Abstract
Emerging evidence has demonstrated that microRNAs (miRNAs/miRs) have various biological functions in the development of human epidermal growth factor receptor 2 (HER2) positive breast cancer. The aim of the present study is to reveal the mechanism of miR-193a-3p inhibiting the progress of HER2 positive breast cancer. The expression of miR-193a-3p was evaluated by quantitative polymerase chain reaction (PCR). The methylation status of miR-193a-3p was evaluated by PCR and pyrosequencing analysis. Overexpression of miR-193a-3p and growth factor receptor bound protein 7 (GRB7) combined with in vitro tumorigenic assays were conducted to determine the carcinostatic capacities of miR-193a-3p in HER2 positive breast cancer cells. The association between miR-193a-3p and GRB7 was determined by luciferase reporter assay. Protein level was evaluated using western blot analysis. miR-193a-3p was down-regulated in HER2 positive breast cancer cells and clinical tissues. Methylation-mediated silencing led to decreased expression of miR-193a-3p in HER2 positive breast cancer. Overexpression of miR-193a-3p could inhibit proliferation, migration and invasion of breast cancer cells. Overexpression of GRB7 could abolish this effect. miR-193a-3p could directly target the 3′ untranslated region of GRB7. miR-193a-3p could directly or indirectly target extracellular signal-regulated kinase 1/2 (ERK1/2) and forkhead box M1 (FOXM1) signaling. In conclusion, it was identified that silencing of miR-193a-3p through hypermethylation can promote HER2 positive breast cancer progress by targeting GRB7, ERK1/2 and FOXM1 signaling. The function of miR-193a-3p in HER2 positive breast cancer implicates its potential application in therapy.
Keywords: breast cancer, human epidermal growth factor receptor 2, microRNA-193a-3p, growth factor receptor bound protein 7
Introduction
Cancer is a principal public health problem globally and breast cancer is one of the most frequently diagnosed types of cancer. It is estimated that there will be 268,670 newly diagnosed cases (2,550 male cases and 266,120 female cases) of breast cancer in the United States in 2018 (1). High-grade types of breast cancer have high aggression and are associated with poor prognosis and shorter survival time (2). The intensive study of molecular mechanisms underlying the progress of breast cancer could aid early diagnosis and treatment. In this respect, the identification of genetic/epigenetic mutations of oncogenes/anti-oncogenes is a potential research direction. At present, a few of immunohistochemistry (IHC) markers as well as clinicopathological variables have become the basis of prognosis prediction and therapy selection for breast cancer (3,4).
Human epidermal growth factor receptor 2 (HER2), estrogen receptor and progesterone receptor are the most commonly used IHC markers for breast cancer. HER2 is an important member of epidermal growth factor receptor family (5,6). In breast cancer clinics, cases associated with HER2 overexpression, which is defined as HER2 positive status, account for ~20% of all patients (5). At the cellular level of breast cancer, HER2 is mainly located in the cell membrane and acts as an oncogene (7,8).
In ~90% of HER2 positive breast cancer cases, HER2 over-expression is caused by HER2 gene amplification (9). Within the 17q12-21 amplicon, HER2 is located at 17q12 (10) and multiple coamplified genes including growth factor receptor bound protein 7 (GRB7), StAR related lipid transfer domain containing 3, DNA topoisomerase II α, protein phosphatase 1 regulatory inhibitor subunit 1B, thyroid hormone receptor α, and retinoic acid receptor α have been identified on the 17q12-21 amplicon (11,12). Coamplified genes on the HER2 amplicon may activate cellular processes that are not directly oncogenic, but have become necessary for the oncogenic state. Therefore, multiple targeted therapies in HER2 positive breast cancer are necessary.
Accumulating evidence has proven that microRNAs (miRNAs/miR) serve important roles in cancer metastasis by reducing the expression of their targets, including mRNA, long noncoding RNA, circular RNA and pseudogenes (13-15). miR-193a-3p has been identified as a key tumor suppressor in cancer (16,17), but little is known about the role of miR-193a-3p in HER2 positive breast cancer.
GRB7 is part of the 17q12-21 amplicon, located close to the HER2 gene (18). Transcript analysis indicates that in breast cancer cells GRB7 RNA expression is always high synchronously with HER2/neuraminidase 1 (Neu) (19). More and more evidence has demonstrated that overexpression of GRB7 is correlated with a metastatic phenotype and deceased survival in breast cancer (18,20). Therefore, GRB7 may have the potential to become a novel therapeutic target in breast cancer.
In order to Figure out better therapies for breast cancer, it is crucial to understand the pathogenesis more thorough. In the present study, insights are provided into the potential effects of miR-193a-3p in HER2 positive breast cancer. As the increase of DNA methylation in the miRNA promoter could reduce the transcription efficiency (21-23), it was identified that during HER2 positive cancer development and progression, miR-193a-3p was silencing by DNA hypermethylation, and the epigenetic silencing of miR-193a-3p made the expression of GRB7 higher and therefore activated the extracellular signal-regulated kinase/forkhead box M1 (ERK/FOXM1) signaling pathway.
Materials and methods
Cell culture and human tissues
The human HER2 positive breast cancer cell lines HCC-1954, 21MT1 and JimT1 and human normal breast cell line MCF-10A were bought from the American Type Culture Collection (Manassas, VA, USA). All of the 4 cell lines were cultured in RPMI-1640 medium (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany), which contained 10% fetal bovine serum (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and incubated at 37°C with 5% CO2. A total of 35 pairs of the clinical HER2 positive breast cancer and adjacent tissues were collected from 35 patients (age, 21 to 58 years old) who received resection surgery in The Third Affiliated Hospital of Kunming Medical University (Kunming, China) from April 2015 to August 2017. All of the human tissues used in the present study were obtained with written informed consent. The present study was approved by the Ethics Committee of The Third Affiliated Hospital of Kunming Medical University.
Plasmid and cell transfection
Synthetic pre-miR-193a-3p (Shanghai GenePharma Co., Ltd., Shanghai, China) was used to transfect cells to overexpress miR-193a-3p as previously described (24). GRB7 overexpression plasmid was constructed as previously described (25). HCC-1954, 21MT1 and JimT1 cells were plated in 6-well plates (2.5×105 cells/well) and were transfected using Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the protocol.
Quantitative polymerase chain reaction (qPCR)
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) was used to extract total RNA from all the 3 cell lines with or without treatment with 5-Aza-dc (5 µM) at room temperature for 4 days and human tissues. Universal cDNA Synthesis kit (Exiqon; Qiagen, Inc., Valencia, CA, USA) was used to synthesize first-strand complementary DNA. miRCURY LNA™ Universal RT microRNA PCR (Exiqon; Qiagen, Inc.) was used to conduct qPCR determining miRNA. cDNA synthesis was conducted at 95°C for 12 min, and the qPCR was conducted with the following thermocycling conditions: 97°C for 5 min, followed by 35 cycles at 95°C for 30 sec, 65°C for 30 sec and 73°C for 1 min, and a final step at 73°C for 10 min; samples were then kept at 4°C until use. U6 was used as an endogenous control. Primers of hsa-miR-193a-3p (product no. 204591) were obtained from Exiqon. The forward primer of miR-193a-3p was 5′-CTGAGGGCTGGGTCTTTGC-3′ and the reverse primer was 5′-GCCGAGAACTGGGACTTTGT-3′. The forward primer and reverse primer of U6 were 5′-CTCGCTTCGGCAGCACA-3′ and 5′-ACGCTTCACGAATTTGCGT-3′, respectively.
Western blotting
Western blotting was conducted as previously described (26). Antibodies against GRB7 (1:2,000; cat. no. sc-13954; Santa Cruz Biotechnology, Inc., Dallas, TX, USA), ERK (1:1,500; cat. no. 4795; Cell Signaling Technology, Inc., Danvers, MA, USA), phosphorylated ERK (1:2,000; cat. no. 4795; Cell Signaling Technology, Inc.), FOXM1 (1:2,000; cat. no. 5436; Cell Signaling Technology, Inc.) and β-actin (1:10,000; cat. no. AC-74; Sigma-Aldrich; Merck KGaA) were used in the present study. The secondary antibody used was horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G (1:1,000; cat. no. sc-2004; Santa Cruz Biotechnology, Inc., Dallas, TX, USA). Semi-quantitative analysis was performed using ImageJ software v1.8.0 (National Institutes of Health, Bethesda, MD, USA).
Cell proliferation assay
Cell Counting Kit-8 (CCK-8) assay (Dojindo Molecular Technologies, Inc., Kumamoto, Japan) was used to measure cell proliferation ability. HCC-1954, 21MT1 and JimT1 cells (3×103 cells/well) were plated in 96-well culture plates for 72 h. The CCK-8 reagent was added to each well and incubated at 37°C for 1 h. Cell viability was assessed by testing the absorbance at 450 nm using Multiskan MS (Thermo Labsystems, Helsinki, Finland).
Colony formation assay
HCC-1954, 21MT1 and JimT1 cells (500 cells/well) were plated in 6-well plates and then cultured in complete media for 10 days. After removing media and being washed by ice-cold PBS 2 times, the colonies were fixed with methanol for 15 min at 4°C and stained with crystal violet for 30 min at room temperature. Images were captured using a digital camera (Canon, Inc., Tokyo, Japan).
Wound healing assay
The wound-healing assay was carried out to measure the migration ability of HER2 positive breast cancer cells. All 3 cell lines (5×105 cells/well) were fused to form a single layer in 6-well plates and a 200-µl sterile pipette tip was used to scratch a single wound on the cell layer. Following 2 rinses with PBS, cells were incubated for another 24 h. The scratch wounds were visualized under an inverted microscope (CKX41; Olympus Corporation, Tokyo, Japan) and the scratch widths were quantified with ImageJ software v1.8.0 (National Institutes of Health).
Cell invasion assay
Transwell invasion assay was performed using Transwell cell invasion assay kits (Corning, Inc., Corning, NY, USA). A total of 3×104 cells were digested and put in the serum-free medium in the upper chamber, with a 2 mg/ml Matrigel-coated membrane containing 8-m pores. The lower Transwell chamber contained Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Invitrogen; Thermo Fisher Scientific, Inc.). Following incubation for 72 h (37°C, 5% CO2), the cells were removed from the upper part of the filters by wiping with a cotton swab. Then, cells on the lower surface of the membrane were fixed with 4% formaldehyde at room temperature for 10 min and stained with 0.5% crystal violet for 15 min at room temperature. Finally, the number of invading cells was imaged and counted at ×200 magnification using an inverted microscope (Nikon Corporation, Tokyo, Japan).
Luciferase reporter assay
The sequence of the GRB7 3′-UTR which is the potential target of miR-193a-3p was ligated into the pmirGLO plasmid (Promega Corporation, Madison, WI, USA). HCC-1954 cells were cotransfected with the pmirGLO-3′-UTR plasmid of the above plasmids or a blank vector using Lipofectamine™ 2000(Invitrogen; Thermo Fisher Scientific, Inc.). The Dual-Luciferase Assay kit (Promega Corporation) was used to conduct luciferase activities, and the transfection efficiency was normalized by co-transfecting with Renilla-luciferase.
Pyrosequencing analysis
A pyrosequencing assay was conducted to detect the percentage of methylation in miR-193a-3p in HER2 positive breast cancer tissues. PSQ Assay Design Software (version 1.0.6; Biotage, Uppsala, Sweden) was used to design the primers used in pyrosequencing analysis.
RNA immunoprecipitation (RIP)
RIP experiments were performed using the Magna RIP™ RNA-Binding Protein Immunoprecipitation kit (EMD Millipore, Billerica, MA, USA) according to the manufacturer's protocol. The co-precipitated RNAs were detected by reverse transcription PCR, as aforementioned. Total RNA (input controls) and normal mouse immunoglobulin G (1:2,000; cat. no. SLM66-0100; Equitech-Bio, Inc., Kerrville, TX, USA) controls were assayed simultaneously to demonstrate that the detected signals were from the RNA that was specifically bound to GRB7 (n=3 for each experiment).
Statistical analysis
All experiments were repeated at least 3 times independently. Data are presented as the mean ± standard deviation. Two-tailed Student's t-test and one-way analysis of variance followed by Dunnett's C were used to calculate statistically significant differences. All statistical analyses were performed using SPSS software (version 20.0; IBM, Corps., Chicago, IL, USA). P<0.05 was considered to indicate a statically significant difference.
Results
miR-193a-3p is downregulated in HER2 positive breast cancer depending on the malignant degree
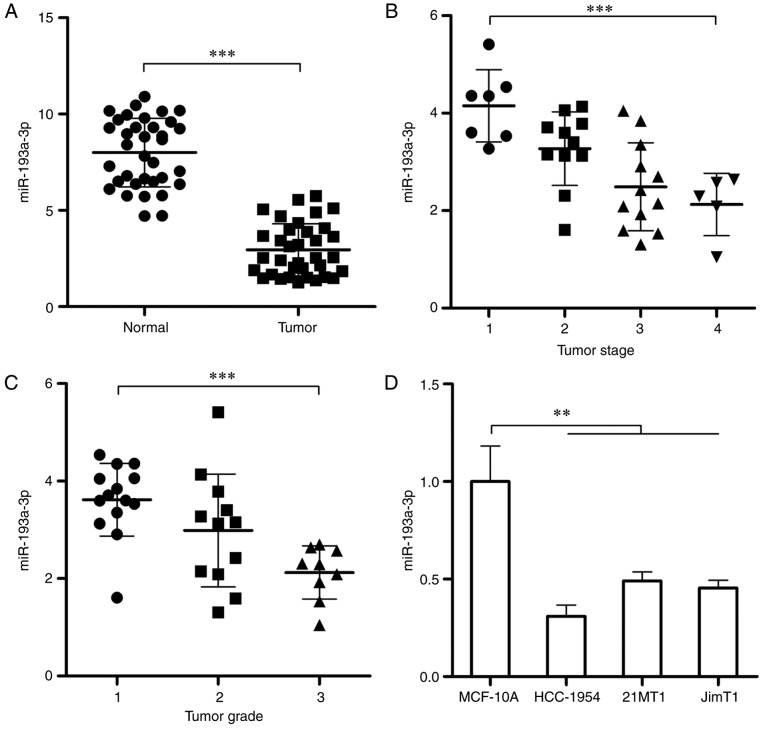
In order to investigate the potential role of miR-193a-3p in HER2 positive breast cancer, the expression of miR-193a-3p in 35 pairs of tumor/adjacent HER2 positive breast cancer tissues were determined and compared. As depicted in Fig. 1A, the expression of miR-193a-3p in tumor tissues was significantly decreased compared with the normal tissues (P<0.001). The expression of miR-193a-3p in HER2 positive breast cancer tissues of different stages and grades was also detected. As presented in Fig. 1B and C, the level of miR-193a-3p decreases significantly with the increase of tumor stage and grade (P<0.001), which means the level of miR-193a-3p in HER2 positive breast cancer is also associated with the malignant degree. Furthermore, the expression of miR-193a-3p in HER2 positive breast cancer cells (3 different cell lines) and normal breast cell was tested by qPCR. The result demonstrated that compared with normal breast cells, miR-193a-3p was significantly downregulated in HER2 positive breast cancer cells (P<0.01; Fig. 1D).
Figure 1.
Expression of miR-193a-3p is decreased in HER2 positive breast cancer and is associated with tumor stage and grade. (A) RT-qPCR was conducted to test the expression of miR-193a-3p in 35 pairs of HER2 positive breast cancer tissues and adjacent tissues. (B) The expression of miR-193a-3p in HER2 positive breast cancer tissues at different stages (Stage 1, n=7; Stage 2, n=11; Stage 3, n=12; Stage 4, n=5). (C) The expression of miR-193a-3p in HER2 positive breast cancer tissues at different grades (Grade 1, n=14; Grade 2, n=12; Grade 3, n=9). (D) RT-qPCR was conducted to determine the expression of miR-193a-3p in normal human breast cells and HER2 positive breast cancer cells. **P<0.01 and ***P<0.001, as indicated. RT-qPCR, reverse transcription-quantitative polymerase chain reaction; HER2, human epidermal growth factor receptor 2; miR, microRNA.
DNA methylation causes the reduction of miR-193a-3p in HER2 positive breast cancer cells
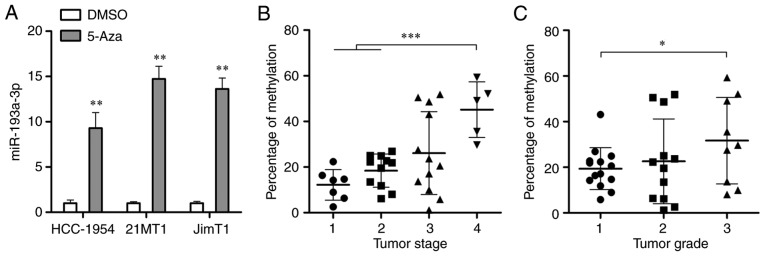
Then the molecular mechanisms underlying the decrease of miR-193a-3p were investigated in HER2 positive breast cancer. According to the usual regulatory mechanism of miRNA in cancer, the alterations of miR-193a-3p expression depending on DNA methylation in HER2 positive breast cancer cells were investigated. After treating with a demethylating agent, 5-Aza-dc (5 µM) for 4 days, qPCR was conducted to detect the expression of miR-193a-3p, which was demonstrated to significantly increase in the 2 tested HER2 positive breast cancer cell lines (P<0.01; Fig. 2A). Subsequently, pyrosequencing analysis demonstrated a significant increase of miR-193a-3p DNA methylation in higher-stage and higher-grade tumors (P<0.01 and P<0.05; Fig. 2B and C, respectively). These results demonstrated that the loss of miR-193a-3p in HER2 positive breast cancer may be caused by DNA hypermethylation.
Figure 2.
Identification of DNA methylation leads to the downregulation of miR-193a-3p in breast cancer cells. (A) Quantitative polymerase chain reaction was carried out to determine the expression of miR-193a-3p in HER2 positive breast cancer cells following 4-days treatment with 5-Aza-dc. **P<0.01 vs. DMSO. (B) Pyrosequencing analysis was conducted to analyze the percentage of methylation in the miR-193-3p promoter in HER2 positive breast cancer tissues at different stages (Stage 1, n=7; Stage 2, n=11; Stage 3, n=12; Stage 4, n=5). (C) Pyrosequencing analysis was conducted to analyze the percentage of methylation in the miR-193-3p promoter in HER2 positive breast cancer tissues at different grades (Grade 1, n=14; Grade 2, n=12; Grade 3, n=9). *P<0.05 and ***P<0.001, as indicated. HER2, human epidermal growth factor receptor 2; miR, microRNA.
Overexpression of miR-193a-3p could inhibit proliferation, migration and invasion of HER2 positive breast cancer cells
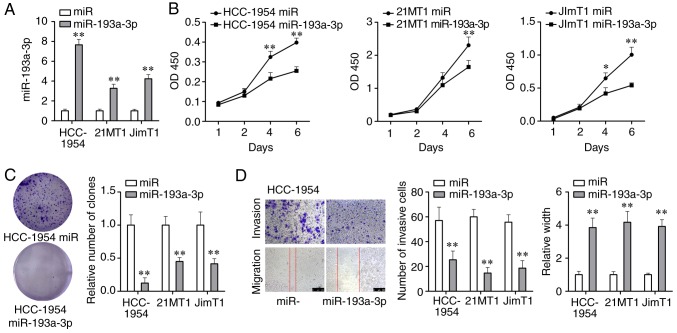
In order to further investigate the role miR-193a-3p serves during the development of HER2 positive breast cancer, the changes of cell viability, colony formation ability, migration ability and invasion ability of HER2 positive breast cancer cells overexpressing miR-193a-3p were tested. miR-193a-3p mimics were transfected into 3 HER2 positive breast cancer cell lines and its expression was significantly upregulated (P<0.01; Fig. 3A). Through the CCK-8 assay, it was demonstrated that cell proliferation was significantly weakened following 4-6 days of the treatment with miR-193a-3p mimics (P<0.01; Fig. 3B). Colony formation assay demonstrated that miR-193a-3p mimics could significantly reduce the number of cancer cell colonies formed (P<0.01; Fig. 3C). The wound-healing and Transwell assays were carried out to investigate the effect of miR-193a-3p mimics on cell migration and invasion abilities. As Fig. 3D displays, cell migration and invasion abilities of HER2 positive breast cancer cells were significantly suppressed by miR-193a-3p mimics (P<0.01). Moreover, overexpression of miR-193a-3p could inhibit proliferation, migration and invasion of another 2 HER2 positive breast cancer cell lines 21MT1 and JimT1 (Fig. S1). In brief, these findings indicated that the overexpression of miR-193a-3p could inhibit proliferation, migration and invasion of HER2 positive breast cancer cells and further inhibit the development of HER2 positive breast cancer.
Figure 3.
Overexpression of miR-193a-3p inhibits proliferation, migration and invasion of HER2 positive breast cancer cells. (A) miR-193-3p mimics were transfected into 3 HER2 positive breast cancer cell lines and the expression of miR-193a-3p was significantly increased compared with the miR group. (B) Cell vitality was significantly downregulated by miR-193a-3p overexpression compared with the miR group. (C) The colony formation capacity was significantly repressed by miR-193a-3p overexpression compared with the miR group. (D) Cell invasion and migration abilities were significantly inhibited by miR-193a-3p overexpression compared with the miR group. *P<0.05 and **P<0.01 vs. the miR group. OD, optical density; miR, microRNA; HER2, human epidermal growth factor receptor 2.
miR-193a-3p could directly repress the expression of GRB7 through binding to its 3′-UTR
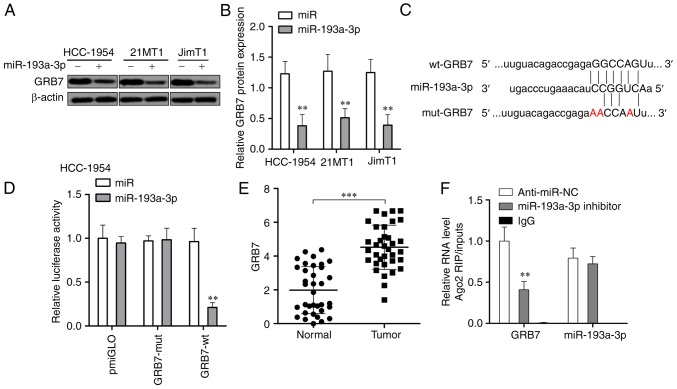
The upregulation of miR-193a-3p significantly downregulated the expression of GRB7 at the protein level in all 3 tested HER2 positive breast cancer cells (P<0.01; Fig. 4A and B). Then, a luciferase reporter assay was carried out to investigate if miR-193a-3p could directly target GRB7. As presented in Fig. 4C and D, miR-193a-3p could specifically downregulate wild-type GRB7 but could not affect the expression of miR-193a-3p with a mutant 3′-UTR in HER2 positive breast cancer cells. GRB7 was significantly upregulated in HER2 positive breast cancer tissues (Fig. 4E). The interaction between GRB7 and miR-193a-3p was further confirmed via RIP assays (Fig. 4F). These results suggested that miR-193a-3p could reduce GRB7 through direct targeting its 3′-UTR.
Figure 4.
miR-193a-3p directly inhibits the expression of GRB7 through targeting its 3′-UTR. (A) Western blotting and (B) statistical analysis of the expression of GRB7, which was significantly reduced by overexpression of miR-193a-3p in 3 HER2 positive breast cancer cell lines. miR-193a-3p could directly target 3′-UTR of GRB7 demonstrated by a (C) Cell Counting Kit-8 and (D) colony formation assay. **P<0.01 vs. the miR group. (E) GRB7 was significantly upregu-lated in 35 pairs of HER2 positive breast cancer tissues. ***P<0.001, as indicated. (F) RNA immunoprecipitation assay demonstrating the association of GRB7 with miR-193a-3p in HCC-1954 cells. **P<0.01 vs. the miR group. miR, microRNA; HER2, human epidermal growth factor receptor 2; UTR, untranslated region; GRB7, growth factor receptor bound protein 7; NC, negative control; OD, optical density.
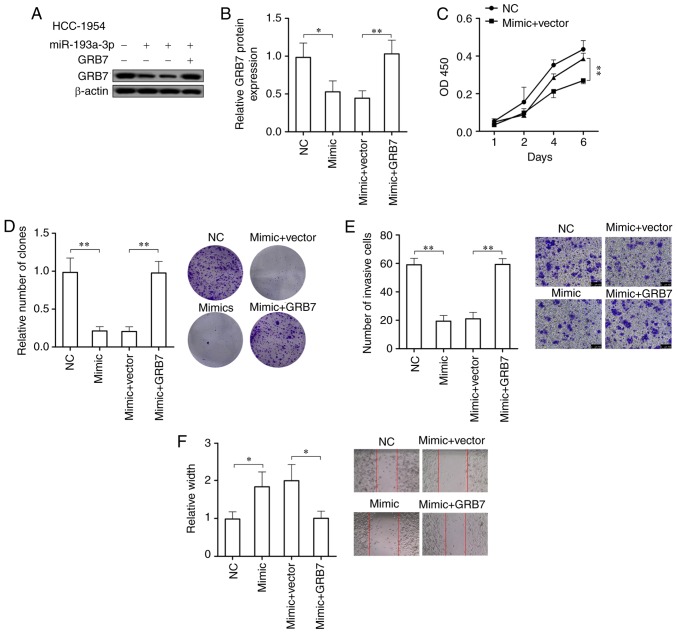
GRB7 overexpression could counteract the inhibitory effect of miR-193a-3p on the oncogenic capacity of breast cancer
In order to prove if miR-193a-3p suppresses the development of HER2 positive breast cancer through targeting GRB7, the cell viability, colony formation ability, migration ability and invasive ability of HER2 positive breast cancer cells overexpressing miR-193a-3p were tested following overexpressing GRB7. After overexpressing miR-193a-3p, GRB7 in HCC-1954 cells was significantly downregulated (P<0.05), while overexpression of GRB7 could significantly abolish the reduction of GRB7 caused by overexpression of miR-193a-3p (P<0.01; Fig. 5A and B). In HCC-1954 cells overexpressing miR-193a-3p, the overexpression of GRB7 significantly promoted the cell viability, colony formation ability, migration ability and invasive ability (P<0.05; Fig. 5C-F). In conclusion, GRB7 overexpression could abolish the effects on HER2 positive breast cancer cells caused by the overexpression of miR-193a-3p and further promote the development of HER2 positive breast cancer.
Figure 5.
GRB7 overexpression abolishes the inhibitory effect of miR-193a-3p on the oncogenic capacity of breast cancer. (A) Western blotting and (B) statistical analysis of the overexpression of GRB7 which abolished the decrease of GRB7 due to overexpression of miR-193a-3p in HER2 positive breast cancer cells. *P<0.05 and **P<0.01, as indicated. (C) Cell vitality was significantly increased. **P<0.01. (D) The colony formation capacity was significantly promoted. **P<0.01, as indicated. (E) Cell invasion and (F) migration abilities were significantly accelerated by GRB7 overexpression. *P<0.05 and **P<0.01. GRB7, growth factor receptor bound protein 7; NC, negative control; OD, optical density; miR, microRNA.
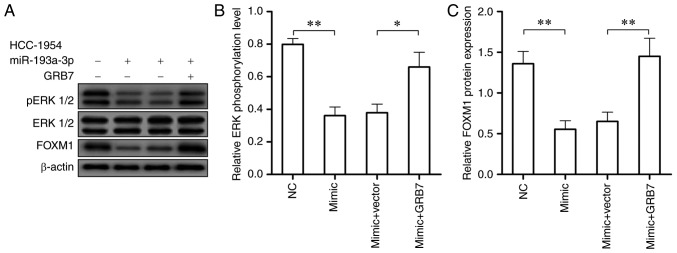
GRB7/ERK/FOXM1 signaling pathway may take part in the effect that miR-193a-3p has on HER2 positive breast cancer
According to the close association between the activation of the ERK/FOXM1 signaling pathway and HER2 positive breast cancer, western blotting was conducted to investigate if miR-193a-3p also represses HER2 positive breast cancer through ERK/FOXM1 signaling pathway. Following the over-expression of miR-193a-3p in HCC-1954 cells, the expression of phosphorylated ERK 1/2 and FOXM1 were significantly reduced (P<0.01; Fig. 6A and B). While the overexpression of GRB7 in HCC-1954 cells overexpressing miR-193a-3p could recover the activity of ERK/FOXM1 signaling pathway, which could accelerate HER2 positive breast cancer tumorigenesis (Fig. 6). These findings demonstrated miR-193a-3p may inhibit HER2 positive breast cancer through downregulating GRB7 and inactivating the ERK/FOXM1 signaling pathway.
Figure 6.
miR-193a-3p may inhibit HER2 positive breast cancer through downregulating GRB7 and inactivating the ERK/FOXM1 signaling pathway. (A) Western blotting and (B) statistical analysis of the protein level of pERK 1/2 and (C) FOXM1 were significantly decreased by GRB7 overexpression in HCC-1954 cells overexpressing miR-193a-3p. *P<0.05 and **P<0.01, as indicated. miR, microRNA; pERK, phosphorylated extracellular signal regulated kinase; FOXM1, forkhead box M1; HER2, human epidermal growth factor receptor 2; GRB7, growth factor receptor bound protein 7.
Discussion
The molecular variation within the 17q12-21 amplicon is one of the major causes of heterogeneity of HER2 positive breast cancer. In 1994, the coexpression of HER2 and GRB7 in human breast cancer cells was first reported by Stein et al (27). Lamy et al (28) analyzed the amplification of 11 genes localized within the 17q12-21 amplicon and demonstrated the frequency of coamplification with HER2 decreases as the distance of the gene from HER2 increases. GRB7 coamplification with HER2 occurred at the greatest frequency, with GRB7 coamplification occurring in 97.7% of HER2 positive breast cancer cases (84 of 86 cases). Several studies have demonstrated that GRB7 can facilitate HER2/Neu-mediated signal transduction and tumor progression (19,29). Therefore, these characteristics of GRB7 make it an attractive therapeutic target for HER2 positive breast cancer. In the present study, it was demonstrated that miR-193a-3p could directly target GRB7 to suppress the tumor. Furthermore, the evidence was also provided that miR-193a-3p could target not only GRB7, but also ERK and FOXM1 signaling in HER2 positive breast cancer cell lines.
miR-193a-3p has been reported to be downregulated in several types of cancer (30,31). The results revealed that miR-193a-3p was decreased in HER2 positive breast cancer and the expression was decreased as the malignant degree of the tumor increased. The present study provides the first evidence to the best of our knowledge, concerning dysregulation of miR-193a-3p in HER2 positive breast cancer. Therefore, attention was focused on understanding the mechanism leading to the downregulation of miR-193a-3p.
In human cancer, epigenetic silencing of tumor suppressors is frequently observed (32). Hypermethylation of the promoter is a major cause of inactivation of tumor suppressors (33). In the present study, hypermethylation of the promoter of miR-193a-3p was also observed in HER2 positive breast cancer and the percentage of methylation of miR-193a-3p was positively associated with the tumor stage and grade. The results of the present study suggest that DNA methylation serves an important role in regulating miR-193a-3p in HER2 positive breast cancer.
In several types of cancer, miR-193a-3p has been demonstrated to be a tumor suppressor (16,31). To further investigate the role miR-193a-3p serves during the development of HER2 positive breast cancer miR-193a-3p was overexpressed in HER2 positive breast cancer cell lines and increased expression of miR-193a-3p could inhibit tumor proliferation, invasion and metastasis.
Through an in silico study, Chen et al (34) revealed that miR-193a-3p was the main target of human GRB7 and miR-193a-3p was frequently downregulated and was inversely correlated with the high expression of GRB7 in ovarian cancer cell lines. In the present study, downregulation of miR-193a-3p and upregulation of GRB7 were also observed in three HER2 positive breast cancer cell lines. Moreover, the result of luciferase reporter assay provided the direct evidence that miR-193a-3p could target GRB7 through binding to its 3′-UTR. To further investigate the role that GRB7 served in the suppressive effect of miR-193a-3p in HER2 positive breast cancer, GRB7 was overexpressed in 3 HER2 positive breast cancer cell lines. Indeed, GRB7 overexpression could counteract the inhibitory effect that miR-193a-3p makes on the oncogenic capacity of breast cancer. Therefore, it was hypothesized that miR-193a-3p suppressed HER2 positive breast cancer through targeting GRB7.
ERK signaling serves a critical role in controlling cancer cell proliferation, survival, metastasis and drug resistance, and abnormal activation of ERK signaling occurs in >85% types of human cancer (35). It is reported that GRB7 can lead to increased ERK1/2 phosphorylation through its interaction with Ras (36). In the present study, it was demonstrated that overexpression of miR-193a-3p could inhibit the phosphorylation of ERK1/2 and overexpression of GRB7 would abolish this effect. FOXM1 is a key transcriptional regulator of the cell cycle, which can be activated by cyclincyclin dependent kinase and ERK mediated phosphorylation (37-39). The activation of FOXM1 can promote nuclear localization to overexpress cell cycle regulators including cell division cycle 25B, baculoviral IAP repeat containing 5 and polo-like kinase 1 (37). In HER2 positive breast cancer, FOXM1 is overexpressed and serves a critical role in tumourigenesis (40). The results of the present study suggested that FOXM1 was a direct or indirect target of miR-193a-3p. The expression of FOXM1 was decreased following overexpression of miR-193a-3p and overexpressing GRB7 could rescue the low expression of FOXM1.
In conclusion, it was determined that miR-193a-3p was downregulated in HER2 positive breast cancer. miR-193a-3p could affect cell proliferation, migration and invasion of HER2 positive breast cancer through affecting different targets. These results reveal the critical role of miR-193a-3p in the progress of HER2 positive breast cancer and implicate its potential application in therapy.
Supplementary Materials
Acknowledgments
Not applicable.
Funding
The present study was supported by the Yunnan Scientific and Technology Committee and Kunming Medical University [Kunming, China; grant no. 2017FE468(-074)].
Availability of data and materials
All data generated or analyzed during this study are included in this manuscript.
Authors' contributions
YT and MW performed the experiments of the study and were responsible for data acquisition. DL conceived and designed the study. YL and SY were responsible for data analysis. YZ and QZ were responsible for statistical analysis. YT and MW were involved in drafting the manuscript. DL revised it critically for important intellectual content. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
All of the human tissues used in the present study were obtained with written informed consent. This study was approved by the Ethics Committee of The Third Affiliated Hospital of Kunming Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Spizzo G, Obrist P, Ensinger C, Theurl I, Dünser M, Ramoni A, Gunsilius E, Eibl G, Mikuz G, Gastl G. Prognostic significance of Ep-CAM AND Her-2/neu overexpression in invasive breast cancer. Int J Cancer. 2002;98:883–888. doi: 10.1002/ijc.10270. [DOI] [PubMed] [Google Scholar]
- 3.Walker RA. Immunohistochemical markers as predictive tools for breast cancer. J Clin Pathol. 2008;61:689–696. doi: 10.1136/jcp.2006.041830. [DOI] [PubMed] [Google Scholar]
- 4.Dunnwald LK, Rossing MA, Li CI. Hormone receptor status, tumor characteristics, and prognosis: A prospective cohort of breast cancer patients. Breast Cancer Res. 2007;9:R6. doi: 10.1186/bcr1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iqbal N, Iqbal N. Human epidermal growth factor receptor 2 (HER2) in cancers: Overexpression and therapeutic implications. Mol Biol Int. 2014;2014:852748. doi: 10.1155/2014/852748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roy AJ, Yankee RA, Brivkalns A, Fitch M. Viability of granulocytes obtained by filtration leukapheresis. Transfusion. 1975;15:539–547. doi: 10.1046/j.1537-2995.1975.15676082228.x. [DOI] [PubMed] [Google Scholar]
- 7.Singh R, Gupta S, Pawar SB, Pawar RS, Gandham SV, Prabhudesai S. Evaluation of ER, PR and HER-2 receptor expression in breast cancer patients presenting to a semi urban cancer centre in Western India. J Cancer Res Ther. 2014;10:26–28. doi: 10.4103/0973-1482.131348. [DOI] [PubMed] [Google Scholar]
- 8.Kim JY, Jung WH, Koo JS. Expression of autophagy-related proteins according to androgen receptor and HER-2 status in estrogen receptor-negative breast cancer. PLoS One. 2014;9:e105666. doi: 10.1371/journal.pone.0105666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Slamon DJ, Godolphin W, Jones LA, Holt JA, Wong SG, Keith DE, Levin WJ, Stuart SG, Udove J, Ullrich A. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244:707–712. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- 10.Thompson SK, Sullivan TR, Davies R, Ruszkiewicz AR. Her-2/neu gene amplification in esophageal adenocarcinoma and its influence on survival. Ann Surg Oncol. 2011;18:2010–2017. doi: 10.1245/s10434-011-1554-1. [DOI] [PubMed] [Google Scholar]
- 11.Sahlberg KK, Hongisto V, Edgren H, Mäkelä R, Hellström K, Due EU, Moen Vollan HK, Sahlberg N, Wolf M, Børresen- Dale AL, et al. The HER2 amplicon includes several genes required for the growth and survival of HER2 positive breast cancer cells. Mol Oncol. 2013;7:392–401. doi: 10.1016/j.molonc.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kauraniemi P, Kallioniemi A. Activation of multiple cancer-associated genes at the ERBB2 amplicon in breast cancer. Endocr Relat Cancer. 2006;13:39–49. doi: 10.1677/erc.1.01147. [DOI] [PubMed] [Google Scholar]
- 13.Valastyan S. Roles of microRNAs and other non-coding RNAs in breast cancer metastasis. J Mammary Gland Biol Neoplasia. 2012;17:23–32. doi: 10.1007/s10911-012-9241-9. [DOI] [PubMed] [Google Scholar]
- 14.Zhang N, Wang X, Huo Q, Sun M, Cai C, Liu Z, Hu G, Yang Q. MicroRNA-30a suppresses breast tumor growth and metastasis by targeting metadherin. Oncogene. 2014;33:3119–3128. doi: 10.1038/onc.2013.286. [DOI] [PubMed] [Google Scholar]
- 15.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 16.Pekow J, Meckel K, Dougherty U, Huang Y, Chen X, Almoghrabi A, Mustafi R, Ayaloglu-Butun F, Deng Z, Haider HI, et al. miR-193a-3p is a key tumor suppressor in ulcerative colitis-associated colon cancer and promotes carcinogenesis through upregulation of IL17RD. Clin Cancer Res. 2017;23:5281–5291. doi: 10.1158/1078-0432.CCR-17-0171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chou NH, Lo YH, Wang KC, Kang CH, Tsai CY, Tsai KW. MiR-193a-5p and -3p play a distinct role in gastric cancer: miR-193a-3p suppresses gastric cancer cell growth by targeting ETS1 and CCND1. Anticancer Res. 2018;38:3309–3318. doi: 10.21873/anticanres.12596. [DOI] [PubMed] [Google Scholar]
- 18.Bivin WW, Yergiyev O, Bunker ML, Silverman JF, Krishnamurti U. GRB7 expression and correlation with HER2 amplification in invasive breast carcinoma. Appl Immunohistochem Mol Morphol. 2017;25:553–558. doi: 10.1097/PAI.0000000000000349. [DOI] [PubMed] [Google Scholar]
- 19.Nadler Y, González AM, Camp RL, Rimm DL, Kluger HM, Kluger Y. Growth factor receptor-bound protein-7 (Grb7) as a prognostic marker and therapeutic target in breast cancer. Ann Oncol. 2010;21:466–473. doi: 10.1093/annonc/mdp346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lesurf R, Griffith OL, Griffith M, Hundal J, Trani L, Watson MA, Aft R, Ellis MJ, Ota D, Suman VJ, et al. Genomic characterization of HER2-positive breast cancer and response to neoadjuvant trastuzumab and chemotherapy-results from the ACOSOG Z1041 (Alliance) trial. Ann Oncol. 2017;28:1070–1077. doi: 10.1093/annonc/mdx048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lujambio A, Ropero S, Ballestar E, Fraga MF, Cerrato C, Setién F, Casado S, Suarez-Gauthier A, Sanchez-Cespedes M, Git A, et al. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 2007;67:1424–1429. doi: 10.1158/0008-5472.CAN-06-4218. [DOI] [PubMed] [Google Scholar]
- 22.Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA, Jones PA. Specific activation of microRNA-127 with down-regulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 23.Yang H, Kong W, He L, Zhao JJ, O'Donnell JD, Wang J, Wenham RM, Coppola D, Kruk PA, Nicosia SV, et al. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targetin PTEN. Cancer Res. 2008;68:425–433. doi: 10.1158/0008-5472.CAN-07-2488. [DOI] [PubMed] [Google Scholar]
- 24.Fan Q, Hu X, Zhang H, Wang S, Zhang H, You C, Zhang CY, Liang H, Chen X, Ba Y. MiR-193a-3p is an important tumour suppressor in lung cancer and directly targets KRAS. Cell Physiol Biochem. 2017;44:1311–1324. doi: 10.1159/000485491. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y, Chan DW, Liu VW, Chiu P, Ngan HY. Differential functions of growth factor receptor-bound protein 7 (GRB7) and its variant GRB7v in ovarian carcinogenesis. Clin Cancer Res. 2010;16:2529–2539. doi: 10.1158/1078-0432.CCR-10-0018. [DOI] [PubMed] [Google Scholar]
- 26.Mak CS, Yung MM, Hui LM, Leung LL, Liang R, Chen K, Liu SS, Qin Y, Leung TH, Lee KF, et al. MicroRNA-141 enhances anoikis resistance in metastatic progression of ovarian cancer through targeting KLF12/Sp1/survivin axis. Mol Cancer. 2017;16:11. doi: 10.1186/s12943-017-0582-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stein D, Wu J, Fuqua SA, Roonprapunt C, Yajnik V, D'Eustachio P, Moskow JJ, Buchberg AM, Osborne CK, Margolis B. The SH2 domain protein GRB-7 is co-amplified, overexpressed and in a tight complex with HER2 in breast cancer. EMBO J. 1994;13:1331–1340. doi: 10.1002/j.1460-2075.1994.tb06386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamy PJ, Fina F, Bascoul-Mollevi C, Laberenne AC, Martin PM, Ouafik L, Jacot W. Quantification and clinical relevance of gene amplification at chromosome 17q12-q21 in human epidermal growth factor receptor 2-amplified breast cancers. Breast Cancer Res. 2011;13:R15. doi: 10.1186/bcr2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bai T, Luoh SW. GRB-7 facilitates HER-2/Neu-mediated signal transduction and tumor formation. Carcinogenesis. 2008;29:473–479. doi: 10.1093/carcin/bgm221. [DOI] [PubMed] [Google Scholar]
- 30.Meng F, Qian L, Lv L, Ding B, Zhou G, Cheng X, Niu S, Liang Y. miR-193a-3p regulation of chemoradiation resistance in oesophageal cancer cells via the PSEN1 gene. Gene. 2016;579:139–145. doi: 10.1016/j.gene.2015.12.060. [DOI] [PubMed] [Google Scholar]
- 31.Nie W, Ge HJ, Yang XQ, Sun X, Huang H, Tao X, Chen WS, Li B. LncRNA-UCA1 exerts oncogenic functions in non-small cell lung cancer by targeting miR-193a-3p. Cancer Lett. 2016;371:99–106. doi: 10.1016/j.canlet.2015.11.024. [DOI] [PubMed] [Google Scholar]
- 32.Cui X, Chen X, Wang W, Chang A, Yang L, Liu C, Peng H, Wei Y, Liang W, Li S, et al. Epigenetic silencing of miR-203 in Kazakh patients with esophageal squamous cell carcinoma by MassARRAY spectrometry. Epigenetics. 2017;12:698–707. doi: 10.1080/15592294.2017.1349045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liang G, Weisenberger DJ. DNA methylation aberrancies as a guide for surveillance and treatment of human cancers. Epigenetics. 2017;12:416–432. doi: 10.1080/15592294.2017.1311434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen K, Liu MX, Mak CS, Yung MM, Leung TH, Xu D, Ngu SF, Chan KK, Yang H, Ngan HY, Chan DW. Methylation-associated silencing of miR-193a-3p promotes ovarian cancer aggressiveness by targeting GRB7 and MAPK/ERK pathways. Theranostics. 2018;8:423–436. doi: 10.7150/thno.22377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Luca A, Maiello MR, D'Alessio A, Pergameno M, Normanno N. The RAS/RAF/MEK/ERK and the PI3K/AKT signalling pathways: Role in cancer pathogenesis and implications for therapeutic approaches. Expert Opin Ther Targets. 2012;16(Suppl 2):S17–S27. doi: 10.1517/14728222.2011.639361. [DOI] [PubMed] [Google Scholar]
- 36.Chu PY, Li TK, Ding ST, Lai IR, Shen TL. EGF-induced Grb7 recruits and promotes Ras activity essential for the tumorigenicity of Sk-Br3 breast cancer cells. J Biol Chem. 2010;285:29279–29285. doi: 10.1074/jbc.C110.114124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laoukili J, Stahl M, Medema RH. FoxM1: At the crossroads of ageing and cancer. Biochim Biophys Acta. 2007;1775:92–102. doi: 10.1016/j.bbcan.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 38.Lüscher-Firzlaff JM, Lilischkis R, Lüscher B. Regulation of the transcription factor FOXM1c by Cyclin E/CDK2. FEBS Lett. 2006;580:1716–1722. doi: 10.1016/j.febslet.2006.02.021. [DOI] [PubMed] [Google Scholar]
- 39.Major ML, Lepe R, Costa RH. Forkhead box M1B transcriptional activity requires binding of Cdk-cyclin complexes for phosphorylation-dependent recruitment of p300/CBP coactiva-tors. Mol Cell Biol. 2004;24:2649–2661. doi: 10.1128/MCB.24.7.2649-2661.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Francis RE, Myatt SS, Krol J, Hartman J, Peck B, McGovern UB, Wang J, Guest SK, Filipovic A, Gojis O, et al. FoxM1 is a downstream target and marker of HER2 overexpression in breast cancer. Int J Oncol. 2009;35:57–68. doi: 10.3892/ijo_00000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this manuscript.