Abstract
This study investigated the effects of dietary supplementation with alternative sources of α-linolenic acid on growth, the composition of rumen microbiota, and the interactions between rumen microbiota and long-chain fatty acid (FA) concentrations, in goat kids. Sixty 4-month-old castrated male Albas white cashmere kids (average BW 18.6 ± 0.1 kg) were randomly allocated among three dietary treatments: (i) basal diet without supplementation (Control), (ii) basal diet supplemented with linseed oil (LSO), (iii) basal diet supplemented with heated linseed grain (HLS). The concentrate:forage ratio was 5:5 and the LSO and HLS treatments provided the kids with similar dietary FA profiles. The diets were fed for 104 d, consisting of 14 d for adaptation followed by 90 d of experimental observation. Treatment did not significantly influence BW, DMI, or bacterial richness or diversity. On the other hand, the relative abundance of bacteria participating in hydrogenation differed significantly among the three groups: the Veillonellaceae and Christensenellaceae were more abundant in LSO kids, Prevotellaceae were more abundant in HLS kids, and the Fibrobacteriaceae were more abundant in Control kids (P < 0.05). Spearman correlation analysis indicated that Ruminobacter, Selenomonas_1, Fretibacterium, Prevotellaceae_UCG-001, Succinimonas, and Ruminococcaceae_NK4A214_group were the genera that participated in hydrogenation of long-chain FAs. HLS-fed kids had a lower relative abundance of Ruminobacter, but a higher abundance of Prevotellaceae_UCG-001 and Fretibacterium than LSO-fed kids. These changes were associated with greater rumen concentrations of C18:3n3 and n-3 PUFA, but lower concentrations of n-6 PUFA and lower n-6/n-3 ratios, in HLS than in LSO-fed kids. In conclusion, feeding kids with HLS increased rumen concentrations of C18:3n3 and n-3 PUFA, but decreased the n-6/n-3 ratio by decreasing the abundance of bacteria that hydrogenate C18:3n3 and increasing the abundance of bacteria that hydrogenate C18:2n6.
Keywords: α-linolenic, biohydrogenation, gut microbiota, lipid metabolism, ruminants
INTRODUCTION
There is an imbalance in the proportion of fatty acids (FAs) in the typical western diet, with a high ratio of n-6 poly-unsaturated FA to n-3 polyunsaturated FA (n-6/n-3 PUFA; Williams, 2000). Consumption of n-3 PUFA has been shown to positively influence immune function, blood pressure, cholesterol and triglyceride levels, and cardiovascular function in humans (Mozaffarian and Wu, 2012). An important source of n-3 PUFA is meat from ruminants. Our previous study has shown that supplementing cashmere kids with a rumen-protected source of α-linolenic acid (C18:3n3), in the form of heated linseed grain (HLS), were more effective in increasing C18:3n3 and n-3 PUFA, while decreasing the n-6/n-3 ratio, in subcutaneous adipose tissue than supplementing with an unprotected source (linseed oil, LSO; Wang et al., 2018). These outcomes suggested that the C18:3n3 in HLS escaped ruminal biohydrogenation, and therefore increased postruminal flow of C18:3n3 and increased its deposition in subcutaneous adipose tissue (Wang et al., 2018).
The ruminal microbiota plays a pivotal role in defining the FA composition of ruminant tissues, and therefore products, by influencing the rumen FA profile – for example, several C18:1 isomers, particularly C18:1 t10 and C18:1 t11, are produced in the rumen and then accumulate in milk and meat (Cremonesi et al., 2018). It has also been shown that the concentration of C18:3n3 in Longissimus thoracis and subcutaneous adipose tissue increases as rumen C18:3n3 concentration increases (Hu et al., 2008). These observations lead to the general hypothesis that the composition of rumen microbiota and the FA metabolites they produce will differ between kids fed HLS and kids fed LSO.
In the present study with cashmere goat kids, we tested whether rumen concentrations of C18:3n3 and n-3 PUFA are higher, and the n-6/n-3 ratio is lower, in kids fed HLS compared with those fed LSO. We also expected a lower abundance of bacteria that hydrogenate C18:3n3 in kids fed HLS than in kids fed LSO.
MATERIALS AND METHODS
Animals, Diets, and Feeding Management
This study was conducted at the Inner Mongolia White Cashmere Goat Breeding Farm, Wulan Town, Etuoke Banner, Ordos City, Inner Mongolia Autonomous Region, China (39°12 N, 107°97 E). The protocol was approved by the Animal Care and Use Committee of Inner Mongolia Agriculture University for the Care and Use of Animals for Experimental and Other Scientific Purposes. Sixty 4-month-old castrated male kids (average BW 18.6 ± 0.1 kg) were selected and randomly allocated among three groups in a randomized block design, with each group comprising four units of five kids. Three dietary treatments were used (Table 1): (i) basal diet without supplementation (Control), (ii) basal diet supplemented with LSO, and (iii) basal diet supplemented with HLS. The LSO and HLS treatments provided the kids with similar dietary FA profiles. The oil-supplemented diets were prepared by manually blending the oil thoroughly into the ground concentrate to ensure a homogenous distribution in the ration. HLS was prepared in a hot air roaster (10 min at 120 °C). Temperature was measured in the heated seeds with a needle thermometer immediately after treatment. After roasting, the linseed grain was air cooled and then stored at 4 °C until it was used; no antioxidant was added. The diets were prepared fresh twice a day and were offered as total mixed ration (concentrate:forage ratio of 5:5) in two equal meals at 0830 and 1630 hours. The kids were given free access to drinking water. The diets were fed for a total of 104 d, consisting of 14 d for adaptation followed by 90 d of measurement. The measurement period was divided into early (1 to 30 d), middle (31 to 60 d), and late periods (61 to 90 d) so levels of nutrition could be increased to meet the changing needs of the kids as they grew (Table 1). FA composition of diets was shown in Table 2. Feed intake was recorded daily for each group of five kids based on the amount of feed offered and refusals. All kids were weighed on days 30, 60, and 90 of the measurement period to determine changes in BW.
Table 1.
Composition and analysis of experimental diets (LSO = linseed oil; HLS = heated linseed)
| 1 to 30 d | 31 to 60 d | 61 to 90 d | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Items | Control | LSO | HLS | Control | LSO | HLS | Control | LSO | HLS |
| Ingredient, % air dry basis | |||||||||
| Alfalfa hay particles | 25 | 25 | 25 | 15 | 15 | 15 | 12.5 | 12.5 | 12.5 |
| Maize straw particles | 5 | 5 | 5 | 20 | 20 | 20 | 25 | 25 | 25 |
| Tall oat grass particles | 20 | 20 | 20 | 15 | 15 | 15 | 12.5 | 12.5 | 12.5 |
| Corn | 28.41 | 23.37 | 23.17 | 30.8 | 30.4 | 29.9 | 31.3 | 29.9 | 29.4 |
| Soybean meal | 11.7 | 10.5 | 11.5 | 9.5 | 11.4 | 11.9 | 8 | 10.4 | 10.9 |
| Distillers dried grains with solubles | 3 | 7.24 | 7.74 | 4 | 0.5 | 0.5 | 4 | 0.5 | 0.5 |
| Flax cake | 4.8 | 4.8 | 0 | 3.5 | 3.5 | 0 | 4.5 | 4.5 | 0 |
| Heated linseed | 0 | 0 | 5.5 | 0 | 0 | 5.5 | 0 | 0 | 7 |
| Linseed oil | 0 | 2 | 0 | 0 | 2 | 0 | 0 | 2.5 | 0 |
| Premix1 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 |
| Calcium carbonate | 0.2 | 0.2 | 0.2 | 0.2 | 0.2 | 0.2 | 0.2 | 0.2 | 0.2 |
| CaHPO4 | 0.2 | 0.2 | 0.2 | 0.2 | 0.2 | 0.2 | 0.2 | 0.2 | 0.2 |
| Sodium chloride | 0.54 | 0.54 | 0.54 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 |
| Sodium bicarbonate | 0.35 | 0.35 | 0.35 | 0.8 | 0.8 | 0.8 | 0.8 | 0.8 | 0.8 |
| Magnesia | 0.3 | 0.3 | 0.3 | 0 | 0 | 0 | 0 | 0 | 0 |
| Chemical composition | |||||||||
| Digestible energy, MJ/kg DM2 | 12.83 | 13.09 | 13.06 | 12.87 | 13 | 12.96 | 12.74 | 13.09 | 13.05 |
| Crude protein, g/kg DM | 188.73 | 188.13 | 188.2 | 162.84 | 158.71 | 159.69 | 153.52 | 151.34 | 151.85 |
| Ether extract, g/kg DM | 29.12 | 53.97 | 53.97 | 28.99 | 45.84 | 46.14 | 26.85 | 48.99 | 49.92 |
| Neutral detergent fibre, g/kg DM | 425.31 | 431.18 | 441.6 | 448.6 | 427.41 | 439.12 | 457.42 | 436.06 | 450.68 |
| Acid detergent fibre, g/kg DM | 232.2 | 231.71 | 243.3 | 242.59 | 235.23 | 248.4 | 247.69 | 240.32 | 256.96 |
| Calcium, g/kg DM | 11.25 | 11.11 | 11 | 10.48 | 10.89 | 10.78 | 10.26 | 10.67 | 10.56 |
| Phosphorus, g/kg DM | 4.65 | 4.67 | 4.78 | 4.5 | 4.44 | 4.56 | 4.31 | 4.22 | 4.33 |
1Provided per kilogram of premix: iron (Fe) 4 g, copper (Cu) 0.8 g, zinc (Zn) 5 g, manganese (Mn) 3 g, iodine (I) 30 mg, selenium (Se) 30 mg, cobalt (Co) 25 mg, vitamin A (VA) 600,000 IU, vitamin D (VD3) 250,000 IU, vitamin E (VE) 1,250 IU, vitamin K (VK3) 180 mg, vitamin B1 (VB1) 35 mg, vitamin B2 (VB2) 850 mg, vitamin B6 (VB6) 90 mg, nicotinic acid 2,200 mg, D-pantothenic acid 1,700 mg, vitamin B12 (VB12) 3 mg, biotin 14 m, folic acid 150 mg.
2Digestible energy is calculated based on the ingredients of the diet and their digestible energy content, not based on the actual dry matter intake.
Table 2.
Fatty acid composition of experimental diets
| Fatty acids (% of total fatty acid)1) | 1 to 30 d | 31 to 60 d | 61 to 90 d | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Control | LSO | HLS | Control | LSO | HLS | Control | LSO | HLS | |
| C16:0 | 16.36 | 10.97 | 12 | 16.25 | 9.27 | 9.99 | 15.83 | 8.49 | 9.16 |
| C18:0 | 3.07 | 3.18 | 3.15 | 3.05 | 3.1 | 3.04 | 3.02 | 3.1 | 3.04 |
| C18:1n9t | 0.47 | 0.39 | 0.26 | 0.46 | 0.39 | 0.24 | 0.44 | 0.37 | 0.21 |
| C18:1n9c | 13.47 | 15.18 | 16.95 | 15.35 | 14.31 | 15.96 | 15.76 | 14.54 | 16.44 |
| C18:2n6t | 0.55 | 0.26 | 0.29 | 0.5 | 0.23 | 0.26 | 0.48 | 0.19 | 0.22 |
| C18:2n6c | 31.54 | 20.55 | 22.39 | 33.89 | 23.95 | 26.11 | 34.06 | 22.65 | 24.84 |
| C18:3n3 | 17.25 | 35.69 | 35.15 | 13.13 | 35.02 | 34.93 | 13.23 | 37.4 | 37.33 |
| C20:5n3 | 0.73 | 0.35 | 0.4 | 0.77 | 0.36 | 0.4 | 0.77 | 0.31 | 0.35 |
| C22:6n3 | 0.76 | 0.36 | 0.4 | 0.91 | 0.42 | 0.46 | 0.96 | 0.38 | 0.42 |
| SFA | 28.29 | 20.3 | 20.18 | 28.26 | 18.52 | 17.95 | 27.68 | 17.38 | 16.52 |
| MUFA | 17.34 | 19.27 | 19.16 | 19.35 | 18.51 | 18.17 | 19.77 | 18.77 | 18.43 |
| PUFA | 54.33 | 60.44 | 60.67 | 52.36 | 62.98 | 63.91 | 52.5 | 63.87 | 65.08 |
| n-3 PUFA | 19.79 | 37.2 | 36.56 | 15.87 | 36.59 | 36.41 | 16 | 38.87 | 39.08 |
| n-6 PUFA | 34.55 | 23.23 | 24.1 | 36.49 | 26.38 | 27.5 | 36.49 | 25.01 | 26 |
| n-3 LCPUFA | 2.54 | 1.51 | 1.42 | 2.73 | 1.58 | 1.48 | 2.77 | 1.47 | 1.33 |
| n-6 LCPUFA | 1.95 | 1.89 | 1.14 | 1.7 | 1.74 | 0.93 | 1.6 | 1.71 | 0.78 |
| P/S | 1.92 | 2.98 | 3.01 | 1.85 | 3.4 | 3.56 | 1.9 | 3.67 | 3.94 |
Total fatty acids = saturated fatty acids (6:0 + 8:0 + 10:0 + 11:0 + 12:0 + 13:0 + 14:0 + 15:0 + 16:0 + 17:0 + 18:0 + 20:0 + 21:0 + 22:0 + 23:0 + 24:0) + monounsaturated fatty acids (14:1 + 15:1 + 16:1 + 17:1 + 18:1t9 + 18:1c9 + 20:1 + 22:1 + 24:1) + n-6 polyunsaturated fatty acids (18:2t6 + 18:2c6 + 18:3n6 + 20:2n-6 + 20:3n6 + 20:4n6 + 22:2n6) + n-3 polyunsaturated fatty acids (18:3n3 + 20:3n3 + 20:5n3 + 22:6n3)
Sampling and Slaughtering Procedures
Residuals were collected and weighed 30 min before each feeding at 0830 hours daily to estimate intake. Samples of total mixed ration were collected at the beginning of each period and stored at –20 °C for chemical analysis. At the end of the experiment, two kids from each unit were randomly selected and slaughtered by exsanguination. Before slaughter, the kids were prevented from consuming food for 24 h and from drinking for 2 h. The rumen was dissected and its contents were squeezed through two layers of cheesecloth; the resulting liquid was snap frozen in liquid N2 and stored at −80 °C until analysis.
Chemical Analysis
Analysis of feed.
Samples of dietary ingredients were analyzed for DM (method 930.15), CP (N × 6.25; method 984.13), ether extract (method 920.39), and calcium and phosphorous (method 935.13), according to AOAC (2000). NDF and ADF were determined according to the methods described by Van Soest et al. (1991) with an Ankom 220 Fiber Analyser (Ankom Co.) and were expressed inclusive of residual ash. Heat stable amylase was not used in the NDF determination.
Measurement of FA.
FA methyl esters were produced from samples of 0.5 g of feed or 1 mL rumen liquid according to the method of O’Fallon et al. (2007), and were analyzed as described previously (Wang et al., 2018).
Metagenomic Analyses
DNA extraction.
Microbial DNA was extracted from rumen samples of five of eight slaughtered kids in each group, using the E.Z.N.A. soil DNA Kit (Omega Biotek, Norcross, GA) according to the manufacturer’s protocols. Once extracted, the DNA was quantified using a BioTek Epoch Spectrophotometer System (Synergy H1, USA Biotec) to measure the absorbance at 260 nm, as well as the A260:A280 ratio. Integrity was evaluated using 1% agarose gel electrophoresis. DNA was diluted to 1 ng/μL using sterile water.
HiSeq sequencing and data analysis.
PCR was used to amplify the V4 region of the bacterial 16S rRNA gene using the universal primers 515F (5-GTGCCAGCMGCCGCGGTAA-3′) and 806R (5-GGACTACHVGGTWTCTAAT-3′; Evans et al., 2014) with the barcode. The forward primer contained 6 base barcode sequences. The reaction was carried out with Phusion High-Fidelity PCR Master Mix (New England Biolabs). The PCR reactions (30 μL) contained 10 μL DNA template, 15 μL of Phusion Master Mix (2×), 1.5 μL of each primer (total 6 μM) and 2 μL of dd H2O. The PCR was performed under the following conditions: 98 °C for 1 min, followed by 30 cycles of 98 °C for 10 s, 50 °C for 30 s, and 72 °C for 30 s, and a final elongation step of 72 °C for 5 min. The same volume of 1× loading buffer (contained SYB green) was mixed with the PCR products and the mixture was subjected to electrophoresis on 2% agarose gel. Samples with a bright main strip between 400 and 450 bp were chosen for further analysis. The PCR products were mixed in equimolar amounts and then purified with a Qiagen Gel Extraction Kit (Qiagen, Germany). Sequencing libraries were generated using TruSeq DNA PCR-Free Sample Preparation Kit (Illumina), following the manufacturer’s recommendations, and index codes were added. Library quality was assessed on the Qubit@ 2.0 Fluorometer (Thermo Scientific) and Agilent Bioanalyzer 2100 system. Finally, the library was sequenced on an Illumina HiSeq 2500 platform and 250 bp paired-end reads were generated.
The generated raw sequences were processed using FLASH and Trimmomatic to merge the paired-end sequences and remove low-quality reads with the following criteria: (i) The reads were truncated at any site receiving an average quality score <20 over a 50 bp sliding window; (ii) primers matching allowed 2-nucleotide mismatching, and reads containing ambiguous bases were removed; (iii) sequences with an overlap longer than 10 bp were merged according to their overlap sequence. Operational taxonomic units (OTUs) were clustered with a 97% similarity cut-off using UPARSE (version 7.1 http://drive5.com/uparse/) and chimeric sequences were identified and removed using UCHIME. The taxonomy of each 16S rRNA gene sequence was analyzed with the Ribosomal Database Project Classifier algorithm (http://rdp.cme.msu.edu/) against the Silva 128 16S rRNA database (Release128 http://www.arb-silva.de) using a confidence threshold of 70%. Bacterial diversity was measured using the QIIME pipeline based on the OTUs (Caporaso et al., 2010).
Taxonomic identification and comparisons were performed at the OTU levels of phylum, family, and species. To eliminate individual variation, so that all samples were compared at the same OTU sequence number, OTU abundance information was normalized using a standard of sequence number corresponding to the sample with the least sequences. Subsequent analysis of α diversity and β diversity were performed on this output-normalized data.
α diversities were calculated according to different microbial diversity metrics (i.e., observed species, Chao1, ACE, Shannon, Simpson, and Good’s coverage index) at a sequence depth of 55,466 and evaluated to determine whether the chosen subset was representative of the overall microbial diversity within each sample and whether there were any differences in terms of microbial diversity between the groups of animals subjected to different diet supplementation.
Statistical Analysis
Spearman correlation was used to correlate ruminal FA with the 20 most relatively abundant bacterial genera using R (pheatmap package). Only correlations with P < 0.05 for the linear model were considered as significant. Principal coordinate analysis (PCoA) was performed using the weighted Unifrac distance using R. Nonparametric multivariate analysis of variance (Adonis) was performed on the weighted Unifrac distances to assess the significance of differences in bacterial community structure among groups. Statistical significance was set at P < 0.05. The ternary plot was created with GGTERN. For all other statistical evaluations, ANOVA and post hoc Tukey honestly significance difference tests were carried out in SAS (S.A.S. Institute, 2002). Student’s t-test was used to analyze differences in diversity indexes. The results are presented as the mean and SEM. Data means were considered significantly different at P < 0.05.
RESULTS
Growth Performance
As shown in Table 3, there were no significant differences in DMI or BW among the treatments, at any stage during the experiment.
Table 3.
Effects of dietary oil supplements (LSO = linseed oil; HLS = heated linseed) on growth performance of cashmere kids
| Items | Control | LSO | HLS | SEM | P-value |
|---|---|---|---|---|---|
| BW, kg | |||||
| Day 30 | 19.8 | 18.5 | 18.4 | 0.59 | 0.353 |
| Day 60 | 22.1 | 21.0 | 19.8 | 0.75 | 0.345 |
| Day 90 | 26.7 | 28.0 | 26.6 | 0.54 | 0.330 |
| DMI, kg/d | |||||
| Days 1 to 30 | 0.64 | 0.63 | 0.63 | 0.04 | 0.994 |
| Days 31 to 60 | 0.76 | 0.79 | 0.67 | 0.06 | 0.344 |
| Days 61 to 90 | 1.28 | 1.16 | 1.20 | 0.08 | 0.436 |
For each value for BW, n = 20; for each DMI, n = 4.
FA Composition
Rumen FA composition is presented in Table 4. Compared with the Control and LSO treatments, the HLS treatment decreased the rumen concentrations of C6:0 (P < 0.0001), C16:0 (P < 0.0001), C21:0 (P = 0.006), C23:0 (P = 0.012), C24:0 (P = 0.010), C17:1 (P = 0.008), C20:1 (P = 0.018), C22:1 (P = 0.012), C20:4n6 (P = 0.006), C22:2n6 (P = 0.006), C20:5n3 (P = 0.008), C22:6n3 (P = 0.008), n-6 PUFA (P = 0.002), n-3 LCPUFA (P = 0.001), n-6 LCPUFA (P = 0.002), and decreased the n-6/n-3 ratio (P < 0.0001), but increased the content of total identified FA methyl esters (P = 0.003), the concentrations of C18:3n3 (P < 0.0001), and n-3 PUFA (P = 0.0004). The rumen concentrations of the FAs listed above did not differ (P > 0.05) between Control and LSO kids. On the other hand, the HLS supplement decreased the rumen concentrations of C8:0 (P = 0.022), C10:0 (P = 0.025), C11:0 (P = 0.021), C22:0 (P = 0.016), C16:1 (P = 0.038), C18:2n6c (P = 0.037), C20:2n6 (P = 0.017), and SFA (P = 0.016), and tended to decrease C24:1 concentration (P = 0.053) compared with Control. The HLS treatment increased the concentration of MUFA compared with Control. Values for LSO kids did not differ (P > 0.05) from those for either Control or HLS kids. The concentration of C18:1n9t was higher (P = 0.0002) in LSO and HLS kids than in Control kids, and did not differ between HLS and LSO kids (P > 0.05). The concentration of C18:1n9c was higher (P = 0.019) in Control than in LSO kids, but values for HLS kids did not differ (P > 0.05) from those for either Control or LSO kids.
Table 4.
Effects of dietary oil supplements (LSO = linseed oil; HLS = heated linseed) on rumen fatty acid profiles in cashmere goat kids. Values are percentages of total identified fatty acid methyl esters.
| Item | Control | LSO | HLS | SEM | P-value |
|---|---|---|---|---|---|
| Total identified fatty acids methyl esters (mg/mL rumen liquid) | 1.44b | 1.50b | 2.06a | 0.11 | 0.003 |
| Saturated fatty acids | |||||
| C6:0 | 2.19a | 2.07a | 1.47b | 0.07 | <.0001 |
| C8:0 | 1.74a | 1.61ab | 1.29b | 0.09 | 0.022 |
| C10:0 | 1.69a | 1.58ab | 1.27b | 0.09 | 0.025 |
| C11:0 | 0.87a | 0.83ab | 0.66b | 0.04 | 0.021 |
| C12:0 | 2.17 | 2.20 | 1.81 | 0.10 | 0.037 |
| C13:0 | 0.94 | 1.02 | 0.97 | 0.04 | 0.399 |
| C14:0 | 3.58 | 3.74 | 3.63 | 0.19 | 0.830 |
| C15:0 | 2.16 | 2.15 | 1.99 | 0.08 | 0.329 |
| C16:0 | 14.80a | 13.65a | 11.96b | 0.28 | <.0001 |
| C17:0 | 2.08 | 1.87 | 1.95 | 0.09 | 0.238 |
| C18:0 | 17.19 | 18.21 | 19.52 | 0.74 | 0.275 |
| C20:0 | 2.34 | 2.36 | 2.12 | 0.06 | 0.521 |
| C21:0 | 0.89a | 0.86a | 0.65b | 0.05 | 0.006 |
| C22:0 | 2.01a | 1.93ab | 1.62b | 0.09 | 0.016 |
| C23:0 | 0.88a | 0.85a | 0.65b | 0.05 | 0.012 |
| C24:0 | 2.07a | 1.97a | 1.62b | 0.09 | 0.010 |
| Monounsaturated fatty acids | |||||
| C14:1 | 1.65 | 1.62 | 1.48 | 0.08 | 0.361 |
| C15:1 | 0.69 | 0.74 | 0.66 | 0.03 | 0.235 |
| C16:1 | 1.96a | 1.79ab | 1.55b | 0.10 | 0.038 |
| C17:1 | 0.97a | 0.90a | 0.68b | 0.05 | 0.008 |
| C18:1n9t | 7.23b | 10.69a | 13.13a | 0.60 | 0.0002 |
| C18:1n9c | 10.97a | 9.35b | 10.55ab | 0.32 | 0.019 |
| C20:1 | 0.95a | 0.94a | 0.73b | 0.05 | 0.018 |
| C22:1 | 0.90a | 0.87a | 0.66b | 0.05 | 0.012 |
| C24:1 | 1.11a | 0.89ab | 0.66b | 0.11 | 0.053 |
| n-6 Polyunsaturated fatty acids | |||||
| C18:2n6t | 0.86 | 0.83 | 0.73 | 0.05 | 0.211 |
| C18:2n6c | 6.31a | 5.65ab | 4.94b | 0.29 | 0.037 |
| C18:3n6 | 0.87 | 0.85 | 0.71 | 0.05 | 0.058 |
| C20:2n6 | 0.86a | 0.82ab | 0.64b | 0.05 | 0.017 |
| C20:3n6 | 0.82 | 0.80 | 0.68 | 0.04 | 0.093 |
| C20:4n6 | 0.90a | 0.86a | 0.67b | 0.04 | 0.006 |
| C22:2n6 | 0.88a | 0.84a | 0.62b | 0.05 | 0.006 |
| n-3 Polyunsaturated fatty acids | |||||
| C18:3n3 | 1.69b | 2.15b | 5.68a | 0.25 | <.0001 |
| C20:3n3 | 0.99 | 0.81 | 0.60 | 0.11 | 0.065 |
| C20:5n3 | 0.89a | 0.82a | 0.64b | 0.05 | 0.008 |
| C22:6n3 | 0.93a | 0.90a | 0.66b | 0.05 | 0.008 |
| Sum and ratio1 | |||||
| SFA | 57.58a | 56.88ab | 53.31b | 0.83 | 0.016 |
| MUFA | 26.42b | 27.79ab | 30.11a | 0.68 | 0.010 |
| PUFA | 16.03 | 15.31 | 15.91 | 0.55 | 0.669 |
| n-3 PUFA | 4.22b | 4.68b | 5.85a | 0.18 | 0.0004 |
| n-6 PUFA | 9.99a | 10.63a | 7.77b | 0.42 | 0.002 |
| n-3 LCPUFA | 2.44a | 2.52a | 1.72b | 0.12 | 0.001 |
| n-6 LCPUFA | 3.19a | 3.31a | 2.35b | 0.15 | 0.002 |
| n-6/n-3 | 2.54a | 2.29a | 1.20b | 0.09 | <.0001 |
| P/S | 0.28 | 0.26 | 0.29 | 0.01 | 0.366 |
a–cMeans within the same row followed by the same superscript letters are not significantly different at P < 0.05. The number of observations for each mean value was eight (n = 8).
1SFA saturated fatty acids (6:0 + 8:0 + 10:0 + 11:0 + 12:0 + 13:0 + 14:0 + 15:0 + 16:0 + 17:0 + 18:0 + 20:0 + 21:0 + 22:0 + 23:0 + 24:0), MUFA monounsaturated fatty acids (14:1 + 15:1 + 16:1 + 17:1 + 18:1n9t + 18:1n9c + 20:1 + 22:1 + 24:1), n-6 PUFA n-6 polyunsaturated fatty acids (18:2n6t + 18:2n6c + 18:3n6 + 20:2n-6 + 20:3n6 + 20:4n6 + 22:2n6), n-3PUFA n-3 polyunsaturated fatty acids (18:3n3 + 20:3n3 + 20:5n3 + 22:6n3), PUFA polyunsaturated fatty acids (n-6 PUFA + n-3 PUFA), n-6LCPUFA n-6 long chain polyunsaturated fatty acids (20:2n6 + 20:3n6 + 20:4n6 + 22:2n6), n-3LCPUFA n-3 long chain polyunsaturated fatty acids (20:3n3 + 20:5n3 + 22:6n3); n-6/n-3 = n-6 long chain polyunsaturated fatty acids/n-3 long chain polyunsaturated fatty acids; P/S = polyunsaturated fatty acids/saturated fatty acids.
Bacterial Diversity
Sequencing coverage and bacterial diversity.
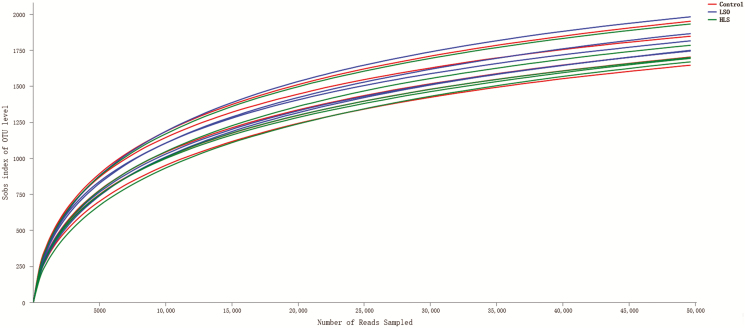
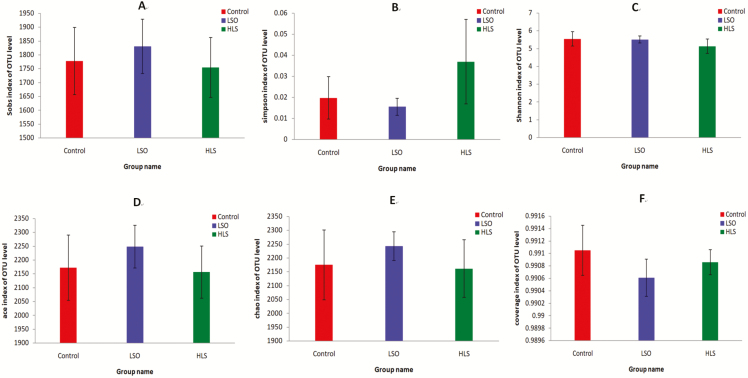
A total of 1,108,740 reads were generated after quality control and chimera removal, which resulted in an average of 73,916 reads per sample, with sequence numbers per sample ranging from 53,249 to 84,831 (median 74,216). A total of 466 unique OTUs that could be taxonomically classified to genus level were identified across all samples. The OTU rarefaction curves of the bacterial communities in the ruminal digesta show that the sampling effort was sufficient to estimate bacterial diversity (Figure 1). α diversity indexes (Figure 2) indicated that supplementation did not exert predominant effects on OTU number, ACE, Chao, Simpson, Shannon, and coverage indexes (P > 0.05).
Figure 1.
The OTU rarefaction curves of the ruminal digesta bacterial communities. Curves were drawn using the least sequenced sample as upper limit for the rarefactions. Each color represents one treatment: The red curves represent kids fed Control diet; the blue curves represent kids fed linseed oil (LSO) diet and the green curves represent kids fed heated linseed grain (HLS) diet.
Figure 2.
Student’s t-test for α diversity indices of ruminal bacteria in kids fed the linseed oil (LSO), heated linseed grain (HLS) and Control diets. (A) Observed species; (B) Simpson index; (C) Shannon index; (D) ACE index; (E) Chao index; (F) Good’s coverage index. For each value, n = 5. Error bars show standard deviation. Each color represents one treatment: The red represents kids fed Control diet; the blue represents kids fed LSO diet and the green represents kids fed HLS diet.
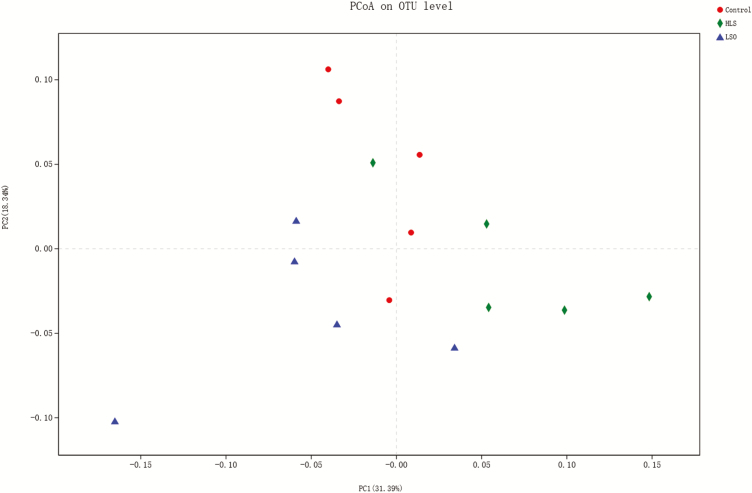
Principal coordinate analysis.
Using the weighted Unifrac similarity metric, the samples clustered according to dietary treatment (Figure 3). Adonis analysis revealed a significant difference (P = 0.005) among treatments in their rumen bacterial communities.
Figure 3.
Principal coordinate analysis (using the weighted Unifrac similarity metric) of bacterial operational taxonomic units (OTU) in the ruminal digesta of goat kids. Each symbol represents one treatment: the red symbols represent kids fed Control diet; the blue symbols represent kids fed linseed oil (LSO) diet and the green symbols represent kids fed heated linseed grain (HLS) diet.
Microbial composition analysis.
Collectively, 29 bacterial phyla, 209 families, 466 genera, and 867 species were identified in the rumen samples. Independent of diets, Bacteroidetes, Firmicutes, Proteobacteria, and Verrucomicrobia were the dominant phyla, as shown in Supplementary Figure 1, totaling about 91.05% on average relative abundance. The most abundant 20 families are listed in Table 5, representing 96% (Control), 89% (LSO), and 94% (HLS) of the total microbiome. The 20 most abundant species at family level are the exactly same among three groups with different abundance.
Table 5.
Effects of dietary oil supplements (LSO = linseed oil; HLS = heated linseed) on relative abundances of the 20 most abundant bacteria (family level) in the rumen of cashmere goat kids
| Phylum | Family | Control | LSO | HLS | SEM | P-values |
|---|---|---|---|---|---|---|
| Bacteroidetes | Prevotellaceae | 0.26b | 0.26b | 0.35a | 0.015 | 0.003 |
| Rikenellaceae | 0.08 | 0.05 | 0.05 | 0.008 | 0.065 | |
| Bacteroidales_BS11_gut_group | 0.09a | 0.04b | 0.04b | 0.01 | 0.007 | |
| Bacteroidales_S24-7_group | 0.03 | 0.03 | 0.02 | 0.005 | 0.321 | |
| Bacteroidales_RF16_group | 0.02 | 0.01 | 0.02 | 0.003 | 0.516 | |
| Unclassified family in Bacteroidetes | 0.02b | 0.01b | 0.05a | 0.006 | 0.002 | |
| Firmicute | Ruminococcaceae | 0.10 | 0.09 | 0.09 | 0.011 | 0.68 |
| Veillonellaceae | 0.03b | 0.11a | 0.04b | 0.008 | <.0001 | |
| Lachnospiraceae | 0.06 | 0.06 | 0.04 | 0.007 | 0.195 | |
| Acidaminococcaceae | 0.01 | 0.02 | 0.01 | 0.004 | 0.105 | |
| Lactobacillaceae | 0.004 | 0.001 | 0.004 | 0.001 | 0.057 | |
| Erysipelotrichaceae | 0.01 | 0.01 | 0.01 | 0.003 | 0.671 | |
| Christensenellaceae | 0.012ab | 0.013a | 0.007b | 0.001 | 0.035 | |
| unclassified_o__Clostridiales | 0.01 | 0.01 | 0.01 | 0.001 | 0.693 | |
| Proteobacteria | Succinivibrionaceae | 0.13 | 0.10 | 0.11 | 0.035 | 0.774 |
| Verrucomicrobia | norank_c__WCHB1-41 | 0.04 | 0.03 | 0.04 | 0.007 | 0.37 |
| Synergistetes | Synergistaceae | 0.01 | 0.02 | 0.01 | 0.003 | 0.641 |
| Fibrobacteres | Fibrobacteraceae | 0.02a | 0.009ab | 0.006b | 0.003 | 0.035 |
| Spirochaetae | Spirochaetaceae | 0.01 | 0.01 | 0.01 | 0.002 | 0.415 |
| Tenericutes | norank_o__Mollicutes_RF9 | 0.01 | 0.01 | 0.01 | 0.001 | 0.518 |
a–cMeans within the same row followed by the same superscript letters are not significantly different at P < 0.05. The number of observations for each mean value was five (n = 5).
Among the top 20 families, an unclassified family was clustered among the families belonging to the Bacteroidetes phylum, and it was labeled “unclassified_k_norank” by the phylogenetic analysis software (Supplementary Figure 2). In the Bacteroidetes, the relative abundances of this unclassified family (P = 0.002) and the Prevotellaceae (P = 0.003) were both greater in HLS kids than in Control or LSO kids, but did not differ (P > 0.05) between Control and LSO kids. Control kids had a greater relative abundance of the Bacteroidales_BS11_gut_group (P = 0.007) than LSO and HLS kids, but relative abundance of this family did not differ (P > 0.05) between HLS and LSO kids. The relative abundance of Veillonellaceae was greater (P < 0.0001) in LSO kids than in Control and HLS kids, but did not differ (P > 0.05) between HLS and Control kids. Compared with HLS kids, the relative abundance of Christensenellaceae (P = 0.035) was greater in LSO kids, but values for Control kids did not differ (P > 0.05) from those in either the LSO or HLS kids. Control kids had a greater (P = 0.035) relative abundance of Fibrobacteraceae than HLS kids, but values for LSO did not differ (P > 0.05) from that for either Control or HLS kids.
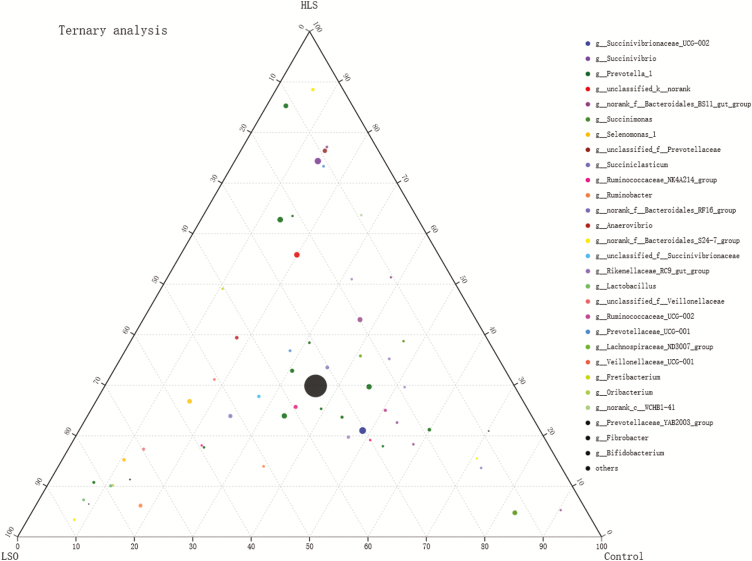
The center of the ternary plot (Figure 4) shows the core microbiome (high-density of circles) that is common to the Control, LSO, and HLS treatments. The OTUs uniquely associated with a specific treatment (where more than 70% of the total abundance of a particular OTU is uniquely associated with only one group) corresponded to the points within the corners of the ternary plot. Most strongly associated with the Control treatment were OTUs that belong to norank_f_Bacteroidales_BS11_gut_group, Succinimonas, norank_f_Bacteroidales_RF16_group, norank_f_Bacteroidales_S24-7_group and Fibrobacter. Most strongly associated with the LSO treatment were OTUs that belong to norank_f_Bacteroidales_S24-7_group, Lactobacillus, Bifidobacterium, Prevotella_1, Oribacterium, Ruminobacter, Prevotellaceae_YAB2003_group, and Selenomonas_1. Most strongly associated with the HLS treatment was OTUs that belong to norank_f_Bacteroidales_S24-7_group, Prevotella_1, norank_f_Bacteroidales_BS11_gut_group, unclassified_f_Prevotellaceae, Succinivibrio, and Prevotellaceae_UCG-001.
Figure 4.
Ternary plot of operational taxonomic units (OTUs) showing the percent of observations for each OTU (>0.5%) present in each dietary group (linseed oil, LSO, heated linseed grain, HLS, Control). The taxonomic list at genus level corresponds to OTU of points. For example, a point positioned within the “70” triangle at the ‘LSO’ corner of the ternary plot indicates that 70% of all observations of that OTU occur within the LSO group. Diameter of plotted points corresponds to relative abundance of the OTU. Compartments of the dotted grid correspond to 10% increments.
Spearman correlation analysis between rumen bacteria abundance and FAs.
The 20 most abundant genera represent 60.6% (Control), 54.6% (LSO), and 64.2% (HLS) of the total bacterial community. Spearman correlation analysis was conducted between the abundance of top 20 bacterial genera and FA ≥ 18 carbon as well as the n-6/n-3 and P/S ratios (Table 6). The threshold |R| > 0.5 is considered as a significant spearman correlation. The results indicated that Succinimonas abundance was positively correlated with concentrations of C21:0, C22:0, C23:0, C24:0, C16:1, C20:1, C22:1, C24:1, C18:3n6, C20:2c6, C20:3n6, C20:4n6, C22:2n6, C20:3n3, C20:5n3 and C22:6n3, but negatively correlated with C18:1n9t concentration. Ruminobacter abundance was positively correlated with the n-6/n-3 ratio, but negatively correlated with C18:3n3 concentration. Succinivibrio abundance was positively correlated with the P/S ratio. Selenomonas_1 abundance was positively correlated with C18:1n9t concentration. Ruminococcaceae_NK4A214_group abundance was positively correlated with C18:0 concentration, but negatively correlated with C18:1n9c concentration. Fretibacterium abundance was negatively correlated with C18:2n6c concentration. norank_f_Bacteroidales_BS11_gut_group abundance was positively correlated with C181n9c concentration. Prevotellaceae_UCG-001 abundance was negatively correlated with C18:2n6c concentration.
Table 6.
Rumen fatty acid showing a significant (P ≤ 0.05) Spearman’s correlation with bacterial microbiome
| Genera | Succinimonas | Ruminobacter | Succinivibrio | Selenomonas_1 | Ruminococcaceae_NK4A214_group | Fretibacterium | norank_f_Bacteroidales_BS11_gut_group | Prevotellaceae_UCG-001 |
|---|---|---|---|---|---|---|---|---|
| C18:0 | 0.654 | |||||||
| C21:0 | 0.568 | |||||||
| C22:0 | 0.607 | |||||||
| C23:0 | 0.586 | |||||||
| C24:0 | 0.571 | |||||||
| C16:1 | 0.682 | |||||||
| C181n9t | −0.7 | 0.521 | ||||||
| C181n9c | −0.693 | 0.536 | ||||||
| C20:1 | 0.625 | |||||||
| C22:1 | 0.586 | |||||||
| C24:1 | 0.575 | |||||||
| C18:2n6c | −0.579 | −0.571 | ||||||
| C18:3n6 | 0.564 | |||||||
| C20:2n6 | 0.536 | |||||||
| C20:3n6 | 0.607 | |||||||
| C20:4n6 | 0.568 | |||||||
| C22:2n6 | 0.568 | |||||||
| C18:3n3 | -0.593 | |||||||
| C20:3n3 | 0.586 | |||||||
| C20:5n3 | 0.568 | |||||||
| C22:6n3 | 0.568 | |||||||
| n-6/n-3 | 0.857 | |||||||
| P/S | 0.525 |
DISCUSSION
Inclusion of HLS in the diet increased the rumen concentrations of C18:3n3 and n-3 PUFA, and decreased the concentration of n-6 PUFA, and therefore decreased the n-6/n-3 ratio, compared with the control and linseed-oil diets. Clearly, supplementation with HLS increases the biohydrogenation of n-6 PUFA but decreases the biohydrogenation of n-3 PUFA, with the outcome being higher concentrations of C18:3n3 and n-3 PUFA in subcutaneous adipose tissue of HLS kids than in LSO kids, supporting the conclusions of our previous study (Wang et al., 2018). The present study adds to our understanding by showing that the PUFA responses to the diets can be explained by the changes in the rumen biota because, in kids fed HLS, there was a lower abundance of bacteria that hydrogenate C18:3n3, and a higher abundance of bacteria that hydrogenate C18:2n6c, than in kids fed LSO.
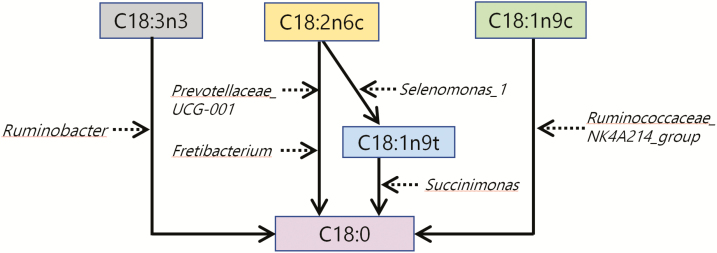
Biohydrogenation of FAs requires hydrogen and energy. The energy would be provided by ATP (Benz et al., 1980) and the hydrogen sources would include H2, water (Rosenfeld and Tove, 1971), NADH, methyl viologen, and electron donors that provide two electrons (Hughes et al., 1980). These energy and hydrogen sources are dependent on the patterns of abundance of the rumen biota, and therefore the diets, thus determining the biohydrogenation pathways that are favored, leading to the PUFA outcomes that we observed as summarized in Figure 5.
Figure 5.
Schema outlining the relationships among bacterial genus that participate in hydrogenation of long-chain fatty acids in the rumen of goat kids. Black solid arrows are used for fatty acids conversion in rumen. Black broken arrows are used for rumen bacteria participate in hydrogenation of fatty acids.
Within the Ruminobacter genus, the principal species are Ruminobacter amylophilus and it can make extensive use of electron transport-linked phosphorylation to generate ATP (Wolin et al., 1997). This is consistent with Ruminobacter abundance being negatively correlated with the concentration of the primary n-3 PUFA, C18:3n3. On the other hand, Ruminobacter cannot use the primary n-6 PUFA, C18:2n6c (Maia et al., 2007), thus explaining the positive correlation between Ruminobacter abundance and the n-6/n-3 ratio.
For C18:2n6c metabolism, the Prevotellaceae are more important (Figure 5). The abundance Prevotellaceae is positively correlated with the concentration of propionic acid (Lyons et al., 2017), and a high abundance of propionate producers is associated with low abundance of methanogens (Van Nevel and Demeyer, 1996; Knapp et al., 2014). Methanogen production and FA biohydrogenation compete for FADH2 and NADH (Knapp et al., 2014), so Prevotellaceae abundance is positively associated with biohydrogenation, explaining the negative correlation between the abundance of Prevotellaceae_UCG-001 and the concentration of C18:2n6c in the present study. The metabolism of C18:2n6c also appears to be controlled by the Fretibacterium genus (Figure 4), a member of the Synergistaceae family that has been reported to be rich in C18:0 (Vartoukian et al., 2013), the terminal product of C18:2n6c hydrogenation (Figure 5). The importance of this pathway is evidenced by the negative correlation between Fretibacterium abundance and C18:2n6c concentration.
Ternary plot analysis revealed different abundances for OTUs that belong to the Ruminobacter, Prevotellaceae_UCG-001, and Fretibacterium in the dietary treatments. The LSO treatment was most strongly associated with a single OTU belonging to Ruminobacter genus. By contrast, the HLS treatment was most strongly associated with one OTU in the Prevotellaceae_UCG-001 genus and 49% of total abundance of one OTU in the Fretibacterium genus. As a consequence, LSO-fed kids had low rumen concentrations of both C18:3n3 and n-3 PUFA, as well as an increased n-6/n-3 ratio, whereas HLS-fed kids had low rumen concentrations of both C18:2n6c and n-6 PUFA. These observations agree with those of He et al. (2017) for the relationships between Ruminobacter amylophilus abundance and n-6/n-3 ratio in the rumen of heifers consuming a diet rich in C18:3n3. They are also consistent with the observation by Cremonesi et al. (2018) that supplementation with linseed grain increases C18:2n6c biohydrogenation in the goat rumen.
The conversion of C18:1n9t (Figure 5) appears to be promoted by a relationship between the Succinimonas and Selenomonas_1 genera. Succinimonas produces succinate that can be decarboxylated by Selenomonas ruminantium to form propionate (Santos and Thompson, 2014), leading to the production of ATP (Wolin et al., 1997). This interaction explains why Succinimonas abundance is negatively correlated with C18:1n9t concentration. The abundance of Succinimonas (Gammaproteobacteria class) was positively correlated with the concentration of many FAs, supporting genomic analyses showing that the class Gammaproteobacteria participates in lipid metabolism (Scully et al., 2014). On the other hand, C18:1n9t is the characteristic intermediate of C18:2n6c biohydrogenation (Cremonesi et al., 2018), so a higher concentration of C18:1n9t implies more biohydrogenation of C18:2n6c. The concentration of C18:1n9t was positively correlated with Selenomonas_1 abundance. We therefore conclude that Selenomonas_1 and Succinimonas act in sequence in the hydrogenation pathway for C18:2n6c (Figure 5).
The LSO treatment was associated with one OTU belonging to Selenomonas_1 and the Control treatment was associated with one OTU belonging to Succinimonas. These relationships probably explain the low concentration of C18:1n9t in Control kids, and why LSO kids had a low concentration of C18:2n6c but a high concentration of C18:1n9t. These observations agree with Dai et al. (2017) who reported that dietary α-linolenic acid increases the relative abundance of Selenomonas ruminantium, and with Polan et al. (1964) who reported that Selenomonas plays an important role in forming C18 monoenoic acid.
As shown in Figure 5, the Ruminococcaceae_NK4A214_group also plays an important role in biohydrogenation of C18:1n9c to C18:0, a suggestion by Huws et al. (2011) and Dai et al. (2017) that is supported by the present results. The abundance of the Ruminococcaceae_NK4A214_group was negatively correlated with the C18:1n9c concentration, but positively correlated with the C18:0 concentration. On the other hand, it has been reported that cellular content of C18:1n9 in Bacteroidales accounts for up to 5.8% of total FA (Ngom, 2018), perhaps explaining why the abundance of Bacteroidales_BS11_gut_group was positively correlated with C18:1n9c concentration in the current study. The ternary plot analysis revealed that 59% of the relative abundance of an OTU in the Ruminococcaceae_NK4A214_group was associated with the LSO treatment, and that two OTUs from the Bacteroidales_BS11_gut_group were associated with the HLS and Control treatments. These observations explain the low concentrations of C18:1n9c in the LSO-fed kids.
Succinivibrio, a genus that ferments cellulose and starch (Liu et al., 2017), was more abundant with the HLS treatment than with the other treatments. This outcome strongly suggests that, when rumen bacteria are presented with rumen-protected fat, rumen bacteria need to source energy from cellulose and starch, in contrast to the situation with the Control and LSO diets where they can utilize dietary fat to produce energy. Fat provides more energy than cellulose and starch so, compared with the Control and LSO diets, the HLS diet provides less energy for the rumen bacteria. However, energy is required for de novo synthesis of saturated FA, so a decrease in energy availability might also explain the positive correlation between Succinivibrio abundance and the P/S ratio. A high (>70%) abundance of one OTU, a member of the Succinivibrio genus was associated with the HLS treatment, so the P/S ratio was a little higher with HLS than with the other diets. We have previously shown that HLS-fed kids have high concentrations of C22:6n3 and n-3 PUFA in adipose tissue (Wang et al., 2018), in agreement with studies in cattle where a high relative abundance of Succinivibrio was correlated with high ear tissue concentrations of C22:6n3 and n-3 PUFA (Liu et al. 2017).
At family level, Veillonellaceae, Christensenellaceae, Prevotellaceae, and Fibrobacteriaceae could provide the hydrogen and energy that are necessary for FA hydrogenation (Vries et al., 1977; Rode et al., 1981; Marounek and Duskova, 1999; Knapp et al., 2014; Zhang et al., 2017; Cremonesi et al., 2018). Bacteroidetes phylum that, according to functional genomics analysis, participates in energy production and conversion (Scully et al., 2014). The LSO-fed kids had the highest relative abundances of Veillonellaceae and Christensenellaceae, the HLS-fed kids had the highest relative abundances of Prevotellaceae and unclassified family in Bacteroidetes, and the Control kids had the highest relative abundance of Fibrobacteriaceae. Together, these observations show that the Veillonellaceae and Christensenellaceae became most important in LSO hydrogenation, while the Prevotellaceae became most important in HLS hydrogenation, and the Fibrobacteriaceae remained most important in the Control diet; and that rumen microorganisms require more energy with the HLS diet. In addition, the relative abundance of the Bacteroidales_BS11_gut_group was reduced with both the LSO and HLS treatments, as reported by Serre et al. (2010). The inclusion of LSO and HLS in the diet changed the community structures of the ruminal bacteria in different ways, but did not influence bacterial richness or diversity, DMI or BW, as reported by other laboratories (Lyons et al., 2017; Li et al., 2017; Vargas et al., 2017). Independently of supplementation, Bacteroidetes, Firmicutes, Proteobacteria, and Verrucomicrobia were the dominant phyla, as seen in other studies with goats and sheep (Lyons et al., 2017; Cremonesi et al., 2018).
In conclusion, compared with the LSO diet, the HLS diet reduced the relative abundance of Ruminobacter and increased the relative abundance of Prevotellaceae_UCG-001 and Fretibacterium, leading to a higher rumen concentration of C18:3n3 but lower rumen concentrations of C18:2n6 and n-6PUFA, and thus a lower n-6/n-3 ratio. More of the dietary PUFA disappeared from the rumen of LSO and HLS kids than from the rumen of the Control kids (evident by comparing the values in Tables 2 and 4). This is consistent with a greater rate of hydrogenation in LSO and HLS kids than in Control kids, and agrees with the observations by Cremonesi et al. (2018). Therefore, the rate of hydrogenation should be considered when using the abundance of hydrogenation bacteria to explain differences in rumen FA concentration between LSO/HLS and Control kids. The findings provide insight into the use of different dietary sources of α-linolenic acid to manipulate FA composition in ruminant products.
Conflict of interest statement. None declared.
Supplementary Material
Footnotes
This study was supported by the National Key R&D Program of China (project no. 2017YFD0500504) and the National Natural Science Foundation of China (project no. 31760685). The authors gratefully acknowledge the staff of the breeding farm of AWCG in Etuoke banner of Ordos in Inner Mongolia and all members of our research group at Inner Mongolia Agriculture University.
LITERATURE CITED
- AOAC 2000. Official Methods of Analysis. 17th ed Arlington, VA:Association of Official Analytical Chemists. [Google Scholar]
- Benz J., Wolf C., and Rüdiger W.. 1980. Chlorophyll biosynthesis: hydrogenation of geranylgeraniol. Plant Sci. Lett. 19:225–230. doi: 10.1016/0304-4211(80)90076-0 [Google Scholar]
- Caporaso J. G., J., Kuczynski J., Stombaugh K., Bittinger F. D., Bushman E. K., Costello N., Fierer A. G., Peña J. K., Goodrich J. I., Gordon, et al. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336. doi:10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremonesi P., G., Conte M., Severgnini F., Turri A., Monni E., Capra L., Rapetti S., Colombini S., Chessa G., Battelli, et al. 2018. Evaluation of the effects of different diets on microbiome diversity and fatty acid composition of rumen liquor in dairy goat. Animal 12:1856–1866. doi:10.1017/S1751731117003433. [DOI] [PubMed] [Google Scholar]
- Dai X., P. J. Weimer K. A. Dill-McFarland V. L. N. Brandao G. Suen, and Faciola A. P.. 2017. Camelina seed supplementation at two dietary fat levels change ruminal bacterial community composition in a dual-flow continuous culture system. Front. Microbiol. 8:2147. doi:10.3389/fmicb.2017.02147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans C. C., K. J., LePard J. W., Kwak M. C., Stancukas S., Laskowski J., Dougherty L., Moulton A., Glawe Y., Wang V., Leone, et al. 2014. Exercise prevents weight gain and alters the gut microbiota in a mouse model of high fat diet-induced obesity. PLoS One 9:e92193. doi:10.1371/journal.pone.0092193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Q., Qiu T., Shao W., Niu C., Xia H., Wang Q., Li Z., Gao Z., Yu H., Su, et al. 2017. Dietary alfalfa and calcium salts of long-chain fatty acids alter protein utilization, microbial populations, and plasma fatty acid profile in Holstein freemartin heifers. J. Agric. Food Chem. 65:10859–10867. doi:10.1021/acs.jafc.7b04173. [DOI] [PubMed] [Google Scholar]
- Hu Q., Fu J. C., and Mu X. D.. 2008. Effects of different oil seeds on the fatty acid composition of ruminal contents and tissue in lambs. J. China Agric. Univ. 13:55–61. doi:10.3724/SP.J.1005.2008.01083. [Google Scholar]
- Hughes P. E., and Tove S. B.. 1980. Identification of an endogenous electron donor for biohydrogenation as alpha-tocopherolquinol. J. Biol. Chem. 255:4447–4452. doi:10.1016/S0082-0784(67)80190-5. [PubMed] [Google Scholar]
- Huws S. A., E. J. Kim M. R. Lee M. B. Scott J. K. Tweed E. Pinloche R. J. Wallace, and Scollan N. D.. 2011. As yet uncultured bacteria phylogenetically classified as prevotella, lachnospiraceae incertae sedis and unclassified bacteroidales, clostridiales and ruminococcaceae may play a predominant role in ruminal biohydrogenation. Environ. Microbiol. 13:1500–1512. doi:10.1111/j.1462-2920.2011.02452.x. [DOI] [PubMed] [Google Scholar]
- Knapp J. R., G. L. Laur P. A. Vadas W. P. Weiss, and Tricarico J. M.. 2014. Invited review: enteric methane in dairy cattle production: quantifying the opportunities and impact of reducing emissions. J. Dairy Sci. 97:3231–3261. doi:10.3168/jds.2013-7234. [DOI] [PubMed] [Google Scholar]
- Li X. Z., C. G. Yan J. Yu Q. S. Gao S. H. Choi J. S. Shin, and Smith S. B.. 2017. Dietary whole and cracked linseed increases the proportion of oleic and α-linolenic acids in adipose tissues and decreases stearoyl-coenzyme A desaturase, acetyl-coenzyme A carboxylase, and fatty acid synthase gene expression in the longissimus thoracis muscle of yanbian yellow cattle. J. Anim. Sci. 95:718–726. doi:10.2527/jas.2016.1050. [DOI] [PubMed] [Google Scholar]
- Liu X. F., Z. Y. Wei C. L. Bai X. B. Ding X. Li G. H. Su L. Cheng L. Zhang H. Guo, and Li G. P.. 2017. Insights into the function of n-3 pufas in fat-1 transgenic cattle. J. Lipid Res. 58:1524–1535. doi:10.1194/jlr.M072983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons T., T. Boland S. Storey, and Doyle E.. 2017. Linseed oil supplementation of lambs’ diet in early life leads to persistent changes in rumen microbiome structure. Front. Microbiol. 8:1656. doi:10.3389/fmicb.2017.01656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maia M. R., L. C. Chaudhary L. Figueres, and Wallace R. J.. 2007. Metabolism of polyunsaturated fatty acids and their toxicity to the microflora of the rumen. Antonie Van Leeuwenhoek 91:303–314. doi:10.1007/s10482-006-9118-2. [DOI] [PubMed] [Google Scholar]
- Marounek M., and Duskova D.. 1999. Metabolism of pectin in rumen bacteria Butyrivibrio fibrisolvens and Prevotella ruminicola. Lett. Appl. Microbiol. 29:429–433. doi: 10.1016/S0030-5898(03)00033-6 [Google Scholar]
- Mozaffarian D., and Wu J. H.. 2012. (N-3) fatty acids and cardiovascular health: are effects of EPA and DHA shared or complementary? J. Nutr. 142:614S–625S. doi:10.3945/jn.111.149633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngom I. I., M., Mailhe D., Ricaboni V., Vitton A., Benezech S., Khelaifia C., Michelle F., Cadoret N., Armstrong A., Levasseur, et al. 2018. Noncontiguous finished genome sequence and description of Mediterranea massiliensis gen. Nov., sp. Nov., a new member of the bacteroidaceae family isolated from human colon. New Microbes New Infect. 21:105–116. doi:10.1016/j.nmni.2017.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Fallon J. V., J. R. Busboom M. L. Nelson, and Gaskins C. T.. 2007. A direct method for fatty acid methyl ester synthesis: application to wet meat tissues, oils, and feedstuffs. J. Anim. Sci. 85:1511–1521. doi:10.2527/jas.2006-491. [DOI] [PubMed] [Google Scholar]
- Polan C. E., J. J. Mcneill, and Tove S. B.. 1964. Biohydrogenation of unsaturated fatty acids by rumen bacteria. J. Bacteriol. 88:1056–1064. doi:10.1111/j.1365-2672.1964.tb05056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rode L. M., B. R. Genthner, and Bryant M. P.. 1981. Syntrophic association by cocultures of the methanol- and CO(2)-H(2)-utilizing species Eubacterium limosum and pectin-fermenting Lachnospira multiparus during growth in a pectin medium. Appl. Environ. Microbiol. 42:20–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld I. S., and Tove S. B.. 1971. Biohydrogenation of unsaturated fatty acids. VI. Source of hydrogen and stereospecificity of reduction. J. Biol. Chem. 246:5025–5030. doi:10.1016/0019-2791(67)90110-3. [PubMed] [Google Scholar]
- Santos E. D. O., and Thompson F.. 2014. The family succinivibrionaceae. In: E. Rosenberg, F. E. Delong, S. Lory, E. Stackebrandt, and F. Thompson, editors, The prokaryotes. Berlin Heidelberg:Springer; p. 639–648. [Google Scholar]
- SAS Institute 2002. STAT user’s guide: Statistics. Version 9.1. Cary, NC:Statistical Analysis System Institute, Inc. [Google Scholar]
- Scully E. D., S. M. Geib J. E. Carlson M. Tien D. McKenna, and Hoover K.. 2014. Functional genomics and microbiome profiling of the Asian longhorned beetle (Anoplophora glabripennis) reveal insights into the digestive physiology and nutritional ecology of wood feeding beetles. BMC Genomics 15:1096. doi:10.1186/1471-2164-15-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serre de La C. B., Ellis C. L., Lee J., Hartman A. L., Rutledge J. C., and Raybould H. E.. 2010. Propensity to high-fat diet-induced obesity in rats is associated with changes in the gut microbiota and gut inflammation. Am. J. Physiol. Gastr. L. 299:G440–G448. doi: 10.1152/ajpgi.00098.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nevel C. J., and Demeyer D. I.. 1996. Influence of pH on lipolysis and biohydrogenation of soybean oil by rumen contents in vitro. Reprod. Nutr. Dev. 36:53–63. doi:10.1051/rnd:19960105. [DOI] [PubMed] [Google Scholar]
- Van Soest P. J., J. B. Robertson, and Lewis B. A.. 1991. Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J. Dairy Sci. 74:3583–3597. doi:10.3168/jds.S0022-0302(91)78551-2. [DOI] [PubMed] [Google Scholar]
- Vargas J. E., S. Andrés T. J. Snelling L. López-Ferreras D. R. Yáñez-Ruíz C. García-Estrada, and López S.. 2017. Effect of sunflower and marine oils on ruminal microbiota, in vitro fermentation and digesta fatty acid profile. Front. Microbiol. 8:1124. doi:10.3389/fmicb.2017.01124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartoukian S. R., J. Downes R. M. Palmer, and Wade W. G.. 2013. Fretibacterium fastidiosum gen. Nov., sp. Nov., isolated from the human oral cavity. Int. J. Syst. Evol. Microbiol. 63(Pt 2):458–463. doi:10.1099/ijs.0.041038-0. [DOI] [PubMed] [Google Scholar]
- Vries de W., Rietveld-Struijk T. R. M., and Stouthamer A. H.. 1977. ATP formation associated with fumarate and nitrate reduction in growing cultures of Veillonella alcalescens. Anton. Leeuw. Int. J. G. 43:153–167. doi: 10.1007/BF00395670 [DOI] [PubMed] [Google Scholar]
- Wang X., Martin G. B., Liu S. L., Shi B. L., Guo X. Y., Zhao Y. L., and Yan S. M.. 2018. The mechanism through which dietary supplementation with heated linseed grain increases n-3 long-chain polyunsaturated fatty acid concentration in subcutaneous adipose tissue of cashmere kids. J. Anim. Sci. 97:385–397. doi:10.1093/jas/sky386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C. M. 2000. Dietary fatty acids and human health. Ann. Zootech. 49:165–180. doi:org/10.1051/animres:2000116 [Google Scholar]
- Wolin M. J., Miller T. L., and Stewart C. S.. 1997. Microbe-microbe interactions. In: P. H. Hobson and C. S. Stewart, editors, The rumen microbial ecosystem. Dordrecht:Springer; p.467–491. [Google Scholar]
- Zhang J., Liu S. C., Li L. L., Ren Y., Feng C. H., Wei C. H., Li Y. P., and Huang Z. L.. 2017. Anaerobic dechlorination of tetrachlorobisphenol a in river sediment and associated changes in bacterial communities. Water Air Soil Pollut. 228:78. doi: 10.1007/s11270-017-3254-3x [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.