Abstract
The physical properties of lipid bilayers can be remodeled by a variety of environmental factors. Here we investigate using molecular dynamics simulations the specific effects of nanoscopic substrates or external contact points on lipid membranes. We expose palmitoyl-oleoyl phosphatidylcholine bilayers unilaterally and separately to various model nano-sized substrates differing in surface hydroxyl densities. We find that a surface hydroxyl density as low as 10% is sufficient to keep the bilayer juxtaposed to the substrate. The bilayer interacts with the substrate indirectly through multiple layers of water molecules, however, despite such buffered interaction, the bilayers exhibit certain properties different from unsupported bilayers. The substrates modify transverse lipid fluctuations, charge density profiles and lipid diffusion rates, although differently in the two leaflets, which creates an asymmetry between bilayer leaflets. Other properties that include lipid cross-sectional areas, component volumes and order parameters are minimally affected. The extent of asymmetry that we observe between bilayer leaflets is well beyond what has been reported for bilayers adsorbed on infinite solid supports. This is perhaps because the bilayers are much closer to our nano-sized finite supports than to infinite solid supports, resulting in a stronger support-bilayer electrostatic coupling. The exposure of membranes to nanoscopic contact points, therefore, cannot be considered as a simple linear interpolation between unsupported membranes and membranes supported on infinite supports. In the biological context, this suggests that the exposure of membranes to non-intercalating proteins, such as those belonging to the cytoskeleton, should not always be considered as passive non-consequential interactions.
INTRODUCTION
The structure, dynamics and organization of lipid bilayers are regulated by a variety of environmental factors. In general, such environmental factors can be categorized as being intrinsic or extrinsic depending on their origin relative to the membrane. Intrinsic factors are those that emanate from within the membrane and include diffusion barriers in the form of membrane proteins,1 preferentially binding lipids such as cholesterol2 and even small molecules such as alcohols.3 In contrast, extrinsic factors are those that originate from the environment surrounding the membrane. These include, for example, ionic strength,4 focal adhesion proteins that latch onto integral membrane proteins,5 non-intercalating proteins that make only external contacts with membranes6 and a variety of different non-biological constructs in the form of solid or polymeric support.7
While considerable knowledge about the effect of some of these environmental factors exists, there are no experiments or computational studies that probe the effect of nanoscopic substrates on membranes. Nanoscopic substrates offer contact points to membranes that are fundamentally different from the type of contact offered by “infinite” solid or polymeric supports. In contrast to infinite supports, the interaction of the nanoscopic substrates with membranes is limited to lateral widths of a few nanometers. In the biological context, such nanoscopic supports can be considered as model representatives to the large family of non-intercalating proteins, such as those belonging to the cytoskeleton or extracellular matrix, that make only external contacts to membranes.6 Here we investigate the hypothesis of whether the interaction of membranes with nanoscopic supports can be considered as a linear interpolation between unsupported membranes and membranes supported on infinite supports.
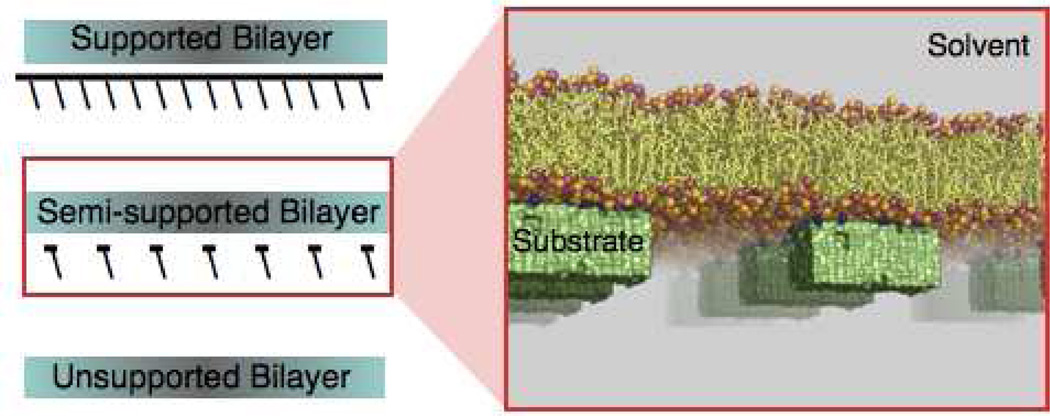
To examine how interactions with such nanoscopic external contact points influence membrane properties, we construct different model membrane configurations and study them using atomistic molecular dynamics (MD) simulations. Model configurations have, in general, been employed on numerous occasions and provide direct insight into underlying mechanisms (for recent examples see Ref. 8). In this study, we expose a single side of a POPC bilayer separately to three different six-nanometer wide substrates. The substrates differ from each other only in the extent of surface hydroxylation. While the surface of one substrate carries no specific functionalization making it essentially hydrophobic, the surfaces of the remaining two substrates carry 10% and 20% hydroxylation. We categorize such membrane configurations under the general family of semi-supported lipid bilayers (Fig. 1).
Figure 1.
Cartoon of a semi-supported POPC bilayer. The phosphate and choline lipid groups are illustrated as pink and orange spheres. The substrates are shown as green solids, and the functional groups on the substrate surface are drawn as blue spheres. The atomic coordinates were taken from the 0.53 microsecond snapshot of a molecular dynamics simulation of a POPC bilayer supported by a substrate carrying 20% hydroxyl surface coverage.
In each of the cases considered, we find that the bilayer interacts with the nano-substrate indirectly through a thin layer of water molecules. Despite such buffered interaction, semi-supported bilayers exhibit properties different from unsupported bilayers. The nano-sized supports modify structural as well as dynamical properties in both leaflets, although the extent of the modification is different in the two leaflets, which creates an asymmetry between bilayer leaflets. The extent of this asymmetry is well beyond what has been reported for bilayers adsorbed on infinite solid supports. Essentially, the properties of semi-supported bilayers are not a linear interpolation between the properties of unsupported bilayers and bilayers supported on infinite supports.
METHODS
To study the effect of nanoscopic substrates on membranes, we carried out four separate MD simulations. First, as a control, an unsupported POPC bilayer containing 64 lipids/leaflet and ~58 waters/lipid was simulated for 0.4 µs. Data from the final 0.3 µs of this simulation was used for analysis. The initial coordinates for this system were taken from an equilibrated ensemble containing 64 lipids/leaflet and ~28 waters/lipid.9
The remaining three simulations are that of a POPC bilayer subjected to different substrates; one substrate with no surface charge density (hydrophobic) and the other two coated partially with hydroxyl groups. A POPC bilayer containing 128 lipids/leaflet and ~94 waters/lipid was first subjected to the hydrophobic substrate. Note that such a lipid-to-water ratio in the unit cell separates the bilayer from its nearest periodic image (along the bilayer normal) by ~ 70 Å. The initial coordinates of the solvated bilayer were taken from the simulation of the unsupported POPC bilayer system. The hydrophobic substrate was constructed by placing a set of uncharged Lennard-Jones (LJ) spheres within a three dimensional grid of size 63×32×25 Å. The LJ parameters for the spheres correspond to the CH4 moiety in the Gromos 43a1 force field.10 This choice of parameters essentially makes the substrate hydrophobic. The LJ spheres were placed at a distance of 4.16 Å from each other; a distance at which the potential energy between two such LJ spheres is minimum. To preserve the topology of the substrate, the LJ spheres were restrained individually using harmonic potentials of force constant 1000 kJ/mol/nm2. In the starting configuration, the shortest distance between the lipid phosphate and the substrate was 10 Å. This semi-supported bilayer system was simulated for 0.45 µs. An additional configuration of this type was also simulated for 0.3 µs in which lateral size of the simulation unit cell was doubled, that is, 256 lipids/leaflet were simulated in the presence of two substrates instead of 128 lipids/leaflet in the presence of one substrate. We did not observe any systematic effects of doubling the unit cell size.
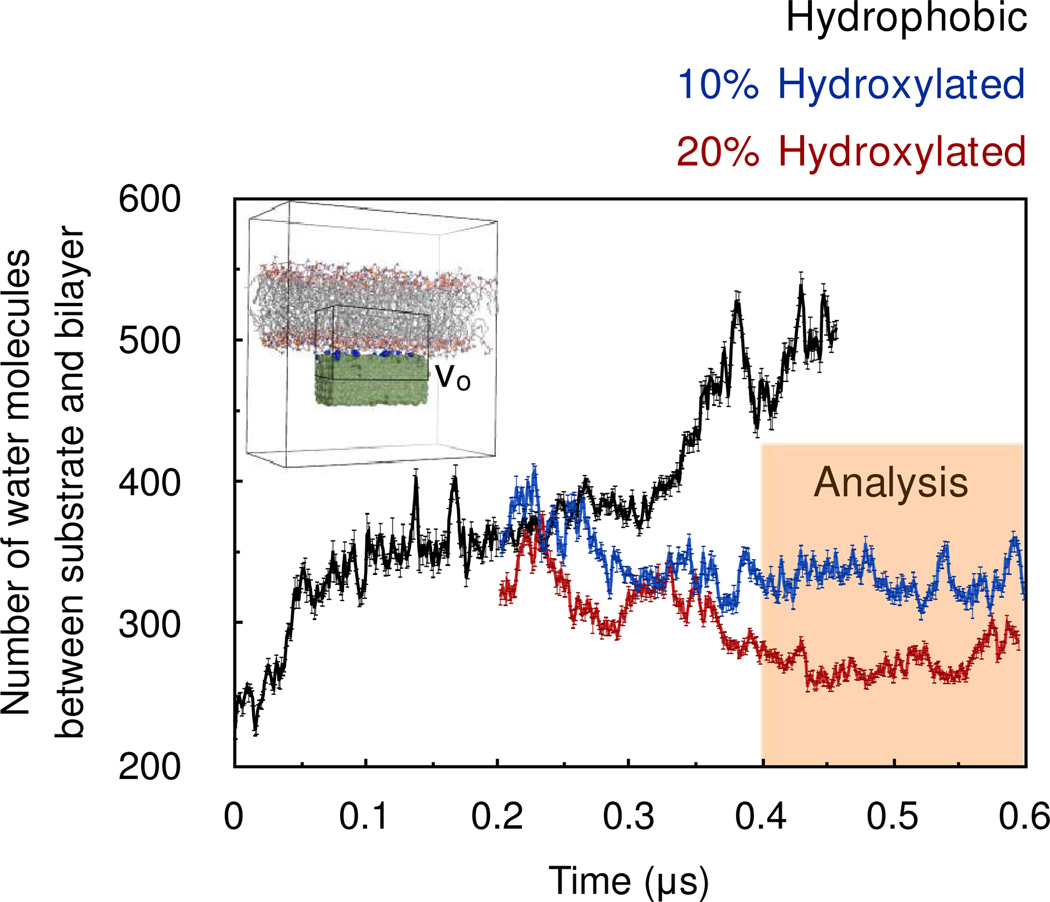
An intermittent point of this trajectory (0.2 µs) was used for setting up the simulations of the bilayers supported on hydroxylated substrates. The rationale behind using an intermittent point is as follows. Since the bilayer is expected to interact more strongly with the hydroxylated substrate than the hydrophobic support, the number of water molecules interleaved between the substrate and the bilayer should decrease upon substrate hydroxylation and level out to smaller numbers. This renders the task of monitoring equilibration easier compared to the scenario in which the simulation was started in parallel with the simulation of the bilayer supported on the hydrophobic substrate. The hydroxylation of the substrate was carried out by substituting randomly a fraction of the CH4 spheres on the surface with CH3OH molecules. In one case ~10% of the CH4 spheres on the surface were substituted with CH3OH molecules, and in another case ~20% of the CH4 spheres on the surface were substituted with CH3OH molecules. Following such substitutions, the CH3 moiety of the CH3OH molecule was restrained harmonically, but no restraints were placed on the hydroxyl group. Each of these systems was subjected to a molecular dynamics simulation of 0.4 µs. The number of water molecules occupying the region between the substrate and the bilayer leveled out in ~ 0.2 µs (Fig. 2), and so the final 0.2 µs of each trajectory was used for analysis. The force field parameters for CH3OH were taken from Walser et al.11
Figure 2.
Time evolution of the number of water molecules between the substrate and the bilayer. The trajectory is divided into 1 ns blocks, and data from each block is represented by an average and a standard deviation. The inset is a representative snapshot from the molecular dynamics trajectory indicates the sub-volume Vo chosen for counting the waters molecules between the substrate and the bilayer.
All molecular dynamics simulations were carried out under isobaric-isothermal conditions. The pressures in the lateral and transverse directions were maintained separately at 1.013 bars using an extended-ensemble approach12 and with a coupling constant of 1 ps and a compressibility of 4.5×10−5 bar−1. An extended ensemble approach13 was also used for maintaining temperature at 303 K, although a shorter coupling constant of 0.2 ps was employed. Electrostatic interactions were computed using the particle mesh Ewald scheme14 with a Fourier grid-spacing of 0.15 nm, a sixth-order interpolation and a direct space cutoff of 10 Å. van der Waals interactions were computed explicitly for interatomic distance up to 16 Å. The bonds in lipid molecules were constrained using the P-LINCS algorithm,15 and the geometries of the water molecules were constrained using SETTLE.16 These constraints permitted use of a large integration time step of 2 fs. The motion of the center of mass was reset every picosecond. The water molecules were described using SPC/E parameters17 and the lipid molecules and lipid-water interactions were described using the recently refined GROMOS 43A1-S3 parameter set,18 which yields lipid component volumes, cross-sectional areas, form factors and electron densities in agreement with experiment.19 All molecular dynamics simulations were carried out using Gromacs 4.0.5.20
RESULTS AND DISCUSSION
To study the effect of nanoscopic external contact points on membranes, we subjected pure POPC bilayers to three different model nano-sized substrates; one carrying no charge density (hydrophobic) while the remaining two coated with hydroxyl groups. As a control, we also simulated an unsupported POPC bilayer.
Figure 2 shows the time evolution of the number of waters between the substrate and bilayer. In the simulation of the bilayer supported on a hydrophobic substrate, the number of water molecules between the substrate and the bilayer increase with simulation time and the bilayer is observed to drift away steadily from the substrate. This is perhaps because the interaction between the substrate and bilayer is weak. The free energy is reduced by the increased entropy of water in this region, resulting in an effective entropic repulsive force between substrate and bilayer. This result does not change even when the lateral size of the simulation unit cell is doubled, that is, when 256 lipids/leaflet are simulated in the presence of two substrates instead of 128 lipids/leaflet in the presence of one substrate. As the separation between the bilayer and substrate increases further, structural properties of the POPC bilayer can be expected to approach those of isolated bilayers. Alternatively, the interaction of the bilayer with a hydrophobic substrate can also unsettle bilayer structure. As observed experimentally,7f templating by periodic micrometer-sized hydrophobic substrates has been shown to give rise to patterned formation of monolayers and bilayers, with monolayers occupying regions directly over the hydrophobic substrate and the bilayers occupying the intermediate unsupported regions.
The introduction of 10% surface hydroxylation on the substrate kept the bilayer juxtaposed to the substrate (Fig. 2). Increasing the surface hydroxyl coverage to 20% also kept the bilayer juxtaposed to the substrate, but it reduced the number of water molecules between the substrate and the bilayer (see also Fig. S1 in supporting information). This suggests that the substrate hydroxyls serve as surrogates for hydrating the lipid bilayer. Note, however, that in neither case were the inner-shell hydration numbers of the head groups altered (Fig. S2 in supporting information). In addition, the lipid head groups did not make any direct contact with the substrate hydroxyls. The radial distributions of the lipid phosphates, carbonyls and choline group around the substrate hydroxyls show that none of these functional groups are within the inner coordination shell of substrate hydroxyls (Fig. S3 in supporting information). At 10% hydroxylation, the lipid phosphates in the lower leaflet fluctuate around an average distance of 7.1 ± 1.6 Å from the substrate and at 20% hydroxylation, this distance reduces to 6.0 ± 1.7 Å. To estimate these average phosphate-substrate distances, only those phosphate groups were considered that lay directly over the substrate. Nevertheless, if this restriction on choosing phosphate groups is lifted the results differ by only 0.1 Å.
The inner coordination shells of the substrate hydroxyls are comprised only of water molecules. Such a buffered interaction between the substrate and the bilayer through water layers also occurs in the case of bilayers supported on infinite substrates. Experiments involving PC bilayers adsorbed on quartz sheets, silicon sheets and mica beads suggest the presence of a 6–30 Å thick layer of interstitial water.21 Recent molecular dynamics simulations employing coarse-grained force fields also suggest presence of a nanometer-thick layer of water between model hydrophilic sheets and PC bilayers.7h It is interesting to note that the interstitial water thickness that we observe in semi-supported membrane simulations is at the lower extreme of values reported in the case of bilayers supported on infinite supports. In other words, it appears that bilayers can be expected to be closer to nano-sized substrates as compared to infinite solid supports, thereby having a stronger electrostatic coupling with the substrate.
But despite the stronger electrostatic coupling between the bilayer and the substrate, we find that most structural properties of the bilayer are not affected. Table 1 lists various structural properties of bilayers computed in the absence and in the presence of substrates. None of these structural properties exhibit any major systematic substrate-induced modifications. The effective volumes of terminal methyls (vCH3), methylenes (vCH2/CH), headgroups (vHG) and glycerol backbones (vGL) were obtained by minimizing the objective function22
| [1] |
In this objective function, ni are the ensemble-average number densities of the different lipid components i{CH2/CH, CH3, HG, GL, H2O}. The outer summation in the function runs over the various slices of the simulation unit cell made along the bilayer normal z, with Δz ~ 0.1 Å. The minimization of the objective function is achieved via enumeration, that is, by systematically scanning over the physiological ranges of vi. The volumes of the hydrocarbon tails, vC, were obtained by summing up the methylene and terminal methyl volumes, that is, vC = 30vCH2/CH + 2vCH3. The hydrocarbon tail thickness, 2dC, is essentially the half-width of the number density of the hydrocarbon core.23 The geometric lipid cross-section areas, AG, were determined by dividing unit cell areas by the number of lipids/leaflets and averaging over the respective trajectories. To be consistent with experimental methods,22 the lipid cross-sectional areas are also estimated by dividing the hydrocarbon tail volumes by their respective thickness, that is, A vC/dC. In addition to the structural properties listed in Table 1, we find no significant changes in the hydrocarbon deuterium order parameters (Fig. S4 in supporting information).
Table 1.
Effect of substrate on the structural properties of POPC bilayer at 303 K.
| Unsupported | Semi-supported (Simulation) | |||
|---|---|---|---|---|
| Expt.a | Simulation | 10% hydroxylated | 20% hydroxylated | |
| vH2O (Å3) | - | 30.3 | 30.4 | 30.5 |
| vCH2/CH (Å3) | 27.6 | 26.6 | 26.8 | 26.7 |
| vCH3 (Å3) | 53.6 | 54.9 | 54.9 | 54.9 |
| vC (Å3) | 924.2 | 909 | 912.6 | 910.8 |
| vHG + vGL (Å3) | 331 | 319.5 | 320.2 | 318.3 |
| vHG + vGL + vC (Å3) | 1256 | 1228.5 | 1232.8 | 1229.1 |
| 2dC (Å) | 27.2 | 27.6 | 28.8 | 27.8 |
| A =vC/dC (Å2) | 68.3 | 65.8 | 63.4 | 65.4 |
| AG (Å2) | - | 64.5 ± 0.4 | 63.8 ± 0.6 | 63.8 ± 0.5 |
Experimental estimate taken from Ref. 19
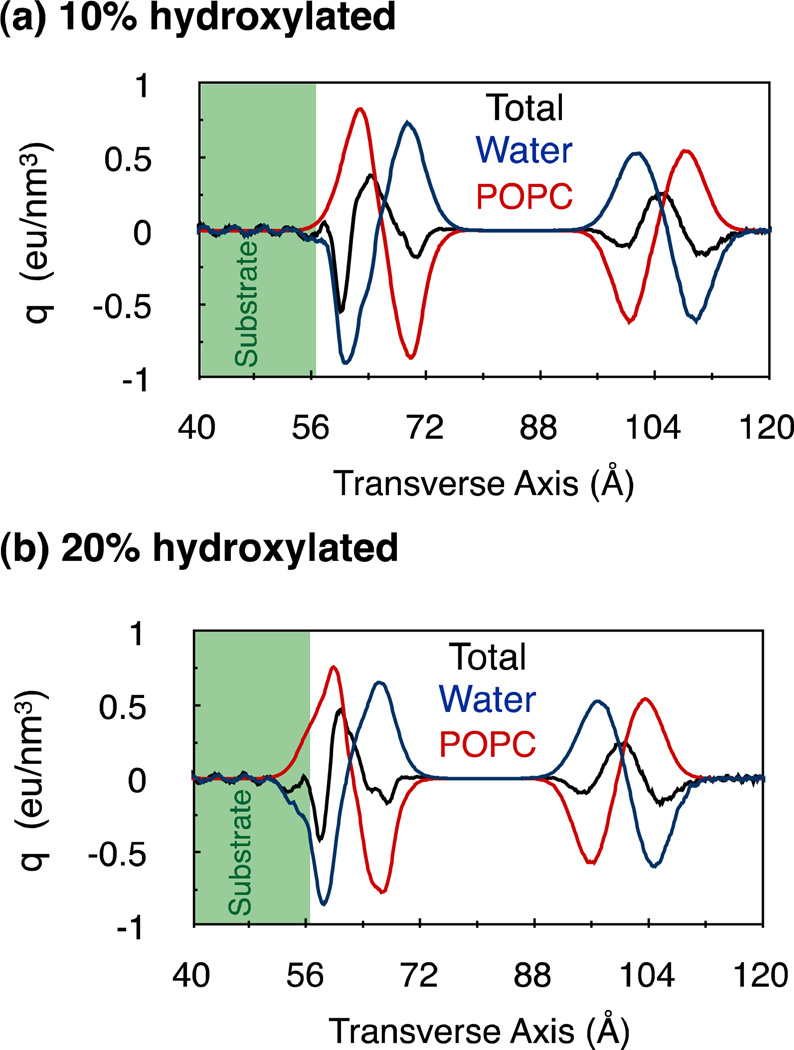
The electrostatic coupling between the substrates and the bilayers, however, significantly modifies the distributions of charges in the hydrated bilayer (Fig. 3). This modification is more pronounced around the lower leaflet in comparison to the complementary upper leaflet, resulting in an overall asymmetry in charge distribution along the bilayer normal. (For a distribution of charges in the unsupported bilayer, see Fig. S5 in supporting information.) We expect that such asymmetry can cause a given external stimuli to interact differently with the bilayer depending on whether it originates closer to the upper leaflet or the lower leaflet. For example, this could directly influence the specific binding of drugs and antimicrobial peptides to membranes,3, 24 the insertion and orientation of integral membrane proteins,25 signal transduction in membrane proteins26 and the selective transport of molecules across membranes,27 ultimately affecting a variety of different physiological processes.28
Figure 3.
Average charge densities in PC bilayer configurations supported on (a) 10% and (b) 20% hydroxylated substrates. The charge densities of the water molecules and the POPC lipids are plotted separately as red and blue lines, respectively. The total charge densities, which are the sums of the charge densities of water molecules, lipids and the substrate-hydroxyl groups, are drawn as solid black lines.
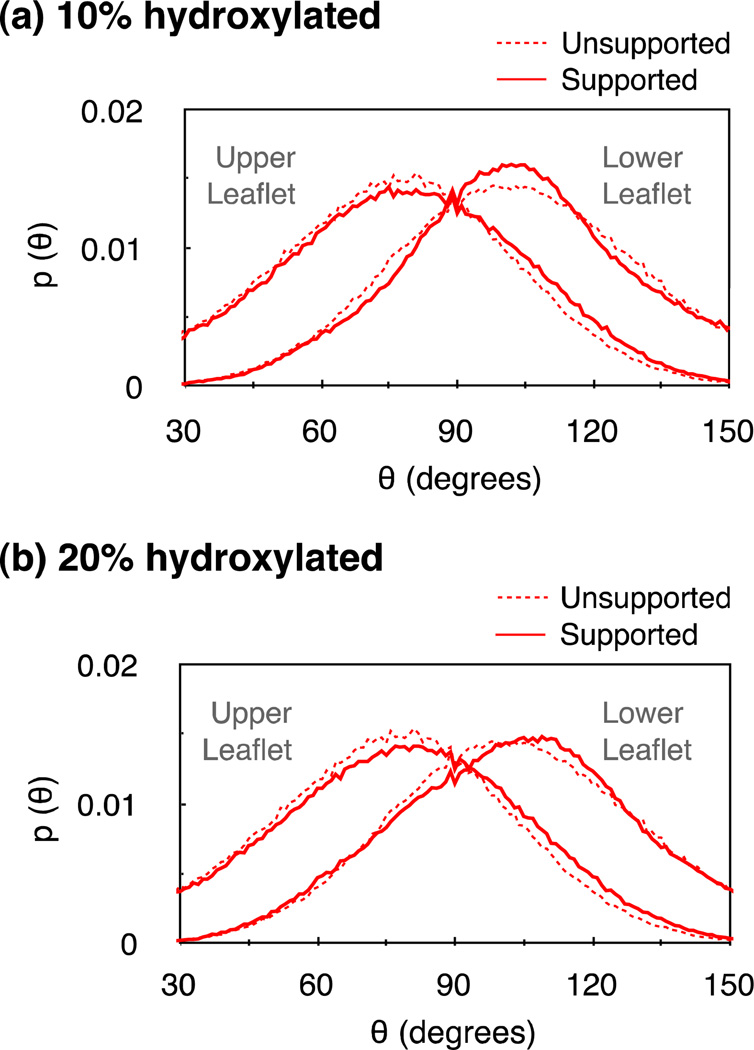
A prominent feature of the substrate-induced modification in charge distribution is an enhancement in peak heights, with compensating reductions in half-widths. The sharpened distribution of water charges suggests an increased alignment of the water dipoles along the bilayer normal. We confirm this explicitly by estimating the effect of the substrate on the density of the transverse component of the water dipoles (see Fig. S6 in supporting information). The modification in the charge densities of lipids can be understood partly by noting that the substrates redistribute the phosphate-choline (PN) dipole orientations. From Fig. 4 we observe that the substrates alter PN angle distributions in both leaflets, although to a greater extent in the lower leaflets. This observation is similar to the one made in simulations of a PC bilayer supported on nano-porous amorphous silica.29 However, we find that such modification in angular distributions has little effect on the average orientations of PN dipoles. In the absence of the substrate, the PN dipoles are aligned at an average angle of 104° ± 27° with respect to the bilayer normal. In the presence of the 10% hydroxylated substrate, the PN dipoles in the lower leaflet align at an angle of 106° ± 27° with respect to the bilayer normal, and this angle is 104° ± 28° in the presence of the 20% hydroxylated substrate.
Figure 4.
Angle distribution p(θ) of phosphate choline (PN) dipoles in bilayers supported on (a) 10% and (b) 20% hydroxylated substrates. θ is the angle between a PN dipole and the bilayer normal. The effect of the substrate on PN angular distributions is more pronounced in the lower leaflet. While a 10% hydroxylation narrows the PN dipole distribution around the natural peak, the 20% hydroxylated substrate shifts the distribution peak by ~10 degrees toward the bilayer normal.
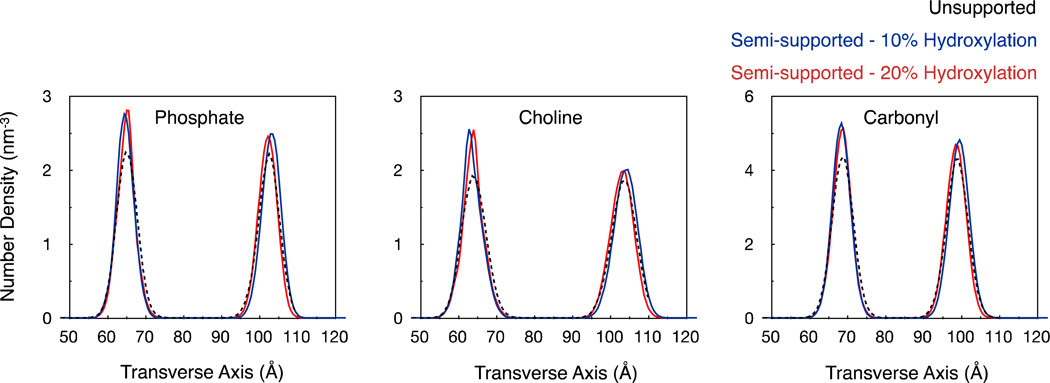
The enhanced sharpness in the lipid charge distribution peaks can also be attributed to substrate-induced reduction in the transverse fluctuations of lipid head groups. Fig. 5 illustrates the number densities of lipid phosphates, cholines and carbonyls along the transverse axis. We find that the substrates sharpen the number distributions, particularly in the lower leaflet, which implies reduced transverse fluctuations of lipid head groups. For a Gaussian distribution of number densities, the statistical (or Shannon) entropy associated with the distribution is proportional to the log of the width of the distribution.30 Thus the observed sharpening of the number densities implies a loss in entropy related to transverse fluctuations. Such a loss in entropy along the transverse direction was also noted in the case of bilayers supported on infinite supports.30 Note, however, that while there is a loss in statistical entropy, it is clearly not large enough to prevent the bilayer from staying juxtaposed to the hydroxylated substrate. Overall, the substrate modifies both head group dipole distributions and their transverse fluctuations, which together result in altered charge distribution profiles.
Figure 5.
Distribution of lipid functional groups along the transverse axis. The distributions from the three bilayer configurations are aligned with each other such that the midpoints between their respective phosphate density peaks coincide.
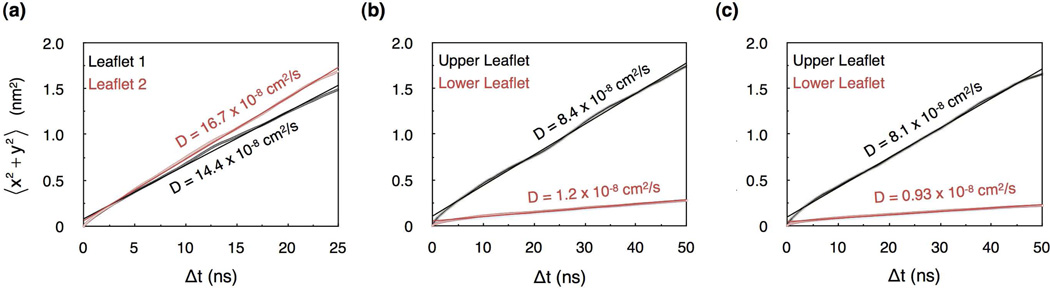
In addition to modifying the transverse fluctuations of lipids, we find that the substrates also modify the lateral motions of lipids significantly. Fig. 6 illustrates the effect of substrates on the lateral mean square displacement (MSD) of lipids. In the absence of a support, the lipids undergo lateral diffusion at the rate of ~15×10−8 cm2/s, which is consistent with the experimental estimate of 9×10−8 cm2/s.31 In the presence of the support, however, lipid diffusion in the lower leaflet slows down by an order of magnitude to values of ~1×10−8 cm2/s. This is a much larger drop in lateral diffusion as noted in experiments and simulations of bilayers supported on infinite supports.7 This difference could be explained by noting that in our simulated cases of semi-supported bilayers, the bilayers are much closer to the substrate as compared to the bilayers supported on infinite supports. The enhanced proximity between the substrate and the bilayer increases the attractive electrostatic force between the substrate-hydroxyls and the lipid dipoles, making lipid diffusion slower. We also find that the substrate slows down lipid diffusion in the upper leaflet, but only slightly in comparison to the modification it introduced in the upper leaflet. Such a decrease in lipid diffusion in the upper leaflet has not been reported in experiments and simulations of bilayers supported on infinite supports. The effect of the substrate on the diffusion of lipids in the upper leaflet could also be attributed to the long-range nature of the electrostatic force between the substrate and the lipids in the upper leaflet. In addition, this could also be attributed to the increased lipid inter-digitation we note in supported bilayers (Fig. S7 in supporting information). Note, however, that the increase in lipid inter-digitation does not affect the hydrocarbon tail thickness (see Table 1).
Figure 6.
Lateral mean square displacement of lipids. (a) unsupported bilayers; (b) bilayers supported on substrates with 10% hydroxyl surface coverage and; (c) bilayers supported on substrates with 20% hydroxyl surface coverage. The straight lines are least square fits, which yield diffusion constants through the Einstein relationship. The attractive electrostatic force between the substrate-hydroxyl’s and the lipid dipoles slows down lipid diffusion, although differently in two leaflets. Lipids in the lower leaflet diffuse at rates almost an order of magnitude slower than those in the upper leaflet.
Taken together, we find that polar nano-substrates are capable of modifying structural as well as dynamical properties of bilayers significantly, and in a manner not reported in the case of bilayers supported on infinite supports. In the case of hydroxylated nano-substrates, several properties, including lipid cross-sectional areas, component volumes and order parameters are minimally affected, however, other properties like transverse lipid fluctuations, charge density profiles and lipid diffusion rates are affected significantly. In addition, the nano-sized substrates introduce a stark asymmetry between bilayer leaflets, an asymmetry that is well beyond what has been seen in the case of bilayers adsorbed on infinite solid supports. The exposure of membranes to nanoscopic substrates, therefore, cannot be considered as a simple linear interpolation between unsupported membranes and membranes supported on infinite supports. This suggests that artificial nano-sized templating could be used for manipulating membrane properties in a manner different from standard templating via infinite supports. While nano-scale templating is undoubtedly challenging, recent advances32 place such technology within grasp in the foreseeable future. In the biological context, these findings suggest that the exposure of cellular membranes to non-intercalating proteins need not always be considered as a passive non-consequential interaction. The very proximity of proteins to membranes can modulate membrane properties, and that too quite significantly. This may very well be a discrete mechanism that nature employs to control membrane function, although more investigation is required.
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge funding from the NIH through their roadmap for medical research and computer time from the Center of Molecular Study of Soft Condensed Matter at the Illinois Institute of Technology. We thank Profs. Eric Jakobsson, Sagar Pandit and Atul Parikh for comments and discussions.
Footnotes
SUUPORTING INFORMATION AVAILABLE
There are seven figures. This information is available free of charge via the Internet at http://pubs.acs.org/.
REFERENCES
- 1.(a) Singer SJ, Nicolson GL. The fluid mosaic model of the structure of cell membranes. Science. 1972;175:720–731. doi: 10.1126/science.175.4023.720. [DOI] [PubMed] [Google Scholar]; (b) Baumgart T, Hammond AT, Sengupta P, Hess ST, Holowka DA, Baird BA, Webb WW. Large-scale fluid/fluid phase separation of proteins and lipids in giant plasma membrane vesicles. Proc. Natl. Acad. Sci. U.S.A. 2007;104:3165–3170. doi: 10.1073/pnas.0611357104. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327(5961):46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 2.(a) Veatch SL, Keller SL. Organization in lipid membranes containing cholesterol. Phys. Rev. Lett. 2002;89:268101. doi: 10.1103/PhysRevLett.89.268101. [DOI] [PubMed] [Google Scholar]; (b) Veatch SL, Keller SL. Miscibility phase diagrams of giant vesicles containing sphingomyelin. Phys. Rev. Lett. 2005;94:148101. doi: 10.1103/PhysRevLett.94.148101. [DOI] [PubMed] [Google Scholar]
- 3.Bruno MJ, Koeppe RE, 2nd, Andersen OS. Docosahexaenoic acid alters bilayer elastic properties. Proc. Natl. Acad. Sci. U.S.A. 2007;104:9638–9643. doi: 10.1073/pnas.0701015104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pandit SA, Bostick D, Berkowitz ML. Molecular dynamics simulation of a dipalmitoylphosphatidylcholine bilayer with NaCl. Biophys. J. 2003;84(6):3743–3750. doi: 10.1016/S0006-3495(03)75102-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.(a) Burridge K, Chrzanowska-Wodnicka M. Focal adhesions, contractility, and signaling. Annu Rev. Cell Dev. Biol. 1996;12:463–518. doi: 10.1146/annurev.cellbio.12.1.463. [DOI] [PubMed] [Google Scholar]; (b) Zaidel-Bar R, Itzkovitz S, Ma'ayan A, Iyengar R, Geiger B. Functional atlas of the integrin adhesome. Nature Cell Biol. 2007;9(8):858–867. doi: 10.1038/ncb0807-858. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Kanchanawong P, Shtengel G, Pasapera AM, Ramko EB, Davidson MW, Hess HF, Waterman CM. Nanoscale architecture of integrin-based cell adhesions. Nature. 2010;468:580–584. doi: 10.1038/nature09621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular Biology of the Cell. 5 ed. Garland Science; 2007. p. 1392. [Google Scholar]
- 7.(a) Tanaka M, Sackmann E. Polymer-supported membranes as models of the cell surface. Nature. 2005;437:656–663. doi: 10.1038/nature04164. [DOI] [PubMed] [Google Scholar]; (b) Castellana ET, Cremer PS. Solid supported lipid bilayers: From biophysical studies to sensor design. Surf. Sci. Rep. 2006;61:429–444. doi: 10.1016/j.surfrep.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Richter RP, Berat R, Brisson AR. Formation of solid-supported lipid bilayers: An integrated view. Langmuir. 2006;22(8):3497–3505. doi: 10.1021/la052687c. [DOI] [PubMed] [Google Scholar]; (d) Troutier AL, Ladaviere C. An overview of lipid membrane supported by colloidal particles. Adv. Colloid Interfac. 2007;133:1–21. doi: 10.1016/j.cis.2007.02.003. [DOI] [PubMed] [Google Scholar]; (e) Kiessling V, Domanska MK, Murray D, Wan C, Tamm LK. Supported Lipid Bilayers: Development and Applications in Chemical Biology. Vol. 4. John Wiley and Sons; 2008. [Google Scholar]; (f) Oliver AE, Parikh AN. Templating membrane assembly, structure, and dynamics using engineered interfaces. Bba-Biomembranes. 2010;1798:839–850. doi: 10.1016/j.bbamem.2009.12.029. [DOI] [PubMed] [Google Scholar]; (g) Chemburu S, Fenton K, Lopez GP, Zeineldin R. Biomimetic Silica Microspheres in Biosensing. Molecules. 2010;15(3):1932–1957. doi: 10.3390/molecules15031932. [DOI] [PMC free article] [PubMed] [Google Scholar]; (h) Xing CY, Faller R. Interactions of lipid bilayers with supports: A coarse-grained molecular simulation study. J. Phys. Chem. B. 2008;112:7086–7094. doi: 10.1021/jp077305l. [DOI] [PubMed] [Google Scholar]
- 8.(a) Baron R, Setny P, McCammon JA. Water in Cavity-Ligand Recognition. J. Amer. Chem. Soc. 2010;132(34):12091–12097. doi: 10.1021/ja1050082. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Varma S, Rogers DM, Pratt LR, Rempe SB. Perspectives on: Ion selectivity Design principles for K(+) selectivity in membrane transport. J Gen. Physiol. 2011;137:479–488. doi: 10.1085/jgp.201010579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pandit SA, Chiu SW, Jakobsson E, Grama A, Scott HL. Cholesterol packing around lipids with saturated and unsaturated chains: A simulation study. Langmuir. 2008;24:6858–6865. doi: 10.1021/la8004135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scott WRP, Hunenberger PH, Tironi IG, Mark AE, Billeter SR, Fennen J, Torda AE, Huber T, Kruger P, van Gunsteren WF. The GROMOS biomolecular simulation program package. J Phys. Chem. A. 1999;103(19):3596–3607. [Google Scholar]
- 11.Walser R, Mark AE, van Gunsteren WF, Lauterbach M, Wipff G. The effect of force-field parameters on properties of liquids: Parametrization of a simple three-site model for methanol. J Chem. Phys. 2000;112:10450–10459. [Google Scholar]
- 12.Parrinello M, Rahman A. Polymorphic Transition in Single Crystals: A New Molecular Dynamics Method. J Appl. Phys. 1981;52:7128–7190. [Google Scholar]
- 13.(a) Nose S. A Molecular-Dynamics Method for Simulations in the Canonical Ensemble. Molecular Physics. 1984;52(2):255–268. [Google Scholar]; (b) Hoover WG. Canonical Dynamics - Equilibrium Phase-Space Distributions. Phys. Rev. A. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 14.Darden T, York D, Pedersen L. Particle Mesh Ewald - an N.Log(N) Method for Ewald Sums in Large Systems. J Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 15.Hess B. P-LINCS: A parallel linear constraint solver for molecular simulation. J Chem. Theory Comput. 2008;4:116–122. doi: 10.1021/ct700200b. [DOI] [PubMed] [Google Scholar]
- 16.Miyamoto S, Kollman P. SETTLE: An Analytical Version of the SHAKE and RATTLE Algorithms for Rigid Water Molecules. J Comp. Chem. 1992;13:952–962. [Google Scholar]
- 17.Berendsen HJ, Grigera JR, Straatsma TP. The missing term in effective pair potentials. J. Phys. Chem. 1987;91:6269–6271. [Google Scholar]
- 18.Chiu SW, Pandit SA, Scott HL, Jakobsson E. An Improved United Atom Force Field for Simulation of Mixed Lipid Bilayers. J. Phys. Chem. B. 2009;113:2748–2763. doi: 10.1021/jp807056c. [DOI] [PubMed] [Google Scholar]
- 19.Kucerka N, Tristram-Nagle S, Nagle JF. Structure of fully hydrated fluid phase lipid bilayers with monounsaturated chains. J Membrane Biol. 2005;208:193–202. doi: 10.1007/s00232-005-7006-8. [DOI] [PubMed] [Google Scholar]
- 20.Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem. Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 21.(a) Bayerl TM, Bloom M. Physical-Properties of Single Phospholipid-Bilayers Adsorbed to Micro Glass-Beads - a New Vesicular Model System Studied by H-2-Nuclear Magnetic-Resonance. Biophys. J. 1990;58(2):357–362. doi: 10.1016/S0006-3495(90)82382-1. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Johnson SJ, Bayerl TM, Mcdermott DC, Adam GW, Rennie AR, Thomas RK, Sackmann E. Structure of an Adsorbed Dimyristoylphosphatidylcholine Bilayer Measured with Specular Reflection of Neutrons. Biophys. J. 1991;59(2):289–294. doi: 10.1016/S0006-3495(91)82222-6. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Koenig BW, Kruger S, Orts WJ, Majkrzak CF, Berk NF, Silverton JV, Gawrisch K. Neutron reflectivity and atomic force microscopy studies of a lipid bilayer in water adsorbed to the surface of a silicon single crystal. Langmuir. 1996;12:1343–1350. [Google Scholar]
- 22.Petrache HI, Feller SE, Nagle JF. Determination of component volumes of lipid bilayers from simulations. Biophys. J. 1997;72:2237–2242. doi: 10.1016/S0006-3495(97)78867-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagle JF, Tristram-Nagle S. Structure of lipid bilayers. BBA-Rev Biomembranes. 2000;1469:159–195. doi: 10.1016/s0304-4157(00)00016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.(a) Brogden KA. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nature Rev. Microbiol. 2005;3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]; (b) Ivankin A, Livne L, Mor A, Caputo GA, DeGrado WF, Meron M, Lin B, Gidalevitz D. Role of the Conformational Rigidity in the Design of Biomimetic Antimicrobial Compounds. Angew Chem Int Edit. 2010;49:8462–8465. doi: 10.1002/anie.201003104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Samartzidou H, Delcour AH. E-coli PhoE porin has an opposite voltage-dependence to the homologous OmpF. EMBO J. 1998;17:93–100. doi: 10.1093/emboj/17.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hille B. Ionic channels of excitable membranes. 3rd ed. Sunderland, Mass.: Sinauer Associates; 2001. p. 607. p. xiii. [Google Scholar]
- 27.Varma S, Sabo D, Rempe SB. K+/Na+ selectivity in K channels and valinomycin: over-coordination versus cavity-size constraints. J. Mol. Biol. 2008;376:13–22. doi: 10.1016/j.jmb.2007.11.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.(a) Manno S, Takakuwa Y, Mohandas N. Identification of a functional role for lipid asymmetry in biological membranes: Phosphatidylserine-skeletal protein interactions modulate membrane stability. Proc. Natl. Acad. Sci. U.S.A. 2002;99:1943–1948. doi: 10.1073/pnas.042688399. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Balasubramanian K, Schroit AJ. Aminophospholipid asymmetry: A matter of life and death. Annu. Rev. Physiol. 2003;65:701–734. doi: 10.1146/annurev.physiol.65.092101.142459. [DOI] [PubMed] [Google Scholar]
- 29.Roark M, Feller SE. Structure and dynamics of a fluid phase bilayer on a solid support as observed by a molecular dynamics computer simulation. Langmuir. 2008;24:12469–12473. doi: 10.1021/la802079h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoopes MI, Deserno M, Longo ML, Faller R. Coarse-grained modeling of interactions of lipid bilayers with supports. J Chem. Phys. 2008;129:175102. doi: 10.1063/1.3008060. [DOI] [PubMed] [Google Scholar]
- 31.Filippov A, Oradd G, Lindblom G. The effect of cholesterol on the lateral diffusion of phospholipids in oriented bilayers. Biophys. J. 2003;84:3079–3086. doi: 10.1016/S0006-3495(03)70033-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.(a) Sanii B, Smith AM, Butti R, Brozell AM, Parikh AN. Bending membranes on demand: fluid phospholipid bilayers on topographically deformable substrates. Nano. Lett. 2008;8:866–871. doi: 10.1021/nl073085b. [DOI] [PubMed] [Google Scholar]; (b) Oliver AE, Kendall EL, Howland MC, Sanii B, Shreve AP, Parikh AN. Protecting, patterning, and scaffolding supported lipid membranes using carbohydrate glasses. Lab Chip. 2008;8:892–897. doi: 10.1039/b800370j. [DOI] [PubMed] [Google Scholar]; (c) Geissler M, Xia YN. Patterning: Principles and some new developments. Adv. Mater. 2004;16:1249–1269. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.