Abstract
The type VI secretion system (T6SS) primarily functions to mediate antagonistic interactions between contacting bacterial cells, but also mediates interactions with eukaryotic hosts. This molecular machine secretes antibacterial effector proteins by undergoing cycles of extension and contraction; however, how effectors are loaded into the T6SS and subsequently delivered to target bacteria remains poorly understood. Here, using electron cryo-microscopy, we analyzed structures of the Pseudomonas aeruginosa effector Tse6 loaded onto the T6SS spike protein VgrG1 in solution and embedded in lipid nanodiscs. Tse6 stability in the absence of membranes requires the chaperone EagT6, two dimers of which interact with the hydrophobic transmembrane domains of Tse6. EagT6 is not directly involved in Tse6 delivery but rather is crucial for its loading onto VgrG1. VgrG1-loaded Tse6 spontaneously enters membranes and its toxin domain translocates across a lipid bilayer indicating that effector delivery by the T6SS does not require puncturing of the target cell inner membrane by VgrG1. Eag chaperone family members from diverse Proteobacteria are often encoded next to putative toxins with predicted transmembrane domains and consequently, we anticipate our findings will be generalizable to numerous T6SS-exported membrane-associated effectors.
INTRODUCTION
Bacteria secrete toxins to compete with other organisms found in their environmental niche. In many species of Gram-negative bacteria, toxic effector proteins are directly delivered into competitor bacteria by a protein secretion apparatus known as the type VI secretion system (T6SS)1–3. The T6SS functions to puncture the outer membrane of adjacent Gram-negative bacteria and release antibacterial effector proteins4.
The T6SS is comprised of a cell envelope-spanning membrane complex and a cytoplasmic assembly that shares structural similarity to the baseplate, sheath, tube and spike components of contractile bacteriophage5,6. The tube component of this assembly is formed by stacks of the hexameric protein Hcp, which is enclosed by a sheath complex that upon T6SS activation is believed to mechanically contract and propel the inner tube outwards from the bacterial cell7,8. An arrowhead-like tip on the Hcp nanotube is formed by a complex between a trimeric protein called VgrG and a proline-alanine-alanine-arginine (PAAR) domain-containing protein. This spike complex likely mediates the first point of contact of the T6SS with the recipient cell9. There is compelling genetic and biochemical evidence indicating that effectors are delivered to the periplasm, whereupon a subset then translocate to the cytoplasm4. Intoxication of effector-producing cells and adjacent sister cells does not occur because each effector is co-expressed with a cognate immunity protein that neutralizes the activity of its associated effector through a direct binding mechanism10.
Likely owing to their differing sizes, effectors have been shown to transit the T6SS via two mechanisms11. Smaller effectors (<40 kDa) bind the lumen of Hcp hexamers and are stabilized by this interaction whereas large, multi-domain effectors bind VgrG12–14. In recent work, genes encoding chaperone proteins have been shown to be required for the association of a subset of effectors with VgrG with three chaperone domain superfamilies having been implicated in this process15,16. One of these families, designated DUF1795, includes EagT6 of Pseudomonas aeruginosa and EagR1 of Serratia marcescens. The precise function of this chaperone family is not known; however, EagT6 and EagR1 are required for the intracellular stability of the PAAR domain-containing effectors, Tse6 (also known as Tne1) and RhsA, respectively17–19. Furthermore, both Eag proteins have been shown to directly bind the N-terminal region of their cognate effectors18,19. Tse6 is unique in that it also requires interaction with Elongation Factor Tu (EF-Tu) for its delivery into recipient bacteria, though the precise role this house-keeping protein plays in the intoxication process is not known19.
Molecular insights into the various T6SS apparatus subcomplexes have been obtained by single particle cryo-EM studies, including the membrane complex20 and the sheath complex in extended and contracted states21,22. In addition, X-ray crystal structures of numerous effector proteins23 as well as secreted structural components such as VgrG have been determined24. However, structures of VgrG in complex with effector-chaperone complexes, which are needed to understand chaperone function, have yet to be determined.
Here, we present the cryo-EM structure of the T6SS effector-chaperone pair Tse6-EagT6 in complex with VgrG1. The structure reveals that two homodimers of the EagT6 chaperone shield the hydrophobic transmembrane domains (TMDs) of the effector Tse6. In biochemical experiments, we show that both interactions are crucial for the specific loading of Tse6 onto VgrG1 and intoxication of target cells. Furthermore, by reconstituting the Tse6-loaded VgrG1 complex into liposomes and nanodiscs we show that Tse6 is able to spontaneously enter membranes and translocate its toxin domain across a lipid bilayer.
RESULTS
Architecture of a “pre-firing” effector complex
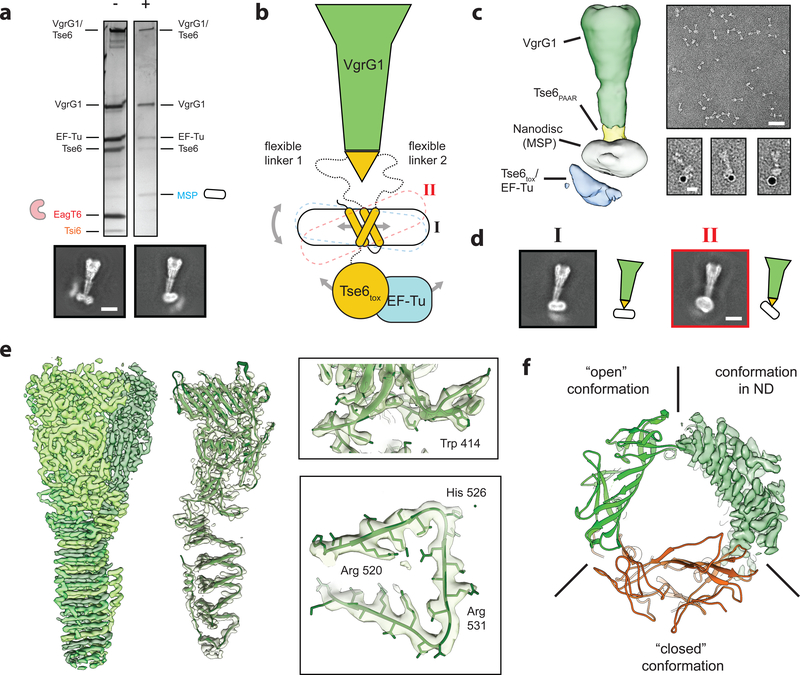
We previously demonstrated that Tse6 is a T6SS-exported effector protein that requires EagT6 for intercellular toxicity19. Furthermore, we determined a low-resolution negative stain EM structure of a complex containing VgrG1-Tse6-EagT6EF-Tu and the Tse6-specific immunity protein Tsi6, which provided initial insights into how these proteins interact19. However, the resolution of this structure limited our understanding of the role EagT6 plays within this assembly.
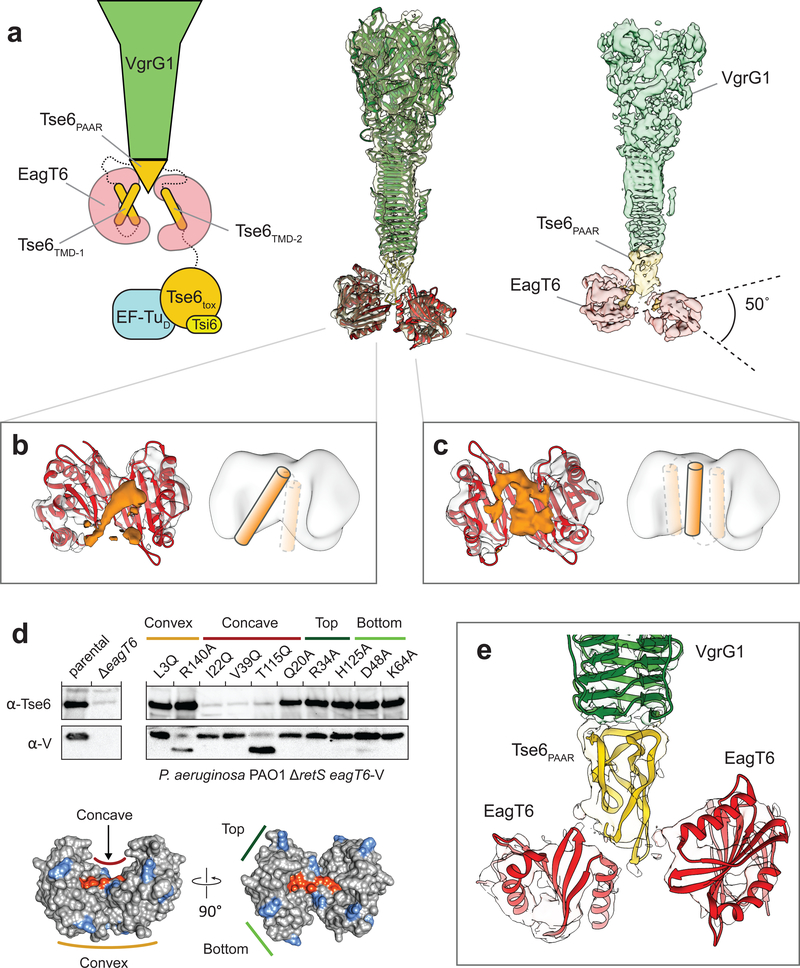
To better understand EagT6 function, we analyzed the structure of the “pre-firing” VgrG1-Tse6-EagT6-EF-Tu-Tsi6 complex (PFC) by cryo-EM and single particle analysis25. The overall resolution of the reconstruction was 4.2 Å and as described below, comprises VgrG1, the N-terminal PAAR and transmembrane domains of Tse6 (hereafter referred to as Tse6PAAR and Tse6TMDs, respectively) and two homodimers of EagT6 (Fig. 1a, Supplementary Fig. 1a–d,2,3,4, Supplementary Table 1). The C-terminal toxin domain of Tse6 (Tse6tox), EF-Tu and Tsi6 could not be resolved indicating that this subcomplex is highly flexible. The clear separation of the β-sheets in the C-terminal triple-stranded β-helix of VgrG1 allowed us to build a refined model of VgrG1 using the deposited crystal structure (pdb:4MTK) as a template (Supplementary Table 1). We assigned the cone-shaped density below VgrG1 to the tapering Tse6PAAR (yellow), which is known to bind to VgrG19. Since the resolution in this region is not sufficient to build an atomic model de novo, we calculated a homology model of Tse6PAAR and fitted it into the density using rigid-body fitting26. Flanking Tse6PAAR at the bottom are two striking horseshoe-shaped densities, the form of which identifies them clearly as two EagT6 dimers. The previously obtained atomic model of EagT6 (pdb:1TU1) fit optimally into these densities. The EagT6 dimers are located directly opposite each other, with the concave openings facing to the center. Interestingly, one of the EagT6 dimers is rotated by ~50°, resulting in different interfaces with Tse6PAAR (Fig. 1e).
Figure 1. Structure of the “pre-firing” VgrG1-Tse6PAAR-Tse6TMD1,2-EagT62 core complex.
(a) Schematic representation (left), fit of available atomic structures into cryo-EM density map (middle) and color-coded segmented cryo-EM density map (right) of the VgrG1-Tse6-EagT62-EF-Tu-Tsi6 complex. Tse6tox, EF-Tu and Tsi6 are not well resolved due to flexibility; see also Fig. 3. The density corresponding to VgrG1 is shown at a higher threshold for visualization. The exact number of transmembrane helices in each TMD is not known. Predictions give a range from one to three helices (see also Supplementary Fig. 5). Illustrated in the schematic representation are the two most likely arrangements for TMD-2 having either one or three (inset) TMHs.
(b,c) Side view on the cavity of the two opposing EagT6 dimers (atomic model in red and density in gray). The additional densities (orange) inside the cavity have a tubular appearance and probably correspond to the transmembrane helices of TMD-1 and TMD-2 of Tse6. In one of the cavities, the enclosed density is less defined indicating a higher degree of flexibility and/or more than one transmembrane helix (c). For corresponding top views, see Supplementary Fig. 1d. Cartoon representations indicate the number of putative transmembrane helices.
(d) Single point mutations at the surface of EagT6 identifying residues interacting with Tse6. Western blot analysis of Tse6 and EagT6 levels in the indicated P. aeruginosa strain (upper panel). Different point mutations were introduced in chromosomally VSV-G (V)-tagged EagT6. The lower panel shows the effect of mutated residues mapped on the surface of EagT6: orange - interacting with Tse6, blue - no effect.
(e) Close-up view on the interfaces between the two EagT6 dimers and Tse6PAAR. See also Supplementary Figures 1–5 and Supplementary Tables 1–3.
Many nonpolar residues are located on the concave surface of EagT6, creating a hydrophobic cavity suited to accommodate the two predicted TMD-containing regions of Tse6 (TMD-1 and TMD-2). The exact number of transmembrane helices in each of the TMDs is not known; however, membrane topology servers consistently predict two transmembrane helices for TMD-1 whereas the predictions for TMD-2 vary from one to three (Supplementary Fig. 5). Consistent with a direct interaction between the TMDs of Tse6 with EagT6, we observed additional density in the concave cavity of both EagT6 dimers (Fig. 1a–c). For one of the EagT6 dimers, a distinct tubular density suggests the binding of a transmembrane helix (Fig. 1b). The density enclosed by the second EagT6 dimer is less well defined, suggesting either a higher degree of flexibility and possibly more than one transmembrane helix (Fig. 1c).
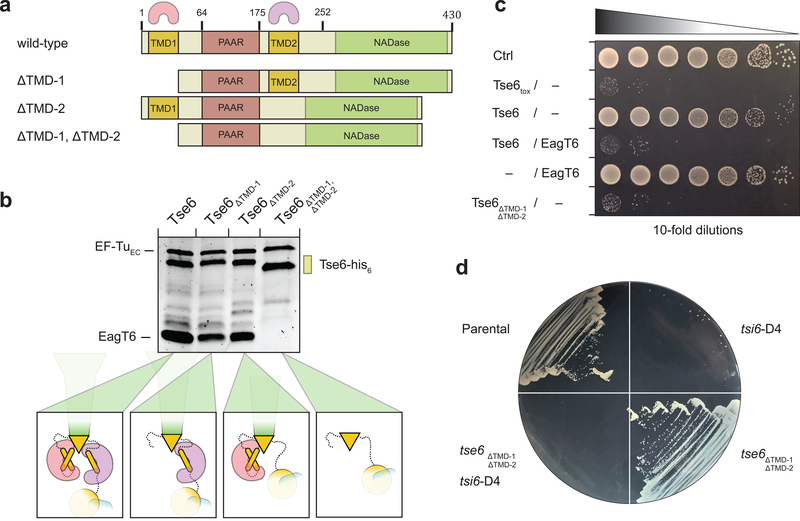
To test the proposed interaction between the TMDs of Tse6 and EagT6, we created TMD-deletion mutants in which each individual TMD region (Tse6ΔTMD-1, Tse6ΔTMD-2) or both TMD regions (Tse6ΔTMD-1, ΔTMD-2) were deleted and performed pull-down assays with EagT6 (Fig. 2a,b). In these experiments, Tse6 co-purified with EagT6 when either of the individual TMD regions were removed but not when both were absent (Fig. 2b). Taken together with our cryo-EM structure, these data demonstrate that each TMD likely interacts with a homodimer of EagT6. We next examined the consequences of removing the TMDs of Tse6 in vivo. Previously, we showed that wild-type Tse6 requires interaction with EagT6 for stability in vivo, perhaps because exposed hydrophobic TMDs result in unstable protein19. However in contrast to wild-type Tse6, we found that Tse6ΔTMD-1, ΔTMD-2 is highly stable in the absence of EagT6 (Supplementary Fig. 6a). To rule out the possibility that deleting the TMDs of Tse6 results in aggregation of misfolded protein, we next assessed the ability of Tse6ΔTMD-1, ΔTMD-2 to inhibit bacterial growth. We found that full-length Tse6 requires co-expression with EagT6 to inhibit the growth of Escherichia coli, presumably due to the stability imparted on Tse6 by EagT6. In contrast, Tse6ΔTMD-1, ΔTMD-2 is highly toxic in the absence of EagT6 (Fig. 2c). Self-intoxication assays in P. aeruginosa with IPTG-induced depletion of Tsi6 produced similar results (Fig. 2d). Together, these data indicate that the TMDs of Tse6 impart instability on the protein, which is prevented through interaction with EagT6.
Figure 2. EagT6 interacts with both TMDs of Tse6.
(a) Primary domain structure of Tse6 and corresponding TMD-deletion mutants. The two EagT6 dimers are indicated by pink and purple cones to highlight the interaction sites on Tse6. TMD-deletion mutants lack either one TMD (Tse6ΔTMD-1 with Δ1–61 and Tse6ΔTMD-2 with Δ180–222) or both TMDs (Tse6 ΔTMD-1, ΔTMD-2 with Δ1–61 and Δ180–222).
(b) Pull-downs of EagT6 and EF-Tu by Tse6wt and TMD-deletion mutants. If one or both of the TMDs are present EagT6 and EF-Tu are pulled down. Absence of both TMDs abolishes EagT6 but not EF-Tu binding.
(c) In E. coli toxicity assays, Tse6ΔTMD-1,ΔTMD-2 shows similar toxicity compared to Tse6toxin and the Tse6/EagT6 complex.
(d) IPTG-inducible depletion of Tsi6-D4 in P. aeruginosa shows that Tse6ΔTMD-1,ΔTMD-2 is as toxic as Tse6wt.
See also Supplementary Figure 6,7 and Supplementary Tables 2 and 3.
The structure of the EagT6 homodimer suggests that a concave cavity and its hydrophobic nature likely mediate its interaction with the TMD regions of Tse6. To test this, we introduced mutations at the eagT6 locus of P. aeruginosa that encode site-specific variants of the EagT6 protein and examined whether mutated EagT6 is still able to stabilize Tse6 in vivo (Fig. 1d). Indeed, substitutions of hydrophobic residues in the concave cavity reduced Tse6 stability whereas mutations in other regions of the EagT6 dimer had no effect (Fig. 1d). For further validation, we tested the ability of each individual TMD fragment of Tse6 to pull down EagT6 (Supplementary Fig. 7). In these experiments, both the TMD-1 and the TMD-2 fragments of Tse6 pulled down EagT6. Furthermore, one of our hydrophobic substitution mutants (V39Q) located within the concave cavity of EagT6, substantially reduced these interactions. Taken together, we conclude that one EagT6 dimer interacts with each of the TMD regions of Tse6 and that these interactions occur via the concave surface of EagT6.
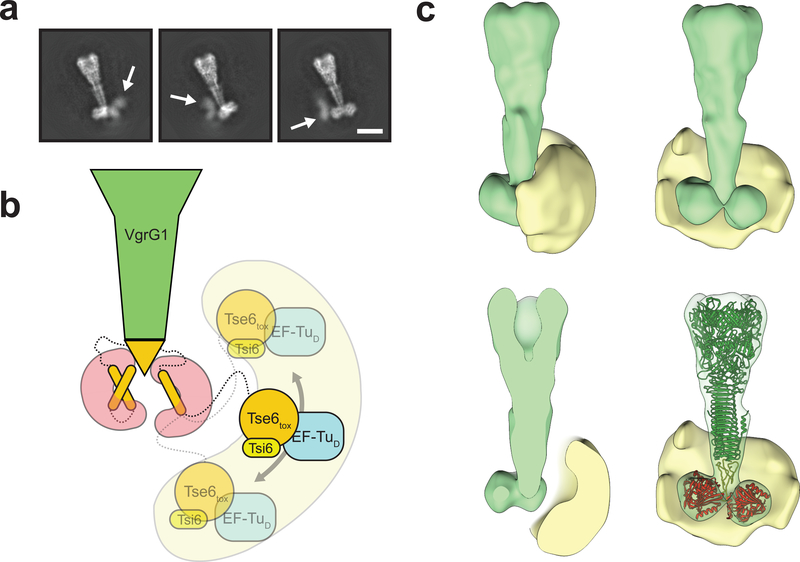
2-D class averages of the PFC indicated that part of the complex is highly flexible, only appearing as diffuse density at its lower part (Fig. 3a, Supplementary Video 1). This density is not resolved in the 3-D reconstruction but based on our prior characterization of Tse6 interaction partners corresponds to the Tse6tox-EF-Tu-Tsi6 subcomplex19. A long linker connects the last resolved domain, Tse6TMD-2, with the first unresolved domain, Tse6tox. The linker likely acts as hinge that allows large movements, explaining the flexibility of the Tse6tox-EF-Tu-Tsi6 subcomplex. The broad range of localization of the subcomplex respective to the core complex becomes more apparent when calculating a 3-D variability map (Fig. 3b,c).
Figure 3. Tse6tox-EF-Tu-Tsi6 subcomplex is highly flexible.
(a) Representative 2-D class averages showing diffuse densities for the Tse6tox-EF-Tu-Tsi6 subcomplex. White arrows indicate flexible region (see also Supplementary Video 1). Scale bar: 10 nm.
(b) Schematic representation of flexibility of the Tse6tox-EF-Tu-Tsi6 subcomplex, which is connected to the more rigid VgrG1-Tse6NTD-EagT62 subcomplex by a linker.
(c) Rotated views and cross-sections of filtered 3-D average (green) and 3-D variability (yellow) densities corresponding to the VgrG1-Tse6-EagT62-EF-Tu-Tsi6 complex. The variability density indicates the range of positions taken by the Tse6tox-EF-Tu-Tsi6 subcomplex. Structures of VgrG1 (green), Tse6PAAR (yellow) and EagT6 (red) are fitted in 3-D average volume for orientation.
See also Supplementary Tables 2 and 3 and Supplementary Video 1.
Interaction with EagT6 is required for Tse6 loading onto VgrG1
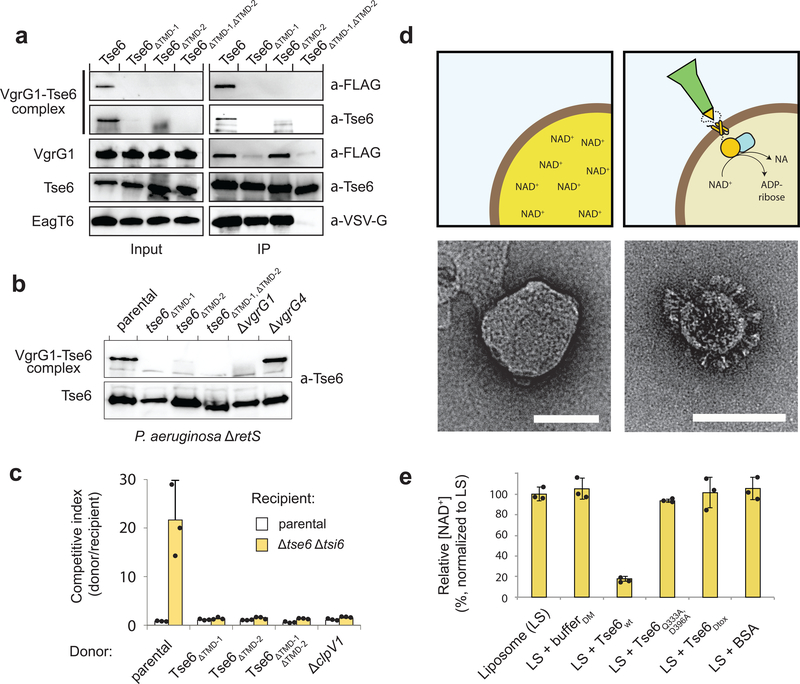
We next sought to determine if EagT6-TMD interactions play any additional role in Tse6 export beyond conferring stability to the effector. The close proximity of the EagT6 homodimers to Tse6PAAR led us to hypothesize that they might also facilitate loading of the effector onto VgrG1. To test this, we performed immunoprecipitation assays on E. coli cells co-expressing Tse6 or each of its TMD-deletion mutants with EagT6 and VgrG1. In these experiments, wild-type Tse6 formed a complex with VgrG1 that was resistant to both SDS and heat denaturation. However, deletion of either of the TMDs abrogated the formation of this highly stable complex (Fig. 4a). Mirroring our in vitro results, we found when expressed from the native tse6 locus in P. aeruginosa, Tse6 variants lacking either TMD region were unable to form SDS-resistant complexes with VgrG1 (Fig. 4b). Mutational inactivation of vgrG1 but not the related vgrG4 gene abrogated complex formation, indicating that the interaction with VgrG1 is specific. From these data, we conclude that the EagT6 interaction is essential for the formation of a highly stable Tse6-VgrG1 complex.
Figure 4. Crucial role of the TMDs of Tse6 in VgrG1 loading and target cell intoxication.
(a) Immunoprecipitation assay of individual and double TMD deletion mutants in E. coli. Only wild-type Tse6 is able to form the detergent-stable Tse6-VgrG1 complex. EagT6 is VSV-G-tagged, VgrG1 has a FLAG-tag.
(b) Western blot analysis of Tse6 levels in the indicated P. aeruginosa strains show that both TMDs are required for high-molecular VgrG1-Tse6 complex formation in vivo. Tse6 only interacts with VgrG1, but not VgrG4, highlighting the specificity of this interaction.
(c) Growth competition experiments between P. aeruginosa donor strains and a parental (Δ retS) or Tse6-susceptible (Δ tse6 Δ tsi6) recipient, showing that only wild-type Tse6 has a fitness advantage, while deletion of one or both TMDs is abolishing this effect. Data are means ± standard deviation from three independent biological measurements (n=3).
(d) Schematic illustration of NAD+-containing liposomes in the presence and absence of the Tse6-loaded VgrG1 complex with corresponding electron micrographs below. NAD+ is converted into ADP-ribose and nicotinamide (NA). Scale bar: 100 nm.
(e) Liposome-based in vitro translocation assay with relative NAD+ levels (normalized to LS), showing degradation of NAD+ after incorporation of the Tse6-loaded VgrG1 complex. DM-containing buffer (bufferDM) showed that the detergent does not cause leakage of liposomes. A catalytically inactive Tse6 mutant (Tse6Q333A,D396A) as well as a Tse6 mutant lacking its C-terminal toxin domain (Tse6Δtox) acted as additional control to exclude perforation of liposomes by the needle-like particles. Data are means ± standard deviation from three independent measurements (n=3).
See also Supplementary Figure 8 and Supplementary Tables 2 and 3.
Tse6 is exported by the H1-T6SS of P. aeruginosa. In addition to Tse6, this pathway is known to secrete a second PAAR-domain containing effector termed Tse511. Consistent with the recent demonstration that at least one PAAR-domain is required for a functional T6SS18, we found when the genes encoding these two H1T6SS delivered PAAR-containing proteins of P. aeruginosa were inactivated, cells were no longer able to secrete the H1-T6SS effector Tse1 (Supplementary Fig. 6b). The same effect was demonstrated when tse5 was deleted in a strain expressing tse6ΔTMD-1, ΔTMD-2 (Supplementary Fig. 6b). This result further highlights the necessity of the TMDs of Tse6 for PAAR-mediated VgrG1-Tse6 complex formation.
To determine whether the SDS-resistant Tse6-VgrG1 complex is necessary for the intoxication of Tse6-susceptible bacteria, we subjected the TMD-deletion mutants to bacterial competition assays (Fig. 4c). The parental strain had the anticipated fitness advantage over a Tse6-susceptible strain; however, strains expressing Tse6 lacking any of its transmembrane regions were unable to outcompete this recipient implying that the highly stable Tse6-VgrG1 complex is essential for intoxication of target cells.
Having established a role for EagT6 in loading Tse6 onto VgrG1, we next wanted to determine whether EagT6 is also involved in effector delivery into recipient bacteria. To test this, we performed secretion assays using P. aeruginosa and found that in contrast to Tse6, EagT6 is not secreted and thus is retained in the bacterium (Supplementary Fig. 6c). These findings are consistent with data from another recently characterized member of the Eag chaperone family from S. marcescens18. Thus EagT6 stabilizes both TMDs of Tse6, which are necessary for the loading of Tse6 onto VgrG1; however, it is not co-secreted with the Tse6-loaded VgrG1 complex. It is therefore crucial for effector loading but not delivery into target bacteria.
Tse6tox is translocated across membranes
The toxin domain of Tse6 acts in the cytoplasm of recipient bacteria where it hydrolyzes the essential dinucleotides NAD+ and NADP+. However, the molecular details of how this domain breaches the inner membrane to reach this cellular compartment are not known19. The presence of two TMD-containing regions within Tse6 suggests that it may be capable of entering membranes and self-translocating its enzymatic domain. To test this hypothesis, we devised a liposome-based assay in which we incubated NAD+-loaded liposomes with the VgrG1-Tse6-EF-Tu-Tsi6 complex (hereafter referred to as Tse6-loaded VgrG1 complex) and measured the levels of NAD+ inside the liposomes (Fig. 4d,e).
In comparison to control liposomes, a significant decrease in NAD+ concentration was observed in the presence of the Tse6-loaded VgrG1 complex, indicating the successful translocation of the Tse6tox domain into the interior of the liposomes (Fig. 4e). To ensure that the needle-shaped particles themselves do not puncture liposomes and cause leakage of NAD+, we also tested complexes containing a catalytically inactive variant of Tse6 (Tse6Q333D,D396A) or with Tse6 lacking its C-terminal toxin domain (Tse6Δtox). We observed no significant decrease in NAD+ levels when incubating the mutant complexes with liposomes, indicating that the integrity of liposomes is not compromised (Fig. 4e, Supplementary Fig. 8a). Negative stain EM of the liposomes clearly showed that the Tse6-loaded VgrG1 complexes decorated the liposomes with Tse6 entering the membrane (Fig. 4d). Nanogold labeling of the his6-tagged Tse6tox further corroborated its successful translocation over the membrane (Supplementary Fig. 8b). However, the complex containing Tse6Δtox did not readily enter liposomes, indicating that the complete Tse6 protein is needed for membrane penetration (Supplementary Fig. 8c). Taken together, these results demonstrate that Tse6 enters the membrane without the need of an additional receptor and translocates its toxin domain across the bilayer.
To obtain a high-resolution structure of this complex embedded in a lipid bilayer, we next exploited the self-insertion capability of the Tse6-loaded VgrG1 complex and reconstituted it in preformed nanodiscs, mimicking the membrane insertion process in situ. We validated the successful insertion of the complex into nanodiscs by negative stain electron microscopy (Supplementary Fig. 9). Either one or two of the complexes were embedded in one nanodisc. The two fractions could be separated by size-exclusion chromatography (Supplementary Fig. 9a) and the peak containing one complex per nanodisc was used for cryo-EM (Supplementary Fig. 1ei).
In agreement with our liposome assay, nanogold labeling of the his6-tagged Tse6tox domain demonstrates that this domain is located on the other side of the bilayer indicating that Tse6tox translocated through the membrane (Fig. 5a,c). Furthermore, we observed that upon reconstitution into membranes, Tsi6 was no longer associated with the Tse6-loaded VgrG1 complex after size exclusion chromatography, indicating that it dissociated from the complex upon its insertion in to the membrane (Fig. 5a).
Figure 5. The toxin domain of Tse6 spontaneously crosses a lipid bilayer.
(a) Silver stained SDS-PAGE of the Tse6-loaded VgrG1 complex in its “pre-firing” conformation (−) and reconstituted in lipid nanodiscs (+). Upon reconstitution, EagT6 and Tsi6 dissociate from the complex and are exchanged by the nanodisc. Scale bar: 10 nm.
(b) Schematic representation of the VgrG1-Tse6-EF-Tu complex reconstituted in lipid nanodiscs. Flexibility caused by (1) lateral movement of the TMDs within the nanodisc, (2) tilting of the nanodisc as well as (3) movement of the Tse6tox-EF-Tu subcomplex impeded structural determination of the bottom part.
(c) Low-resolution cryo-EM reconstruction of VgrG1-Tse6-EF-Tu complex embedded in lipid nanodiscs (left) and representative negatively stained electron micrograph areas of the complex (right). Scale bar: 50 nm. Right panel shows three examples of VgrG1-Tse6-EF-Tu complexes in nanodiscs, labeled with 5 nm NTA-coated nanogold to label his-tagged Tse6. Scale bar: 10 nm.
(d) Two representative class averages of the VgrG1-Tse6-EF-Tu complex in side view (black I) and tilted view (red II), corresponding to the conformations shown in Fig. 5b. Scale bar: 10 nm.
(e) 3.2 Å cryo-EM reconstruction of VgrG1 obtained from the same dataset and applying C3 symmetry. Subunits of trimeric VgrG1 are colored in different green hues (left), fit of atomic model in single subunit (middle), as well as close-ups showing side chain densities (right) of the VgrG1 trimer.
(f) Comparison between atomic VgrG1 structures in ‘open’ and ‘closed’ conformations, showing that VgrG1 within the VgrG1-Tse6-EF-Tu complex in nanodiscs adopts an ‘open’ conformation.
See also Supplementary Figures 1–3 and 9 and Supplementary Tables 1–3 and Supplementary Video 2.
Using cryo-EM and single-particle analysis we determined the structure of the VgrG1-Tse6-EF-Tu complex embedded in lipid nanodiscs (Fig. 5b,c, Supplementary Fig. 1e–i,2,3). In this structure, density for VgrG1 and the nanodisc is clearly recognizable while additional density corresponding to Tse6tox and EF-Tu is also apparent (Fig. 5c). Although 2-D class averages clearly show the outlines of the nanodisc as well as secondary structure elements in VgrG1 (Fig. 5a, Supplementary Fig. 1e,2c), the resolution of this reconstruction was limited to ~ 11 Å (Supplementary Fig. 1g). Attempts to increase the resolution were impeded by several difficulties. First, the regions of Tse6 connecting its PAAR domain to its TMDs induce substantial flexibility, which allows the TMDs embedded in the nanodisc to wobble (Fig. 5b,d, Supplementary Fig. 1h). Second, lateral movements of the TMDs inside the nanodisc, as well as the dynamic nature of the Tse6tox-EF-Tu subcomplex (Fig. 5b, Supplementary Fig. 1h,i) further aggravated image processing.
Nevertheless, we were able to resolve the region of our complex containing VgrG1 to a final resolution of 3.2 Å by imposing C3 symmetry (Fig. 5e, Supplementary Video 2). The resulting atomic model was almost identical to that of VgrG1 prior to membrane insertion (Supplementary Table 1). Two conformations of VgrG1 have been observed in crystal structures and referred to as ‘open’ and ‘closed’ states24. In the ‘closed’ conformation, there is a large loop that protrudes into the bowl-shaped cavity of VgrG1, restricting its diameter. In this conformation, several C-terminal residues adopt a fold that is not favorable for PAAR interaction. When comparing our cryo-EM structure of VgrG1 before and after membrane insertion, it becomes evident that both structures are in the ‘open’ conformation (Fig. 5f). The obtained density map correlated even on the side-chain level with the crystal structure of VgrG1 in the ‘open’ conformation (Fig. 5f, Supplementary Table 1). We therefore propose that the ‘open’ conformation is the physiological relevant form of VgrG1.
Conserved N-terminal loading and translocation region in Proteobacteria
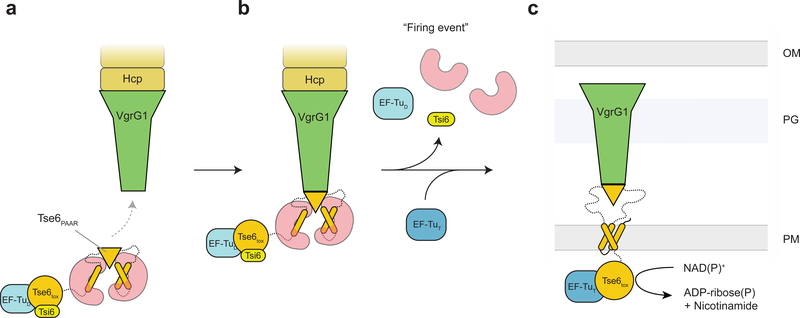
The PAAR domain and its flanking TMDs are comprised of the ~250 N-terminal amino acids of Tse6 (Supplementary Fig. 10a). Having established that the entirety of this region is required for Tse6 loading onto VgrG1 we next wanted to examine the conservation of this domain arrangement within Proteobacteria (Supplementary Fig. 10a,11). Interestingly, we found this N-terminal domain arrangement to be present in many predicted T6SS effectors from diverse Gram-negative bacteria (Supplementary Fig. 10a). Furthermore, the majority of these TMD-containing effectors also have predicted chaperone genes in their immediate genetic neighborhood (Supplementary Fig. 10b). Based on our findings presented herein, we designate the conserved region of these effectors the ‘N-terminal loading and translocation region’ (NLTR). In line with previously published informatic studies27, we found the C-terminus of NLTR containing proteins to be highly sequence divergent and are predicted to encode toxins with a variety of enzymatic activities (Supplementary Fig. 10a). Given the conservation of the NLTR among Proteobacteria (Supplementary Fig. 11), we believe that the mechanism of effector loading and translocation mediated through the NLTR is paradigmatic for all Eag-associated effectors. Our observations support a generalizable function of Eag proteins in the stabilization and loading of effectors onto their cognate VgrG proteins (Fig. 6).
Figure 6. Model for Tse6 effector loading and delivery.
(a) EagT6 (red) binds to the hydrophobic TMDs of Tse6 (orange). This prevents the protein from aggregating and ensures the correct folding of the PAAR domain. The PAAR domain specifically recognizes VgrG1 and mediates the loading of Tse6 onto VgrG1. EagT6 is therefore crucial for the efficient assembly of the Tse6-VgrG1 complex. The binding of EF-Tu (light blue) and Tsi6 (yellow) completes the T6S effector/chaperone complex. (b) Prior to firing EagT6, EF-Tu and Tsi6 dissociate from the complex activating the Tse6toxin. (c) The T6SS punctures the outer membrane of the target cell, forcefully bringing Tse6 into the periplasm. Tse6 spontaneously enters the inner membrane and translocates the Tse6toxin domain across the membrane. On the cytosolic side of the membrane, Tse6toxin binds to EF-Tu and acts as glycohydrolase depleting the cytosolic NAD(P)+ pool. OM – outer membrane, PG – peptidoglycan, IM – inner membrane, D – donor, T – target.
DISCUSSION
Our cryo-EM reconstruction of the PFC core, comprising VgrG1, Tse6PAAR, Tse6TMDs, and EagT62, represents the first structural characterization of a T6SS chaperone-effector pair loaded onto its cognate VgrG. Together with our biochemical data, we also uncover the role of EagT6 in Tse6-mediated interbacterial killing. We show that the hydrophobic TMDs of Tse6 are shielded from the hydrophilic environment of the cytoplasm by EagT6 in order to prevent aggregation and degradation of Tse6. Furthermore, we demonstrate that the association of EagT6 with the TMDs of Tse6 allows its PAAR domain to interact with VgrG1, which is then loaded into the T6SS apparatus.
The requirement of EagT6 for Tse6 recruitment to the T6SS shares some similarities to membrane protein export by the host-cell targeting bacterial type III secretion system (T3SS). The T3SS translocon is a pore formed at the tip of the T3SS apparatus that allows soluble effectors to access the host cell cytoplasm. The membrane protein that forms the translocon is itself a T3SS substrate and requires a cognate chaperone for solubility in the bacterial cytoplasm prior to its loading into the T3SS apparatus. Structural studies of the AcrH T3SS chaperone from Aeromonas hydrophila show that like EagT6, it directly binds and stabilizes the transmembrane domains of the AopB translocon prior to its export from the cell28.
We have also shown that Tse6 is capable of entering membranes and that its toxin domain is able to self-translocate across a lipid bilayer. Based on these observations, we propose that the T6SS delivers the Tse6-VgrG1 complex into the periplasm and spontaneous membrane insertion allows Tse6tox to translocate across the inner membrane into the cytoplasm where it exerts toxicity. Membrane translocation has recently been shown for the VgrG3 and TseL effectors of Vibrio cholerae29. Both effectors target periplasmic substrates. Interestingly, they exerted their toxic activity in the periplasm even though they were produced in the cytosol. The authors suggest that the effectors are translocated across the inner membrane by an intrinsic membrane-penetrating activity or by an as yet unknown active transport mechanism. TseL and VgrG3 are not predicted to contain TMDs; therefore, their mechanism of membrane translocation likely differs from that of Tse6.
The role of EF-Tu binding in the interbacterial delivery of Tse6 is still not understood. We previously proposed that EF-Tu present in the recipient cell might act as a molecular ratchet by preventing the toxin domain of Tse6 from translocating back into the periplasm once it enters the cytoplasm19. In our in vitro translocation assay, EF-Tu is only present at the outside but not at the inside of NAD+-loaded liposomes. Therefore, we can exclude that EF-Tu on the other side of the membrane is required for the translocation of the toxin domain of Tse6 across a lipid bilayer. Whether EF-Tu functions to anchor the toxin domain of Tse6 within the cytoplasm after its translocation across the inner membrane remains to be determined and will be the subject of future study.
The ability of Tse6 to translocate its toxin domain across membranes without the help of accessory factors is remarkable and poses the question of how this task is accomplished. The TMDs of Tse6 are predicted to possess up to five total transmembrane helices, which are likely unable to form a pore large enough to accommodate the 17 kDa toxin domain in its folded state. We therefore suggest a scenario in which the TMDs of Tse6 form a small translocation pore that allows unfolded Tse6tox to thread through and ultimately refold in the cytosol of the target cell. This unfolding might be induced by the interaction of the transmembrane helices with the inner membrane and translocation could be catalyzed by refolding in the cytoplasm. In line with this, we have shown that the toxin domain of Tse6 readily refolds in vitro19. This model shares similarity with the current model for membrane translocation by Corynebacterium diphtheriae diphtheria toxin. This toxin is capable of translocating its enzymatic A fragment through a pore created by two membrane-inserted α-helical hairpins belonging to the translocation domain of the B fragment30.
This study provides the first detailed insight into both the mechanism of effector loading onto VgrG proteins and the delivery of the Tse6 toxin across the membrane of the target cell. It not only enhances our knowledge about how the T6SS machinery functions, but also lays the foundation for a mechanistic understanding of the vital role of T6SS in interbacterial killing within bacterial communities. Ultimately, these insights could lead to the development of novel treatments for drug-resistant pathogens.
METHODS
Bacterial strains, plasmids and growth conditions
All P. aeruginosa strains generated were derived from the sequenced strain PAO1 (Supplementary Table 2)31. P. aeruginosa was grown in Luria-Bertani (LB) media at 37°C or 30°C supplemented with 30 μg/mL gentamicin, 25 μg/mL irgasan, 40 μg/mL X-gal, 5% (w/v) sucrose and 0.5 mM IPTG as required. In-frame deletions, chromosomal fusions and chromosomal point mutations were generated as previously described using the pEXG2 suicide vector1,32. pPSV38 was used for inducible protein expression in P. aeruginosa. E. coli strains required for this study included XL1-Blue for plasmid maintenance, SM10 for conjugal transfer of allelic exchange plasmids into P. aeruginosa, and BL21 Codon Plus (Novagen) for protein overexpression and toxicity experiments. E. coli strains were grown in media at 37°C supplemented with 50 μg/mL kanamycin, 150 μg/mL carbenicillin, 30 μg/mL chloramphenicol, 200 μg/mL trimethoprim, 0.1% (w/v) L-rhamnose and the indicated concentrations of IPTG as required. Plasmids used for inducible protein expression in E. coli were pETDuet-1, pSCrhaB2-CV, pPSV35-CV and pRSFDuet-1.
Protein expression and purification
Expression of the VgrG1-Tse6-EagT62-EF-Tu-Tsi6 complex was performed as described previously19. Briefly, 12 L of E. coli BL21 CodonPlus cells harboring pETDuet-1::tse6-his8::tsi6 and pRSFDuet-1::vgrG1::eagT6 were grown at 37°C to mid-log phase and protein expression was induced by the addition of 1mM IPTG. Following incubation at 30°C for an additional 5–6 h, cells were pelleted and lysed in either detergent-free buffer or buffer containing 0.2% β-D-decylmaltopyranoside (depending on the downstream application). VgrG1-Tse6-EagT62-EF-Tu-Tsi6 complexes were then further purified by Ni-NTA chromatography and size exclusion chromatography as described previously19. For the detergent-bound complex, a second purification step using a Superose 6 Increase 5/150 column was performed.
Reconstitution into nanodiscs
VgrG1-Tse6-EF-Tu-Tsi6 solubilized in detergent (β-D-decylmaltopyranoside) was mixed with preformed nanodiscs (Cube Biotech), containing MSP1D1-ΔH5 and POPC, in a molar ratio of 1:4 and dialyzed against ND-buffer (20 mM Tris-HCl, pH 8.0, 300 mM NaCl) for 96 h at RT. ND-buffer was exchanged against fresh one after 24 h. Subsequently, size-exclusion chromatography with a Superose 6 Increase 10/300 GL column (GE Healthcare) was used to separate aggregates and/or empty nanodiscs, as well as non-reconstituted complex.
Co-purification and co-immunoprecipitation assays
Full-length Tse6 and its truncated variants Tse6ΔTMD-1, Tse6ΔTMD-2 and Tse6ΔTMD-1, ΔTMD-2 were cloned into MCS-1 of pETDuet-1 using the NcoI/HindIII restriction sites and a 3’ primer that fuses a His8-tag to the C-terminus of the protein. To neutralize effector cytotoxicity during overexpression in E. coli, these Tse6 variants were cloned into pETDuet-1 already containing the tsi6 gene in MCS-2 as described previously19. The resulting plasmids contain C-terminal His8-tagged Tse6 variants and untagged Tsi6. To elucidate EagT6 binding sites on Tse6, the above pETDuet-1 plasmids were co-transformed with pPSV35-CV::eagT6 in E. coli BL21 (DE3) Codon Plus cells. Protein expression was performed using similar conditions as was used for expression of the VgrG1-Tse6-EagT62-EF-Tu-Tsi6 complex except that 100 mL culture volumes were used. Pelleted cells were resuspended in 4 mL of 50 mM Tris-HCl pH 8.0, 300 mM NaCl, 5mM imidazole prior to lysis by sonication. Cleared lysates were purified using Ni-NTA agarose (Qiagen) as per the instructions of the manufacturer. Presence or absence of an interaction between EagT6 and the Tse6 variants was assessed by SDS-PAGE followed by TGX staining (Bio-Rad). To assess VgrG1 binding, E. coli BL21 (DE3) cells were transformed with pETDuet-1::vgrG1-FLAG plasmid. Stationary phase overnight cultures were sub-inoculated in 300 mL of LB and protein expression was induced as described for the VgrG1-Tse6-EagT62-EF-Tu-Tsi6 complex. 75 mL of culture was co-pelleted with 75 mL of each of the BL21 (DE3) CodonPlus strains co-expressing EagT6 with either Tse6 or the various Tse6 truncations. Following cell lysis by sonication, 50 μL aliquots constituting the input fraction were mixed 1:1 with SDS-PAGE sample loading buffer and boiled for 10 min. The remainder of the lysate was incubated with Ni-NTA agarose beads for 1.5 hours, washed extensively with lysis buffer, and eluted using lysis buffer supplemented with 400 mM imidazole. Elution samples were prepared in a similar manner to the input fraction and all samples were analyzed by SDS-PAGE and western blotting.
Tsi6 depletion assays
A sequence encoding the C-terminal DAS+4 degradation tag was fused to the 3’ end of the native tsi6 locus in P. aeruginosa strains lacking the native sspB gene and expressing either wild-type Tse6 or Tse6ΔTMD-1, ΔTMD-2. An IPTG-inducible plasmid containing sspB was used to stimulate controlled degradation of Tsi6-DAS+4 (Tsi6D4). The SspB protein recognizes DAS+4 tagged proteins and delivers them to the ClpXP protease for degradation. Strains harboring this plasmid were streaked on LB agar supplemented with 500 μM IPTG.
Secretion assays
Overnight cultures of P. aeruginosa strains were used to inoculate 2 ml of LB at a ratio of 1:500. Cultures were grown at 37°C with shaking to mid-log phase, and cell and supernatant fractions were prepared as described previously3.
Western blotting
Western blot analyses of protein samples were performed as described previously3 using rabbit α-VSV-G (diluted 1:5000, Sigma), mouse α-FLAG (diluted 1:5000, Sigma) rabbit α-Tse1 (diluted 1:2000), rabbit α-Tse6 (diluted 1:3000) or mouse α-RNAP (diluted 1:5000, Sigma) and detected with α-rabbit or α-mouse horseradish peroxidase-conjugated secondary antibodies (diluted 1:5000, Sigma). Western blots were developed using chemiluminescent substrate (Clarity Max, Bio-Rad) and imaged with the ChemiDoc Imaging System (Bio-Rad).
Competition assays
Recipient P. aeruginosa strains contained lacZ insertion at the neutral phage attachment site (attB) to differentiate these from an unlabeled donor. Overnight cultures of donor and recipient strains were mixed in a 1:1 (v/v) ratio and diluted 1:2 (v/v) in LB. Starting ratios of donor and recipient were enumerated by plating on LB agar containing 40 μg/mL X-gal. Five microlitres of each competition mixture was then spotted in triplicate on a 0.2 μm nitrocellulose membrane overlaid on a 3% LB agar plate and incubated face up at 37°C for 18–20 h. Competitions were then harvested by resuspending cells in LB and enumerating cfu by plating on LB agar containing 40 μg/mL X-gal. The final donor/recipient cfu were normalized to the starting ratio of donor and recipient strains.
E. coli toxicity assays
Plasmids containing Tse6ΔTMD-1, ΔTMD-2, under non-inducing conditions were not tolerated. Therefore, SOE pcr was used to generate a variant (D396A) that we previously demonstrated reduces but does not abolish the toxic NADase activity of Tse619. To allow for pairwise comparison of toxicity levels between strains, this amino acid substitution was also introduced into the plasmids expressing Tse6 and Tse6tox (Supplementary Table 3). E. coli BL21 Codon Plus cells were co-transformed with pSCrhaB2-CV or pSCrhaB2-CV expressing full-length Tse6, Tse6tox or Tse6ΔTMD-1, ΔTMD-2 and pPSV35-CV or pPSV35-CV expressing EagT6. Overnight cultures of these cells were diluted 106 in 10-fold increments and each dilution was spotted onto LB agar plates containing 0.1% (w/v) L-rhamnose, 0.1mM IPTG and the appropriate antibiotics. Photographs were taken after overnight growth at 37°C.
Negative-stain electron microscopy
4 μL of sample at a concentration of 0.01 mg/mL was applied onto glow-discharged carbon-coated copper grids. After 60 s of incubation at RT, excess liquid was blotted away with Whatman No. 4 filter paper, followed by two wash steps with purification buffer and subsequently stained with 0.75 % (w/v) uranyl formate. Images were recorded manually with a JEOL JEM-1400 microscope, equipped with a LaB6 cathode and 4k x 4k CMOS detector F416 (TVIPS), operating at 120 kV.
Sample vitrification
3 μL of VgrG1-Tse6-EagT62-EF-Tu-Tsi6 complex at a concentration of 0.015 mg/mL was applied on freshly glow-discharged Quantifoil 2/1 (with 2 nm additional carbon layer) cryo-EM grids, automatically blotted and plunged in liquid ethane using a CryoPlunge3 (Gatan) at a humidity between 90 and 100 %. For the Tse6-loaded VgrG1 complex in nanodiscs, 3 μL of 0.02 mg/mL was applied on freshly glow-discharged QF 2/1 (2 nm additional carbon) grids, and subsequently blotted and plunged using a Vitrobot (FEI) at 100% humidity. To improve ice quality and distribution 0.01 % Tween-20 was added during this step. Grid quality was assessed before data collection using a JEOL JEM-1400 (see above) or JEOL JEM-3200FSC equipped with a FEG and an in-column energy filter, operating at 300 kV. Long-term storage of grids was done in liquid nitrogen.
Cryo EM and image processing
Two cryo-EM data sets for the PFC complex were collected on a Titan Krios electron microscope (FEI) equipped with a Cs-Corrector, operated at 300 kV acceleration voltage. Micrographs were recorded on a Falcon II direct electron detector (FEI) at a magnification of 122,807 (equals nominal magnification of 59,000), corresponding to a pixel size of 1.14 Å. 24 frames with 62.5 ms time intervals per frame were collected during each exposure, resulting in a total exposure time of 1.5 s and a total electron dose of 60 e-/Å2. Using the automated data collection software EPU (FEI) two datasets, with a defocus range of 1.7 – 4.2 μm, were automatically collected featuring 5822 and 5820 micrographs, respectively. The 24 frames were aligned and summed with the help of MotionCor2 (3 × 3 patches) 33,34. Furthermore, dose-weighted and unweighted full-dose images were calculated. Data processing was performed using the software package SPHIRE/EMAN225. Un-weighted full-dose images were used for defocus and astigmatism determination with CTER (SPHIRE). After merging both data sets and visual inspection of the integrated images, 8744 micrographs were selected for subsequent processing. A combination of manual and automated particle selection, as well as several rounds of 2D classification, yielded a number of 137,906 “clean” dose-weighted and drift-corrected particles that were extracted with a final window size of 360 × 360 pixel (Supplementary Fig. 2a,c). 2D classification was performed using the iterative stable alignment and clustering (ISAC) implemented in SPHIRE. The particle stack was subjected to sxmeridien (3D refinement in SPHIRE) with imposed C3 symmetry, resulting in a 3.6 Å map of the C3 symmetric VgrG1 (top) part of the complex, estimated by the ‘gold standard’ criterion of FSC = 0.143. The determined three-dimensional projection parameters for each particle were subsequently used to create a symmetrized particle stack. This new stack contained three copies of each original particle with projection parameters rotated by 120° along the (C3-) symmetry axis. The ensuing 3D classification resulted in three volumes that were rotated by 120° to each other (Supplementary Fig. 4). As anticipated, the three copies of the original particle were evenly distributed to these classes. However, given that classification procedures are not perfect in reality, we further confirmed that not more than one copy of the original particle is present in each class. Finally, one of the classes, containing 55,000 particles, was selected and subjected to a new local 3D refinement without imposing symmetry. This resulted in the 4.2 Å density map of the PFC, whereas the resolution of the EM density decreases towards the periphery of the map (Supplementary Fig. 1b). Global resolutions were calculated between two independently refined half maps at the 0.143 FSC criterion, local resolution calculated using sxlocres of SPHIRE. Final densities were filtered to estimated average resolution. To visualize local resolution gradients within the map, it was colored according to the local resolution in Chimera35. Graphical rendering of 3D average and variability was calculated using sx3dvariability of the SPHIRE software package and filtered for illustrative purposes. The final electron density map allowed for placement of crystal structures of EagT6 (PDB:1TU1) and the homology model of Tse6PAAR domain (Phyre2 web server)26 using the Rigid-body Fit-in-Map tool of Chimera. Rosetta was used to perform a relaxation of the known crystal structure of VgrG1 (PDB:4MTK) into the obtained cryo-EM density map36.
The cryo EM data set for the Tse6-loaded VgrG1 complex in nanodiscs was collected on the same Titan Krios microscope as described above, using a K2 direct electron detector (Gatan) with in-column energy filter and a magnification of 80,893 (nominal magnification of 59,000), yielding a pixel size of 1.1 Å. In total 100 frames (each 150 ms) were recorded, resulting in a total exposure time of 15 s and a total electron dose of 91 e-/Å2. 1873 micrographs were automatically collected using EPU software, with a defocus range of 0.5 to 3.2 μm and energy filter width of 15 eV. Frame alignment and summing was done with MotionCor2 (3 × 3 patches) and un-weighted full-dose-, dose-weighted full-dose- and low-dose- (15e- without 1st frame/25e- without 1st frame) images were generated. After visual inspection 1358 micrograph were selected for further processing and un-weighted full-dose images were used for CTF estimation with CTER. 114,879 particles were manually selected using EMAN2 boxer and extracted from dose-weighted full-dose images with a final box size of 360 × 360 pixel. 2D classification (ISAC) and visual sorting yielded a total number of 72,190 particles that were subjected to 3D refinement with imposed C3 symmetry (Supplementary Fig. 2b,c). The obtained final reconstruction had an average resolution of 3.33 Å. To further improve the resolution, the last iterations of the 3D refinement were performed in continuing mode with extracted particles of the 25e− without 1st frame-low-dose images, resulting in a 3.25 Å electron density map. Rosetta was subsequently used to perform a relaxation of the known crystal structure of VgrG1 (PDB:4MTK) into the cryo-EM density map. Local resolution was calculated and visualized as described above. The low-resolution map of the Tse6-loaded VgrG1 complex with an overall resolution of 10.7 Å was obtained by using only 2-D classes where the nanodisc was almost perpendicular to VgrG1 (in total 11,000 particles, C1 refinement) (Supplementary Fig. 2b,c).
Angular distribution plots for all structures as well as beautified 2D class averages were calculated using SPHIRE (Supplementary Fig. 2c,3).
Nanogold staining
For detection of His-tagged proteins, samples were incubated for 1 hour with a 5 nm Ni-NTA Nanogold solution (Nanoprobes) on ice at a 9 : 1 ratio (v/v) and subsequently negatively stained according to the procedure described above.
Fluorescence-based liposome assay
POPC liposomes encapsulating NAD were prepared by sonication and freeze-thaw cycles. POPC (Avanti Polar lipids) was added to LSNAD-assay buffer (20 mM Tris-HCl, pH=8.0, 150 mM NaCl, 50 mM NAD) to obtain a final concentration of 10 mg/mL. After 30 min sonication, followed by 4 cycles of freezing and thawing (−196°C and 40°C, respectively), liposomes were separated from non-encapsulated NAD via two rounds of PD midiTrap G-25 columns equilibrated with LS-assay buffer (20 mM Tris-HCl, pH=8.0, 150 mM NaCl) following the manufacturer’s instruction (only first 0.8 mL of 1.5mL total elution was used for subsequent steps to ensure absence of non-encapsulated NAD).
Liposome assays were performed in 96-well plate format. 20 μL of liposomes containing 50 mM NAD+ were mixed with 10 μL sample (Tse6wt: 0.7 mg/mL Tse6-complex in LS-assay buffer containing 0.03 % n-Decyl-β-D-Maltoside (DM, Anatrace), bufferDM: LS-assay buffer + 0.03 % DM, Tse6Q333A, D396A: 0.7 mg/mL catalytically inactive mutant complex in LS-assay buffer containing 0.03 % DM, Tse6Δtox: 0.7 mg/mL complex of Tse6 mutant lacking its C-terminal domain in LS-assay buffer containing 0.03 % DM, BSA: 0.5 mg/mL BSA in LS-assay buffer + 0.03 % DM) and filled up to final volume of 100 μL with LS-assay buffer. After 16 h of incubation at 37 °C (300 rpm), samples were heated for 15 min at 96 °C to stop the reaction and 15 μL 7.66 % (v/v) of Triton X-100 was added (final conc.: 1 % (v/v)) and incubated for 10 min to permeabilize liposomes and release NAD. After addition of 35 μL of 8.57 M NaOH (final conc.: 2 M) reactions were incubated for 30 min in the dark to allow for the development of the base-catalyzed fluorescent breakdown product of NAD. Subsequently, plates were analyzed using a POLARstar Omega plate reader with the following optical settings: Excitation wavelength of 355 nm and Emission of 400–10 nm. All reactions were measured in triplicates.
Bioinformatic tools
Multiple Sequence Alignments were carried out with the Clustal Omega server (EMBL-EBI, https://www.ebi.ac.uk/Tools/msa/clustalo/). Sequence logos were generated using WebLogo3 server (https://weblogo.berkeley.edu/logo.cgi). Vizualization of sequence conservation was done with Jalview37. For prediction of membrane topology the TOPCONS server38, as well as the TopGraph server were used39. Visualization, analysis and figure preparation was done with Chimera (UCSF)35.
Supplementary Material
Acknowledgements
We thank O. Hofnagel and D. Prumbaum for their valuable assistance in electron microscopy and C. Gatsogiannis, M. Stabrin, as well as T. Moriya for the lively exchange regarding image processing. We thank the SPHIRE developer team, in particular Pawel A. Penczek for developing the cryo-EM image processing software used in this study. D.Q. is a fellow of Fonds der Chemischen Industrie. This work was supported by the Max Planck Society (to S.R.), the European Council under the European Union’s Seventh Framework Programme (FP7/ 2007–2013) (grant no. 615984) (to S.R.), the National Institutes of Health (R01-AI080609) (to J.D.M.), the Howard Hughes Medical Institute (to J.D.M.), start-up funds from McMaster University (to J.C.W.), a project grant from the Canadian Institutes of Health Research (to J.C.W.) and equipment provided by the Michael DeGroote Institute for Infectious Disease Research (to J.C.W.).
Footnotes
Competing interests
The authors declare no competing financial interests.
Data availability
The cryo-EM density maps of the PFC and the VgrG1-Tse6-EF-Tu complex in nanodisc are deposited into the Electron Microscopy Data Bank with the accession numbers EMD-0135 and EMD-0136, respectively. The refined models of VgrG1 in the PFC and the VgrG1-Tse6-EF-Tu complex in nanodisc have the PDB entry IDs 6H3L and 6H3N, respectively. Relevant data and details of plasmids and strains are available from the corresponding author upon request.
Statistics and Reproducibility
Digital electron micrographs were recorded multiple times with similar results, and representative areas are shown in the figures (Fig. 4d, 5c, Supplementary Fig. 1a,e,2c, 8, 9). 2-D class averages were generated using the ‘Iterative Stable Alignment and Clustering’ (ISAC) algorithm implemented in SPHIRE40, that is based on reproducible K-means classification. It determines validated and homogenous subsets of images by repeatedly testing the stability and reproducibility of the calculated solution (Fig. 3a,5a,d, Supplementary Fig. 1a,e,2c). All gels as well as western blots were repeated at least twice with similar results (Fig. 1d,2b,4a,b,5a, Supplementary Fig. 5a,6,7,8a,12).
References
- 1.Mougous JD et al. A Virulence Locus of Pseudomonas aeruginosa Encodes a Protein Secretion Apparatus. Science 312, 1526–1530 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pukatzki S et al. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. PNAS 103, 1528–1533 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hood RD et al. A Type VI Secretion System of Pseudomonas aeruginosa Targets a Toxin to Bacteria. Cell Host & Microbe 7, 25–37 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Russell AB et al. Type VI secretion delivers bacteriolytic effectors to target cells. Nature 475, 343–347 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang YW, Rettberg LA, Ortega DR & Jensen GJ In vivo structures of an intact type VI secretion system revealed by electron cryotomography. EMBO reports e201744072 (2017). doi: 10.15252/embr.201744072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Silverman JM, Brunet YR, Cascales E & Mougous JD Structure and Regulation of the Type VI Secretion System. 10.1146/annurev-micro-121809-151619 66, 453–472 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lossi NS et al. The HsiB1C1 (TssB-TssC) complex of the Pseudomonas aeruginosa type VI secretion system forms a bacteriophage tail sheathlike structure. J. Biol. Chem. 288, 7536–7548 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Basler M, Pilhofer M, Henderson GP, Jensen GJ & Mekalanos JJ Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 483, 182–186 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shneider MM et al. PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature 500, 350–353 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li M et al. Structural Basis for Type VI Secretion Effector Recognition by a Cognate Immunity Protein. PLOS Pathogens 8, e1002613 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whitney JC et al. Genetically distinct pathways guide effector export through the type VI secretion system. Molecular Microbiology 92, 529–542 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Silverman JM et al. Haemolysin Coregulated Protein Is an Exported Receptor and Chaperone of Type VI Secretion Substrates. Molecular Cell 51, 584–593 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flaugnatti N et al. A phospholipase A1 antibacterial Type VI secretion effector interacts directly with the C‐terminal domain of the VgrG spike protein for delivery. Molecular Microbiology 99, 1099–1118 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Pukatzki S, Ma AT, Revel AT, Sturtevant D & Mekalanos JJ Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. PNAS 104, 15508–15513 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Unterweger D, Kostiuk B & Pukatzki S Adaptor Proteins of Type VI Secretion System Effectors. Trends in Microbiology 25, 8–10 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Bondage DD, Lin J-S, Ma L-S, Kuo C-H & Lai E-M VgrG C terminus confers the type VI effector transport specificity and is required for binding with PAAR and adaptor–effector complex. PNAS 113, E3931–E3940 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang JY, Bullen NP, Ahmad S & Whitney JC Diverse NADase effector families mediate interbacterial antagonism via the type VI secretion system. J. Biol. Chem. 293, 1504–1514 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cianfanelli FR et al. VgrG and PAAR Proteins Define Distinct Versions of a Functional Type VI Secretion System. PLOS Pathogens 12, e1005735 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whitney JC et al. An Interbacterial NAD(P)+ Glycohydrolase Toxin Requires Elongation Factor Tu for Delivery to Target Cells. Cell 163, 607–619 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Durand E et al. Biogenesis and structure of a type VI secretion membrane core complex. Nature 523, 555–560 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Ge P et al. Atomic structures of a bactericidal contractile nanotube in its pre- and postcontraction states. Nature Structural & Molecular Biology 22, 377–382 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kudryashev M et al. Structure of the Type VI Secretion System Contractile Sheath. Cell 160, 952–962 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang H, Gao Z-Q, Su X-D & Dong Y-H Crystal structure of type VI effector Tse1 from Pseudomonas aeruginosa. FEBS Letters 586, 3193–3199 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Spínola-Amilibia M et al. The structure of VgrG1 from Pseudomonas aeruginosa, the needle tip of the bacterial type VI secretion system. Acta Crystallogr Sect D Struct Biol 72, 22–33 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Moriya T et al. High-resolution Single Particle Analysis from Electron Cryo-microscopy Images Using SPHIRE. J Vis Exp e55448–e55448 (2017). doi: 10.3791/55448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kelley LA, Mezulis S, Yates CM, Wass MN & Sternberg MJE The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10, 845–858 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang D, de Souza RF, Anantharaman V, Iyer LM & Aravind L Polymorphic toxin systems: Comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biol. Direct 7, 18 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nguyen VS et al. Structure of AcrH-AopB Chaperone-Translocator Complex Reveals a Role for Membrane Hairpins in Type III Secretion System Translocon Assembly. Structure 23, 2022–2031 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Ho BT, Fu Y, Dong TG & Mekalanos JJ Vibrio cholerae type 6 secretion system effector trafficking in target bacterial cells. Proc. Natl. Acad. Sci. U.S.A. 114, 9427–9432 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murphy JR Mechanism of Diphtheria Toxin Catalytic Domain Delivery to the Eukaryotic Cell Cytosol and the Cellular Factors that Directly Participate in the Process. Toxins (Basel) 3, 294–308 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stover CK et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406, 959–964 (2000). [DOI] [PubMed] [Google Scholar]
- 32.Rietsch A, Vallet-Gely I, Dove SL & Mekalanos JJ ExsE, a secreted regulator of type III secretion genes in Pseudomonas aeruginosa. PNAS 102, 8006–8011 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng SQ et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nature Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang G et al. EMAN2: an extensible image processing suite for electron microscopy. Journal of Structural Biology 157, 38–46 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Pettersen EF et al. UCSF Chimera—A visualization system for exploratory research and analysis. Journal of Computational Chemistry 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Wang RY-R et al. Automated structure refinement of macromolecular assemblies from cryo-EM maps using Rosetta. eLife Sciences 5, 352 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Waterhouse AM, Procter JB, Martin DMA, Clamp M & Barton GJ Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsirigos KD, Peters C, Shu N, Käll L & Elofsson A The TOPCONS web server for consensus prediction of membrane protein topology and signal peptides. Nucleic Acids Res. 43, W401–7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Elazar A, Weinstein JJ, Prilusky J & Fleishman SJ Interplay between hydrophobicity and the positive-inside rule in determining membrane-protein topology. Proc. Natl. Acad. Sci. U.S.A 113, 10340–10345 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang Z, Fang J, Chittuluru J, Asturias FJ & Penczek PA Iterative Stable Alignment and Clustering of 2D Transmission Electron Microscope Images. Structure 20, 237–247 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.