Summary
In Vibrio species, quorum sensing controls gene expression for numerous group behaviors, including bioluminescence production, biofilm formation, virulence factor secretion systems, and competence. The LuxR/HapR master quorum sensing regulators activate expression of hundreds of genes in response to changes in population densities. The mechanism of transcription activation by these TetR-type transcription factors is unknown, though LuxR DNA binding sites that lie in close proximity to the −35 region of the promoter are required for activation at some promoters. Here, we show that Vibrio harveyi LuxR directly interacts with RNA polymerase to activate transcription of the luxCDABE bioluminescence genes. LuxR interacts with RNA polymerase in vitro and in vivo and specifically interacts with both the N- and C-terminal domains of the RNA polymerase α-subunit. Amino acid substitutions in the RNAP interaction domain on LuxR decrease interactions between LuxR and the α-subunit and result in defects in transcription activation of quorum sensing genes in vivo. The RNAP-LuxR interaction domain is conserved in Vibrio cholerae HapR and is required for activation of the HapR-regulated gene hapA. Our findings support a model in which LuxR/HapR bind proximally to RNA polymerase to drive transcription initiation at a subset of quorum sensing genes in Vibrio species.
Keywords: Quorum sensing, transcriptional activation, Vibrio, DNA-directed RNA polymerase
Abbreviated Summary
Cell-cell communication called quorum sensing regulates numerous behaviors in bacteria, including bioluminescence, biofilm formation, and virulence. At the epicenter of quorum sensing pathways are the transcriptional regulators that respond to changes in bacterial cell density and accordingly alter gene expression. Here we show that the master quorum sensing transcription factors LuxR/HapR in Vibrio harveyi and Vibrio cholerae activate quorum sensing genes via direct interactions with RNA polymerase in vivo.

Introduction
Bacterial communication is central to the regulation of various developmental processes, including biofilm formation, virulence factor secretion, competence, and bioluminescence (Rutherford & Bassler, 2012). Intra- and interspecies communication, called quorum sensing, directs population-wide changes in gene expression through release and detection of signaling molecules called autoinducers. Autoinducers convey the number and type of cells within their vicinity; as the concentration of autoinducers changes, bacteria alter gene expression to coordinate behaviors.
In many Vibrio species, autoinducers are detected via membrane-bound histidine-kinase receptors that initiate a phosphorylation cascade upon autoinducer binding (Ng & Bassler, 2009, Hawver et al., 2016). This signaling pathway culminates in expression of the master quorum sensing regulator, which is called LuxR in Vibrio harveyi (Ball et al., 2017). LuxR homologs are highly conserved in vibrios and all act as the master quorum sensing regulators, including HapR in Vibrio cholerae, SmcR in Vibrio vulnificus, OpaR in Vibrio parahaemolyticus, VanT in Vibrio anguillarum, and VtpR in Vibrio tubiashii. At low cell densities, autoinducer concentrations are low, resulting in low expression of the master quorum sensing regulator, LuxR. At high cell densities, autoinducer concentrations are high, resulting in maximal expression of LuxR. LuxR expression increases 10-fold between low cell density and high cell density, producing a global change in the quorum sensing gene network via activation and repression of downstream genes ranging 2- to 200-fold (van Kessel et al., 2013b, Waters & Bassler, 2006).
Although the V. harveyi master transcription regulator is named LuxR, it has a distinct structure and function compared to the LuxR proteins that bind autoinducers in typical LuxI/LuxR quorum sensing systems of Gram-negative bacteria (e.g., the LuxR present in Vibrio fischeri) (Ball et al., 2017). The master quorum sensing factor LuxR in V. harveyi, suffering from confusing nomenclature, is actually a member of the TetR family of transcription factors and is distinct from V. fischeri LuxR at the molecular, genetic, and biochemical levels (Pompeani et al., 2008, van Kessel et al., 2013b). V. fischeri, which encodes a canonical LuxI/LuxR system, also has a LuxR/HapR homolog called LitR that functions upstream of LuxR in its quorum sensing system (Fidopiastis et al., 2002, Studer et al., 2008). V. harveyi LuxR-type proteins are structurally conserved among vibrios and share high amino acid identity with the master regulators in other species: 71% with HapR (V. cholerae), 96% with OpaR (V. parahaemolyticus), 93% with SmcR (V. vulnificus), and 60% with LitR (V. fischeri). LuxR proteins function as dimers and have a similar structure to TetR but are distinct from most other TetR proteins in their function (Ramos et al., 2005). LuxR family proteins can both activate and repress hundreds of QS regulated genes, whereas TetR-type proteins typically act only as repressors and regulate 1 or 2 genes. In V. harveyi, the LuxR regulon is 625 genes, and LuxR binds to multiple binding sites within the promoter regions of genes it directly regulates (van Kessel et al., 2013a, van Kessel et al., 2013b), unlike many characterized TetR-type proteins that regulate their promoters by binding to a single binding site. For example, there are eight LuxR binding sites in the luxCDABE locus, which drives the expression of the bioluminescence genes, and two LuxR binding sites in the betIBA-proXWV promoter, which controls the expression of the osmotic stress response genes (Chaparian et al., 2016, van Kessel et al., 2015). The luxCDABE locus is one of the most highly activated by LuxR, and its expression is increased >100-fold in the presence of LuxR (van Kessel et al., 2013a, van Kessel et al., 2013b). Because few TetR-type proteins have been found to act as activators, the mechanism of activation and the reason that multiple binding sites are necessary for activation at this and other LuxR regulated promoters remains unknown.
One proposed mechanism for LuxR transcription activation is that multiple binding sites enable LuxR interactions with other proteins. Importantly, two LuxR binding sites in the luxCDABE promoter in V. harveyi lie in close proximity to the −35 promoter recognition region of the promoter (Chaparian et al., 2016, Pribnow, 1975), suggesting that LuxR binding to these sites may facilitate an interaction with RNA polymerase (RNAP). In addition, we previously showed that LuxR DNA binding at yet another site in the luxCDABE promoter enables synergistic DNA binding with Integration Host Factor (IHF), which drives proper timing of bioluminescence gene expression during quorum sensing (Chaparian et al., 2016). Related to this finding, IHF also plays a role in transcription activation of the vvpE promoter controlled by SmcR in V. vulnificus (Jeong et al., 2010). In this study by Jeong et al., IHF was shown to bind to two sites between the SmcR binding site and the −10/−35 in the vvpE promoter region. An in vivo assay demonstrated that the presence of IHF enabled co-immunoprecipitation of DNA bound by both RNAP and SmcR, whereas a strain lacking IHF did not immunoprecipitate both DNA fragments. From this experiment, the authors concluded that IHF bends the DNA to facilitate a direct interaction between SmcR and RNAP and that this interaction directs transcription activation. However, because this immunoprecipitation assay was performed in vivo, the proposed interaction between SmcR and RNAP may be direct or may be bridged by other proteins. Thus, these previous studies are suggestive of a role for LuxR-type proteins in interactions with RNAP to promote transcription activation, but none have conclusively demonstrated that this interaction directly occurs or whether it impacts transcription activation.
In this study, we demonstrate that LuxR directly interacts with RNAP both in vitro and in vivo in V. harveyi. Biochemical analyses show that LuxR interacts specifically with both the N-terminal and C-terminal domains of the α-subunit of RNAP in vitro. Mutations in specific interaction regions of LuxR decrease α-subunit binding affinity in vitro and abrogate transcription activation of quorum sensing genes in vivo. Finally, we extend our findings to the closely related LuxR homolog HapR in V. cholerae and show that mutations in the analogous RNAP-interaction domain on HapR disrupt activation of quorum sensing genes.
Results
LuxR interacts with RNAP in vivo
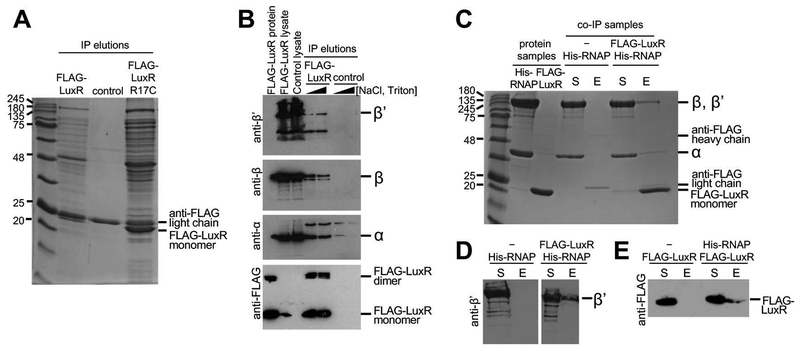
To identify potential proteins that interact with LuxR, an in vivo immunoprecipitation (IP) was performed using LuxR with a 1x-FLAG epitope tag (FLAG-LuxR) that has been shown to be fully functional in vivo (van Kessel et al., 2013b). Lysates from a V. harveyi ΔluxR strain containing FLAG-tagged LuxR (expressed via its native promoter on an extrachromosomal plasmid) or control strains were incubated with anti-FLAG beads, and proteins that remained bound to the beads through washing steps were analyzed by mass spectrometry (Fig. 1A, Tables S4–6). The control strain tested was a ΔluxR strain containing an empty vector, and the empty vector control sample was also analyzed by mass spectrometry (Fig. 1A, Table S7). We also tested for proteins that interacted with the LuxR R17C mutant, which has been shown to be completely defective for DNA binding in vitro and in vivo (Fig. 1A) (Svenningsen et al., 2008, van Kessel et al., 2013b). Our analysis of proteins bound to anti-FLAG beads revealed numerous proteins (Fig. 1A), the majority of which were retained through high stringency buffer incubations (Fig. S1). LuxR was the most prominent band and several other expected proteins were also present (Fig. 1A). In addition, IHF was present in the list of proteins identified by mass spectrometry, which has been previously shown to interact with LuxR in vitro (Chaparian et al., 2016). Catabolite Activator Protein (CAP) was also identified, which is required for transcription activation at some LuxR-activated promoters (Chatterjee et al., 2002). We observed that some of the most prominent proteins in the LuxR IP were subunits of RNAP. We identified the core enzyme subunits α, β, and β’, as well as several sigma factors (σ70, σ38, σ54; Tables S4–6). Importantly, RNAP holoenzyme proteins were also identified in IPs performed with the DNA binding mutant FLAG-LuxR R17C (Fig. 1A; data not shown). In IPs with the FLAG-LuxR R17C mutant protein, the LuxR band is more prevalent because LuxR autorepresses its expression (Chatterjee et al., 1996). Thus, the DNA binding mutant cannot bind DNA to repress its own expression and FLAG-LuxR R17C has higher levels of LuxR expression and immunoprecipitates more proteins. The R17C DNA binding mutant shows that the proteins are being pulled down by a LuxR-protein interaction and not simply by LuxR binding DNA and pulling down proteins that are also bound to that segment of DNA. The results of these experiments suggested that LuxR interacts with RNAP in vivo, and this interaction is independent of DNA binding by LuxR. To corroborate the mass spectrometry results, western blot analyses were performed with antibodies against RNAP α, β, and β’. These RNAP subunits were observed in IP elutions only in the strain containing FLAG-LuxR and not in a control strain (Fig. 1B). Thus, we conclude that LuxR interacts with RNAP in vivo in V. harveyi.
Figure 1. LuxR interacts with RNAP in vivo and in vitro.
(A) Elutions from IP reactions with a ΔluxR V. harveyi strain (KM669) containing plasmids expressing either FLAG-LuxR (pAP116), an empty vector control plasmid (pSLS3), or FLAG-R17C (pST012). The bands corresponding to the FLAG-LuxR protein and the light chain antibody to the FLAG epitope are indicated. (B) Western blot analyses of lysates and elutions from IP reactions with a ΔluxR V. harveyi strain (KM669) containing either a plasmid expressing FLAG-LuxR (pAP116) or an empty vector control plasmid (pSLS3). Purified FLAG-LuxR protein (1 μg) was included in lane 1 as a positive control for the FLAG western. For each strain, two IP reactions were tested with differing stringencies in the wash steps: 1) 100 mM NaCl, 0.1% Triton-X, and 2) 500 mM NaCl, 1% Triton-X. Antibodies used are indicated on the left of each panel. (C, D) SDS-PAGE (C) and western blot (D) analyses of co-IP experiments with purified FLAG-LuxR and His-RNAP holoenzyme incubated with anti-FLAG resin. Purified E. coli His-RNAP (216 pmol) and purified FLAG-LuxR (54 pmol) were loaded in lanes 2 and 3, respectively, in panel C. Samples were taken of the supernatants (S) after incubation with the anti-FLAG resin and samples of the elution (E) were collected from the resin after elution. The western blot was probed with antibodies against β’. (E) Western blot analysis of reciprocal co-IP experiments performed with FLAG-LuxR and His-RNAP incubated with nickel-NTA resin. Samples were taken of the supernatants (S) after incubation with the nickel-NTA resin and samples of the elution (E) were collected from the resin after elution. The western blot was probed with antibodies against the FLAG epitope.
LuxR directly interacts with RNAP in vitro
To test if LuxR and RNAP directly interact, both proteins were purified and a co-immunoprecipitation (co-IP) experiment was performed. V. harveyi FLAG-LuxR was the bait protein to test whether it could pull down E. coli His-RNAP using anti-FLAG beads. We chose to use purified E. coli RNAP for these assays for three reasons: 1) we have shown that LuxR-activated genes (e.g., luxCDABE) are expressed in E. coli in the presence of LuxR (van Kessel et al., 2013b), 2) V. harveyi RNAP subunits share 82 – 91% amino acid identity with E. coli RNAP subunits, and 3) expression plasmids for E. coli RNAP purification were readily available. Following the co-IP, His-RNAP was observed to only be present in the elutions of reactions that also contained FLAG-LuxR (Fig. 1C), and this result was confirmed by western blot analysis (Fig. 1D). Reciprocal reactions were performed in which His-RNAP acted as the bait protein using nickel-NTA resin. FLAG-LuxR was only observed to be present in the elutions of the reactions that also contained His-RNAP (Fig. 1E). These results show that LuxR directly interacts with RNAP in vitro.
LuxR binds to two sites proximal to the −10/−35 region of the luxCDABE promoter
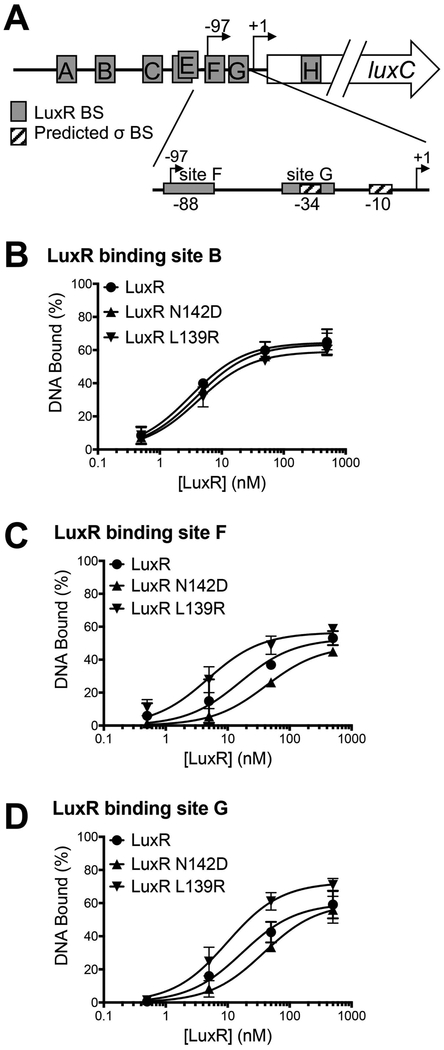
To determine whether LuxR interactions with RNAP are relevant to LuxR activity as a transcriptional regulator in V. harveyi, we examined promoters in which LuxR binds proximally to RNAP. Although LuxR binding site locations in the V. harveyi genome have been determined by ChIP-seq (van Kessel et al., 2013b), transcription start sites are not known for most promoters in V. harveyi. Thus, LuxR binding site positions relative to transcription start sites are not known for most genes. However, previous work has identified many LuxR binding sites in the promoter of the luxCDABE operon, two of which that lie in close proximity to the defined primary transcription initiation site (Fig. 2A) (Chaparian et al., 2016). There are two known transcription start sites at this promoter; the upstream low cell density start site (Fig. 2A, labeled at −97) is weak and repressed by LuxR, and the downstream high cell density start site (Fig. 2A, labeled at +1) is the primary site for high levels of transcription initiation during quorum sensing (Swartzman & Meighen, 1993, Swartzman et al., 1992). LuxR binding sites A-G are all necessary for transcription activation at the primary start site to varying degrees; scrambling the LuxR binding sequence at these binding sites eliminates expression of the luxCDABE genes (Chaparian et al., 2016). Specifically, site F is centered at −88 upstream of the primary transcription start site, and site G is centered at −34, directly over the predicted −35 site. Because the F and G LuxR binding sites are both candidates for LuxR positioning near RNAP for interaction at this promoter, we examined the binding affinity for LuxR at these sites relative to a site distantly located from the transcription start site (site B). Site B is a highly studied binding site for LuxR at this promoter, and LuxR is known to have a high affinity to site B (van Kessel et al., 2013b). Thus, we wanted to compare LuxR binding at site B with the relatively newly discovered LuxR binding sites F and G. Electrophoretic mobility shift assays (EMSAs) were performed with LuxR and DNA containing sites B, F, or G from the luxCDABE promoter (Fig. 2, Fig. S2). The binding affinity for LuxR to site B was relatively high at 3.2 nM and is similar to previously published results (Fig. 2B, Fig. S2A; previously calculated at 14.7 nM (van Kessel et al., 2013b)). We calculated the binding affinity for LuxR to sites F and G to be 16.0 nM and 17.1 nM, respectively (Fig. 2C, Fig. S2B, Fig. 2D, Fig. S2C). Thus, the binding affinities for sites F and G are in a similar range as site B (van Kessel et al., 2013b). We conclude that LuxR binds with specificity to sites F and G that are in close proximity to RNAP during transcription initiation at the luxCDABE promoter.
Figure 2. LuxR DNA binding to PluxC promoter binding sites.
(A) Diagram of the luxCDABE promoter region. Gray boxes indicate LuxR binding sites (BS) and hatched boxes are predicted σ BSs. The primary transcription start site is indicated by a black arrow at +1, and the upstream low cell density transcription start site is indicated by a black arrow at −97. (B, C, D) Electrophoretic mobility shift assays were conducted with dsDNA substrates corresponding to LuxR binding sites B (panel B; oligos JCV617, JCV363), F (panel C; oligos AB190, AB191), or G (panel D; oligos AB188, AB189) with purified proteins LuxR, LuxR N142D, and LuxR L139R at concentrations ranging from 0.5 to 500 nM. The percentage of DNA bound is graphed for each protein. A two-way analysis of variance (ANOVA) followed by Tukey’s multiple-comparison test (n=3) was performed on all data points, and no statistical differences were observed (p < 0.001).
LuxR directly interacts with the RNAP α-subunit N- and C-termini
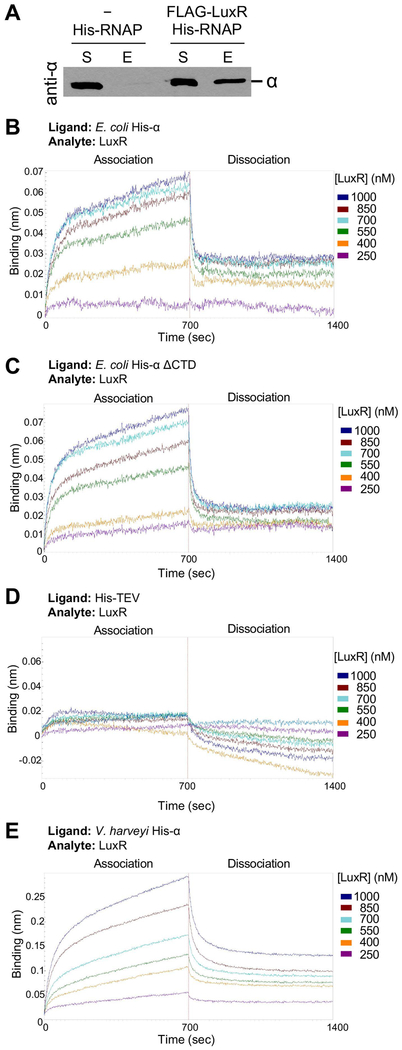
To identify which subunit of RNAP interacts with LuxR, E. coli His-α was purified (Tang et al., 1995). The α-subunit was chosen first for these experiments for two reasons: 1) It is the only subunit that can fold correctly in the absence of the other RNAP core enzyme subunits and can therefore be purified in the absence of the other subunits (Tang et al., 1995), and 2) many activator proteins that are known to interact with RNAP, such as Fis and CAP, have been shown to interact with the α-subunit (Bokal et al., 1995, Heyduk & Heyduk, 1993). Co-IP experiments were performed with FLAG-LuxR and His-α. Using FLAG-LuxR as the bait, the α-subunit was found to only be present in the reactions containing FLAG-LuxR (Fig. 3A). From these data, we conclude that LuxR directly interacts with the α-subunit. However, this finding does not rule out the possibility of additional interactions with other subunits of RNAP.
Figure 3. LuxR interacts with the N-terminal domain of RNAP α subunit.
(A) Western blot analysis of co-IP experiments with purified FLAG-LuxR and E. coli His-α incubated with anti-FLAG resin. Samples were taken of the supernatants (S) after incubation with the anti-FLAG resin and samples of the elution (E) were collected from the resin after elution. The western blot was probed with antibodies against α. (B, C, D, E) BLI analyses of LuxR (analyte) interactions with various ligands (each at 200 nM): E. coli His-α (B), E. coli His-α ΔCTD (C), His-TEV protease (D), or V. harveyi His-α (E). LuxR concentrations are indicated, and the association and dissociation curves are presented.
Several specific interaction interfaces have been identified on the α-subunit of RNAP (Savery et al., 2002). Most proteins interact with the C-terminus of α (Bell et al., 1990, Niu et al., 1996, Igarashi et al., 1991, Ishihama, 1992), though CAP is known to interact with both the N-terminal domain (NTD) and C-terminal domain (CTD) of α at class II promoters (Niu et al., 1996). To further assess the interaction between LuxR and the α-subunit, we used Bio-Layer Interferometry (BLI) to measure the relative dissociation constant (Kd) between these proteins in an in vitro assay. In this assay, one protein, the ‘ligand’, is immobilized on a biosensor tip and incubated with various concentrations of a second protein, the ‘analyte’. The purpose of this assay is to provide a relative quantitative measurement to compare proteins in vitro, but is not intended to indicate the in vivo binding affinity between proteins. In our initial experiments, E. coli His-α was used as the ligand and immobilized with Ni-NTA biosensor tips, and native LuxR served as the analyte. The Kd between LuxR and His-α was determined to be 150 nM (Fig. 3B, Table 1). We next purified two mutant versions of the His-α protein: ΔCTD and ΔNTD. When we performed a BLI experiment with LuxR and His-α ΔCTD, we found the Kd to be 256 nM, which is statistically different (p = 0.007) than the Kd between LuxR and His-α (Fig. 3C, Table 1). Unfortunately, the ΔNTD protein did not stably associate with the BLI sensor tip, and thus the BLI experiment could not be performed with this mutant protein (data not shown). Our result with the ΔCTD protein suggests that the N-terminal domain of α is not sufficient for maximal interaction with LuxR. However, because there was still a definite observed interaction between LuxR and His-α ΔCTD with a moderate affinity, we conclude that there are binding interaction sites between LuxR and the NTD of α. As a negative control for this experiment, we performed BLI with purified His-tagged Tobacco Etch Virus Nla proteinase (His-TEV protease) as the ligand and LuxR as the analyte (Blommel & Fox, 2007). Because the mass spectrometry data from the LuxR IP experiment indicated that LuxR has the ability to interact with many proteins in vivo, we chose to test a negative control protein for its ability to interact with LuxR in the BLI assay. We chose His-TEV protease because 1) we had an active batch of TEV protein purified, 2) it is a protein that is not natively expressed in V. harveyi, and 3) our preparation contained a His-tag for use in western blots and BLI experiments. Neither an association nor a dissociation were observed for experiments with His-TEV and LuxR, and a Kd could not be calculated (Fig. 3D, Table 1). Thus, we conclude that LuxR specifically interacts with the α-subunit of RNAP in vitro and the N-terminal domain of α contributes to some, but not all, of this interaction, requiring the C-terminal domain for maximal interaction. In addition, we purified the α-subunit of RNAP from V. harveyi and determined that it has a Kd of 139 nM for binding to LuxR, which is not significantly different from the binding affinity of LuxR to E. coli α (Fig. 3E, Table 1, p = 0.999).
Table 1.
BLI analyses of LuxR-protein interactions.
| Ligand | Analytea | Kd (nM) | R2 |
|---|---|---|---|
| His-α E. coli | LuxR | 150 ± 3 | 0.891 |
| His-α ΔCTD E. coli * | LuxR | 255 ± 37 | 0.922 |
| His-α V. harveyi | LuxR | 139 ± 19 | 0.898 |
| His-TEV protease | LuxR | ND | ND |
| His-α V. harveyi | LuxR N142D * | 376 ± 24 | 0.901 |
| His-α V. harveyi | LuxR L139R | 128 ± 17 | 0.981 |
| His-α V. harveyi | LuxR S76A * | 250 ± 51 | 0.997 |
| His-α V. harveyi | LuxR I4A | 198 ± 24 | 0.970 |
| His-α V. harveyi | LuxR S76A/N142D * | 327 ± 36 | 0.964 |
An asterisk (*) indicates that the mutant protein is significantly different than the wild-type protein counterpart (p < 0.01; one-way analysis of variance (ANOVA), followed by Tukey’s multiple-comparison test; n = 3).
Identification of the interaction interface on LuxR that interacts with the RNAP α-subunit
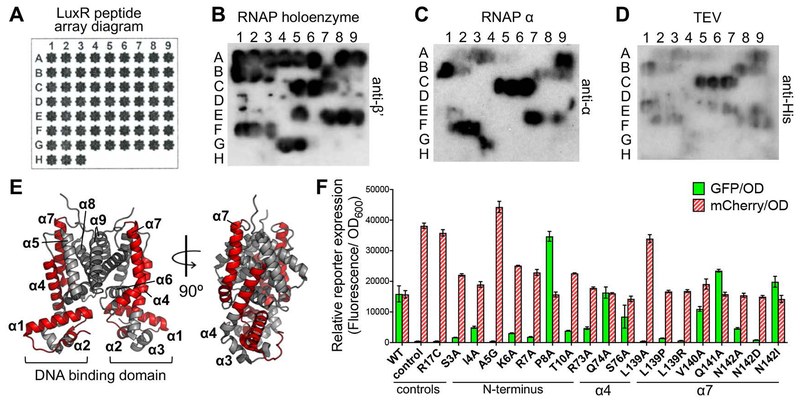
Though we demonstrated that the LuxR-α interaction is mediated through both the NTD and CTD of α, it was unknown where the interaction occurs on the LuxR dimer. To identify LuxR amino acids in the interface of the LuxR-α interaction, we developed a peptide array assay to probe the LuxR protein sequence. A synthetic peptide array of the LuxR protein primary structure was synthesized using ten amino acid peptides spotted on a membrane, with seven amino acid overlaps in sequence between neighboring peptides (Fig. 4A, Fig. S3A). We incubated this peptide membrane with purified E. coli His-RNAP and probed for RNAP using an anti-β’ antibody. Spots on the array clearly identified where His-RNAP interacted with LuxR peptides (Fig. 4B). We mapped these onto the crystal structure of the LuxR homolog SmcR to visualize the relative arrangement of these peptides near the DNA binding domain (helices 2 and 3), dimerization domain (helices 8 and 9), and the ligand binding pocket (helices 4–8) (Fig. 4E) (Kim et al., 2010, Kim et al., 2018). We observed that RNAP interacts with peptides corresponding to alpha helices 1, 4, and 7, as well as an N-terminal region near (but not part of) the helices in the DNA binding domain (Fig. 4E). None of the peptide hits were in the dimerization domain. Among the peptides that interact with RNAP are two residues that were previously studied: L139 and N142. Substitutions at these LuxR residues (L139P, L139R, and N142D) result in a LuxR activation defect in which multiple promoters (including luxCDABE) have decreased or no transcription activation, with no apparent effect on LuxR repression at any tested promoter (van Kessel et al., 2013b).
Figure 4. The LuxR-RNAP interaction domain on LuxR revealed by peptide arrays and in vivo assays.
(A) Diagram of LuxR peptide array layout; corresponding peptides presented in Figure S2A. (B, C, D) LuxR peptide array probed with His-RNAP (panel B, western blot against β’), E. coli His-α (panel C, western blot against α), or His-TEV protease (panel D, western blot against 6xHis epitope). (E) Crystal structure of V. vulnificus SmcR (PDB: 3KZ9, gray); image generated by PyMol. The LuxR peptides identified in the peptide array that interact with RNAP are highlighted in red. (F) GFP and mCherry expression from E. coli strains containing plasmids expressing LuxR (pJV239), LuxR R17C (pAB38), empty vector (pJV036), or LuxR with amino acid substitutions as listed (see Table S2 for all plasmid names). Relative fluorescence expression was calculated by dividing the fluorescence value by the OD600 value. The locations of amino acid substitutions in LuxR are indicated (N-terminus, α-helix 4, or α-helix 7).
We also probed the LuxR peptide array with V. harveyi RNAP subunit α (Fig. 4C). We observed that the α subunit binds a specific subset of the regions of LuxR that the RNAP holoenzyme was found to bind: helices 1, 4, and 7. As a negative control, we probed the LuxR peptide array with His-TEV protease (Fig. 4D). The purpose of this control was to identify non-specific “sticky” regions of the LuxR peptide array and differentiate specific regions of LuxR required for protein-protein interactions. Only three spots had a strong signal on this control peptide array with His-TEV protease. This result demonstrates that only a single region of LuxR on alpha helix 4 is a “sticky” area that enabled binding of all three proteins tested in this assay, though this region may still be important for LuxR-protein interactions. We conclude that RNAP holoenzyme and RNAP α bind to specific regions of LuxR in helices 1, 4, and 7 and in the unstructured N-terminus.
Mutations in the LuxR interaction interface disrupt the LuxR-α interaction
Because RNAP interacts with peptides from multiple regions of LuxR, we used a genetic approach to determine the key residues required for the LuxR-α interaction from among those identified in the peptide array. We made alanine substitutions in various amino acid residues that were prevalent in the peptides bound by RNAP in the peptide array, specifically in the N-terminal loop and helices 4 and 7 (Fig. 4E). Using a previously established assay in E. coli (van Kessel et al., 2013b), we measured transcription activation and repression driven by expression of luxR mutant alleles using fluorescent reporters: activation is measured using a PluxCDABE-gfp reporter and repression is measured using a P05222-mCherry reporter. For example, wild-type LuxR activates transcription of PluxCDABE (high GFP levels) and represses transcription of P05222 (low mCherry levels) compared to the empty vector control strain (Fig. 4F). Conversely, a DNA-binding defective LuxR R17C mutant does not activate transcription of PluxCDABE (low GFP levels) and does not repress transcription of P05222 (high mCherry levels) (Fig. 4F). As shown previously, L139P, L139R, and N142D have wild-type levels of repression but have major activation defects (Fig. 4F). We also observed activation-defective phenotypes in most of the tested LuxR mutant proteins, such as for R73A and S76A (Fig. 4F). To test whether mutations in different helices had combinatorial effects in this assay, we constructed double mutant alleles. The phenotype of a strain expressing a LuxR allele with a combination of the L139P and N142D mutations was not additive (Fig. S3B), nor was combination of the N142D and S76A mutations (Fig. S3C), suggesting that the activation defect for each mutant allele is likely linked to the same interaction interface. We also observed some mutant proteins that have an increased activation phenotype (P8A, Q141A, and N142I).
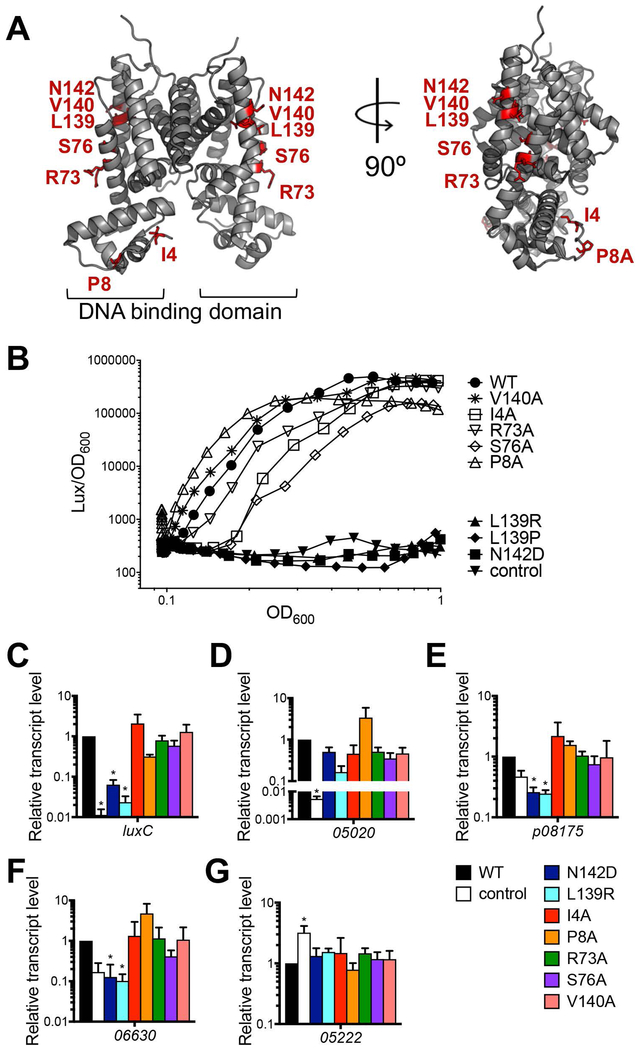
To further assess the phenotypes of the luxR mutants identified in the E. coli reporter screen, we constructed V. harveyi ΔluxR strains containing wild-type or mutant luxR alleles expressed by induction on extrachromosomal plasmids and assayed for activation of the luxCDABE promoter by LuxR and mutant LuxR proteins. Because we used an IPTG-inducible promoter, LuxR production was not regulated by quorum sensing but rather by addition of IPTG. Thus, we performed a bioluminescence time-course to observe LuxR regulation of bioluminescence following induction. We chose to focus on eight mutants with the most dramatic activation-specific phenotypes in the E. coli assay and mapped these onto the SmcR crystal structure (Fig. 5A): I4A, P8A, R73A, S76A, V140A, L139P, L139R, and N142D (van Kessel et al., 2013b). The mutants were determined to be to have activation-specific phenotypes if they retained WT levels of repression. All of the mutants but LuxR P8A and V140A had a decreased and/or delayed bioluminescence phenotype (Fig. 5B). The L139P, L139R, and N142D luxR mutations completely abrogated bioluminescence activation and were comparable to the empty vector control strain (Fig. 5B). LuxR S76A displayed a delayed onset of bioluminescence as well as lower levels of bioluminescence throughout the curve. LuxR P8A had an accelerated onset of bioluminescence but ultimately failed to reach WT levels of bioluminescence. LuxR V140A behaved similarly to wild-type LuxR and did not show a phenotype in this assay. Western blot analysis showed that all LuxR proteins were produced at similar levels, indicating that phenotypic differences were not due to protein level (Fig. S4).
Figure 5. Amino acid substitutions in the RNAP interaction domain on LuxR alter gene expression.
(A) Crystal structure of V. vulnificus SmcR (PDB: 3KZ9, gray); image generated by PyMol. The amino acids for which substitutions are assayed in panel B are indicated by red sticks. (B) Bioluminescence assay with V. harveyi ΔluxR strains (KM699) containing plasmids expressing wild-type LuxR (WT; pJV239), empty vector (control; pJV036), LuxR N142D (pJV261), LuxR L139R (pJV260), LuxR L139P (pJV242), LuxR I4A (pAB17), LuxR P8A (pAB21), LuxR R73A (pAB25), LuxR S76A (pAB28), or LuxR V140A (pAB33). Bioluminescence is calculated as light units (lux) per OD600. Data points represent mean values for three independent experiments. (C, D, E, F, G) Relative transcript levels of V. harveyi genes were determined by qRT-PCR from the same V. harveyi ΔluxR strains and plasmids expressing LuxR alleles as described in panel B for genes luxC (C), VIBHAR_05020 (D), VIBHAR_p08175 (E), VIBHAR_06630 (F), and VIBHAR_05222 (G). The legend for panels C-G denotes the LuxR allele expressed on a plasmid for each strain. Asterisks indicate significant differences compared to the WT luxR phenotype ((C, D): p < 0.0001; one-way ANOVA, followed by Tukey’s multiple comparisons test of log-transformed data; n = 3); (E): p < 0.05; one-way ANOVA, followed by Dunnett’s multiple comparisons test; n = 3); (F,G): p < 0.05; one-way ANOVA, followed by Dunnett’s multiple comparisons test of log-transformed data; n = 3)
We also analyzed expression of several quorum sensing genes that are directly activated by LuxR: luxC, VIBHAR_05020, VIBHAR_p08175, and VIBHAR_06630. Our qRT-PCR analyses comparing strains expressing wild-type LuxR versus mutant LuxR show that the LuxR N142D and L139R alleles were significantly decreased in activation at all tested LuxR-activated promoters except VIBHAR_05020 (Fig. 5C–F). As we observed with the bioluminescence data, L139P and L139R have the same phenotype (Fig. 5B, data not shown). Conversely, strains expressing the LuxR I4A, P8A, R73A, and V140A mutant proteins had varied phenotypes at activated promoters with differing levels of activation but none were significantly different from wild-type LuxR (Fig. 5C–F). All mutant proteins displayed wild-type levels of repression at VIBHAR_05222, a repressed gene in the LuxR regulon (Fig. 5G) (van Kessel et al., 2013a, van Kessel et al., 2013b). These results suggest that the N142D, L139P, and L139R mutations disrupt the interaction between LuxR and RNAP at most activated promoters. However, the variable phenotypes of the other mutant alleles suggest that there may be other effects of these mutations at each promoter.
We next assayed the effects of LuxR amino acid substitutions on the interaction between LuxR and V. harveyi α in vitro using BLI. Surprisingly, we observed that the Kd between LuxR L139R and His-α was not significantly different from the Kd between wild-type LuxR and His-α (128 nM and 139 nM, respectively), suggesting that this substitution alone does not disrupt the interaction measured in vitro (Table 1, Fig. S5A). Conversely, the Kd between LuxR N142D and His-α was calculated to be 376 nM, which is a significant 2.7-fold decrease in binding affinity compared to wild-type LuxR (Table 1, Fig. S5B). We assayed the DNA binding affinity of LuxR mutant proteins L139R and N142D to LuxR binding sites F and G from the luxCDABE promoter. The binding affinities for LuxR L139R at sites B, F, and G were similar to wild-type LuxR and not significantly different at any point assayed (Fig. 2B, 2C, 2D, 4.2 nM, 4.8 nM, and 9.5 nM, respectively; p < 0.001). The binding affinities for LuxR N142D at sites B, F, and G were also similar to wild-type LuxR and not significantly different (Fig. 2B, 2C, 2D, 3.9 nM, 41.9 nM, and 38.1 nM, respectively; p < 0.001). From these results, we conclude that the L139R and N142D substitutions do not affect DNA binding at any site proximal to RNAP. The N142D substitution causes a decrease in interaction with RNAP α, which likely results in the observed defects in transcription activation in vivo.
BLI experiments were also performed with LuxR mutant proteins I4A and S76A because the strains containing these mutant alleles displayed a decrease in activation at several promoters (Fig 5B, 5C). The Kd between V. harveyi His-α and LuxR I4A or LuxR S76A was calculated to be 198 nM and 250 nM, respectively (Table 1, Fig. S5C, S5D). Although LuxR I4A appears to have a slight binding defect compared to wild-type LuxR binding with the α subunit (139 nM), it is not significant (p = 0.222). However, LuxR S76A has a significant 1.8-fold decrease (p = 0.005) in binding affinity compared to wild-type LuxR (Table 1, Fig. S5D). This indicates that the activation deficiency displayed by LuxR S76A is likely due to a decrease in binding with RNAP α.
Mutation of the RNAP α interaction domain of HapR disrupts transcription activation in V. cholerae
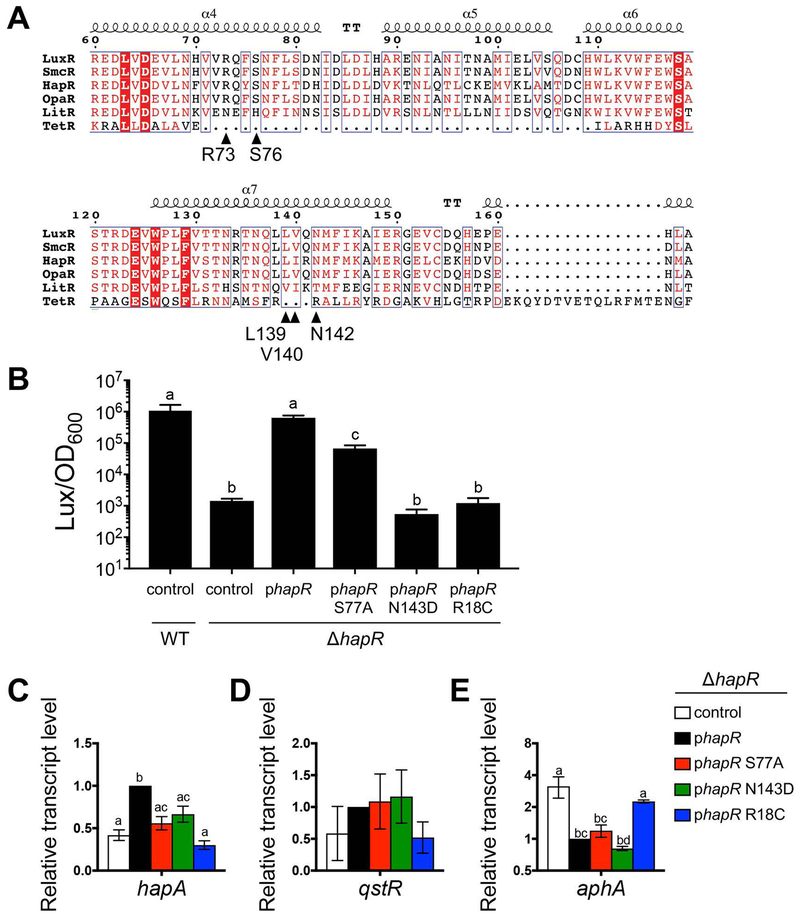
LuxR homologs in Vibrio species share high percent identities, particularly in the DNA binding domain (van Kessel et al., 2013b). However, the amino acid identity is less well-conserved in the region near alpha helices 4–7 (Fig. 6A). We examined the conservation of amino acid residues at the key LuxR interaction positions identified by our in vitro experiments and observed that these residues are highly conserved between LuxR, SmcR, HapR, and OpaR. Conversely, these residues are strikingly absent in the alignment with E. coli TetR. In addition, LitR has no identity with LuxR at these residues, though LitR encodes amino acids that may have functional conservation (e.g., valine at position 139 in place of the leucine in LuxR, or histidine in position 76 in place of the serine in LuxR). Given the high identity between LuxR proteins in these Vibrio species, we hypothesized that these residues are important for RNAP interactions in these species as well. To test this, we examined the effects of making substitutions in V. cholerae HapR in the residues analogous to LuxR N142 and S76 (which correspond to N143 and S77 in HapR, respectively). We also constructed the analogous R17C DNA-binding deficient substitution in HapR (which corresponds to R18C in HapR). The hapR wild-type and mutant alleles were cloned onto extrachromosomal plasmids under control of the IPTG-inducible Ptac promoter in a ΔhapR V. cholerae strain background and compared to a wild-type V. cholerae strain. We first tested the bioluminescence phenotype of strains expressing these mutant proteins using a low-copy plasmid containing the luxCDABE locus (operon and native promoter) from V. harveyi. This experiment with hapR alleles was performed with 1 μM IPTG, which produced comparable levels of bioluminescence in a ΔhapR strain containing the hapR-expression plasmid compared to the wild-type strain (Fig. 6B).
Figure 6. Amino acid substitutions in HapR alter gene expression.
(A) Alignment of predicted RNAP interaction domains of LuxR homologs from V. harveyi (LuxR, AAA27539), V. vulnificus (SmcR, AAF72582), V. cholerae (HapR; ABD24298), V. parahaemolyticus (OpaR, NP_798895), and V. fischeri (LitR, YP_205560) aligned to E. coli TetR (P0ACT4). Sequence alignments were assembled using the Clustal Omega software program (Sievers et al., 2011) and viewed using the ESPript program (Robert & Gouet, 2014). Black triangles indicate the location of amino acids shown. The predicted alpha helices are shown above the alignment. (B) Endpoint bioluminescence assay with V. cholerae wild-type and ΔhapR strains (AC53 and SAD793, respectively) containing plasmids expressing wild-type hapR (WT; pSLS1096), empty vector (control; pEVS143), hapR S77A (pJV377), hapR N143D (pJV378), or hapR R18C (pJV379). Bioluminescence is calculated as light units per OD600 (relative light units, RLU). Different letters indicate significant differences (p < 0.0001; one-way ANOVA, followed by Tukey’s multiple comparisons test of log-transformed data; n = 3). (C, D, E) Relative transcript levels of V. cholerae genes were determined by qRT-PCR from V. cholerae ΔhapR strains containing plasmids expressing hapR (pSLS1096), empty vector (control; pEVS143), hapR S77A (pJV377), hapR N143D (pJV378), or hapR R18C (pJV379). Different letters indicate significant differences (p < 0.05; one-way ANOVA, followed by Dunnett’s multiple comparisons test (C) or Tukey’s multiple comparisons test (D, E) of log2-transformed data; n = 3).
Bioluminescence production exhibited significant decreases of 10-fold and 1177-fold in strains expressing hapR S77A and N143D, respectively, compared to the strain expressing wild-type hapR (Fig. 6B). As expected, bioluminescence production was completely abrogated in the strain expressing the hapR R18C DNA binding mutant. Interestingly, the defective phenotypes of hapR N143D and S77A were restored to wild-type bioluminescence levels upon over-expression with 10 μM IPTG, whereas the R18C mutant was not changed (Fig. S6). From these results, we conclude that the N143D and S77A HapR residues are required for maximal expression of the luxCDABE genes in V. cholerae under conditions of endogenous hapR levels. Because these substitutions in HapR result in similar phenotypes to the analogous substitutions in the RNA polymerase interaction domain of LuxR, the data suggest that HapR interacts with RNA polymerase via the same interaction domain on HapR as on LuxR.
We next assayed transcript levels of quorum sensing genes in V. cholerae that have previously been shown to be directly activated by HapR: hapA and qstR (Lo Scrudato & Blokesch, 2013, Jobling & Holmes, 1997, Tsou et al., 2009, Zheng et al., 2011, Metzger et al., 2016). As a control for HapR transcription repression activity, we assayed transcript levels of aphA. As expected, only the hapR R18C DNA binding mutant was unable to repress, indicating there is no defect in the DNA binding ability of hapR S77A and N143D. In strains containing the hapR S77A and N143D mutants, transcript levels significantly decreased ~2 fold for hapA compared to wild-type hapR (Fig. 6C). Conversely, none of the mutants exhibited significant differences in qstR transcript levels compared to wild-type hapR (Fig. 6D). This is a similar finding to our results in V. harveyi where some, but not all, LuxR-activated genes were affected by the S76A and N142D substitutions. From these qRT-PCR and bioluminescence data, we conclude that substitutions in HapR N143D and S77A decrease transcription at specific promoters in V. cholerae. This suggests that the RNAP interaction domain is conserved in HapR.
Discussion
At tightly controlled promoters with sub-optimal sigma-factor binding site consensus sequences, activators are often required to recruit, stabilize, or aid in open complex formation during transcription activation. In this study, we identified the interaction between the V. harveyi master regulator, LuxR, and RNAP. We found that LuxR and RNAP directly interact, and this interaction is likely necessary for the proper expression of some QS regulated genes. LuxR interacts with RNAP at both the NTD and CTD of the α subunit, though our data have not ruled out the possibility that LuxR may also interact with other subunits of RNAP holoenzyme. The likelihood that LuxR interacts with other subunits of RNAP is supported by the results of our experiments with LuxR peptide arrays. Both the RNAP holoenzyme and the α subunit alone interacted with sites on the peptide array with the majority of these sites overlapping. However, the RNAP holoenzyme displayed more interaction sites on LuxR peptides than α alone. These extra interaction sites between RNAP and LuxR could be mediated by the other subunits of RNAP. The interaction between LuxR and the α-NTD is especially interesting because few proteins, such as CAP, have been shown to interact in that region of the α subunit (Niu et al., 1996, Rhodius et al., 1997, Lee et al., 2012). It is likely that both α-CTD and α-NTD interactions are required at LuxR binding sites that overlap the −35, as both are required at similar CAP class II promoters.
Another important similarity to note between LuxR and CAP is that CAP is capable of binding to promoters in almost exactly the same positions relative to RNAP as LuxR does at PluxCDABE in V. harveyi. At class I CAP activated promoters, CAP binds at a site centered at −62 to −103 bp upstream of the transcription start site and interacts with the α-CTD, while at class II CAP activated promoters, CAP binds to a site centered at −41bp that overlaps with the −35 element and replaces it while it interacts with the α-CTD, α-NTD, and σ70 (Niu et al., 1996, Gaston et al., 1990, Zhou et al., 1994a, Niu et al., 1994, Rhodius et al., 1997, Savery et al., 1998, Rhodius & Busby, 2000a, Rhodius & Busby, 2000b, Zhou et al., 1994b). At PluxCDABE, LuxR binds to a site centered at −88 (site F) and a site centered at −34 (site G), and we have also shown an interaction between LuxR and the α-NTD and α-CTD. Another TetR-type protein, DhaS in Lactococcus lactis, had been found to be an activator instead of a canonical TetR repressor. DhaS has been found to interact with another protein, DhaQ, to activate the dha operon. Not only was this TetR-type protein found to interact with another protein, like LuxR, but it also has a binding site in the promoter of the dha operon that overlaps with the −35 binding site of RNAP (Christen et al., 2006). This information, coupled with our data showing that LuxR interacts with RNAP and has a binding site in the same location, indicates that this interaction and method of activation may be consistent in the small subset of TetR-type proteins that can act as activators. These similarities have led us to propose a mechanism for LuxR activation at PluxCDABE and other QS regulated promoters that is similar to the mechanisms of CAP activation at class I and II promoters (Fig. 7): at class I promoters, CAP recruits RNAP, and at class II promoters, CAP both recruits and aids in open complex formation (Niu et al., 1996, Rhodius & Busby, 2000a, Rhodius & Busby, 2000b, Rhodius et al., 1997). Also of note, CAP has been shown to be able to interact with RNAP in the absence of promoter DNA with similar affinity as LuxR to RNAP (Heyduk et al., 1993). The ability of LuxR to bind RNAP in the absence of DNA, much like CAP (Fig. 1), indicates that LuxR likely has the ability to recruit RNAP to the promoter. Further, the positioning of LuxR binding site G as well as the LuxR-α NTD interaction indicates that LuxR may participate in open complex formation. Further investigation of the function of LuxR at both sites will serve to determine if our model is correct.
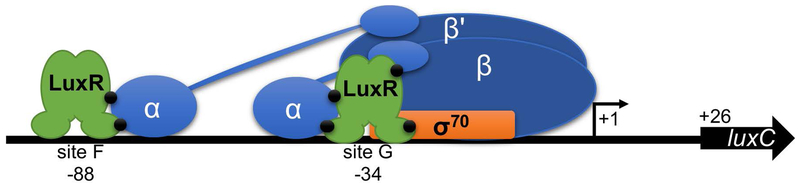
Figure 7. Model of LuxR interaction with RNAP holoenzyme at the luxCDABE promoter in V. harveyi.
LuxR binds to sites F and G in the luxCDABE promoter, which are centered at −88 and −34 relative to the primary transcription start site (small black arrow), respectively. LuxR binding facilitates recruitment of RNAP via interactions with the α subunit, both at the C-terminal domain (sites F and G) and N-terminal domain (site G). Putative sites of interaction between LuxR and RNAP are indicated by black circles. LuxR binding site G is positioned on top of the −35 site for σ70, which may result in an interaction between LuxR and σ at this site.
The PluxCDABE promoter is undoubtedly complex in regard to transcription regulation. Although LuxR binding sites A-G are all required for full activation at PluxCDABE, it is unclear what roles the majority of the binding sites are playing. We have shown that LuxR binding at site C drives an interaction with IHF and perhaps aids in IHF bending the DNA. LuxR binding to site E appears to block activation from the LCD transcription start site at high LuxR concentrations (Chaparian et al., 2016, Swartzman & Meighen, 1993). In addition to the seven LuxR binding sites, CAP was found to bind at PluxCDABE. In strains deleted for cap, bioluminescence expression decreased 1000-fold (Chatterjee et al., 2002). This is especially interesting because CAP was another protein pulled down by LuxR in the FLAG-LuxR IP (Fig. 1A, Table S6), indicating it may also interact with LuxR. However, the exact role of CAP in the activation of PluxCDABE remains to be investigated. Further, the order of LuxR binding to each of the binding sites is unknown, as this may be driven by factors in addition to LuxR binding affinity for the site. There is no evidence of oligomerization of LuxR at any promoter, thus it appears that each binding site is independently bound by LuxR. At HCD, there are approximately 6,500 dimers of LuxR in the cell, a sufficient amount to bind all 1,165 LuxR binding sites identified in V. harveyi (van Kessel et al., 2013b). During the transition from LCD to HCD when the concentration of LuxR in the cell is increasing (~10-fold), the concentration of LuxR required to interact with proteins such as RNAP and IHF is not known. The delay of, or inability to, reach WT levels of bioluminescence expression by many of the luxR mutants in this study indicates that the LuxR interaction with RNAP is critical at this promoter. Our results show that these specific mutants do not have DNA binding defects at sites F and G (Fig. 2C, 2D). Thus, collectively our data support the conclusion that LuxR interacts with RNAP via binding to sites F and G, placing LuxR in close proximity to RNAP.
Our data have also shown that RNAP interacts with multiple surface exposed α-helices of LuxR, and mutating the amino acids within these α-helices affects the ability of RNAP to bind to LuxR. Most mutations that we introduced in the LuxR interaction interface either had no effect or decreased LuxR’s ability to activate at some promoters. This suggests that these regions of LuxR are important for the function of LuxR in transcription activation. In particular, LuxR N142D and S76A were shown to decrease the ability of LuxR to activate at PluxCDABE and other QS regulated genes, as well as have a decreased ability to bind α in vitro (Fig. 4F, Fig. 5B, Fig. 5D–F, Table 1), indicating that the decrease in LuxR activation is likely caused by an interruption of the LuxR-α interaction. Thus, we propose that the major interaction region of LuxR-RNAP is on LuxR α-helix 7 (L139 and N142), with another contributing region on α-helix 4 (S76) (Fig. 5A, Fig. 7). We also note that numerous mutations in the LuxR N-terminal loops and in α-helix 1 near the DNA binding domains yielded activation defects. Although the effects of these mutations on RNAP interactions are difficult to tease apart from possible interruptions of DNA binding, some of these mutations (e.g., I4A) show wild-type levels of repression in vivo, suggesting that they are not affecting DNA binding at all sites. We propose that the LuxR-RNAP interaction interface also includes this N-terminal region and that this may play a role in interaction with the α CTD (Fig. 5A, Fig. 7).
The key residues 139 and 142 in the major LuxR-RNAP interaction region are conserved in vibrios (Fig. 6A) and are surface-exposed in the crystal structures of both HapR and SmcR (De Silva et al., 2007, Kim et al., 2010). Notably, none of these residues are involved in coordinating the ligand binding pockets (Kim et al., 2018). LuxR P8A, Q141A, and N142I all displayed an increase in PluxCDABE activation, with LuxR Q141A and N142I displaying a moderate increase. The result from this experiment is intriguing because LuxR N142D displays a completely abrogated activation phenotype, indicating that the positively charge asparagine may be important at this interaction site, yet LuxR N142I lost this positive charge and displays an increased activation phenotype. In LuxR Q141A, the polar and hydrophobic amino acid, glutamine, is replaced with the non-polar and hydrophilic amino acid, alanine. When considering both these variants, if an amino acid in this area that likely plays either no role or an opposing role in this interaction is changed to an amino acid that contributes to this protein-protein interaction or is more hydrophobic, the mutant LuxR protein could interact more strongly with RNAP. Regarding the LuxR P8A mutation, it displayed an increased ability to activate at PluxCDABE compared to wild-type LuxR in the beginning stages of LuxR expression. As LuxR P8A induction progresses, the relative expression of bioluminescence decreases compared to wild-type LuxR and remains lower than wild-type levels at the end of the curve (Fig 5B). It is possible that the amino acid structural change in LuxR P8A is advantageous for binding to a more ‘naked’ promoter early in the transition from LCD to HCD but eventually becomes a hindrance to activation.
Notably, our data shows that the LuxR-RNAP interaction is conserved across vibrios. When the analogous LuxR S76A and N142D mutations were made in the V. cholerae LuxR homolog, HapR, an activation-deficient phenotype was observed not only at PluxCDABE but also on the V. cholerae QS regulated gene hapA. This indicates that the LuxR/HapR-RNAP interaction is necessary for QS gene expression in not only V. harveyi but also in V. cholerae and likely across the Vibrio genus.
It is important to note that the activation-defective phenotypes of the LuxR mutant proteins could be caused by something in addition to an alteration in their ability to interact with RNAP. For example, it has been shown that LuxR and IHF synergistically bind to the PluxCDABE promoter, IHF is required for full activation of PluxCDABE, and LuxR interacts with IHF in vitro (Chaparian 2016). Our LuxR IP also identified an interaction between LuxR and IHF in vivo. Thus, it is possible that a LuxR activation-deficient mutant could retain a wild-type level of interaction with RNAP and instead be altered in its ability to interact with other proteins such as IHF (e.g., LuxR I4A and L139R which were not significantly altered in interaction with α in BLI experiments).
Our current model for LuxR-RNAP interaction-mediated activation at the PluxCDABE promoter is presented in Figure 7. We propose that LuxR interacts with the α-CTD at promoters in which there are LuxR binding sites >60 but <100 bp upstream of the transcription start site. We also propose that LuxR interacts with the α-CTD, α-NTD, and possibly σ70 at promoters in which there are LuxR binding sites that overlap with the −35 element. While this fits the biochemical and genetic data for the PluxCDABE promoter, not all LuxR activated promoters fit these promoter architecture descriptions, indicating there is likely a RNAP interaction-independent mode of LuxR activation. The mechanism for how LuxR activates at promoters where its binding sites are not within range of interaction with the α-subunit of RNAP remains to be investigated.
Surprisingly, despite stringent washes and conditions, our in vivo FLAG-LuxR IP indicated that LuxR interacts with hundreds of other proteins (Fig. 1A, Tables S4–6). This high volume of potential LuxR interacting proteins may be due to that “sticky” area on LuxR alpha helix 4 discovered in the control peptide array (Fig. 4D). Even so, many of the identified proteins seem like legitimate LuxR interaction partners. As we have shown, LuxR interacts with RNAP and in this experiment every subunit of RNAP was pulled down, including many sigma factors that may interact with LuxR or simply have been pulled down via their interaction with the core enzyme. Many of the proteins pulled down in association with LuxR are transcription factors, and direct interactions between transcription factors that aid in activation or repression is not uncommon. Several of the proteins pulled down were nucleoid-associated proteins including IHF, which has been validated as an interaction partner with LuxR (Chaparian et al., 2016). Whether or not many of these proteins are bona fide LuxR interaction partners and their possible role in transcription regulation remains to be determined.
Experimental Procedures
Bacterial Strains and Media
The E. coli strains S17–1λpir and DH10B were used for cloning purposes, and the E. coli BL21 (DE3) strain was used for protein expression. E. coli strains were grown at 30°C or 37°C shaking at 275 RPM in Lysogeny Broth (LB) medium supplemented with 40 μg/ml kanamycin, 100 μg/ml ampicillin, and 10 μg/ml chloramphenicol as required. The strain of Vibrio harveyi is BB120 (BAA-1116), which was recently reclassified as Vibrio campbellii BAA-1116 (Lin et al., 2010). For consistency with the plethora of literature with BB120, we have referred to this strain as V. harveyi throughout the manuscript. BB120 and derivatives were grown at 30°C in Luria Marine (LM) medium supplemented with 250 μg/ml kanamycin and either 5 μg/ml or 10 μg/ml chloramphenicol as required. Plasmids were transferred from E. coli to V. harveyi strains by conjugation on LB plates, and exconjugants were selected on LM plates with polymyxin B at 50 U/ml and the appropriate selective antibiotic. V. cholerae strain AC53 and derivatives were grown at 30°C in LB medium supplemented 40 μg/ml kanamycin and 5 μg/ml or 10 μg/ml tetracyclin (liquid and agar medium, respectively) as required. The SAD793 strain containing the mutation in hapR was constructed as previously described using natural transformation of splicing-by-overlap extension PCR products by natural transformation (Dalia et al., 2013). All strains were verified by PCR and sequencing.
Molecular Methods and Statistical Analyses
PCR was performed using Phusion HF polymerase (New England Biolabs). Restriction enzymes and enzymes for isothermal DNA assembly (Gibson et al., 2009) (T5 exo, Q5 polymerase, Taq ligase) were purchased from New England Biolabs. Phusion HF polymerase was used for site-directed mutagenesis PCR following the QuikChange mutagenesis (Stratagene) protocol. All oligonucleotides were ordered from Integrated DNA Technologies. Sequencing of plasmids/PCR products was conducted at ACGT or Eurofins Scientific. Plasmid cloning procedures and PCR primer sequences are available upon request. DNA samples were resolved on 1% agarose gels, and protein samples were resolved on 12%, 15%, or 4–20% gradient SDS-PAGE gels. RNA extraction and qRT-PCR experiments were performed as described with cells collected at OD600 = 1.0 (Chaparian et al., 2016). The ΔΔCT method was used to analyze data normalized to hfq as the internal standard gene for V. harveyi or rpoB for V. cholerae.
Unless otherwise noted, data are plotted for triplicate independent experiments. Symbols on graphs represent the mean values, and error bars are standard deviations. Statistical analyses were performed with GraphPad Prism version 7.0c. Additional information about statistical analyses is included in the figure legends.
Fluorescence and Bioluminescence Assays
Bioluminescence and fluorescence assays were performed in strains containing plasmids with luxR expressed under control of the IPTG-inducible Ptac promoter and lacIq system. For bioluminescence assays, overnight cultures of V. harveyi or V. cholerae were diluted to OD600 = 0.05 in fresh LM media or LB media, respectively, containing selective antibiotic and 10 μM IPTG (V. harveyi) or 1 μM IPTG (V. cholerae) in 200 μl in 96-well plates. The plate was incubated shaking at 30°C for 24 h in a BioTek Synergy H1 plate reader with bioluminescence and OD600 readings taken every 30 minutes. For fluorescence assays to measure GFP and mCherry expression, E. coli cultures containing LuxR expression plasmids were grown shaking at 30°C for 16 h, and 200 μl of the cultures was measured in 96-well plates for GFP, mCherry, and OD600 in a BioTek Synergy H1 plate reader.
Immunoprecipitation Assays
LuxR immunoprecipitation (IP) assays were performed with a ΔluxR V. harveyi strain (KM669) containing plasmids expressing either FLAG-LuxR (pAP116) from the native luxR promoter, FLAG-LuxR R17C (pST012) from the native luxR promoter, or empty vector (pSLS3). The strain was inoculated in 25 ml LM at a dilution of 1:25 and grown shaking at 30°C for 16 hours (hr) overnight. To prepare lysates, 50 OD600 units of cells were pelleted, 1 ml lysis buffer (1 ml BugBuster® (Millipore Sigma), 1x protease inhibitors (Sigma-Aldrich), 0.15 mg/ml lysozyme, 0.01% Triton X-100, 1 mM PMSF, 100 mM NaCl, 50 mM Hepes-KOH pH 7.5, 10 mM MgCl2, 0.08 mg/ml DNase) was added, and this mixture was incubated rotating at room temperature (RT) for 20 min. Lysates were clarified by centrifuging at 13,000 x g for 10 min. ANTI-FLAG® M2 Affinity Gel (Sigma-Aldrich) was washed 3 times with IP buffer. At RT, 400 μl IP buffer (50 mM Hepes-KOH pH 7.5, 0.5 mM EDTA, 0.1% Triton X-100, 500 mM NaCl, 1 mM PMSF) and 10 μl ANTI-FLAG® M2 Affinity Gel were added to 100 μl of lysate and incubated 1 hr rotating at RT. The affinity gel was washed with 500 μl IP buffer while rotating for 5 minutes 3 times. To elute proteins from the affinity gel, 25 μl of 6x Sample Buffer (0.5 M Tris-HCl pH 6.8, 6.4% SDS, 6% β-mercaptoethanol, 12% glycerol, 6 mg Bromophenol blue) was added to the beads and boiled at 95°C for 5 min. Proteins were resolved on a 15% SDS-PAGE gel, and select gel slices containing sample were analyzed by the Laboratory for Biological Mass Spectrometry at Indiana University. The control sample lane was sent in its entirety, whereas the FLAG-LuxR sample lane was cut into three segments for processing (top, middle, bottom; see Tables S4–S6).
Mass Spectrometry
Gel bands were diced into 1 mm cubes and incubated for 45 min at 57°C with 2.1 mM dithiothreitol to reduce cysteine residue side chains. These side chains were then alkylated with 4.2 mM iodoacetamide for 1 h in the dark at 21°C. A solution containing 1 μg trypsin in 25 mM ammonium bicarbonate was added and the samples were digested for 12 hours at 37°C. The resulting peptides were desalted using a ZipTip (Millipore, Billerica MA). The samples were dried down and injected into an Eksigent HPLC coupled to a LTQ Velos mass spectrometer (Thermo Fisher Scientific, Waltham MA) operating in “top five” data dependent MS/MS selection. The peptides were separated using a 75 micron, 15 cm column packed in-house with C18 resin (Michrom Bioresources, Auburn CA) at a flow rate of 300 nl/min. A one hour gradient was run from Buffer A (2% acetonitrile, 0.1% formic acid) to 60% Buffer B (100% acetonitrile, 0.1% formic acid).
Co-Immunoprecipitation Assays
FLAG-LuxR and His-RNAP were incubated in a 4:1 ratio at 7.2 μM: 1.8 μM, respectively, with 333 nM RNAP binding DNA substrate (substrate generated by PCR with oligos AB052, AB053) in 30 μl reactions in co-IP buffer (50 mM HEPES pH 7.5, 200 mM NaCl, 0.5% Triton X-100). The reactions were incubated for 30 min at RT. Subsequently, 10 μl of ANTI-FLAG® M2 Affinity Gel (Sigma-Aldrich) (to immunoprecipitate FLAG-LuxR) or HIS-Select® Nickel Affinity Gel (Sigma) (to immunoprecipitate His-RNAP) were washed 3 times in co-IP buffer and then added to the reactions and incubated for 30 min at RT rotating. The reactions were washed 3 times in co-IP for 5 min rotating. To elute proteins from the gel, 20 μl of 2% SDS was added and boiled for 5 min at 95°C. Samples were analyzed on 12% SDS-PAGE gels. These gels were either stained with coomassie brilliant blue stain or further analyzed by western blot.
Western blots
Proteins from SDS-PAGE gels were transferred to a 0.45 μm nitrocellulose membrane by wet transfer in Transfer Buffer (48 mM Tris Base, 39 mM Glycine, 0.037% SDS, 20% methanol) for 60–70 min at 500 mAmps. Membranes were blocked overnight in a 5% milk/TBS-T solution (5 g nonfat dry milk per 100 ml buffer, 25 mM Tris-Cl pH 8.0, 125 mM NaCl 0.1% Tween 20). The membrane was washed 3 times with TBS-T rocking for 5 min and subsequently incubated with anti-FLAG-HRP primary antibody (Sigma, 1:5,000), anti-His-HRP primary antibody (Sigma, 1:3,000), primary antibodies against RNAP subunits (α 1:10,000; β, β’, σ, 1:1,000; all from Neoclone), or primary antibodies against LuxR (1:3,000) for 1 hr at RT. All primary incubations were performed in TBS-T except incubations with anti-LuxR antibodies, which were incubated with 5% milk in TBS-T. Membranes were washed 3 times with TBS-T rocking for 5 min and, when necessary, incubated with anti-mouse or anti-rabbit HRP-conjugated secondary antibody (1:10,000, Promega) for 30 mins at RT. Membranes were washed 3 times with TBS-T rocking for 5 min, treated with Pierce® ECL Western Blotting Substrate (Thermo Scientific) for 5 min and exposed to film.
Protein purification
The His-RNAP core enzyme purification plasmid pAI900 was a generous gift from the Richard Gourse lab. It expresses α, β, β’, and ω subunits with a C-terminal H10x His-tag on β’. An overnight culture of BL21(DE3) cells containing pAI900 was diluted 1:100 into 1L fresh LB with 100 μg/ml ampicillin medium and grown at 30°C until OD600 reached 0.5–0.6. Expression of His-RNAP was induced with 1 mM IPTG, followed by growth for 3 hr at 30°C. Cells were pelleted at 4000 x g for 10 min at 4°C, and the pellet was resuspended in 2 ml lysis buffer per gram of pellet (1 ml BugBuster® (Millipore Sigma), 1x protease inhibitors, 0.15 mg/ml lysozyme, 0.01% Triton X-100, 1 mM PMSF, 100 mM NaCl, 50 mM Hepes-KOH pH 7.5, 10 mM MgCl2, 0.08 mg/ml DNase) and incubated at 4°C rocking for 30 min. The lysate was clarified at 14,000 x g for 20 min at 4°C. The lysate was applied to a HisTrap™ column (GE Healthcare) on an Akta Pure L, washed with 20 column volumes of HisTrap RNAP Buffer A (50 mM Na2HPO4, 0.3 M NaCl, 30 mM imidazole, 0.1% Tween 20, 5% ethanol, 5 mM β-mercaptoethanol), and eluted with HisTrap RNAP Buffer B (50 mM Na2HPO4, 0.3 M NaCl, 300 mM Imidazole, 0.1% Tween 20, 5% Ethanol, 5 mM β-mercaptoethanol). The eluted fractions were pooled and applied to a HiTrap™ Heparin HP column (GE Healthcare), washed with 20 column volumes of RNAP Heparin Buffer A (10 mM Tris-HCl pH 8, 0.1 mM EDTA, 5% glycerol, 0.2 M NaCl), and eluted with RNAP Heparin Buffer B (10 mM Tris-HCl pH 8, 0.1 mM EDTA, 5% glycerol, 0.6 M NaCl). The fractions were pooled and dialyzed overnight at 4°C in RNAP Storage Buffer (10 mM Tris-HCl pH 8.0, 100 mM KCl, 10 mM MgCl2, 0.1 mM EDTA, 50% Glycerol, 0.5 mM DTT). Purified RNAP was flash frozen on liquid N2 and stored at −80°C.
The His-α purification plasmid pRLG3538 (N-terminal 6x His-tagged WT α subunit) and pRLG3657 His-α Δ235 (N-terminal 6xHis-tagged with a C-terminal truncation, expressing α 1–234) were also generous gifts from the Gourse lab. An overnight culture of BL21(DE3) cells containing pRLG3538 or pRLG3657 were diluted 1:100 into 1L fresh LB medium with 100 μg/ml ampicillin and grown at 37°C until OD600 reached 0.5–0.6. Expression of His-α or His-α Δ235 was induced with 1 mM IPTG and incubated shaking 3 hr at 37°C. The cells were pelleted at 4000 x g for 10 min at 4°C. The pellet was resuspended in 2 ml lysis buffer per gram of pellet or lysed using an Avestin EmulsiFlex-C3 and incubated at 4°C rocking for 30 min. The lysate was clarified by centrifuging at 14000 x g for 20 min at 4°C. The lysate was applied to a HisTrap™ column (GE Healthcare), washed with 20 column volumes of Alpha Buffer A (20mM Tris pH 8.0, 500mM NaCl 5mM imidazole), and eluted with Alpha Buffer B (20mM Tris pH 8.0, 500mM NaCl 500mM imidazole). The eluted fractions were pooled and applied to a HiPrep™ Sephacryl S200 (16/60) column (GE Healthcare) using gel filtration buffer (10 mM Tris-HCl pH 8.0, 100 mM KCl, 10 mM MgCl2, 0.1 mM EDTA). The fractions were pooled and dialyzed overnight at 4°C in RNAP storage buffer. Aliquots of purified His-α and His-α Δ235 were flash frozen on liquid N2 and stored at −80°C.
LuxR, His-LuxR, and FLAG-LuxR were purified as previously described (Chaparian et al., 2016) from BL21(DE3) cells containing either pJV079 (wild-type LuxR), pJV080 (His-LuxR), or pJV143 (FLAG-LuxR).
Purification of His-TEV protease was performed using plasmid pMHT238Δ (Blommel & Fox, 2007), which is an engineered TEV protease lacking C-terminal residues 238–242. Plasmid pMHT238Δ was transformed into B834(DE3)::pRIL cells, and colonies were inoculated into 1 ml LB media with kanamycin and chloramphenicol grown shaking at 37°C for 3 hr. These cultures were added to 25 ml of fresh selective medium and grown shaking at 37°C for 2 hr. Finally, the two 25-ml cultures were used to inoculate two 1-L cultures of fresh selective medium and grown shaking at 37°C for 3 hr. The culture was moved to 18°C shaking for 30 min, 0.5 mM IPTG added, and incubated shaking at 18°C for 15.5 hr. The cells were pelleted and resuspended in lysis buffer (25 mM Na2HPO4, 25 mM NaH2PO4, pH 7.8, 1M NaCl, 30 mM imidazole, 20 μg/ml DNaseI, and 1X protease inhibitors (Sigma)). Fastbreak (Promega) was added at 1X concentration for 15 min stirring at room temperature. Insoluble material was pelleted at 15,000 RPM for 30 min at 4°C. The lysate was applied to a HisTrap™ FF column (GE Healthcare) equilibrated with column buffer (25 mM Na2HPO4, 25 mM NaH2PO4, pH 7.8, 1M NaCl, 30 mM imidazole), washed with 20 column volumes of column buffer, and eluted with 5 column volumes of elution buffer (25 mM Na2HPO4, 25 mM NaH2PO4, pH 7.8, 1M NaCl, 0.5M imidazole). Fractions containing His-TEV protease were collected, dialyzed into final buffer (25 mM Na2HPO4, 25 mM NaH2PO4, pH 7.8, 1M NaCl, 2 mM DTT, 20% glycerol), flash frozen on liquid N2, and stored at −80°C.
Electrophoretic Mobility Shift Assays (EMSAs)
EMSAs were performed as described (van Kessel et al., 2013b). For analyses of LuxR protein binding affinities, the amount of probe shifted was quantified compared to unbound probe using ImageJ software. Background density (determined by reactions lacking LuxR protein) was subtracted from the densities of shifted probe bands. Percent DNA bound was calculated as the fraction of shifted probe divided by the total density from shifted and unshifted probe. Binding affinities and statistical analyses were performed on GraphPad Prism version 7.0c.
Bio-layer Interferometry
Bio-layer interferometry was performed on an Octet® K2 System using Dip and Read™ Ni-NTA (NTA) Biosensors (FortéBio®). In a 96-well plate, the following reactions were prepared: 1) a reference well containing BLI Buffer (1x Kinetics Buffer™ (FortéBio®) diluted in PBS (Sigma)), 200 mM NaCl, 0.05% Tween 20), 2) a ligand-loading well containing 200 nM E. coli α, E. coli ΔCTD α, V. harveyi α, or tobacco etch virus TEV protease (i.e., the ligand) in BLI buffer, and 3) an association well containing the V. harveyi LuxR or various LuxR mutant proteins (i.e., the analyte) at various concentrations (1000, 850, 700, 550, 400, 250, and 0 nM). The method on the Octet® K2 System data acquisition software was set to perform a 30-sec sensor check in the reference well, 600 sec ligand-loading step in the ligand-loading well, 60 sec baseline step in the reference well, 700 sec association step in the association well, and 700 sec dissociation step in the reference well. All steps were performed shaking at RT for each concentration of analyte. The data were analyzed using Octet® K2 System data analysis software.
LuxR Peptide Array
A LuxR peptide array was designed and produced by Kinexus Bioinformatics Corp. The array consisted of the entire primary amino acid sequence of LuxR synthesized in 10 amino acid segments each shifted by 3 amino acids with 7 amino acids of overlap between segments. The peptide array membrane was hydrated with 100% methanol for 10 min, washed in TBS for 5 min 3 times, and blocked in 5% milk/TBS-T rocking overnight at 4°C. The membrane was washed once in TBS-T for 5 min rocking at RT. The membrane was incubated with 5 μg/ml E. coli His-RNAP, 0.8 μg/ml V. harveyi α subunit, or 0.8 μg/ml tobacco etch virus TEV protease in TBS-T for 3 hr. The membrane was washed 3 times in TBS-T for 5 min with rocking. The membrane was incubated with 5ml TBS-T/anti-β’ antibody (1:1,000) or 10ml TBS-T/anti-α antibody (1:10,000) for 1.5 hr or 15 ml TBS-T/anti-His antibody for 2 hrs, washed 3 times in TBS-T for 5 min, incubated with the anti-mouse secondary (1:1,000) for 1 hr when applicable, and washed 3 times in TBS-T for 5 min. The membrane was incubated with Pierce® ECL Western Blotting Substrate (Thermo Scientific) for 2 min and then exposed to film.
Supplementary Material
Acknowledgments
The authors wish to acknowledge Trevor Douglas for allowing us to use the Octet system and Sundhar Subramanian for assistance with BLI experiments and data analysis. We also thank Jonathan Trinidad and the Indiana University Laboratory for Biological Mass Spectrometry for analysis of protein samples. We gratefully thank Richard Gourse and Wilma Ross for generously providing RNA polymerase expression plasmids, strains, and guidance on experiments. We thank Jared Cochran for sharing the His-TEV protease plasmid-containing E. coli strain and Susanne Ressl and Jane Newman for assistance with PyMol image generation. The V. cholerae strains and rpoB primers were generously provided by Ankur Dalia. We thank Chelsea Simpson for excellent technical assistance and Clay Fuqua and Matt Bochman for comments on the manuscript. This work was supported by National Institutes of Health (NIH) grant 1R35GM124698–01 to JVK.
Footnotes
Conflict of Interest Statement
The authors declare that that they have no conflicts of interest to declare.
References
- Ball AS, Chaparian RR & van Kessel JC, (2017) Quorum sensing gene regulation by LuxR/HapR master regulators in vibrios. J Bacteriol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell A, Gaston K, Williams R, Chapman K, Kolb A, Buc H, Minchin S, Williams J & Busby S, (1990) Mutations that alter the ability of the Escherichia coli cyclic AMP receptor protein to activate transcription. Nucleic Acids Res 18: 7243–7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blommel PG & Fox BG, (2007) A combined approach to improving large-scale production of tobacco etch virus protease. Protein expression and purification 55: 53–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokal A.J.t., Ross W & Gourse RL, (1995) The transcriptional activator protein FIS: DNA interactions and cooperative interactions with RNA polymerase at the Escherichia coli rrnB P1 promoter. J Mol Biol 245: 197–207. [DOI] [PubMed] [Google Scholar]
- Chaparian RR, Olney SG, Hustmyer CM, Rowe-Magnus DA & van Kessel JC, (2016) Integration host factor and LuxR synergistically bind DNA to coactivate quorum-sensing genes in Vibrio harveyi. Mol Microbiol 101: 823–840. [DOI] [PubMed] [Google Scholar]
- Chatterjee J, Miyamoto CM & Meighen EA, (1996) Autoregulation of luxR: the Vibrio harveyi lux-operon activator functions as a repressor. Mol Microbiol 20: 415–425. [DOI] [PubMed] [Google Scholar]
- Chatterjee J, Miyamoto CM, Zouzoulas A, Lang BF, Skouris N & Meighen EA, (2002) MetR and CRP bind to the Vibrio harveyi lux promoters and regulate luminescence. Mol Microbiol 46: 101–111. [DOI] [PubMed] [Google Scholar]
- Christen S, Srinivas A, Bahler P, Zeller A, Pridmore D, Bieniossek C, Baumann U & Erni B, (2006) Regulation of the Dha operon of Lactococcus lactis: a deviation from the rule followed by the Tetr family of transcription regulators. J Biol Chem 281: 23129–23137. [DOI] [PubMed] [Google Scholar]
- Dalia AB, Lazinski DW & Camilli A, (2013) Characterization of undermethylated sites in Vibrio cholerae. J Bacteriol 195: 2389–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva RS, Kovacikova G, Lin W, Taylor RK, Skorupski K & Kull FJ, (2007) Crystal structure of the Vibrio cholerae quorum-sensing regulatory protein HapR. J Bacteriol 189: 5683–5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidopiastis PM, Miyamoto CM, Jobling MG, Meighen EA & Ruby EG, (2002) LitR, a new transcriptional activator in Vibrio fischeri, regulates luminescence and symbiotic light organ colonization. Mol Microbiol 45: 131–143. [DOI] [PubMed] [Google Scholar]
- Gaston K, Bell A, Kolb A, Buc H & Busby S, (1990) Stringent spacing requirements for transcription activation by CRP. Cell 62: 733–743. [DOI] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA 3rd & Smith HO, (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6: 343–345. [DOI] [PubMed] [Google Scholar]
- Hawver LA, Jung SA & Ng WL, (2016) Specificity and complexity in bacterial quorum-sensing systems. FEMS Microbiol Rev 40: 738–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyduk E & Heyduk T, (1993) Physical studies on interaction of transcription activator and RNA-polymerase: fluorescent derivatives of CRP and RNA polymerase. Cell Mol Biol Res 39: 401–407. [PubMed] [Google Scholar]
- Heyduk T, Lee JC, Ebright YW, Blatter EE, Zhou Y & Ebright RH, (1993) CAP interacts with RNA polymerase in solution in the absence of promoter DNA. Nature 364: 548–549. [DOI] [PubMed] [Google Scholar]
- Igarashi K, Hanamura A, Makino K, Aiba H, Aiba H, Mizuno T, Nakata A & Ishihama A, (1991) Functional map of the alpha subunit of Escherichia coli RNA polymerase: two modes of transcription activation by positive factors. Proc Natl Acad Sci U S A 88: 8958–8962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihama A, (1992) Role of the RNA polymerase alpha subunit in transcription activation. Mol Microbiol 6: 3283–3288. [DOI] [PubMed] [Google Scholar]
- Jeong HS, Kim SM, Lim MS, Kim KS & Choi SH, (2010) Direct interaction between quorum-sensing regulator SmcR and RNA polymerase is mediated by integration host factor to activate vvpE encoding elastase in Vibrio vulnificus. J Biol Chem 285: 9357–9366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobling MG & Holmes RK, (1997) Characterization of hapR, a positive regulator of the Vibrio cholerae HA/protease gene hap, and its identification as a functional homologue of the Vibrio harveyi luxR gene. Mol Microbiol 26: 1023–1034. [DOI] [PubMed] [Google Scholar]
- Kim BS, Jang SY, Bang YJ, Hwang J, Koo Y, Jang KK, Lim D, Kim MH & Choi SH, (2018) QStatin, a Selective Inhibitor of Quorum Sensing in Vibrio Species. mBio 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Kim BS, Park YJ, Choi WC, Hwang J, Kang BS, Oh TK, Choi SH & Kim MH, (2010) Crystal structure of SmcR, a quorum-sensing master regulator of Vibrio vulnificus, provides insight into its regulation of transcription. J Biol Chem 285: 14020–14030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DJ, Minchin SD & Busby SJ, (2012) Activating transcription in bacteria. Annu Rev Microbiol 66: 125–152. [DOI] [PubMed] [Google Scholar]
- Lin B, Wang Z, Malanoski AP, O’Grady EA, Wimpee CF, Vuddhakul V, Alves N Jr, Thompson FL, Gomez-Gil B & Vora GJ, (2010) Comparative genomic analyses identify the Vibrio harveyi genome sequenced strains BAA-1116 and HY01 as Vibrio campbellii. Environ Microbiol Rep 2: 81–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Scrudato M & Blokesch M, (2013) A transcriptional regulator linking quorum sensing and chitin induction to render Vibrio cholerae naturally transformable. Nucleic Acids Res 41: 3644–3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger LC, Stutzmann S, Scrignari T, Van der Henst C, Matthey N & Blokesch M, (2016) Independent Regulation of Type VI Secretion in Vibrio cholerae by TfoX and TfoY. Cell Rep 15: 951–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng WL & Bassler BL, (2009) Bacterial quorum-sensing network architectures. Annu Rev Genet 43: 197–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu W, Kim Y, Tau G, Heyduk T & Ebright RH, (1996) Transcription activation at class II CAP-dependent promoters: two interactions between CAP and RNA polymerase. Cell 87: 1123–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu W, Zhou Y, Dong Q, Ebright YW & Ebright RH, (1994) Characterization of the activating region of Escherichia coli catabolite gene activator protein (CAP). I. Saturation and alanine-scanning mutagenesis. J Mol Biol 243: 595–602. [DOI] [PubMed] [Google Scholar]
- Pompeani AJ, Irgon JJ, Berger MF, Bulyk ML, Wingreen NS & Bassler BL, (2008) The Vibrio harveyi master quorum-sensing regulator, LuxR, a TetR-type protein is both an activator and a repressor: DNA recognition and binding specificity at target promoters. Mol Microbiol 70: 76–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pribnow D, (1975) Nucleotide sequence of an RNA polymerase binding site at an early T7 promoter. Proc Natl Acad Sci U S A 72: 784–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos JL, Martinez-Bueno M, Molina-Henares AJ, Teran W, Watanabe K, Zhang X, Gallegos MT, Brennan R & Tobes R, (2005) The TetR family of transcriptional repressors. Microbiol Mol Biol Rev 69: 326–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodius VA & Busby SJ, (2000a) Interactions between activating region 3 of the Escherichia coli cyclic AMP receptor protein and region 4 of the RNA polymerase sigma(70) subunit: application of suppression genetics. J Mol Biol 299: 311–324. [DOI] [PubMed] [Google Scholar]
- Rhodius VA & Busby SJ, (2000b) Transcription activation by the Escherichia coli cyclic AMP receptor protein: determinants within activating region 3. J Mol Biol 299: 295–310. [DOI] [PubMed] [Google Scholar]
- Rhodius VA, West DM, Webster CL, Busby SJ & Savery NJ, (1997) Transcription activation at class II CRP-dependent promoters: the role of different activating regions. Nucleic Acids Res 25: 326–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert X & Gouet P, (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42: W320–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford ST & Bassler BL, (2012) Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harbor perspectives in medicine 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savery NJ, Lloyd GS, Busby SJ, Thomas MS, Ebright RH & Gourse RL, (2002) Determinants of the C-terminal domain of the Escherichia coli RNA polymerase alpha subunit important for transcription at class I cyclic AMP receptor protein-dependent promoters. J Bacteriol 184: 2273–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savery NJ, Lloyd GS, Kainz M, Gaal T, Ross W, Ebright RH, Gourse RL & Busby SJ, (1998) Transcription activation at Class II CRP-dependent promoters: identification of determinants in the C-terminal domain of the RNA polymerase alpha subunit. EMBO J 17: 3439–3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD & Higgins DG, (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer SV, Mandel MJ & Ruby EG, (2008) AinS quorum sensing regulates the Vibrio fischeri acetate switch. J Bacteriol 190: 5915–5923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenningsen SL, Waters CM & Bassler BL, (2008) A negative feedback loop involving small RNAs accelerates Vibrio cholerae’s transition out of quorum-sensing mode. Genes Dev 22: 226–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartzman E & Meighen EA, (1993) Purification and characterization of a poly(dA-dT) lux-specific DNA-binding protein from Vibrio harveyi and identification as LuxR. J Biol Chem 268: 16706–16716. [PubMed] [Google Scholar]
- Swartzman E, Silverman M & Meighen EA, (1992) The luxR gene product of Vibrio harveyi is a transcriptional activator of the lux promoter. J Bacteriol 174: 7490–7493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Severinov K, Goldfarb A & Ebright RH, (1995) Rapid RNA polymerase genetics: one-day, no-column preparation of reconstituted recombinant Escherichia coli RNA polymerase. Proc Natl Acad Sci U S A 92: 4902–4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou AM, Cai T, Liu Z, Zhu J & Kulkarni RV, (2009) Regulatory targets of quorum sensing in Vibrio cholerae: evidence for two distinct HapR-binding motifs. Nucleic Acids Res 37: 2747–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kessel JC, Rutherford ST, Cong JP, Quinodoz S, Healy J & Bassler BL, (2015) Quorum sensing regulates the osmotic stress response in Vibrio harveyi. J Bacteriol 197: 73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kessel JC, Rutherford ST, Shao Y, Utria AF & Bassler BL, (2013a) Individual and Combined Roles of the Master Regulators AphA and LuxR in Control of the Vibrio harveyi Quorum-Sensing Regulon. J Bacteriol 195: 436–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kessel JC, Ulrich LE, Zhulin IB & Bassler BL, (2013b) Analysis of activator and repressor functions reveals the requirements for transcriptional control by LuxR, the master regulator of quorum sensing in Vibrio harveyi. mBio 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters CM & Bassler BL, (2006) The Vibrio harveyi quorum-sensing system uses shared regulatory components to discriminate between multiple autoinducers. Genes Dev 20: 2754–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Ho B & Mekalanos JJ, (2011) Genetic analysis of anti-amoebae and anti-bacterial activities of the type VI secretion system in Vibrio cholerae. PLoS One 6: e23876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Merkel TJ & Ebright RH, (1994a) Characterization of the activating region of Escherichia coli catabolite gene activator protein (CAP). II. Role at Class I and class II CAP-dependent promoters. J Mol Biol 243: 603–610. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Pendergrast PS, Bell A, Williams R, Busby S & Ebright RH, (1994b) The functional subunit of a dimeric transcription activator protein depends on promoter architecture. EMBO J 13: 4549–4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.