Abstract
Astaxanthin is a high-value carotenoid used as a pigmentation source in fish aquaculture. In addition, a beneficial role of astaxanthin as a food supplement for humans is becoming evident. The unicellular green alga Haematococcus pluvialis seems to be a suitable source for natural astaxanthin. Astaxanthin accumulation in H. pluvialis occurs in response to environmental stress such as high light and salt stress. Here, the isolation of the H. pluvialis carotenoid biosynthesis gene phytoene synthase is reported. Furthermore, the expression of phytoene synthase and carotenoid hydroxylase, two key enzymes in astaxanthin biosynthesis, was investigated at the transcriptional level. The application of environmental stress resulted in increased steady-state mRNA levels of both genes. High-light intensity led to a transient increase in carotenoid hydroxylase mRNA followed by moderate astaxanthin accumulation. In contrast, salt stress in combination with high light resulted in a sustained increase in both transcripts. The addition of compounds inducing reactive oxygen species did not influence transcript levels of phytoene synthase and carotenoid hydroxylase. The application of an inhibitor of photosynthesis, 3-(3, 4-dichlorophenyl)-1,1-dimethylurea, indicated that the light-induced expression of these carotenoid biosynthesis genes may be under photosynthetic control.
The ketocarotenoid astaxanthin (3,3′-dihydroxy-4,4′-diketo-β-carotene) is a high-value carotenoid used as a feed supplement for fish aquaculture and as pigmentation source for egg yolk (Boussiba et al., 1992; Lorenz and Cysewski, 2000). As well as this use, a beneficial role of astaxanthin as a food supplement for humans is becoming evident. Thus, it was shown that astaxanthin possesses a higher anti-oxidant activity when compared with β-carotene and α-tocopherol and reveals a strong activity as inhibitor of lipid peroxidation (Miki, 1991; Mortensen et al., 1997). Beneficial effects of astaxanthin, such as the reduction of gastric inflammation and bacterial load in H. pylori-infected mice and humans, the prevention of age-related macular degeneration, the reduction of risk of arteriosclerosis, and the prevention of carcinogenesis are currently under examination (Tanaka et al., 1994; Bennedsen et al., 1999; Lorenz and Cysewski, 2000).
Astaxanthin biosynthesis has been observed in a limited number of organisms, e.g. in some marine bacteria, in the yeast Phaffia rhodozyma, and in some green algae (Johnson and Schroeder, 1995). The unicellular green alga Haematococcus pluvialis reveals the highest astaxanthin accumulation (up to 4% by dry weight) and seems to be the most suitable source for natural astaxanthin (Boussiba, 2000). The physiology of astaxanthin accumulation in H. pluvialis, which occurs in response to various environmental stress conditions such as high-light intensities, nitrogen and phosphate limitations, and salt stress has been intensively studied (Kobayashi et al., 1993; Boussiba et al., 1999; Boussiba, 2000). The biosynthesis of astaxanthin is normally accompanied by a morphological change of the vegetative cells into non-motile cyst cells in which astaxanthin was shown to accumulate in the cytoplasm (Santos and Mesquita, 1984). At present, the role of astaxanthin accumulation in H. pluvialis is not well understood and various beneficial effects such as photoprotection and protection against oxidative stress have been discussed (Yong and Lee, 1991; Kobayashi et al., 1997).
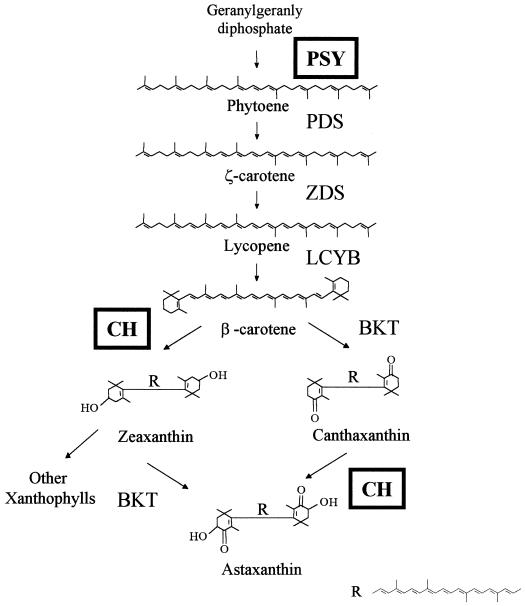
The biosynthesis of astaxanthin starts with the condensation of two geranylgeranyl diphosphate molecules to form phytoene (Fig. 1; for review, see Cunningham and Gantt, 1998). Four desaturation reactions lead to the synthesis of lycopene followed by two cyclization reactions for the biosynthesis of β-carotene. The conversion of β-carotene into astaxanthin in H. pluvialis is carried out by two enzymes, β-carotene ketolase and carotenoid hydroxylase. The H. pluvialis genes coding for β-carotene ketolase and carotenoid hydroxylase were isolated, and the gene products have already been studied to some extent (Kajiwara et al., 1995; Lotan and Hirschberg, 1995; Breitenbach et al., 1996; Linden, 1999).
Figure 1.
Carotenoid biosynthetic pathway of astaxanthin in H. pluvialis. Several intermediates were omitted for the sake of simplification. The carotenoid biosynthesis enzymes phytoene synthase (PSY), phytoene desaturase (PDS), ζ-carotene desaturase (ZDS), lycopene cyclase (LCYB), β-carotene ketolase, and carotenoid hydroxylase (CH) are indicated. The gene expression of phytoene synthase and carotenoid hydroxylase (boxed) have been studied in the present publication.
Although some of the biosynthesis genes have been cloned, the molecular basis of astaxanthin biosynthesis regulation in H. pluvialis has not been thoroughly investigated to date. In two recent studies, the expression of several carotenoid genes during the induction of astaxanthin biosynthesis by light was examined (Sun et al., 1998; Grünewald et al., 2000). However, in both studies the induction of gene expression was examined in flagellate cells, whereas massive accumulation of astaxanthin occurs during cyst cell formation.
The aim of the present study was to gain insight into the molecular basis of stress-induced astaxanthin accumulation in H. pluvialis. The gene expression of two key enzymes of astaxanthin biosynthesis in H. pluvialis was investigated. The application of various environmental stress conditions resulted in increased steady-state levels of both phytoene synthase and carotenoid hydroxylase mRNAs. We conclude that H. pluvialis is capable of responding to stress conditions by the differential regulation of mRNA steady-state levels of carotenoid biosynthesis genes.
RESULTS
Isolation and Amino Acid Sequence of Phytoene Synthase from H. pluvialis
To examine the expression of a carotenoid biosynthesis enzyme involved in the first specific step of carotenogenesis, an H. pluvialis phytoene synthase (EMBL GenBank accession no. AF305430) cDNA was isolated by functional complementation in Escherichia coli. Thus, the plasmid DNAs from a cDNA library prepared from red cyst cells were introduced into E. coli carrying plasmid pACCAR25ΔcrtB (Misawa et al., 1990). The plasmid harbored several carotenoid biosynthesis genes from Erwinia uredovora but lacked a functional phytoene synthase gene. Upon cotransformation with the H. pluvialis cDNA library, three yellow colonies were identified out of approximately 70,000 colonies that revealed an E. coli color. The corresponding plasmids were isolated, and it was shown by DNA sequencing that all three cDNA inserts represented the same gene (data not shown).
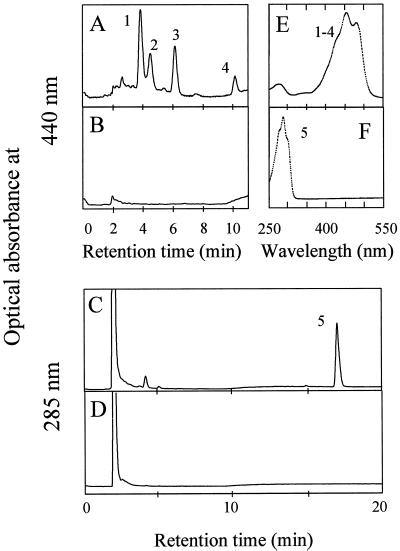
The longest cDNA was used further for complementation experiments and DNA sequencing. HPLC analysis of carotenoid pigments from the yellow transformant was carried out (Fig. 2). The yellow transformant accumulated zeaxanthin and several zeaxanthin glycosides, whereas the control carrying only plasmid pACCAR25ΔcrtB did not reveal any colored carotenoids (Fig. 2, A, B, and E). Cotransformation of the cDNA with plasmid pACCRT-E, which carried the geranylgeranyl diphosphate synthase gene from E. uredovora, resulted in the accumulation of phytoene (Fig. 2, C and F). Sequence analysis of the entire cDNA insert was carried out, and one open reading frame was identified (data not shown). An alignment of the predicted open reading frame with other known phytoene synthases revealed high overall sequence similarity to the higher plant enzymes and the phytoene synthase of the cyanobacterium Synechocystis PCC 6803 (51%–54% identity; data not shown). When compared with bacterial phytoene synthases, the H. pluvialis enzyme revealed an N-terminal extension indicating the presence of a chloroplast targeting sequence.
Figure 2.
Heterologous complementation of H. pluvialis phytoene synthase in E. coli. The HPLC analyses of E. coli carotenoids following the cotransformation of H. pluvialis phytoene synthase gene together with either pACCAR25ΔcrtB or plasmid pACCRT-E are shown in A and C, respectively. HPLC separations of carotenoid pigments extracted from E. coli cells carrying either the complementation plasmids pACCAR25ΔcrtB or pACCRT-E are shown in B and D. In addition, the absorption spectra of peaks 1 through 4 (zeaxanthin and zeaxanthin glycosides) as well as the spectrum of peak 5 (phytoene) are shown in E and F.
Phytoene Synthase and Carotenoid Hydroxylase Show Higher Steady-State mRNA Levels in Response to Various Stress Conditions
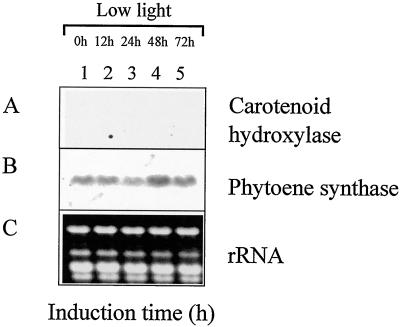
The expression of the phytoene synthase and carotenoid hydroxylase genes was examined by northern-blot analysis using the respective cDNAs as probes. To gain information about the post-translational effects of the various stress conditions, the amount of accumulated astaxanthin was determined. In addition, the percentage of cyst cell formation was monitored by microscopic examination (Table I). No transcript for carotenoid hydroxylase could be detected after growth for 4 d under a dark/light cycle and low-light conditions (Fig. 3A, lane 1). In contrast, the phytoene synthase was shown to be expressed at low levels under these conditions (Fig. 3B, lane 1). When growth was continued for an additional 72 h under the same conditions, no increase in steady-state mRNA levels was observed for either gene (Fig. 3, A and B, lanes 2–5). In addition, neither astaxanthin accumulation nor cyst cell formation was observed (Table I).
Table I.
Gene expression of phytoene synthase and carotenoid hydroxylase, formation of H. pluvialis cyst cells, and astaxanthin accumulation after growth under various stress conditions
| Experiment No. | Growth and Stress Conditions | Phytoene Synthase Expression | Hydroxylase Expression | Cyst Cells | Astaxanthina |
|---|---|---|---|---|---|
| (%) | (mg g−1 dry wt) | ||||
| 1 | Standard growth conditions | Low | n.d.b | <10 | 0 |
| 2 | Sodium acetate, Fe2+, high light | High | High | >80 | 13.5 ± 0.5 |
| 3 | Sodium acetate, low light | High, delayed | High, delayed | ∼25 | 5.7 ± 0.8 |
| 4 | Sodium acetate, Fe2+, low light | High, delayed | High, delayed | ∼25 | 4.7 ± 0.2 |
| 5 | Sodium acetate, high light | High | High | >80 | 9.5 ± 0.1 |
| 6 | Sodium chloride, high light | High | High | >80 | 7.8 ± 0.2 |
| 7 | High light | Temporary increase | Transient | <10 | 6.1 ± 0.5 |
| 8 | High light, methyl viologen | Temporary increase | Transient | <10 | 6.5 ± 0.2 |
| 9 | High light, Fe2+ | Temporary increase | Transient | <10 | 5.2 ± 0.1 |
| 10 | Sodium acetate, Fe2+, high light, Cycloheximide | High | High | <10 | 0.4 ± 0.04 |
| 11 | High light, DCMU | Low | n.d. | <10 | 0 |
Typical results of at least three independent experiments are shown.
Astaxanthin was quantified after 72 h of induction.
n.d., Not detected.
Figure 3.
Expression of carotenoid hydroxylase and phytoene synthase under standard growth conditions. RNA was isolated from H. pluvialis cells harvested after 4 d of growth (lane 1) and after additional growth under standard culture conditions for 12 (lane 2), 24 (lane 3), 48 (lane 4), and 72 (lane 5) h. For northern-blot analysis, the H. pluvialis carotenoid hydroxylase (A) and phytoene synthase (B) cDNAs were used as specific probes. For comparison, total RNA was stained with ethidium bromide (C).
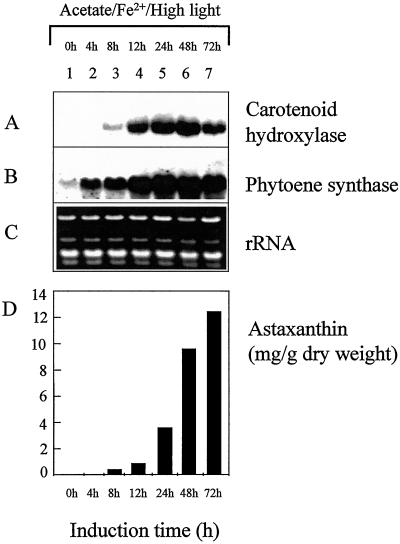
Addition of sodium acetate and ferrous sulfate and increasing the light intensity resulted in a strong increase in steady-state mRNA levels of both the carotenoid hydroxylase and phytoene synthase (Fig. 4, A and B). The induction of phytoene synthase transcript was already detectable after 4 h, whereas carotenoid hydroxylase transcripts became detectable 8 h after the onset of stress conditions (Fig. 4A, lane 3; Fig. 4B, lane 2). The highest transcript levels of carotenoid hydroxylase were found at 24 to 48 h, and the steady-state mRNA levels decreased at 72 h after induction (Fig. 4A, lanes 5–7). A similar pattern was observed for phytoene synthase, although the higher transcript levels were sustained from 24 to 72 h (Fig. 4B, lanes 5–7). At the same time astaxanthin started to accumulate, reaching 13.5 mg g−1 dry weight after induction for 72 h (Fig. 4D). At this time most of the cells were present as cysts cells (Table I). When the cells were grown in the presence of sodium acetate and ferrous sulfate under low illumination, the increase in steady-state levels of phytoene synthase and carotenoid hydroxylase transcripts was delayed, and highest transcript levels were found at 48 h after induction (Table I). The lower light intensities also resulted in a lower astaxanthin accumulation of 4.7 mg g−1 dry weight and a decreased percentage of cyst cell formation (approximately 25%). In the presence of sodium acetate only and under low illumination, a high but delayed expression of phytoene synthase and carotenoid hydroxylase genes was observed (Table I).
Figure 4.
The expression of carotenoid hydroxylase and phytoene synthase during the induction of astaxanthin biosynthesis. The biosynthesis of astaxanthin was induced by high light and by addition of sodium acetate and FeSO4. The H. pluvialis cells used for the isolation of RNA were harvested after 4 d of growth (lane 1) and after additional growth under astaxanthin-inducing conditions for 12 (lane 2), 24 (lane 3), 48 (lane 4), and 72 (lane 5) h. For northern-blot analysis, the H. pluvialis carotenoid hydroxylase (A) and phytoene synthase (B) were used as specific probes. For comparison, total RNA was stained with ethidium bromide (C). In addition, the accumulation of astaxanthin was examined (D).
Addition of either sodium acetate or sodium chloride and growth under high-light intensities resulted in a strong increase in steady-state mRNA levels of both genes and in the accumulation of astaxanthin of 9.5 and 7.8 mg g−1 dry weight, respectively (Table I). Growth in the presence of either sodium chloride or sodium acetate also resulted in encystment with more than 80% of cyst cells formed after 72 h of induction.
Phytoene Synthase and Carotenoid Hydroxylase Show Increased Gene Expression in Response to High Illumination
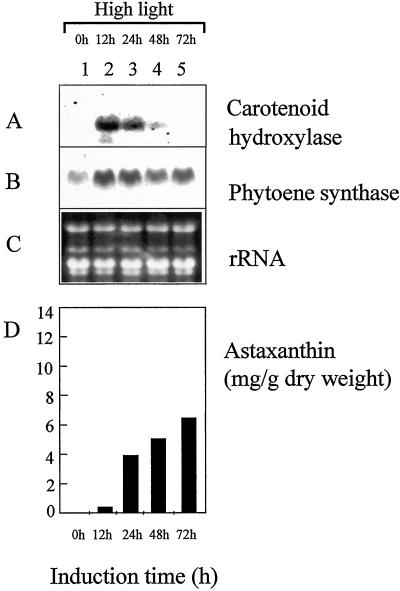
The regulation of transcript levels of carotenoid hydroxylase and phytoene synthase were examined following the induction by higher light intensities (125 μmol m−2 s−1, Fig. 5). For carotenoid hydroxylase, a maximal induction was found at 12 h following the onset of high light (Fig. 5A, lane 2). The induction pattern was transient and the carotenoid hydroxylase mRNA was not detectable after 72 h of high-light illumination (Fig. 5A, lanes 3–5). Phytoene synthase only revealed a minor increase with highest transcript levels after 12 h of high light (Fig. 5B, lane 2). The mRNA levels always seemed to be elevated thereafter when compared with transcript levels prior to high-light exposure (Fig. 5B, lanes 1–5). The induction by high light resulted in an astaxanthin production of approximately 6 mg g−1 dry weight after 72 h of high light (Fig. 5D). The high-light treatment did not lead to the formation of non-motile cyst cells (Table I).
Figure 5.
Expression of carotenoid hydroxylase and phytoene synthase genes in response to increased illumination. The H. pluvialis cells used for the preparation of RNA were harvested after 4 d of growth (lane 1) and after additional growth under high-light conditions for 12 (lane 2), 24 (lane 3), 48 (lane 4), and 72 (lane 5) h. For northern-blot analysis, the H. pluvialis carotenoid hydroxylase (A) and phytoene synthase (B) were used as specific probes. For comparison, total RNA was stained with ethidium bromide (C). In addition, the accumulation of astaxanthin was examined (D).
The Involvement of Reactive Oxygen Species in the Up-Regulation of Carotenoid Biosynthesis Genes
To examine a possible effect of ROS on the expression of phytoene synthase and carotenoid hydroxylase, methyl viologen was added to H. pluvialis cultures grown under high-light conditions (Table I). The expression pattern of carotenoid hydroxylase and phytoene synthase induction reflected the kinetics observed following the induction by high light only (Fig. 5). In addition, the accumulation of astaxanthin, with a maximum at approximately 6.5 mg g−1 dry weight, was similar in both experiments (Table I). Whereas methyl viologen leads to the formation of the superoxide anion radical, Fe2+ seems to result mainly in the formation of the hydroxyl radical (Halliwell and Gutteridge, 1989). Nevertheless, the same results were obtained for the steady-state mRNA kinetics using Fe2+ (Table I). Furthermore, neither the addition of methyl viologen nor the supplementation with Fe2+ led to the formation of cyst cells (Table I).
Up-Regulation of Carotenoid Hydroxylase and Phytoene Synthase Is Independent of De Novo Protein Biosynthesis
When the protein biosynthesis inhibitor cycloheximide was added prior to the application of stress conditions, the induction of carotenoid hydroxylase and phytoene synthase was still detected (Table I). The expression of both genes revealed similar kinetics and transcript quantities for the first 24 h of induction when compared with the induction observed without the addition of cycloheximide (Fig. 4, A and B). However, astaxanthin biosynthesis and the formation of cyst cells were inhibited under these conditions (Table I).
The effect of the photosynthetic electron transport inhibitor 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU) on gene expression was investigated. Following the addition of DCMU, the light induction of carotenoid hydroxylase was abolished and basal expression levels were observed for phytoene synthase (Table I).
DISCUSSION
Here we report the isolation of a new carotenoid biosynthesis gene from the unicellular alga H. pluvialis. It was shown by sequence similarities with phytoene synthases from other organisms, as well as by functional complementation in E. coli, that this gene codes for phytoene synthase (Fig. 2). Whereas phytoene synthase is involved in the early steps of the general carotenoid biosynthetic pathway, the previously described carotenoid hydroxylase gene is involved in the final steps of astaxanthin synthesis (Linden, 1999). With these two gene probes at hand, we addressed several important questions concerning the stress-induced astaxanthin biosynthesis in H. pluvialis.
First of all, we showed that the mRNAs of both carotenoid hydroxylase and phytoene synthase are up-regulated in response to various stress conditions. Together with published reports on the up-regulation of phytoene desaturase, β-carotene ketolase and isopentenyl diphosphate isomerase, these results suggest that the regulation of carotenoid gene transcript levels plays an important role in the stress response of H. pluvialis (Sun et al., 1998; Grünewald et al., 2000). A second question addressed was the involvement of the various stress factors in the up-regulation of transcript levels of the two carotenoid biosynthesis genes examined. The addition of sodium acetate and Fe2+ and growth under high light, which was reported to bring about the highest astaxanthin production, also led to a strong induction of steady-state mRNA levels for both genes (Fig. 4). Sodium acetate could be replaced by sodium chloride in the induction of carotenoid genes (Table I). This finding indicated that the effect of sodium acetate on carotenoid gene expression is the result of salt stress and is not due to an increased carbon/nitrogen ratio as suggested previously (Kakizono et al., 1992). The application of higher light intensities only resulted in a moderate induction of gene expression, which revealed a transient induction pattern in the case of carotenoid hydroxylase (Fig. 5). The same induction patterns were identified following the addition of methyl viologen and Fe2+ under high-light conditions (Table I). It has previously been observed that ROS-generating compounds such as Fe2+, methyl viologen, and methylene blue resulted in increased astaxanthin accumulation, which led to the hypothesis that the stress response in H. pluvialis may be mediated by ROS (Kobayashi et al., 1993; Fan et al., 1998; Boussiba, 2000). However, the results presented here suggest that ROS generators are not involved in the transcriptional regulation of phytoene synthase and carotenoid hydroxylase. In corroboration of this finding, previous reports showed that the effect of Fe2+ on astaxanthin accumulation is independent of de novo protein biosynthesis, and the authors suggested a function of ROS at the post-translational level (Kobayashi et al., 1993).
Another important feature is the interrelation between cyst cell formation, accumulation of astaxanthin, and the up-regulation of the carotenoid biosynthesis genes. Astaxanthin accumulation occurred in flagellate cells in response to higher light intensities, which seems to be a consequence of the moderate and transient up-regulation of carotenoid biosynthesis genes under these conditions (Table I, Fig. 5; Grünewald et al., 1997). This induction pattern can be interpreted as an acclimation process to higher light conditions, which occurs within 1 d of the increase in irradiance (Hagen et al., 2000). However, increased production of astaxanthin was coupled with the formation of non-motile cyst cells (Table I). Growth under illumination with high light only, or high light plus either Fe2+ or methyl viologen, did not support the formation of cyst cells. On the other hand, high light in combination with salt stress seemed to be indispensable for the formation of cyst cells, whereas the application of salt stress only led to a moderate encystment. Under the latter conditions, the up-regulation of carotenoid hydroxylase and phytoene synthase transcript levels was shown to be delayed in comparison with the induction by sodium acetate and high light (Table I). Therefore, the strong up-regulation of mRNA levels in response to high light and salt stress seems to result from the additive effects of the respective stress conditions (Fig. 4). In addition, higher levels of expression of the two carotenoid biosynthesis genes in response to stress were shown to be independent of de novo protein biosynthesis. In contrast, the inhibition of photosynthesis abolished the high-light-induced up-regulation of carotenoid hydroxylase and phytoene synthase (Table I). This result indicates that the light-induced expression of these carotenoid biosynthesis genes may be under photosynthetic control.
In conclusion, H. pluvialis appears to be capable of responding to various stress conditions in different ways. Whereas high light leads to a transient response and to moderate accumulation of astaxanthin, the combination of various stress conditions such as high light and salt stress is obligatory for encystment and the strong up-regulation of carotenoid genes.
MATERIALS AND METHODS
Hematococcus pluvialis Strain, Growth Conditions, and Supplements
H. pluvialis Flotow NIES-144 was obtained from the National Institute for Environmental Studies (Tsukuba, Japan). The basal medium (pH 6.8) for growth of H. pluvialis contained 1.2 g of sodium acetate, 2.0 g of yeast extract, 0.4 g of l-Asn, 0.2 g of MgCl2 × 6H2O, 0.01 g of FeSO4 × 7H2O, and 0.02 g of CaCl2 × 2H2O per liter (Kobayashi et al., 1993). H. pluvialis was grown at 22°C under a dark/light cycle of 12 h of low light (20 μmol m−2 s−1, provided by universal-white lamps Osram L65W/25S) and 12 h dark for 4 d (final cell density approximately 4 × 105 cells per mL). A cell density of approximately 6 × 105 cells per mL was determined after additional growth for 72 h under standard culture conditions. Growth was performed in 200 mL of basal medium in 500 mL of Erlenmeyer flasks without aeration, and cultures were shaken manually once a day. For induction of astaxanthin biosynthesis, various supplements were added and cultures were shaken continuously (Kobayashi et al., 1993). Sodium acetate and FeSO4 were used at a final concentration of 45 mm and 450 μm, respectively. The translational inhibitor cycloheximide (final concentration 100 ng mL−1) as well as the inhibitor of photosynthesis DCMU (final concentration 20 μm) were added 2 h prior to the induction of astaxanthin biosynthesis. The reactive-oxygen-generating reagent methyl viologen was used at a final concentration of 10−11 m. For high-light treatment, growth of H. pluvialis was performed at 125 μmol m−2 s−1 of continuous light according to Kajiwara et al. (1995).
For analysis of carotenoids, cultures of Escherichia coli JM101 containing different plasmids were grown in Luria-Bertani medium at 28°C for 48 h and ampicillin (50 μg mL−1), chloramphenicol (30 μg mL−1), and isopropyl-β-d-thio-galactopyranosid (0.5 mm) were added as required (Sambrook et al., 1989).
H. pluvialis cDNA Expression Libraries, Plasmids, Screening, and DNA Sequencing
The construction of H. pluvialis cDNA libraries from cyst cells was described previously (Linden, 1999). After in vivo excision using the ExAssist/SOLR system (Stratagene, La Jolla, CA), the cDNA libraries were further used for complementation experiments. E. coli strain JM101 was used as a host for screening and complementation experiments with plasmids pACCAR25ΔcrtB and pACCRT-E. Plasmid pACCAR25ΔcrtB harbors the carotenoid biosynthesis genes crtE, crtI, crtY, crtZ, and crtX from Erwinia uredovora (Misawa et al., 1990). Plasmid pACCRT-E carries the crtE gene from E. uredovora and resulted in the accumulation of geranylgeranyl diphosphate (Misawa et al., 1995). The screening for phytoene synthase was carried out by the heterologous complementation procedure reported previously using pACCAR25ΔcrtB as complementation plasmid (Linden et al., 1993). The nucleotide sequences of H. pluvialis phytoene synthase cDNAs were determined for both strands using the Abi Prism Dye Terminator Cycle Sequencing Ready Reaction Kit (Perkin-Elmer Applied Biosystems, Foster City, CA). The analysis of nucleotide and derived amino acid sequences was carried out using the PCGENE program (Intelligenetics, Oxford Molecular Group, Oxford).
Northern-Blot Analysis
After 4 d of growth, the H. pluvialis cells were collected by centrifugation either directly or after varying induction times of astaxanthin biosynthesis. The cells were frozen and subsequently powdered under liquid nitrogen using a mortar and pestle. RNA was then isolated according to the miniprep RNA extraction procedure described by Sokolowsky et al. (1990). For northern-blot analysis, total RNA (10 μg) was denatured in formaldehyde, electrophoresed on a 1% (w/v) agarose gel containing 6% (v/v) formaldehyde, transferred to positively charged nylon membrane (Boehringer Mannheim/Roche, Basel), and hybridized in the presence of 50% (v/v) formamide. Probe labeling and hybridization were carried out according to the instructions in the DIG Nonradioactive Nucleic Acid Labeling and Detection System (Boehringer Mannheim/Roche).
Carotenoid Extraction and HPLC Analysis
For the isolation of carotenoids (carotenes and hydroxylated products) from E. coli, cells were harvested by centrifugation, frozen in liquid nitrogen, and dried in a freeze dryer (Alpha, Christ, Osterode, Germany) under vacuum. Subsequently, the cells were extracted twice with acetone at 55°C for 15 min. The combined extracts were then partitioned into diethylether/petrol (boiling point 35°C–80°C; 1:9, v/v) and evaporated to dryness. Carotenoid extracts were separated on an ODS-1 column (Maisch, Ammerbuch, Germany) at 1.4 mL min−1 starting with acetonitrile:methanol:0.1 m Tris-HCl buffer (74:12:4, v/v) as eluent. After 4 min, a linear gradient to methanol:hexane (4:1, v/v) was used (Gilmore and Yamamoto, 1991). Spectra were recorded directly from elution peaks using a 994 diode array detector (Waters, Milford, MA).
Quantification of astaxanthin and astaxanthin esters from H. pluvialis cells was carried out by modifying a procedure by Boussiba et al. (1992). The freeze-dried cells were powdered, resuspended in a solution containing 5% (v/v) KOH and 30% (v/v) methanol, and heated in a water bath (70°C) for 5 min. After centrifugation the supernatant, which contained the chlorophylls, was discarded. The pellet was extracted twice with dimethyl sulfoxide at 70°C for 5 min. To allow the quantification of astaxanthin and astaxanthin esters separately from other carotenoids, the absorbance of the combined extracts was determined at 550 nm. The values were subsequently multiplied by 3.2, a factor determined by measuring the absorbance of a purchased astaxanthin standard (Sigma, St. Louis) at two different wavelength (A492/A550). The amount of astaxanthin was then calculated applying an absorption coefficient for astaxanthin in dimethyl sulfoxide of 2,220 according to Boussiba et al. (1992).
ACKNOWLEDGMENTS
This work was only possible due to the generous support from Prof. Peter Böger (Konstanz, Germany). We are grateful to Silvia Kuhn for excellent technical assistance. Due thanks are expressed to Dr. Nirihiko Misawa, (Kirin Brewery Co., Yokohama, Japan) for the gift of the plasmids for complementation in E. coli. We thank Dr. Susanne Römer for critically reading the manuscript. The authors are grateful to Edel O'Halloran for help in the preparation of the manuscript.
LITERATURE CITED
- Bennedsen M, Wang X, Willen R, Wadstrom T, Andersen LP. Treatment of H. pylori infected mice with antioxidant astaxanthin reduces gastric inflammation, bacterial load and modulates cytokine release by splenocytes. Immunol Lett. 1999;70:185–189. doi: 10.1016/s0165-2478(99)00145-5. [DOI] [PubMed] [Google Scholar]
- Boussiba S. Carotenogenesis in the green alga Haematococcus pluvialis: cellular physiology and stress response. Physiol Plant. 2000;108:111–117. [Google Scholar]
- Boussiba S, Fan L, Vonshak A. Enhancement and determination of astaxanthin accumulation in green alga Haematococcus pluvialis. Methods Enzymol. 1992;213:386–391. [Google Scholar]
- Boussiba S, Wang B, Yuan PP, Zarka A, Chen F. Changes in pigments profile in the green alga Haematococcus pluvialis exposed to environmental stresses. Biotechnol Lett. 1999;21:601–604. [Google Scholar]
- Breitenbach J, Misawa N, Kajiwara S, Sandmann G. Expression in Escherichia coli and properties of the carotene ketolase from Haematococcus pluvialis. FEMS Microbiol Lett. 1996;140:241–246. doi: 10.1016/0378-1097(96)00187-5. [DOI] [PubMed] [Google Scholar]
- Cunningham FX, Gantt E. Genes and enzymes of carotenoid biosynthesis in plants. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:557–583. doi: 10.1146/annurev.arplant.49.1.557. [DOI] [PubMed] [Google Scholar]
- Fan L, Vonshak A, Zarka A, Boussiba S. Does astaxanthin protect Haematococcus against light damage? Z Naturforsch. 1998;53:93–100. doi: 10.1515/znc-1998-1-217. [DOI] [PubMed] [Google Scholar]
- Gilmore AM, Yamamoto HY. Zeaxanthin formation and energy-dependent fluorescence quenching in pea chloroplasts under artificially mediated linear and cyclic electron transport. Plant Physiol. 1991;96:635–643. doi: 10.1104/pp.96.2.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grünewald K, Eckert M, Hirschberg J, Hagen C. Phytoene desaturase is localized exclusively in the chloroplast and up-regulated at the mRNA level during accumulation of secondary carotenoids in Haematococcus pluvialis (Volvocales, chlorophyceae) Plant Physiol. 2000;122:1261–1268. doi: 10.1104/pp.122.4.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grünewald K, Hagen C, Braune W. Secondary carotenoid accumulation in flagellates of the green alga Haematococcus lacustris. Eur J Phycol. 1997;32:387–392. [Google Scholar]
- Hagen C, Grünewald K, Schmidt S, Müller J. Accumulation of secondary carotenoids in flagellates of Haematococcus pluvialis is accompanied by increase in chlorophyll productivity of photosynthesis. Eur J Phycol. 2000;35:75–82. [Google Scholar]
- Halliwell B, Gutteridge JMC. Free radicals in biology and medicine. Oxford: Clarendon Press; 1989. [Google Scholar]
- Johnson EA, Schroeder WA. Microbial carotenoids. Adv Biochem Eng Biotechnol. 1995;53:119–178. doi: 10.1007/BFb0102327. [DOI] [PubMed] [Google Scholar]
- Kajiwara S, Kakizono T, Saito T, Kondo K, Ohtani T, Nishio N, Nagai S, Misawa N. Isolation and functional identification of a novel cDNA for astaxanthin biosynthesis from Haematococcus pluvialis, and astaxanthin synthesis in Escherichia coli. Plant Mol Biol. 1995;29:343–352. doi: 10.1007/BF00043657. [DOI] [PubMed] [Google Scholar]
- Kakizono T, Kobayashi M, Nagai S. Effect of carbon/nitrogen ratio on encystment accompanied with astaxanthin formation in a green alga, Haematococcus pluvialis. J Ferment Bioeng. 1992;74:403–405. [Google Scholar]
- Kobayashi M, Kakizono T, Nagai S. Enhanced carotenoid biosynthesis by oxidative stress in acetate-induced cyst cells of a green unicellular alga, Haematococcus pluvialis. Appl Environ Microbiol. 1993;59:867–873. doi: 10.1128/aem.59.3.867-873.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M, Kakizono T, Nishio N, Nagai S, Kurimura Y, Tsuji Y. Antioxidant role of astaxanthin in the green alga Haematococcus pluvialis. Appl Microbiol Biotechnol. 1997;48:351–356. [Google Scholar]
- Linden H. Carotenoid hydroxylase from Haematococcus pluvialis: cDNA sequence, regulation and functional complementation. Biochim Biophys Acta. 1999;1446:203–212. doi: 10.1016/s0167-4781(99)00088-3. [DOI] [PubMed] [Google Scholar]
- Linden H, Vioque A, Sandmann G. Isolation of a carotenoid biosynthesis gene coding for ζ-carotene desaturase from Anabaena PCC 7120 by heterologous complementation. FEMS Microbiol Lett. 1993;106:99–104. [Google Scholar]
- Lorenz RT, Cysewski GR. Commercial potential for Haematococcus microalgae as a natural source of astaxanthin. Trends Biotechnol. 2000;18:160–167. doi: 10.1016/s0167-7799(00)01433-5. [DOI] [PubMed] [Google Scholar]
- Lotan T, Hirschberg J. Cloning and expression in Escherichia coli of the gene encoding β-C- 4-oxygenase, that converts β-carotene to the ketocarotenoid canthaxanthin in Haematococcus pluvialis. FEBS Lett. 1995;364:125–128. doi: 10.1016/0014-5793(95)00368-j. [DOI] [PubMed] [Google Scholar]
- Miki W. Biological functions and activities of animal carotenoids. Pure Appl Chem. 1991;63:141–146. [Google Scholar]
- Misawa N, Nakagawa M, Kobayashi K, Yamano S, Izawa Y, Nakamura K, Harashima K. Elucidation of the Erwinia uredovora carotenoid biosynthetic pathway by functional analysis of gene products expressed in Escherichia coli. J Bacteriol. 1990;172:6704–6712. doi: 10.1128/jb.172.12.6704-6712.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misawa N, Satomi Y, Kondo K, Yokoyama A, Kajiwara S, Saito T, Ohtani T, Miki W. Structure and functional analysis of a marine bacterial carotenoid biosynthesis gene cluster and astaxanthin biosynthetic pathway proposed at the gene level. J Bacteriol. 1995;177:6575–6584. doi: 10.1128/jb.177.22.6575-6584.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen A, Skibsted LH, Sampson J, Rice-Evans C, Everett SA. Comparative mechanims and rates of free radical scavenging by carotenoids antioxidants. FEBS Lett. 1997;418:91–97. doi: 10.1016/s0014-5793(97)01355-0. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Santos MF, Mesquita JF. Ultrastructural study of Haematococcus lacustris (Girod.) Rostafinsky (Volvocales): I. Some aspects of carotenogenesis. Cytologia. 1984;49:215–228. [Google Scholar]
- Sokolowsky V, Kaldenhoff R, Ricci M, Russo VEA. Fast and reliable mini-prep RNA extraction from Neurospora crassa. Fungal Genet Newslett. 1990;36:41–43. [Google Scholar]
- Sun Z, Cunningham FX, Gantt E. Differential expression of two isopentenyl pyrophosphate isomerases and enhanced carotenoid accumulation in a unicellular chlorophyte. Proc Natl Acad Sci USA. 1998;95:11482–11488. doi: 10.1073/pnas.95.19.11482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Morishita Y, Suzui M, Kojima T, Okumura A, Mori H. Chemoprevention of mouse urinary bladder carcinogenesis by the naturally occurring carotenoid astaxanthin. Carcinogenesis. 1994;15:15–19. doi: 10.1093/carcin/15.1.15. [DOI] [PubMed] [Google Scholar]
- Yong YYR, Lee Y-K. Do carotenoids play a photoprotective role in the cytoplasm of Haematococcus lacustris (Chlorophyta)? Phycologia. 1991;30:257–261. [Google Scholar]