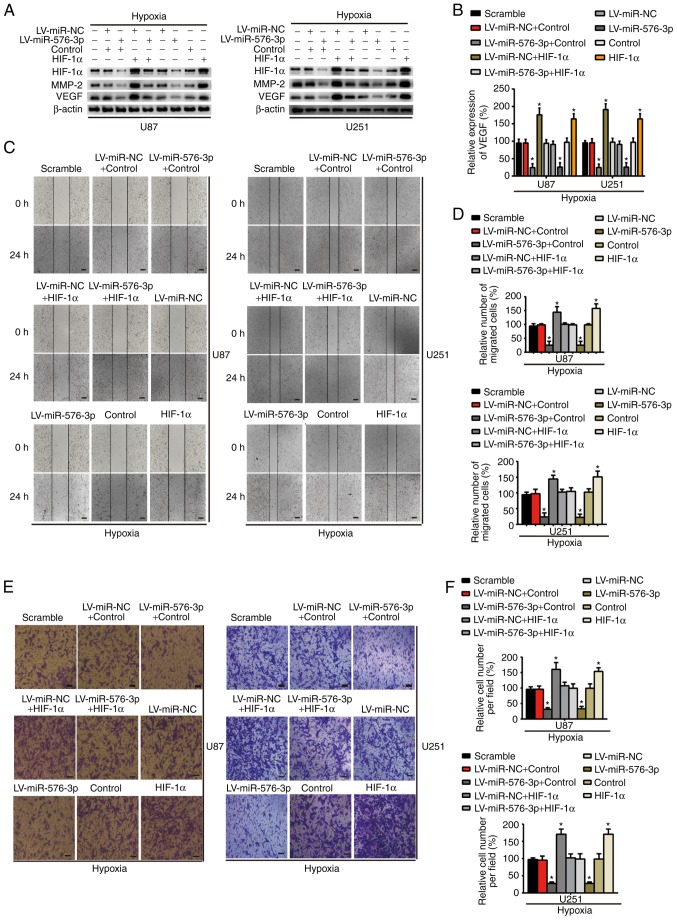
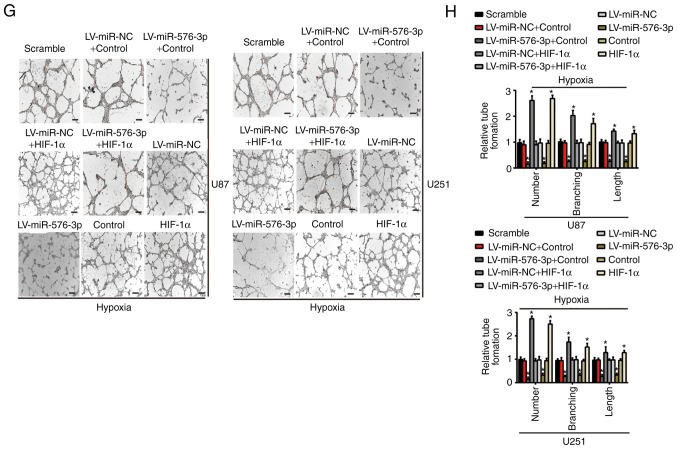
Reintroduction of HIF-1α attenuates miR-576-3p-induced inhibition of cell migration and angiogenesis
As presented in Fig. 5A, compared with the scramble group, the LV-miR-NC group, the control group or the co-transfection group by transfection of LV-miR-NC + control, a reduction of the HIF-1α expression in the co-transfection of LV-miR-576-3p and control group was rescued by HIF-1α overexpression in the co-transfection of LV-miR-576-3p and HIF-1α group. Notably, the expression of MMP-2 and VEGF (Fig. 5A), in addition to the corresponding VEGF expression levels in the conditioned media (P<0.05; Fig. 5B), also changed in a similar manner to the HIF-1α expression levels. Therefore, the reduction in MMP-2 and VEGF expression caused by the overexpression of miR-576-3p may be abolished by upregulating HIF-1α. Wound healing, Transwell invasion and tube formation assays were performed to validate whether HIF-1α is a pivotal target of miR-576-3p in U87 and U251 cells subsequent to hypoxia treatment for 24 h. As presented in Fig. 5C-H, HIF-1α was reintroduced into the hypoxia-treated glioma cells following transfection with miR-576-3p and it significantly abolished miR-576-3p inhibition of cell migration and proangiogenic effects compared with the scramble group, the LV-miR-NC group, the control group or the co-transfection group by transfection of LV-miR-NC + control (P<0.05). No significant difference in the cell migration and proangiogenic effects were detected between the scramble group, the LV-miR-NC group, the control group and the co-transfection group by transfection of LV-miR-NC + control. The results of the present study indicate that the malignant progression of glioma cells is affected by miR-576-3p through its regulation of HIF-1α.
Figure 5.
HIF-1α overexpression rescues the effects of miR-576-3p in vitro. (A) Western blotting of HIF-1α, MMP-2 and VEGF in the indicated cells (U87 and U251) under hypoxic conditions subsequent to the corresponding transfection. (B) Expression of VEGF in the conditioned medium of U87 and U251 cells treated with hypoxia for 24 h was determined using an enzyme-linked immunosorbent assay. Effect of HIF-1α or control in the presence or absence of miR-576-3p overexpression or miR-NC expression on the migration and pro-angiogenesis of glioma cells after hypoxic incubation for 24 h, as determined using (C) a wound healing (scale bar, 200 µm) and (D) quantified, (E) a Transwell invasion (scale bar, 100 µm) and (F) quantified. Effect of HIF-1α or control in the presence or absence of miR-576-3p overexpression or miR-NC expression on the migration and pro-angiogenesis of glioma cells after hypoxic incubation for 24 h, as determined using (G) a tube formation assay (scale bar, 300 µm) and (H) quantified. Data are presented as the mean ± standard deviation of three replicates. *P<0.05 vs. compared with the scramble group, the LV-miR-NC group, the control group or the co-transfection group by transfection of LV-miR-NC + control. miR, microRNA; NC, negative control; LV, lentivirus; HIF-1α, hypoxia-inducible factor-1α; MMP, matrix metalloproteinase; VEGF, vascular endothelial growth factor.
Discussion
An increasing number of studies have reported the presence of deregulated miRNA in various human cancer types (24-26), including in glioblastoma (27,28). Furthermore, studies have demonstrated that miRNA-576-3p is an inhibitor of bladder cancer through reducing cyclin D1 (29) and may be a prognostic biomarker for breast cancer subsequent to the development of chemotherapy resistance (30). However, the functional impact of miR-576-3p on glioma has not yet been reported to the best of our knowledge.
In the present study, the expression of miR-576-3p in glioma was investigated using 19 glioma specimens and 7 NBTs, and a number of glioma cell lines. RT-qPCR was used to detect the levels of miR-576-3p in glioma tissues and NBTs; miR-576-3p was significantly elevated in the non-cancerous brain tissue compared with in glioma specimens, and the downregulation of miR-576-3p was observed in the hypoxia-induced glioma cells. Previous studies have revealed that glioma, a solid tumor that is rich in blood vessels, has the histological properties of necrosis and capillary endothelial cell proliferation, which affect multiple processes including cell proliferation and invasion and angiogenesis (8,31). The present study investigated the effect of miR-576-3p on the migration and proangiogenic abilities of hypoxia-treated glioma cells and the results revealed that the overexpression of miR-576-3p results in the notable inhibition of these abilities compared with the control cells.
In order to investigate how miR-576-3p exerts its effects on glioma, an in-depth analysis of miR-576-3p targets was performed using a number of computational methods. miRWalk, miRanda and TargetScan online tools were used to predict candidate targets for miR-576-3p. Using a luciferase reporter assay and western blotting analysis, it was demonstrated that the protein expression of HIF-1α in glioma cells under hypoxia may be reduced through miR-576-3p overexpression by directly targeting its 3′-UTR.
A previous study confirmed that hypoxia-induced angiogenesis serves a notable function in the prognosis of patients with cancer (32). The mechanism has been investigated extensively in previous years and HIF-1α is a necessary regulator. HIF-1 is a heterodimeric protein composed of HIF-1α and HIF-1β subunits (33). Under normoxic conditions, the HIF-1α protein is ubiquitinated and quickly degraded by the proteasome pathway (34). In a low-oxygen environment, the HIF-1α protein accumulates and then is translocated to the nucleus, constructs a heterodimeric protein complex with HIF-1β and activates a variety of notable genes (35,36). HIF-1α has been reported to be involved in numerous types of tumor (37,38), including glioma, and is closely associated with tumor invasion and metastasis (39,40). HIF-1α expression has been demonstrated to be involved in the invasiveness of glioblastoma and is closely associated with grade and vascular density (8). To validate whether miR-576-3p affects hypoxia-treated glioma by regulating the levels of HIF-1α, lentiviruses carrying shRNA-HIF-1α were applied to knockdown the expression of HIF-1α in the present study. The results demonstrated that HIF-1α knockdown under hypoxic conditions is as effective as miR-576-3p overexpression in inhibiting the migration and angiogenesis of hypoxia-induced glioma cells. Notably, hypoxia-induced VEGF expression is the main driving force behind neovascular maturation during tumor progression and embryonic development (41,42). VEGF may be produced in the cystic fluid of patients with GBM, where the levels of VEGF are higher compared with those in the serum (43). As one of the main mediators of tumor angiogenesis, VEGF may be stimulated by hypoxia through the major hypoxia-responsive transcriptional activator HIF-1α (44). In addition, MMPs are necessary for ECM degradation. MMP-mediated ECM digestion lays the foundation for the migration of proliferating endothelial cells and their invasion of the matrix, which is a necessary step in neovascular maturation (45). Furthermore, MMPs may degrade ECM macromolecules, which enables cell infiltration and invasion and results in the failure of conventional treatment of brain tumor types (46). In glial brain tumor types, MMP-2 has been demonstrated to be a downstream regulator of HIF-1α (47). In the present study, western blot analysis and an ELISA revealed that the downregulation of HIF-1α results in a substantial reduction in the expression of MMP-2 and VEGF compared with control cells. Conversely, the restored expression of HIF-1α in LV-miR-576-3p-transfected glioma cells abolished the effect of miR-576-3p on cell migration and angiogenesis under hypoxia. The western blotting results additionally revealed the corresponding activation of VEGF and MMP-2. Therefore, these results confirm that miR-576-3p-induced resistance to glioma cell migration and angiogenesis may be achieved by targeting HIF-1α and may result in the decline of VEGF and MMP-2 expression.
In conclusion, the present study demonstrates that miR-576-3p is markedly downregulated in hypoxia-induced glioma cells compared with those in a normoxic environment, and that the overexpression of miR-576-3p in turn reduces the migration and proangiogenic abilities of hypoxia-treated glioma cells by attenuating HIF-1α expression. Based on these results, the targeting of miR-576-3p is a novel therapeutic strategy for patients with glioma. However, in vivo studies should be performed to confirm that miR-576-3p is a promising candidate for the treatment of gliomas. In addition, further studies are required to determine the detailed mechanism of miR-576-3p downregulation during glioma progression.
Acknowledgments
Not applicable.
Funding
The present study was supported by the Natural Science Foundation of Jiangxi Province (grant no. 20171ACB20035), the National Natural Science Foundation (grant nos. 81760446 and 81660420), the Foreign Science and Technology Cooperation Plan of Jiangxi Province (grant no. 20151BDH80009) and the Construction Plan of the Superior Science and Technology Innovation Team of Jiangxi Province (grant no. 20152BCB24009).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
SL and XZ conceived the experiments. QH and FL wrote the manuscript. QH, FL and TY conducted the experiments. MW, MY and GS analyzed the data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was ethically approved by the Ethics Committee of the Second Affiliated Hospital of Nanchang University and written informed consent was obtained from all patients.
Patient consent for publication
Patients provided consent for publication.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 3.Van Meir EG, Hadjipanayis CG, Norden AD, Shu HK, Wen PY, Olson JJ. Exciting new advances in neuro-oncology: The avenue to a cure for malignant glioma. CA Cancer J Clin. 2010;60:166–193. doi: 10.3322/caac.20069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Senger D, Cairncross JG, Forsyth PA. Long-term survivors of glioblastoma: Statistical aberration or important unrecognized molecular subtype. Cancer J. 2003;9:214–221. doi: 10.1097/00130404-200305000-00009. [DOI] [PubMed] [Google Scholar]
- 5.Brandes AA, Franceschi E, Gorlia T, Wick W, Jacobs AH, Baumert BG, van den Bent M, Weller M, Stupp R, European Organisation for Research and Treatment of Cancer Brain Tumour Group Appropriate end-points for right results in the age of antiangiogenic agents: Future options for phase II trials in patients with recurrent glioblastoma. Eur J Cancer. 2012;48:896–903. doi: 10.1016/j.ejca.2011.10.027. [DOI] [PubMed] [Google Scholar]
- 6.Hou LC, Veeravagu A, Hsu AR, Tse VC. Recurrent glioblastoma multiforme: A review of natural history and management options. Neurosurg Focus. 2006;20:E5. doi: 10.3171/foc.2006.20.4.2. [DOI] [PubMed] [Google Scholar]
- 7.Soda Y, Myskiw C, Rommel A, Verma IM. Mechanisms of neovascularization and resistance to anti-angiogenic therapies in glioblastoma multiforme. J Mol Med (Berl) 2013;91:439–448. doi: 10.1007/s00109-013-1019-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zagzag D, Zhong H, Scalzitti JM, Laughner E, Simons JW, Semenza GL. Expression of hypoxia-inducible factor 1alpha in brain tumors: Association with angiogenesis, invasion, and progression. Cancer. 2000;88:2606–2618. doi: 10.1002/1097-0142(20000601)88:11<2606::AID-CNCR25>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 9.Kakudo N, Morimoto N, Ogawa T, Taketani S, Kusumoto K. Hypoxia enhances proliferation of human adipose-derived stem cells via HIF-1a activation. PLoS One. 2015;10:e0139890. doi: 10.1371/journal.pone.0139890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bae WY, Choi JS, Kim JE, Jeong JW. Cinnamic aldehyde suppresses hypoxia-induced angiogenesis via inhibition of hypoxia-inducible factor-1α expression during tumor progression. Biochem Pharmacol. 2015;98:41–50. doi: 10.1016/j.bcp.2015.08.095. [DOI] [PubMed] [Google Scholar]
- 11.Antica M, Kusic B, Hranilovic D, Dietz AB, Vuk-Pavlovic S. Cloning the cDNA for murine U2 snRNP-A' gene and its differential expression in lymphocyte development. Immunol Lett. 2002;82:217–223. doi: 10.1016/S0165-2478(02)00064-0. [DOI] [PubMed] [Google Scholar]
- 12.Li HS, Zhou YN, Li L, Li SF, Long D, Chen XL, Zhang JB, Li YP, Feng L. WITHDRAWN: Mitochondrial targeting of HIF-1α inhibits hypoxia-induced apoptosis independently of its transcriptional activity. Free Radic Biol Med. 2018 doi: 10.1016/j.freeradbiomed.2018.04.568. [DOI] [PubMed] [Google Scholar]
- 13.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 14.Esquela-Kerscher A, Slack FJ. Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 15.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: Tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 17.Shenouda SK, Alahari SK. MicroRNA function in cancer: Oncogene or a tumor suppressor. Cancer Metastasis Rev. 2009;28:369–378. doi: 10.1007/s10555-009-9188-5. [DOI] [PubMed] [Google Scholar]
- 18.Hossian AKMN, Sajib MS, Tullar PE, Mikelis CM, Mattheolabakis G. Multipronged activity of combinatorial miR-143 and miR-506 inhibits lung cancer cell cycle progression and angiogenesis in vitro. Sci Rep. 2018;8:10495. doi: 10.1038/s41598-018-28872-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Ouyang M, Wang Q, Jian Z. MicroRNA-142-3p inhibits hypoxia/reoxygenation-induced apoptosis and fibrosis of cardiomyocytes by targeting high mobility group box 1. Int J Mol Med. 2016;38:1377–1386. doi: 10.3892/ijmm.2016.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li L, Sun P, Zhang C, Li Z, Zhou W. MiR-98 suppresses the effects of tumor-associated macrophages on promoting migration and invasion of hepatocellular carcinoma cells by regulating IL-10. Biochimie. 2018;150:23–30. doi: 10.1016/j.biochi.2018.04.016. [DOI] [PubMed] [Google Scholar]
- 21.Rainov NG, Heidecke V. Clinical development of experimental therapies for malignant glioma. Sultan Qaboos Univ Med J. 2011;11:5–28. [PMC free article] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Miletic H, Niclou SP, Johansson M, Bjerkvig R. Anti-VEGF therapies for malignant glioma: Treatment effects and escape mechanisms. Expert Opin Ther Targets. 2009;13:455–468. doi: 10.1517/14728220902806444. [DOI] [PubMed] [Google Scholar]
- 24.Cheng RF, Wang J, Zhang JY, Sun L, Zhao YR, Qiu ZQ, Sun BC, Sun Y. MicroRNA-506 is up-regulated in the development of pancreatic ductal adenocarcinoma and is associated with attenuated disease progression. Chin J Cancer. 2016;35:64. doi: 10.1186/s40880-016-0128-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hong-Yuan W, Xiao-Ping C. miR-338-3p suppresses epithelial-mesenchymal transition and metastasis in human nonsmall cell lung cancer. Indian J Cancer. 2015;52(Suppl 3):E168–E171. doi: 10.4103/0019-509X.186569. [DOI] [PubMed] [Google Scholar]
- 26.Wu JH, Wang YH, Wang W, Shen W, Sang YZ, Liu L, Chen CM. MiR-18b suppresses high-glucose-induced proliferation in HRECs by targeting IGF-1/IGF1R signaling pathways. Int J Biochem Cell Biol. 2016;73:41–52. doi: 10.1016/j.biocel.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 27.Zhou Y, Liu Y, Hu C, Jiang Y. MicroRNA-16 inhibits the proliferation, migration and invasion of glioma cells by targeting Sal-like protein 4. Int J Mol Med. 2016;38:1768–1776. doi: 10.3892/ijmm.2016.2775. [DOI] [PubMed] [Google Scholar]
- 28.Li Q, Zhou L, Wang M, Wang N, Li C, Wang J, Qi L. MicroRNA-613 impedes the proliferation and invasion of glioma cells by targeting cyclin-dependent kinase 14. Biomed Pharmacother. 2018;98:636–642. doi: 10.1016/j.biopha.2017.12.044. [DOI] [PubMed] [Google Scholar]
- 29.Liang Z, Li S, Xu X, Xu X, Wang X, Wu J, Zhu Y, Hu Z, Lin Y, Mao Y, et al. MicroRNA-576-3p inhibits proliferation in bladder cancer cells by targeting cyclin D1. Mol Cells. 2015;38:130–137. doi: 10.14348/molcells.2015.2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xia Lv J, Xu K, Sun P, Ma E, Gao J, Zhou S, Zhang Q, Wang M, Chen FF, et al. miRNA expression patterns in chemoresistant breast cancer tissues. Biomed Pharmacother. 2014;68:935–942. doi: 10.1016/j.biopha.2014.09.011. [DOI] [PubMed] [Google Scholar]
- 31.Sun B, Zhang D, Zhang S, Zhang W, Guo H, Zhao X. Hypoxia influences vasculogenic mimicry channel formation and tumor invasion-related protein expression in melanoma. Cancer Lett. 2007;249:188–197. doi: 10.1016/j.canlet.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 32.Liao D, Johnson RS. Hypoxia: A key regulator of angiogenesis in cancer. Cancer Metastasis Rev. 2007;26:281–290. doi: 10.1007/s10555-007-9066-y. [DOI] [PubMed] [Google Scholar]
- 33.Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci USA. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang LE, Gu J, Schau M, Bunn HF. Regulation of hypoxia-inducible factor 1alpha is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc Natl Acad Sci USA. 1998;95:7987–7992. doi: 10.1073/pnas.95.14.7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 36.Papandreou I, Cairns RA, Fontana L, Lim AL, Denko NC. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 2006;3:187–197. doi: 10.1016/j.cmet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 37.Liu X, Chen S, Tu J, Cai W, Xu Q. HSP90 inhibits apoptosis and promotes growth by regulating HIF-1α abundance in hepatocellular carcinoma. Int J Mol Med. 2016;37:825–835. doi: 10.3892/ijmm.2016.2482. [DOI] [PubMed] [Google Scholar]
- 38.Shiau AL, Shen YT, Hsieh JL, Wu CL, Lee CH. Scutellaria barbata inhibits angiogenesis through downregulation of HIF-1 α in lung tumor. Environ Toxicol. 2014;29:363–370. doi: 10.1002/tox.21763. [DOI] [PubMed] [Google Scholar]
- 39.Tang JH, Ma ZX, Huang GH, Xu QF, Xiang Y, Li N, Sidlauskas K, Zhang EE, Lv SQ. Downregulation of HIF-1a sensitizes U251 glioma cells to the temozolomide (TMZ) treatment. Exp Cell Res. 2016;343:148–158. doi: 10.1016/j.yexcr.2016.04.011. [DOI] [PubMed] [Google Scholar]
- 40.Shi H, Zheng B, Wu Y, Tang Y, Wang L, Gao Y, Gong H, Du J, Yu R. Ubiquitin ligase Siah1 promotes the migration and invasion of human glioma cells by regulating HIF-1α signaling under hypoxia. Oncol Rep. 2015;33:1185–1190. doi: 10.3892/or.2014.3695. [DOI] [PubMed] [Google Scholar]
- 41.Cui H, Wang Y, Huang H, Yu W, Bai M, Zhang L, Bryan BA, Wang Y, Luo J, Li D, et al. GPR126 protein regulates developmental and pathological angiogenesis through modulation of VEGFR2 receptor signaling. J Biol Chem. 2014;289:34871–34885. doi: 10.1074/jbc.M114.571000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gupta B, Chiang L, Chae K, Lee DH. Phenethyl isothiocyanate inhibits hypoxia-induced accumulation of HIF-1α and VEGF expression in human glioma cells. Food Chem. 2013;141:1841–1846. doi: 10.1016/j.foodchem.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 43.Takano S, Yoshii Y, Kondo S, Suzuki H, Maruno T, Shirai S, Nose T. Concentration of vascular endothelial growth factor in the serum and tumor tissue of brain tumor patients. Cancer Res. 1999;56:2185–2190. [PubMed] [Google Scholar]
- 44.Forsythe JA, Jiang BH, Iyer NV, Agani F, Leung SW, Koos RD, Semenza GL. Activation of vascular endothelial growth factor gene transcription by hypoxia-inducible factor 1. Mol Cell Biol. 1996;16:4604–4613. doi: 10.1128/MCB.16.9.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Osenkowski P, Toth M, Fridman R. Processing, shedding, and endocytosis of membrane type 1-matrix metalloproteinase (MT1-MMP) J Cell Physiol. 2004;200:2–10. doi: 10.1002/jcp.20064. [DOI] [PubMed] [Google Scholar]
- 46.Schenk S, Hintermann E, Bilban M, Koshikawa N, Hojilla C, Khokha R, Quaranta V. Binding to EGF receptor of a laminin-5 EGF-like fragment liberated during MMP-dependent mammary gland involution. J Cell Biol. 2003;161:197–209. doi: 10.1083/jcb.200208145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fujiwara S, Nakagawa K, Harada H, Nagato S, Furukawa K, Teraoka M, Seno T, Oka K, Iwata S, Ohnishi T. Silencing hypoxia-inducible factor-1alpha inhibits cell migration and invasion under hypoxic environment in malignant gliomas. Int J Oncol. 2007;30:793–802. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.