Abstract
Merkel cells are mechanosensory cells involved in tactile discrimination. Merkel cells have been primarily studied in the murine back skin, where they are found in specialized structures called touch domes located around primary hair follicles. Yet, little is known about the morphogenesis of Merkel cells in areas of the skin devoid of hair, such as the glabrous paw skin. Here, we describe Merkel cell formation in the glabrous paw skin during embryogenesis. We first found in the glabrous paw skin that Merkel cells were specified at E15.5, 24 hours later, compared to in the back skin. Additionally, by performing lineage-tracing experiments, we found that unlike in the back skin, SOX9(+) cells do not give rise to Merkel cells in the glabrous paw skin. Finally, we compared the transcriptomes of Merkel cells in the back and the glabrous paw skin and showed that they are similar. Genetic and transcriptome studies showed that the formation of Merkel cells in both regions was controlled by similar regulators. Among them was FGFR2, an upstream factor of MAPK signaling that was reported to have a critical function in Merkel cell formation in the back skin. Here, we showed that FGFR2 is also required for Merkel cell development in the glabrous paw skin. Taken together, our results demonstrate that Merkel cells in the murine back skin and glabrous paw skin are similar, and even though their formation is controlled by a common genetic program, their precursor cells might differ.
Keywords: Merkel cells, glabrous skin, hair follicle, FGFR2
1. INTRODUCTION
Merkel cells were first described by Friedrich Merkel in 1875 as cells that may serve as “touch cells”1. It took over 100 years to prove that Merkel cells have a function in skin sensation2. Merkel cells are mechanosensory cells, which are innervated by afferent neurons and play a key role in the tactile discrimination of the shape and texture of objects2,3. Functional studies in mutant mice, in which Merkel cells were not formed, have demonstrated that the animals were unable to discriminate between different textures when performing behavioral tasks4. In humans, Merkel cells are most abundant on the fingertips, lips and face5–7. In mice, they are found mainly in the back skin, paws, and whiskers8.
Most of our knowledge about Merkel cells comes from studies focused on the murine back skin, where these cells are found in specialized structures called touch domes which are located exclusively around primary hair follicles9. Recently, we discovered that a subset of SOX9 expressing cells located inside of developing hair follicles are Merkel cell progenitors that give rise to Merkel cells at embryonic day (E) 14.510. We also found that fibroblast growth factor receptor 2 (FGFR2) plays an essential function in Merkel cell specification in the back skin, as loss of Fgfr2 leads to drastic decreases in the number of Merkel cells but does not affect the formation of SOX9 positive (+) cells10. To further our investigations, we sought to determine whether our recent findings in the murine back skin apply to other types of skin in the mouse such as the glabrous skin. When do Merkel cells first appear in the glabrous skin? Do they share common progenitors with the Merkel cells in the hairy skin? How similar are the Merkel cells amongst these different anatomical regions? These are some of the unanswered questions we wanted to take on.
In this study, we characterized Merkel cell formation in the glabrous paw skin during embryogenesis and determined how known Merkel cell regulators in the back skin affect Merkel cell formation in the glabrous paw skin to uncover common regulators of Merkel cells.
2. Methods
2.1. Mice
All mice were housed in the Center for Comparative Medicine and Surgery (CCMS) at Icahn School of Medicine at Mount Sinai (ISMMS) in accordance with the Institutional Animal Care and Use Committee (IACUC) approved protocol LA11–0020. At least three animals from independent litters were used for each analysis. Fgfr2flox mice were generously provided by Dr. Philippe Soriano11. Sox9-CreER mice were described in12. R26-mT/mG (stock number: 007676), Atoh1-GFP (stock number: 013593), Gli1-CreER (stock number: 007913) and Krt14-Cre (stock number: 004782) mice were obtained from Jackson Laboratories. For Tamoxifen treatment, pregnant females carrying Sox9-CreER; R26-mT/mG or Gli1-CreER; R26-mT/mG embryos were injected with Tamoxifen (Sigma-Aldrich; St. Louis, MO) doses totaling 40 μg/g body weight at E13.5 and E14.5. Pups were collected at E17 and P0 for further analysis. Mice were genotyped mice by PCR using DNA extracted from tail skin.
2.2. Immunofluorescence staining and microscopy
For immunofluorescence staining, tissues were collected and embedded into OCT (Tissue-Tek; Torrance, CA) and subsequently cut into 10μm sections using a Leica Cryostat. Slides were fixed for 10 minutes in 4% paraformaldehyde (PFA; Electron Microscopy Sciences) in PBS and blocked for one hour at room temperature or overnight at 4°C in PBS with 1% Triton X-100, 1% BSA and 0.25% normal donkey serum (NDS). Primary antibodies were diluted in blocking solution and incubations were carried out for one hour at room temperature or overnight at 4°C, followed by incubation in secondary antibodies for one hour at room temperature. Slides were counterstained with DAPI, mounted using antifade mounting media and then imaged using a Leica DM5500 upright slide microscope using 10x, 20x or 40x objectives.
2.3. Antibodies
The following antibodies were used: KRT14 (generously gifted by Dr. Julie Segre of the National Human Genome Research Institute, USA, 1/20,000), KRT8 (Developmental Studies Hybridoma Bank, TROMA-1, 1/500), KRT20 (Dako, M7019, 1/70), SOX2 (Stemgent, 09–0024, 1/150), GFP (Abcam, ab13970, 1/1000), SOX9 (Abcam, ab185966, 1/500) and FGFR2 (Cell Signaling, 23328, 1/150). For immunofluorescence staining, secondary antibodies were coupled with Alexa 488, 549 or 649 from Jackson Laboratories (1/1000).
2.4. Quantifications
Merkel cells were quantified by counting the number of KRT8(+) cells per millimeter (mm) of skin. Briefly, the length of each section was measured, and the number of positively stained cells was counted. Typical section lengths were between 7–14 mm and at least 100 mm of skin was counted for each condition. A large number of Merkel cells were counted in control conditions (> 200 KRT8(+) cells) and subsequently the number of Merkel cells in a similar length of back and paw skin were counted for the knockout condition. Comparisons and statistics were performed between matching knockout and control littermates.
2.5. Statistics
In column bar graphs in Figure 4D and4E, mean value ± one standard deviation was presented. Box-and-whisker plots show first to third quartiles around the median, with whiskers showing a 5%–95% range and outliers presented as individual data points. To determine the significance between two groups, a Mann-Whitney test was performed. For all statistical tests, p < 0.05 was considered for statistical significance, and furthermore, the actual p-values (to four decimal places) are provided in the figure legends. Significance levels were defined as *p < 0.05, **p < 0.01, ***p < 0.001. n.s., not significant. For statistical analyses, GraphPad Prism 5 was used.
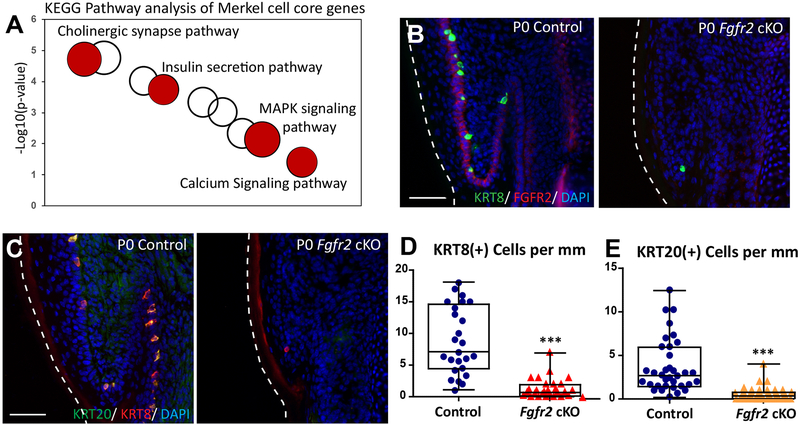
Figure 4. FGFR2 is critical for Merkel cell formation in the glabrous paw skin.
(A). KEGG pathway analysis of Merkel cell core genes from the back skin and glabrous paw skin. Selected KEGG terms are in red. The size of a cycle indicates the number of genes in a KEGG term. (B). Immunofluorescence analysis of Merkel cell markers KRT8 (green) and FGFR2 (red) in P0 control and Fgfr2 cKO glabrous paw skin. Note that FGFR2 (red) is completely gone in P0 Fgfr2 cKO. (C). Immunofluorescence analysis of Merkel cell markers KRT20 (green) and KRT8 (red) in P0 control and P0 Fgfr2 cKO glabrous paw skin. (D). Quantification of KRT8 (+) Merkel cells in P0 control and P0 Fgfr2 cKO, p < 0.0001, n = 4. (E). Quantification of KRT20(+) Merkel cells in P0 control and P0 Fgfr2 cKO, p < 0.0001, n = 4. White dotted lines indicate the edges of the tissue. Scale = 50μm in (B) and (C). The data presented in box plots (D, E) show the median with 25th and 75th percentile borders. Whiskers extend from minimum to maximum. ***p < 0.001 (per Mann-Whitney test).
2.6. Fluorescence-activated cell sorting (FACS)
Interfollicular epidermis (IFE) and Merkel cells were isolated and pooled from multiple Atoh1-GFP newborn mice from more than 3 litters (about 50–100 newborn mice) using FACS. Briefly, P0 Atoh1-GFP back skins were collected and incubated for 4–6 hours in 1.26U/mL dispase (Invitrogen) at 4°C. The epidermis was then gently peeled from the underlying dermis, dissociated by 0.25% Trypsin with 1mM EDTA (Corning Cellgro; Manassas, Virginia, USA) and washed with 1x PBS. To collect glabrous skin, P0 Atoh1-GFP paws were collected and incubated in 1.26U/mL dispase (Invitrogen) and 0.3% type 1 collagenase (Worthington) overnight at 4°C. The paw epidermis was then separated from the dermis, dissociated in 0.25% Trypsin with 1mM EDTA (Corning Cellgro; Manassas, Virginia, USA) for 15 min at 37° C and washed with 1x PBS.
The cell suspension was stained in HBSS+2%FBS with 1:100 SCA1-PerCP-Cy5.5 (Biolegend) and 1:500 EPCAM-APC (Biolegend) for 20 minutes at room temperature and washed with 1x HBSS prior to cell sorting in HBSS with DAPI. IFE was sorted as ATOH1-GFP(−), EPCAM(+), and SCA1(+). Merkel cells were sorted as ATOH1-GFP(+), EPCAM(+), and SCA1(−). Note that alpha6-integrin was not used in the flow sorting scheme to isolate Merkel cells, but alpha6-integrin(+) cells were included in FACS-purification of hair follicle cells13. All cell isolations were performed on a BD FACSAria II instrument (BD, Franklin Lakes, New Jersey, USA) in the Flow Cytometry Core Facility at ISMMS.
2.7. RNA purification, RT-qPCR, and library preparation
FACS-purified cells were collected directly into RLT Plus buffer (QIAGEN), and RNA was purified from these sorted cells with the RNeasy Plus Micro Kit (QIAGEN) according to manufacturer’s instructions. To perform RT-qPCR, complimentary DNA was reverse-transcribed from total RNA using qScript cDNA SuperMix (Quanta Biosciences, Gaithersburg, Maryland, USA). Prior to library construction, sample quality was measured using an Agilent Bioanalyzer. Only samples with RNA integrity numbers > 8 were used for library preparation. Briefly 10ng of RNA was first reverse-transcribed and amplified using the Ovation RNA-seq System V2 (Nugen). Libraries from 100ng of sonicated cDNA (Covaris) were then constructed using the Ovation Ultra-Low DR Multiplex system (Nugen). All RNA Libraries were sequenced on an Illumina HiSeq platform at GENEWIZ.
2.8. RNA-Seq analysis and data visualization
RNA-seq reads were aligned to the mouse reference genome (mm10) using Tophat (v2.0.13)14. The mouse gene models of Refgene were downloaded from the UCSC genome browser (genome.uscs.edu) on March 13, 2017. FPKM (fragments per kilobase of transcript per million mapped reads) values were generated using cufflinks (v2.2.1)15. Lowly expressed genes (mean FPKM values < 2) were excluded from differential expression analysis. For each gene, read counts were obtained using the counts feature in the subread package (v1.4.6)16. Differentially expressed genes were identified with absolute fold change > 5 and adjusted p-value < 0.01 by DESeq217. The sample PCA plot was generated using the plotPCA function in DESeq2 package after a variance stabilizing transformation of the count data by the variance Stabilizing Transformation function in DESeq2 package. The heatmap was generated using the heatmap.2 function in gplots package with normalized counts from the DESeq2 package scaled by row.
2.9. Gene ontology enrichment analysis
Significantly over-represented functional categories were identified using DAVID Bioinformatics Resources 6.818. Selected Gene ontology (GO) terms and KEGG pathways were considered significant with p values < 0.05 and are showed in Fig. 4 and Table S1–S5.
2.10. Data Availability
RNA-seq data of back skin/paw glabrous skin Merkel cells and IFE at P0 were uploaded to the GEO database (GSE122598; this study). RNA-seq data of hair follicle cells at P0 was obtained from the previous publication13 and in the GEO database (GSM3069360, GSM3069361 and GSM3069362). Additional data that support the findings of this study is available from the corresponding author upon request.
3. Results
3.1. Merkel cells are formed at embryonic day 15.5 in the glabrous paw skin
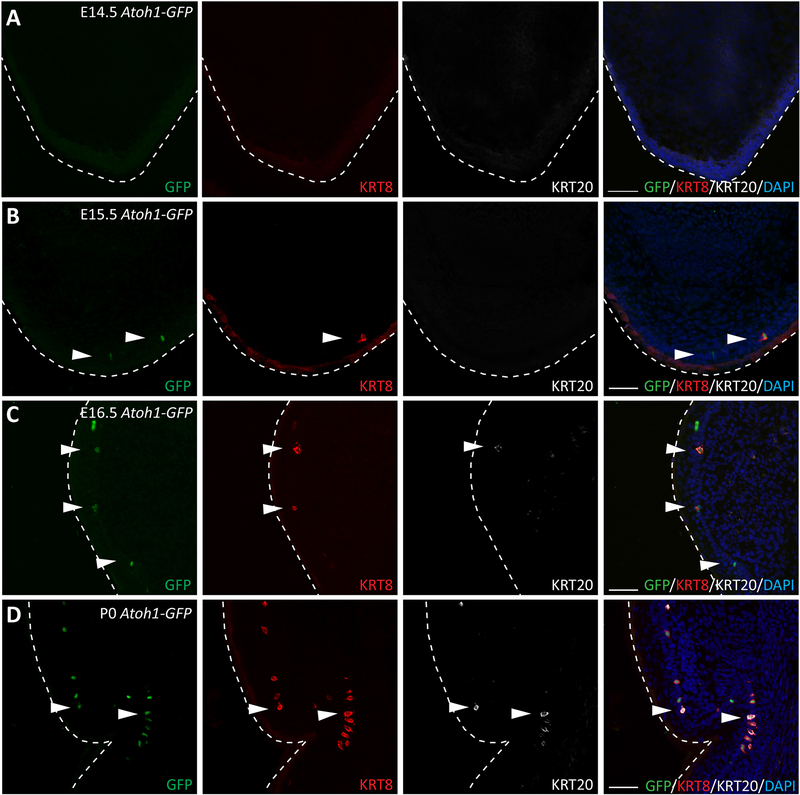
While Merkel cells in the back skin, which contains hair follicles, have been extensively studied, little is known about the development of Merkel cells in glabrous skin, which is devoid of hair follicles. To characterize Merkel cell formation in glabrous skin, we used Atoh1-GFP mice, in which an enhanced green fluorescent protein (GFP) is fused to the 3′-end of the atonal homolog 1 gene (Atoh1)19. We collected paws from Atoh1-GFP pups at embryonic day (E) 14.5, E15.5, E16.5 and postnatal day (P) 0 and performed immunofluorescence analyses of ATOH1-GFP and the keratin intermediate filaments 8 and 20 (KRT8 and KRT20, respectively) that are expressed in more mature Merkel cells20,21. At E14.5, when Merkel cells were already formed inside of the hair placode of the back skin10,21, we could not detect any ATOH1-GFP(+), KRT8(+) or KRT20(+) Merkel cells in the mouse paw (Fig. 1A). At E15.5, a few ATOH1-GFP(+) and KRT8(+) Merkel cells were observed in the mouse paw (Fig. 1B). At E16.5, we detected more cells positive for ATOH1-GFP and KRT8, as well as a few cells that were also positive for KRT20 (Fig. 1C). At P0, the number of Merkel cells positive for ATOH1-GFP, KRT8, and KRT20 drastically increased (Fig. 1D). Similar to in the back skin21, induction of expression of ATOH1, KRT8 and KRT20 markers was temporally regulated in the glabrous paw skin as out of more than 200 Merkel cells analyzed, no KRT20(+)/KRT8(−) cells were observed and only a small number of KRT8(+)/ATOH1-GFP(−) cells were detected (Fig. S1A, B). Thus, similar to in the back skin, the formation of Merkel cells in the glabrous paw skin is a temporally regulated process, although the timing of the induction of Merkel cell formation in the glabrous paw skin occurs one day later compared to in the back skin.
Figure 1. Merkel Cells are formed at Embryonic day 15.5 in the glabrous paw skin.
(A–D). Immunofluorescence analysis of early, intermediate, and late Merkel cell differentiation markers ATOH1-GFP (green), KRT8 (red), and KRT20 (white), respectively, in the glabrous paw skin of Atoh1-GFP mice 14.5 (A), E15.5 (B), E16.5 (C) and P0 (D). White arrows indicate Merkel cells. White dotted lines indicate the edges of the tissue. Note that ATOH1-GFP(+) Merkel cells and KRT8(+) Merkel cells were first observed at E15.5, KRT20(+) Merkel cells were first observed at E16.5. Scale = 50μm in all panels.
3.2. SOX9(+) cells do not give rise to Merkel cells in the glabrous paw skin at E14.5.
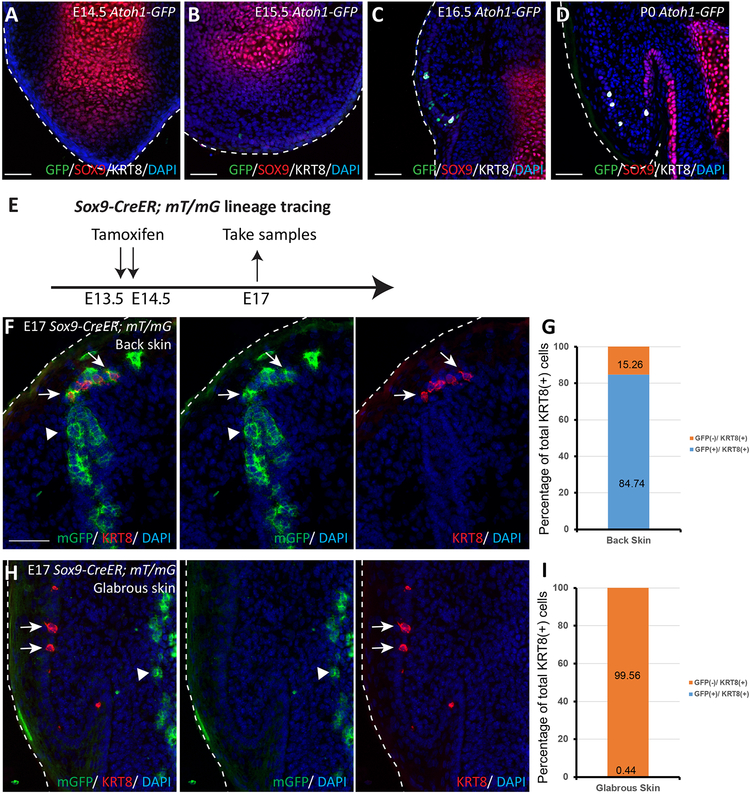
In the back skin, we previously found that Merkel cells formed from SOX9(+) cells present in primary hair placodes10. Therefore, we sought to determine whether Merkel cells in glabrous skin are also formed from SOX9(+) cells. By performing immunofluorescence studies at previously examined time points E14.5-E16.5 and P0, we observed in the glabrous paw skin that Merkel cells were not near SOX9(+) cells (Fig. 2A–D), suggesting that SOX9(+) cells may not be a common precursor cell population for Merkel cells in all of the different regions of the body.
Figure 2. During embryogenesis, SOX9(+) cells do not give rise to Merkel cells in the glabrous paw skin at E14.5.
(A–D). Immunofluorescence analysis of SOX9 (red), Merkel cell markers ATOH1-GFP (green), and KRT8 (white) at E14.5 (A), E15.5 (B), E16.5 (C), and at P0 (D) in the glabrous paw skin of Atoh1-GFP mice. White dotted lines indicate the edges of the tissue. Note that ATOH1-GFP(+) and KRT8(+) Merkel cells do not overlap with SOX9(+) cells. (E). Experimental design for Sox9-CreER; mT/mG lineage tracing experiment. (F). Sox9-CreER; mT/mG lineage tracing in the back skin. White arrows point to Merkel cells; arrow heads point to SOX9-traced cells. SOX9-traced cells are GFP(+) (green). KRT8(+) Merkel cells are stained in red. Note the overlap between SOX9 progenies and KRT8 staining. (G). Quantification of Merkel cells positive for KRT8 and GFP in Sox9-CreER; mT/mG back skin sections at E17. n = 6. Data is presented as a percentage of the total 401 cells quantified. The bar graph shows the percentage of KRT8(+) cells, which are co-labelled with GFP (blue) or not co-labelled with GFP (orange). (H) Sox9-CreER; mT/mG lineage tracing in the glabrous paw skin. White arrows point to Merkel cells; white arrow heads point to SOX9-traced cells. SOX9-traced cells are GFP(+) (green). KRT8(+) Merkel cells are stained in red. Right side panels show separated color channels. (I). Quantification of Merkel cells positive for KRT8 and GFP in Sox9-CreER; mT/mG glabrous paw skin sections at E17. n = 6. Data is presented as percentages of the total 325 cells quantified. The bar graph shows the percentage of KRT8(+) cells that are co-labelled with GFP (blue) or not co-labelled with GFP (orange). White dotted lines indicate the edges of the tissue. Scale bar is 50μm in (A-D), (F) and (H).
To test this hypothesis, we used Sox9-CreER; R26-mT/mG mice to fate map cells originating from SOX9(+) cells. We treated embryos with Tamoxifen at E13.5-E14.5, when Merkel cells had not yet been specified and collected paws and back skins at E17.5 (Fig. 2E). Immunofluorescence analysis showed that GFP staining was found in approximately 85% of KRT8(+) Merkel cells in the back skin (Fig. 2F, G), which is consistent with our previous report that showed that Merkel cells originate from SOX9(+) precursor cells in the back skin10. In contrast, KRT8(+) Merkel cells were not GFP(+) in the glabrous paw skin (Fig. 2H, I; Fig. S1C). In fact, GFP(+) cells were negative for E-Cadherin, a marker of epithelial cells (Figure S1D). These fate-mapping studies show that at E14.5, when SOX9(+) cells give rise to Merkel cells in the back skin, SOX9(+) cells do not give rise to Merkel cells in the glabrous paw skin.
3.3. Differential requirement of SHH for Merkel cell formation in the back and glabrous paw skin
In the back skin, Merkel cells are located around primary hair follicles and during development originate from SOX9(+) cells located in developing hair follicles9,10. In contrast, our data showed that Merkel cells in the glabrous paw skin do not originate from SOX9(+) cells. We therefore tested whether the formation of Merkel cells in the back and glabrous skin is similarly affected in mouse mutants with impaired hair follicle formation and loss of SOX9(+) cells. SHH and Smoothened (SMO), two critical factors of the SHH signaling pathway, are essential for hair follicle formation and specification of SOX9(+) cells22,23. In mouse mutants that lack Shh or Smo genes in the skin epithelium, Merkel cells are not formed in the back skin22,23. However, in the glabrous paw skin, loss of Shh or Smo had much less of an effect on Merkel cell formation as these cells were still formed, and their total count was only slightly reduced, compared to control23 (Table 1). This lesser effect on Merkel cell formation was also found in the glabrous paw skin when coupling the loss of Smo with the loss of EED, a component of the Polycomb repressive complex (PRC) 2 that temporally restricts Merkel cell formation23,24. Loss of EED leads to an ectopic expansion of Merkel cells in both the back and glabrous paw skin23,24. Concurrent loss of EED and Smo in the skin epithelium depleted Merkel cells in the back skin, however, it leads to a slight increase in the number of Merkel cells in the glabrous paw skin23 (Table 1).
Table 1.
Table of genes encoding for the critical regulator of Merkel cell formation in the back skin but not in the glabrous paw skin.
| Gene Symbol(s) | Mouse models used to study the gene loss of function effects | Merkel Cell phenotype in the back skin | Merkel Cell phenotype in the glabrous skin |
|---|---|---|---|
| Shh |
ShhEGFPcre/EGFPcre Krt14-Cre; Shhflox/flox |
Merkel Cells are not formed (Perdigoto et al., 2016; Xiao et al., 2016) |
Reduced number of Merkel Cells (Perdigoto et al., 2016) |
| Smo |
Krt14-Cre; Smoflox/flox Krt5-tTA; TRE-Cre; Smoflox/flox |
Merkel Cells are not formed (Perdigoto et al., 2016; Xiao et al., 2016) |
Reduced number of Merkel Cells (Perdigoto et al., 2016) |
| Eed, Smo | Krt14-Cre; Eedflox/flox; Smoflox/flox | Reduced number of Merkel Cells (Perdigoto et al., 2016) |
Increased number of Merkel Cells (Perdigoto et al., 2016) |
These observations encouraged us to test if SHH-responding cells, known to serve as Merkel cell precursors in the back skin22, are also the precursors of Merkel cells in glabrous skin. By performing lineage-tracing experiments using Gli1-CreER; mT/mG reporter mice, we demonstrated that at E14.5, GLI1(+) cells do not give rise to Merkel cells in the glabrous paw skin (Fig. S2A–C). In fact, GFP(+) cells were negative for E-cadherin suggesting that they are not of epithelial origin (Figure S2D). Thus during development SHH signaling is important for Merkel cell formation in the back skin but to a lesser extent in the glabrous paw skin. Together, these results show that in the back skin, SHH signaling is required for hair follicle formation, SOX9(+) cell specification and Merkel cell development, however, in the glabrous paw skin, SHH signaling makes minor contributions to Merkel cell development.
3.4. Transcriptome profiles of Merkel cells isolated from the back and glabrous paw skin are similar.
To look further into the mechanisms of Merkel cell formation, we performed transcriptional profiling of Merkel cells isolated from the back and glabrous skin regions. By performing FACS, we purified Merkel cells from the back and glabrous paw skins of P0 Atoh1-GFP mice and collected IFE cells from corresponding regions. We next isolated total RNAs from these FACS-purified cells and subjected them to high-throughput RNA sequencing (RNA-seq). We also included in the analysis hair follicle cells from the back skin for comparative reference purposes13. Principal component analysis (PCA) of the top 5,000 most variable genes (across all samples) showed that Merkel cell populations from back and glabrous paw skins clustered near each other but away from the epidermal and hair follicle populations (Fig. 3A). Hierarchical clustering of the significantly differentially expressed genes between Merkel cells and IFE cells also showed that Merkel cell populations from the back skin and glabrous skin clustered together and that they are distinct from the epidermis and hair follicle cells (Fig. 3B). It should be noted that since the RNA-seq data of purified hair follicle cell populations is obtained from a previous study13, there could be a batch effect in the PCA and hierarchical clustering analysis.
Figure 3. RNA-seq transcriptional profiling of Merkel cells isolated from back and glabrous paw skins revealed common core Merkel cell genes.
(A). Principal component analysis based on the top 5000 most variable genes in the back skin and the glabrous paw skin. (B). Hierarchical clustering of the significantly differentially expressed genes between Merkel cells and IFE in either the back skin or glabrous paw skin (Fig. S3), indicating distinct clusters of genes with either shared or unique gene expression in all isolated skin cell types. (C). List of core Merkel cell genes. Selected signature genes from the 514 genes that were commonly upregulated in Merkel cells of back and the glabrous paw skin regions, compared to IFE of the corresponding skin region and selected by these criteria: FPKM > 2, fold-change > 5 and adjusted p-value < 0.01, are organized according to functional categories. (D). Differential expression analysis of genes expressed in Merkel cells in the glabrous paw skin versus the back skin. Genes with mean FPKM > 2, absolute fold-change > 2 and adjust p-value < 0.01 were considered as differentially expressed genes.
Seeking to determine which genes are specifically expressed in the back skin and glabrous skin, we first identified 1151 genes that were significantly upregulated in Merkel cells in the back skin compared to adjacent IFE cells and 753 genes that were significantly upregulated in Merkel cells in the glabrous paw skin compared to adjacent IFE cells, by fold change > 5 and adjusted p-value < 0.01 (Fig. S3, Table S1 and Table S2). By performing KEGG pathway analysis of genes enriched in Merkel cells of the back skin and the glabrous paw skin, we observed common enrichment for genes of the cholinergic synapse, synaptic vesicle cycle, and MAPK signaling pathways (Table S1 and Table S2). Gene ontology (GO) term analysis also showed similar enrichment for genes of ion transport, nervous system development and synaptic transmission in both back skin and glabrous paw skin Merkel cell populations (Table S1 and Table S2).
In line with our PCA and hierarchical clustering analyses which showed similarity in the Merkel cell populations of the back skin and glabrous paw skin, we observed a large overlap between up-regulated genes in Merkel cells in the back and glabrous paw skins, in comparison to their corresponding IFE. We identified 514 up-regulated genes in common and classified them as core Merkel cell genes (Fig. 3C and Table S3). Among them were Atoh1, Isl1, Sox2, Krt20 and Piezo2, genes with known roles in Merkel cells of the back and glabrous skin19,21,25–27 (Fig. 3C and Table 2).
Table 2.
Table of genes encoding critical regulators of Merkel Cell formation in both back and the glabrous paw skin.
| Gene Symbol(s) | Mouse models used to study the gene loss of function effects | Merkel Cell phenotype in the back skin | Merkel Cell phenotype in the glabrous skin |
|---|---|---|---|
| Atoh1 |
Hoxb1Cre; Atoh1flox/flox Krt14-Cre; Atoh1flox/flox |
Merkel Cells are not formed (Maricich et al., 2009; Perdigoto et al., 2014; Van Keymeulen et al., 2009) |
Merkel Cells are not formed (Maricich et al., 2009; Van Keymeulen et al., 2009) |
| Sox2 | Krt14-Cre; Sox2flox/flox | Reduced number of Merkel Cells (Lesko et al., 2013; Bardot et al., 2013) |
Reduced number of Merkel Cells (Bardot et al., 2013) |
| Ezh1/2 | Krt14-Cre; Ezh1/2flox/flox | Increased number of Merkel Cells (Bardot et al., 2013) |
Increased number of Merkel Cells (Bardot et al., 2013) |
| Eed | Krt14-Cre; Eedflox/flox | Increased number of Merkel Cells (Dauber et al., 2016; Perdigoto et al., 2016) |
Increased number of Merkel Cells (Dauber et al., 2016; Perdigoto et al., 2016) |
| Suz12 | Krt14-Cre; Suz12flox/flox | Increased number of Merkel Cells (Dauber et al., 2016) |
Increased number of Merkel Cells (Dauber et al., 2016) |
We next performed differential expression analysis between Merkel cells in the glabrous paw skin and those in the back skin using FDR < 0.01 and fold-change cut off > 2. The analysis showed 198 genes that were more highly expressed in glabrous paw skin Merkel cells and 168 genes that were more highly expressed in back skin Merkel cells, out of more than 11,000 genes expressed in these cells (Fig. 3D; Table S4 and S5). We also observed expression of Hox family genes, exclusive to the back skin or glabrous paw skin. Among these were Hoxb3, Hoxb7 and Hoxb5 in back skin Merkel cells and Hoxc9 and Hoxc10 in glabrous paw skin Merkel cells. Additionally, KEGG pathway and GO term analyses of genes expressed higher in the glabrous paw skin Merkel cell showed enrichment of genes pertaining to focal adhesion, cell division and cell cycle pathways (Table S5), while genes expressed higher in the back skin Merkel cells showed enrichment of genes pertaining to focal adhesion and cell adhesion pathways (Table S4). We did not observe any enrichment for the SHH signaling pathway or its components, which could explain the differing requirement of SHH signaling of Merkel cell formation in the glabrous paw skin and the back skin. Together, this result suggests that Merkel cells in the glabrous skin and back skin are very similar and differences in gene expression between these two populations are likely due to their distinct localizations within the body such as Hox genes.
3.5. FGFR2 is critical for Merkel cell formation in both back and glabrous paw skin.
As described above, KEGG pathway analysis of Merkel cell core genes revealed enrichment for genes of MAPK signaling pathways (Fig. 4A and Table S3). Interestingly, we recently reported that FGFR2-mediated signaling is required for Merkel cell specification in the back skin10, and it is known that the MAPK signaling pathway is a downstream effector of FGFR2-mediated signaling28. To test whether FGFR2 signaling also controls Merkel cell formation in the glabrous paw skin, we analyzed Fgfr2 cKO mice in which Fgfr2 is conditionally ablated in the skin epithelium10. Immunofluorescence analysis of mouse glabrous paw skin confirmed the loss of FGFR2 in P0 Fgfr2 cKO mice compared to control (Fig. 4B) and revealed significant reductions in the numbers of KRT8(+) and KRT20(+) Merkel cells (Fig. 4C–E). Thus, FGFR2–mediated signaling is required for Merkel cell formation in both back and glabrous paw skin.
4. Discussion
Merkel cells are important epidermal mechanoreceptors involved in tactile recognition of shapes and textures of objects4,29. In the body, Merkel cells are innervated by afferent neurons and located in touch-sensitive areas, which in mice are located on their back skin, whisker pads, and footpads. Merkel cells in the back skin are located in touch domes around primary hair follicles and are innervated by two types of neurofilament fibers, NFH+/TrkC+ and Ret+/TrkA+ fibers30. In contrast to in the back skin, Merkel cells in the glabrous paw skin are mainly innervated by NFH+/TrkC+ fibers30.
The majority of studies on Merkel cells have been done on the murine back skin. Our recent study revealed that Merkel cells within the back skin arise from SOX9(+) precursors located in hair placodes10. In contrast, there is little understanding of the mechanisms that control Merkel cell formation in areas of the skin devoid of hair, such as the glabrous paw skin.
In this paper, we performed a comparative analysis of Merkel cells located in the back skin and the glabrous paw skin. Our study shows that in the glabrous paw skin, the formation of Merkel cells occurs one day later than it does in the back skin, but it follows the same maturation process characterized by progressive expression of early (Atoh1), mid (Krt8) and late (Krt20) differentiation markers21.
We performed transcriptional profiling of Merkel cells isolated from back and glabrous paw skins of newborn mice and identified that these cells share similar transcriptomes. Consistent with previous studies, among commonly expressed genes were transcription factors Atoh1 and Sox231,32, which have been shown to be essential for Merkel cell formation in glabrous and back skin19,21,26,27. In epidermal progenitors, Atoh1 and Sox2 are regulated by the epigenetic repressor PRC2. The loss of PRC2 by knocking out their critical components Ezh1/2, Eed or Suz12 in the skin epithelium leads to activation of Atoh1 and Sox2 expression, which results in the formation of ectopic Merkel cells23,26. As in the back skin, we observed a similar phenotype in the glabrous paw PRC2-null skin epithelium23,24,26. To gain further insight into the general regulators of Merkel cell control, we performed KEGG pathway analysis of commonly upregulated genes and identified enrichment for genes in the MAPK signaling pathway. One of the pathways that activates the MAPK kinase cascade is the FGF signaling pathway28. As our recent study shows that FGFR2 is critical for Merkel cell formation in the back skin10, we thus examined FGFR2’s role in the glabrous paw skin and demonstrated that the loss of Fgfr2 leads to a drastic decrease in the number of Merkel cells, highlighting a general requirement for the FGFR2 signaling pathway in Merkel cell formation. Thus, Merkel cells in newborn back skin and glabrous paw skin are similar and share some common regulators that control Merkel cell formation.
Our studies also uncover the differences between Merkel cell development in both back and glabrous skin. Although SOX9(+) cells were recently shown to give rise to Merkel cells in the back skin at E14.510, our fate-mapping experiments showed that SOX9(+) cells do not give rise to Merkel cell in the glabrous paw skin at a similar time point. Furthermore, we found that SHH signaling, which is essential for Merkel cell formation in the back skin22,23, is important but has minor roles in Merkel cell development in the glabrous skin.
How can we reconcile between these differences? In the back skin, SHH signaling is essential for proper hair follicle development, including SOX9(+) cell specification22,23. Because glabrous skin lacks hair follicles, the mechanisms regulating Merkel cell formation through control of hair follicle development should have no effect on Merkel cell formation in this region. Because SOX9(+) cells do not give rise to Merkel cells in the glabrous paw skin, there could be a different precursor population in this region. Alternatively, the back and glabrous paw skins might share a common Merkel cell progenitor population, with SOX9 expression within this progenitor population occurring only in the back skin. In support of the latter hypothesis, previous studies have shown that the loss of Sox9 in the skin epithelium does not affect Merkel cell formation in the back skin, suggesting that even in this region, SOX9 is not required for Merkel cell development and serves simply as a marker10. Future work on uncovering a population of Merkel cell precursors in glabrous skin will reconcile these possibilities.
Future studies on the morphogenesis of Merkel cells are important not only for our understanding of the biology of the skin as a sensory organ but also for uncovering processes that lead to human pathologies. Hyper- or hypo-sensitivity to touch have long been reported in patients with diabetes, inflammation, or autism33–36. Recent studies have shown that Merkel cells can inhibit alloknesis, a disease state where mechanical light touch stimuli provoke itching37. Finally, these discoveries might help us uncover which mechanisms control Merkel cell carcinoma, a deadly skin disease with no effective treatment38–41.
Supplementary Material
ACKNOWLEDGMENTS
For help, critical suggestions, reagents, and experimental input, we are grateful to Dr. Ya-Chieh Hsu, Dr. Julie Segre and Sergei Ezhkov. We would like to thank Dr. Philippe Soriano for the Fgfr2 floxed mice. We also thank the personnel of the Microscope Facility at ISMMS. Research reported in this publication was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under award number R01 AR063724 (to E.E.). We also thank the NCI P30 Cancer Center Support Grant (to E.E.) for their support, as well as the NIH/NHBLI grant, the R01HL133120 (to D.Zheng.). This content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Health.
Footnotes
CONFLICT OF INTERESTS
The authors have declared no conflicting interests.
REFERENCES
- 1.Winkelmann RK, Breathnach AS. The Merkel cell. J Invest Dermatol. 1973;60(1):2–15. [DOI] [PubMed] [Google Scholar]
- 2.Haeberle H, Lumpkin EA. Merkel Cells in Somatosensation. Chemosens Percept. 2008;1(2):110–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lumpkin EA, Marshall KL, Nelson AM. The cell biology of touch. J Cell Biol. 2010;191(2):237–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maricich SM, Morrison KM, Mathes EL, Brewer BM. Rodents rely on Merkel cells for texture discrimination tasks. J Neurosci. 2012;32(10):3296–3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iggo A, Muir AR. The structure and function of a slowly adapting touch corpuscle in hairy skin. J Physiol. 1969;200(3):763–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boot PM, Rowden G, Walsh N. The distribution of Merkel cells in human fetal and adult skin. Am J Dermatopathol. 1992;14(5):391–396. [DOI] [PubMed] [Google Scholar]
- 7.Lacour JP, Dubois D, Pisani A, Ortonne JP. Anatomical mapping of Merkel cells in normal human adult epidermis. Br J Dermatol. 1991;125(6):535–542. [DOI] [PubMed] [Google Scholar]
- 8.Moll I, Paus R, Moll R. Merkel cells in mouse skin: intermediate filament pattern, localization, and hair cycle-dependent density. J Invest Dermatol. 1996;106(2):281–286. [DOI] [PubMed] [Google Scholar]
- 9.Halata Z, Grim M, Bauman KI. Friedrich Sigmund Merkel and his “Merkel cell”, morphology, development, and physiology: Review and new results. Anat Rec Part A. 2003;271a(1):225–239. [DOI] [PubMed] [Google Scholar]
- 10.Nguyen MB, Cohen I, Kumar V, et al. FGF signalling controls the specification of hair placode-derived SOX9 positive progenitors to Merkel cells. Nat Commun. 2018;9(1):2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Molotkov A, Mazot P, Brewer JR, Cinalli RM, Soriano P. Distinct Requirements for FGFR1 and FGFR2 in Primitive Endoderm Development and Exit from Pluripotency. Dev Cell. 2017;41(5):511–526 e514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu Z, Wang W, Jiang K, et al. Embryonic attenuated Wnt/beta-catenin signaling defines niche location and long-term stem cell fate in hair follicle. Elife. 2015;4:e10567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohen I, Zhao D, Bar C, et al. PRC1 Fine-tunes Gene Repression and Activation to Safeguard Skin Development and Stem Cell Specification. Cell Stem Cell. 2018;22(5):726–739 e727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg S. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biology. 2013;14(4):R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trapnell C, Roberts A, Goff L, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protocols. 2012;7(3):562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30. [DOI] [PubMed] [Google Scholar]
- 17.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. [DOI] [PubMed] [Google Scholar]
- 19.Maricich SM, Wellnitz SA, Nelson AM, et al. Merkel cells are essential for light-touch responses. Science. 2009;324(5934):1580–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moll I, Kuhn C, Moll R. Cytokeratin 20 is a general marker of cutaneous Merkel cells while certain neuronal proteins are absent. J Invest Dermatol. 1995;104(6):910–915. [DOI] [PubMed] [Google Scholar]
- 21.Perdigoto CN, Bardot ES, Valdes VJ, Santoriello FJ, Ezhkova E. Embryonic maturation of epidermal Merkel cells is controlled by a redundant transcription factor network. Development. 2014;141(24):4690–4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiao Y, Thoresen DT, Miao LL, et al. A Cascade of Wnt, Eda, and Shh Signaling Is Essential for Touch Dome Merkel Cell Development. Plos Genetics. 2016;12(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perdigoto CN, Dauber KL, Bar C, et al. Polycomb-Mediated Repression and Sonic Hedgehog Signaling Interact to Regulate Merkel Cell Specification during Skin Development. PLoS Genet. 2016;12(7):e1006151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dauber KL, Perdigoto CN, Valdes VJ, Santoriello FJ, Cohen I, Ezhkova E. Dissecting the Roles of Polycomb Repressive Complex 2 Subunits in the Control of Skin Development. J Invest Dermatol. 2016;136(8):1647–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Keymeulen A, Mascre G, Youseff KK, et al. Epidermal progenitors give rise to Merkel cells during embryonic development and adult homeostasis. Journal of Cell Biology. 2009;187(1):91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bardot ES, Valdes VJ, Zhang JS, et al. Polycomb subunits Ezh1 and Ezh2 regulate the Merkel cell differentiation program in skin stem cells. Embo J. 2013;32(14):1990–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lesko MH, Driskell RR, Kretzschmar K, Goldie SJ, Watt FM. Sox2 modulates the function of two distinct cell lineages in mouse skin. Dev Biol. 2013;382(1):15–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ornitz DM, Itoh N. The Fibroblast Growth Factor signaling pathway. Wires Dev Biol. 2015;4(3):215–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Owens DM, Lumpkin EA. Diversification and specialization of touch receptors in skin. Cold Spring Harb Perspect Med. 2014;4(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Niu J, Vysochan A, Luo W. Dual innervation of neonatal Merkel cells in mouse touch domes. Plos One. 2014;9(3):e92027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haeberle H, Fujiwara M, Chuang J, et al. Molecular profiling reveals synaptic release machinery in Merkel cells. Proc Natl Acad Sci U S A. 2004;101(40):14503–14508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoffman BU, Baba Y, Griffith TN, et al. Merkel Cells Activate Sensory Neural Pathways through Adrenergic Synapses. Neuron. 2018;100(6):1401–1413 e1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blakemore SJ, Tavassoli T, Calo S, et al. Tactile sensitivity in Asperger syndrome. Brain Cognition. 2006;61(1):5–13. [DOI] [PubMed] [Google Scholar]
- 34.Tomchek SD, Dunn W. Sensory processing in children with and without autism: A comparative study using the short sensory profile. Am J Occup Ther. 2007;61(2):190–200. [DOI] [PubMed] [Google Scholar]
- 35.Richards KF, Guastafierro A, Shuda M, Toptan T, Moore PS, Chang Y. Merkel cell polyomavirus T antigens promote cell proliferation and inflammatory cytokine gene expression. J Gen Virol. 2015;96(12):3532–3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang W, Kanda H, Ikeda R, Ling J, DeBerry JJ, Gu JG. Merkel disc is a serotonergic synapse in the epidermis for transmitting tactile signals in mammals. Proc Natl Acad Sci U S A. 2016;113(37):E5491–5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feng J, Luo J, Yang P, Du J, Kim BS, Hu H. Piezo2 channel-Merkel cell signaling modulates the conversion of touch to itch. Science. 2018;360(6388):530–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Samimi M, Touze A. Merkel cell carcinoma: The first human cancer shown to be associated with a polyomavirus. Presse Med. 2014;43(12 Pt 2):e405–411. [DOI] [PubMed] [Google Scholar]
- 39.Thakuria M, LeBoeuf NR, Rabinowits G. Update on the biology and clinical management of Merkel cell carcinoma. Am Soc Clin Oncol Educ Book. 2014:e405–410. [DOI] [PubMed] [Google Scholar]
- 40.Boyse K, Foley EH, Bradley V, Scarborough D. Merkel cell carcinoma: a case report with treatment summary and updates. Cutis. 2004;74(6):350–356. [PubMed] [Google Scholar]
- 41.Porceddu SV, Veness MJ, Guminski A. Nonmelanoma Cutaneous Head and Neck Cancer and Merkel Cell Carcinoma: Current Concepts, Advances, and Controversies. J Clin Oncol. 2015;33(29):3338–+. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data of back skin/paw glabrous skin Merkel cells and IFE at P0 were uploaded to the GEO database (GSE122598; this study). RNA-seq data of hair follicle cells at P0 was obtained from the previous publication13 and in the GEO database (GSM3069360, GSM3069361 and GSM3069362). Additional data that support the findings of this study is available from the corresponding author upon request.