Abstract
A ureidoglycolate-degrading activity was analyzed in different organs of chickpea (Cicer arietinum). Activity was detected in all the tissues analyzed, but highest levels of specific activity were found in pods, from which it has been purified and characterized. This is the first ureidoglycolate-degrading activity that has been purified to homogeneity from any photosynthetic organism. Only one ureidoglycolate-degrading activity was found during the purification. The enzyme was purified 1,500-fold, and specific activity for the pure enzyme was 8.6 units mg−1, which corresponds with a turnover number of 1,600 min−1. The native enzyme has a molecular mass of 180 kD and consists of six identical or similar-sized subunits of 31 kD each. The enzyme exhibited hyperbolic, Michaelian kinetics for (−) ureidoglycolate with Km values of 6 and 10 μm in the presence or absence of Mn2+, respectively. Optimum pH was between 7 and 8 and maximum activity was found at temperatures above 70°C, the enzyme being extremely stable and resistant to heat denaturation. The activity was inhibited by EDTA and enhanced by several bivalent cations, thus suggesting that the enzyme is a metalloprotein. This enzyme has been characterized as a ureidoglycolate urea-lyase (EC 4.3.2.3), which catalyzes the degradation of (−) ureidoglycolate to glyoxylate and urea. This is the first time that such an activity is detected in plant tissues. A possible function for this activity and its implications in the context of nitrogen mobilization in legume plants is also discussed.
Nitrogen availability often limits plant growth because nitrogen is the most limiting element in plant nutrition after carbon (Schubert, 1986). Leguminous plants establish a symbiotic association with certain soil bacteria such as Rhizobium sp., which are capable of fixing atmospheric nitrogen, and plants assimilate this nitrogen fixed by the bacteria. This organic nitrogen can be transported to the upper parts of the plant either as amides (Asn and Gln) or as ureides (allantoin and allantoate), so that legumes are classified as amide or ureide exporters according to the compounds used for nitrogen mobilization.
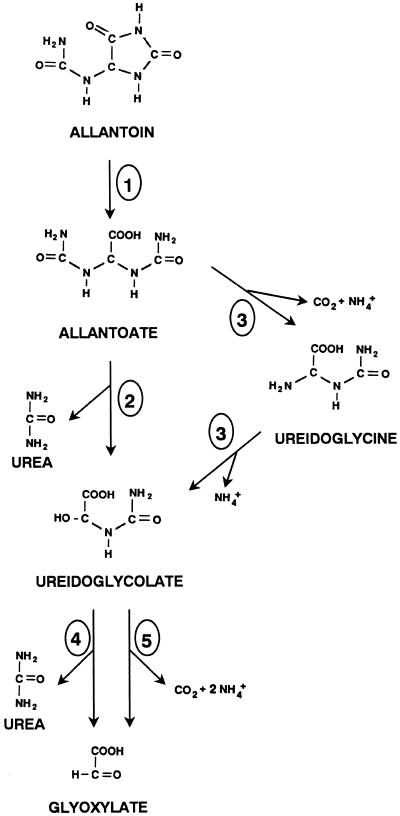
The major route for ureide biogenesis is the enzymatic oxidation of purine to allantoin, a pathway that is now being fully understood in plants (Schubert and Boland, 1990), and the doubts about the final step of urate oxidation and allantoin synthesis have been recently resolved (Sarma et al., 1999). The conversion of ureides into useful metabolic intermediates involves the release of nitrogen and possibly a two-carbon molecule (Schubert and Boland, 1990). Possible pathways for the catabolism of ureides in plants are shown in Figure 1. Allantoin is degraded to allantoate by allantoinase (EC 3.5.2.5), which has been well characterized from plant extracts (Wells and Lees, 1992; Webb and Lindell, 1993; Bell and Webb, 1995). According to the studies done with bacteria, fungi, algae, and animals, allantoate degradation can be catalyzed either by allantoate amidohydrolase (EC 3.5.3.9) or by allantoate amidinohydrolase (EC 3.5.3.4). Both allantoate-degrading enzymes produce ureidoglycolate, which is metabolized to glyoxylate by either ureidoglycolate urea-lyase (EC 4.3.2.3) or ureidoglycolate amidohydrolase (EC 3.5.3.19). Those activities differ in the nature of the nitrogen compound that is released: Allantoate and ureidoglycolate amidohydrolases yield ammonium, whereas allantoate amidinohydrolase and ureidoglycolate urea-lyase release urea.
Figure 1.
Putative pathways for ureide degradation in plants. The enzymatic activities represented are: allantoinase (1), allantoate amidinohydrolase (2), allantoate amidohydrolase (3), ureidoglycolate urea-lyase (4), and ureidoglycolate amidohydrolase (5).
The exact pathway for ureide degradation in plants was suggested only on the basis of physiological studies. One study showed urea accumulation in soybean (Glycine max) leaf pieces incubated in the presence of allantoin and a urease inhibitor (Shelp and Ireland, 1985). However, urease inhibitors could be inhibiting ureide-degrading enzymes as well (Winkler et al., 1985). Studies of ureide catabolism using urease-deficient mutant soybean plants (Stebbins and Polacco, 1995) or others based on the Ni2+ requirement of urease (Polacco et al., 1982) support the idea of ureide degradation without any detectable urea production. In addition, labeled ureides are degraded to ammonium without detectable urea (Winkler et al., 1987). Stahlhut and Widholm (1989a, 1989b) presented evidence for the ammonium pathway in soybean cell suspension cultures. Thus, it seems to be evidence for ureide degradation by allantoate amidohydrolase and ureidoglycolate amidohydrolase, rather than by urea-releasing enzymes, although both pathways could work in parallel. In fact, it has been reported that soybean degrades ureides mainly by the amidohydrolase pathway producing ammonium, although with a small portion of ureide degradation through urea (Stebbins and Polacco, 1995).
Allantoate- or ureidoglycolate-degrading activities have been determined in just a few cases in plants (Singh, 1968; Winkler et al., 1985; Wells and Lees, 1991; Lukaszewski et al., 1992) and green algae (Piedras et al., 1998). Allantoicase from the alga Chlamydomonas reinhardtii is the only allantoate-degrading activity purified from a photosynthetic organism (Piedras et al., 2000). Only one ureidoglycolate-degrading enzyme has been partially purified, from common bean developing pods (Wells and Lees, 1991), and this enzyme has been reported to be a ureidoglycolate amidohydrolase. There is no clear evidence of ureidoglycolate urea-lyase, and this activity has never been detected in plant extracts. In contrast, most of the ureidoglycolate-degrading activities detected in microorganisms and animals have been reported as lyases (Trijbels and Vogels, 1967; Takada and Noguchi, 1986; Takada and Tsukiji, 1987; Fujiwara and Noguchi, 1995).
In our approach to understand ureide catabolism in plants, we have studied ureidoglycolate degradation in the legume chickpea (Cicer arietinum), a plant species that uses Asn for nitrogen export from the root nodules to the aerial organs (Atkins, 1991). We have detected a single activity in our crude extracts, which was purified to electrophoretic homogeneity. This enzyme is a (−) ureidoglycolate urea-lyase that catalyzes the degradation of (−) ureidoglycolate to glyoxylate and urea; this is the first time that such an activity has been reported in plant extracts.
RESULTS
Purification of the Ureidoglycolate-Degrading Activity
We analyzed ureidoglycolate-degrading activity in cotyledons, seedlings, leaves, and developing pods of chickpea. Activity was detected in all the tissues examined, but with highest specific activity (up to 6 milliUnits [U] mg−1) in developing pods (data not shown). Therefore, this tissue was chosen as starting material for further analysis. During the purification only one peak of activity was detected, suggesting that a single ureidoglycolate-degrading enzyme is the dominant activity.
The enzyme responsible for this activity was purified to electrophoretic homogeneity, using the procedure listed in Table I, and a purification factor of 1,530 was obtained, suggesting that ureidoglycolase could represent as little as 0.06% of the total protein present in the crude extract. The purification procedure had a 5.9% yield and the specific activity of purified ureidoglycolase was 8.6 U mg−1 protein (Table I).
Table I.
Purification procedure for ureidoglycolate-degrading activity from chickpea
| Step | Total Activity | Total Protein | Specific Activity | Purification Factor | Yield |
|---|---|---|---|---|---|
| mU | mg | mU mg−1 | % | ||
| Crude extract | 9,261 | 1,650 | 5.6 | – | 100 |
| Ammonium sulfate | 5,393 | 498 | 10.8 | 1.9 | 58.2 |
| Gel filtration | 4,224 | 323 | 13.1 | 2.3 | 45.6 |
| Heat treatment | 2,713 | 36 | 75.8 | 13.5 | 29.3 |
| Ion exchange | 1,249 | 3 | 416.2 | 74.3 | 13.5 |
| Electrophoresis | 549 | 0.064 | 8,573.4 | 1,531 | 5.9 |
Substrate Stereoselectivity and Reaction Products
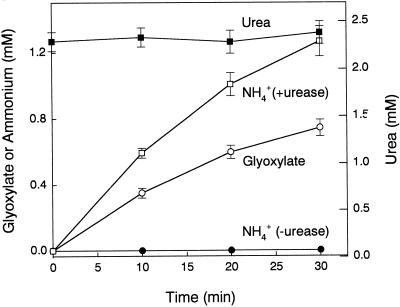
To distinguish between the two types of ureidoglycolate-degrading activities (Fig. 1), reaction products were determined for the enzyme from chickpea. As shown in Figure 2, the production of glyoxylate was not associated with the production of ammonium. However, when aliquots from the same reactions were later incubated with urease, ammonium was detected at levels doubling the glyoxylate concentration (Fig. 2). Urea production was determined also by the diacetyl monoxime method. In this method, there is a step consisting of a boiling treatment of the samples, which nonenzymatically degrades the remaining ureidoglycolate to glyoxylate and urea. If the enzymatic degradation of ureidoglycolate was associated with ammonium production, the concentration of urea would decrease as the glyoxylate concentration increased. On the contrary, if the enzyme produced urea, its concentration would keep constant during the reaction. The concentration of urea during the reaction catalyzed by the enzyme from chickpea remained constant (Fig. 2), indicating that the nitrogenous compound released is urea.
Figure 2.
Reaction products of ureidoglycolase purified from chickpea pods. Enzymatic activity was performed in 100 mm Tea (triethanolamine) buffer (pH 7.8), 0.5 mm MnSO4, 3 mm phenylhydrazine, 2.5 mm ureidoglycolate, and purified enzyme (urease-free) and incubated at 30°C. At the indicated times, aliquots were taken and analyzed for glyoxylate (white circles), urea (black squares), and ammonium (black circles). Ammonium production was also monitored in the presence of urease (white squares). Urea values represent free urea plus urea derived from ureidoglycolate .
We have purified an allantoicase activity from C. reinhardtii enzyme that catalyzes the degradation of allantoate to (−) ureidoglycolate (Piedras et al., 2000). The enzyme from chickpea was able to use this (−) ureidoglycolate generated by C. reinhardtii allantoicase as substrate, catalyzing its degradation to glyoxylate. To assess if the enzyme can use (+) ureidoglycolate as substrate as well, we performed activity assays both with the (−) ureidoglycolate produced by C. reinhardtii allantoicase and with commercial ureidoglycolate (which is a racemic mixture of both isomers; Table II). Substrate concentrations (6 μm) lower than the Km (12 μm for racemic ureidoglycolate) were used to obtain first-order kinetics, and therefore the reaction rate would depend on substrate concentration. The activity was approximately double with (−) ureidoglycolate (Table II), indicating that the amount of substrate was double. As a consequence, this result indicates that (+) ureidoglycolate is not a substrate for ureidoglycolase from chickpea. Therefore, the purified enzyme catalyzes the degradation of (−) ureidoglycolate to glyoxylate and urea, so it is a (−) ureidoglycolate urea-lyase (EC 4.3.2.3; Enzyme Nomenclature Recommendations, 1992).
Table II.
Glyoxylate formation by chickpea ureidoglycolate urea-lyase with (±) ureidoglycolate and (−) ureidoglycolate as substrates
| Substrate (6 μm)a | Glyoxylate
Produced
|
|
|---|---|---|
| Heatb | Enzymec | |
| μm | ||
| (±) Ureidoglycolate | 5.92 | 0.72 |
| (−) Ureidoglycolated | 6.02 | 1.38 |
Standard reaction mixtures were performed with 6 μm of either (±) ureidoglycolate or (−) ureidoglycolate.
This substrate concentration was obtained by using the appropriate volume of a stock solution in which the actual concentration was determined after the transformation of ureidoglycolate to glyoxylate by boiling for 10 min.
To confirm the initial ureidoglycolate concentration in the reaction mixtures, glyoxylate was determined in an aliquot of these mixtures after transformation of ureidoglycolate to glyoxylate by heat treatment (10 min boiling).
Glyoxylate formed enzymatically after 30-min incubation at 30°C. Data for this enzymatic production are corrected by subtracting the nonenzymatic degradation.
(−) Ureidoglycolate was obtained by incubating 7 mm allantoate, 0.5 mm MnSO4, and pure allantoicase from C. reinhardtii in 50 mm TES (pH 7.8).
Enzyme Properties
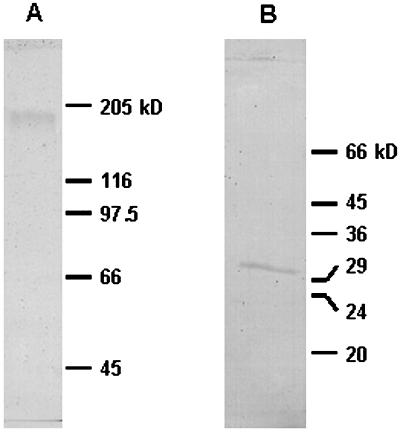
A molecular mass of about 180 kD was found by gel filtration (see “Materials and Methods” and Table III). A similar value was determined by electrophoresis in the presence of SDS (Fig. 3A). However, when the pure enzyme was subjected to electrophoresis under reducing conditions, a single band with a molecular mass of 31 kD was obtained (Fig. 3B). These data suggest that the enzyme is a hexamer of about 186 kD consisting of six identical or similar-sized subunits of 31 kD each, bound by disulfide bridges.
Table III.
Physicochemical and molecular properties of ureidoglycolate urea-lyase from chickpea
| Stokes' radius (nm) | 5.6 |
| Molecular mass, native enzyme | |
| Gel filtration | 180 |
| SDS-PAGE | 184 |
| Molecular mass, denatured enzyme | 31 |
| Number of monomers | 6 |
| Type of subunits | 1 |
| Optimum pH | 7–8 |
| Optimum temperature (°C) | >70 |
| Activation energy (kJ mol−1) | 57 |
| Increase in rate of process produced by raising temperature by 10°C (40°C to 50°C) | 2.2 |
| Km (μM) | 12 |
| Turnover number (min−1) | 1,600 |
Figure 3.
SDS-PAGE of the purified ureidoglycolase from chickpea. A, The sample was mixed with loading buffer containing SDS, and loaded in SDS-PAGE with 6% (w/v) acrylamide. B, The sample was mixed with loading buffer containing SDS and dithiothreitol, boiled for 5 min, and loaded in SDS-PAGE with acrylamide concentration of 10% (w/v). Both gels were stained with Coomassie Brilliant Blue R-250.
Ureidoglycolase from chickpea showed hyperbolic kinetics for ureidoglycolate, with a Km value of 12 μm when commercial (±) ureidoglycolate was used, in the presence of 0.5 mm manganese. Because only (−) ureidoglycolate is the substrate, the Km would be 6 μm. This Km increased to 20 μm without Mn2+. The Vmax for the reaction catalyzed by the ureidoglycolate lyase from chickpea was influenced by Mn2+, with the Vmax approximately double in the presence of Mn2+. These data suggest a catalytic role for the metal, which is corroborated by the fact that the activity was completely inhibited by incubation with the chelator EDTA (data not shown). In addition, when this preparation was dialyzed to remove EDTA, the activity was recovered to about 50% of the initial level, suggesting that ions are tightly bound to the protein and are not fully removed during EDTA treatment. This less active preparation was incubated with several cations to test their effect. Mn2+, Zn2+, and Mg2+ recovered the activity to the initial levels or even higher, whereas other ions such as Fe2+ and Ni2+ strongly inactivated the enzyme (not shown).
Allantoate is a structural analog of ureidoglycolate. Although the purified enzyme from chickpea does not degrade allantoate to glyoxylate, we tested the possible inhibitory effect of allantoate, but no inhibition was observed. Other compounds that did not inhibit the enzyme were urea, ammonium, albiziin, and citruline. All the inhibitors were used at a final concentration of 2.5 mm (equimolar with ureidoglycolate).
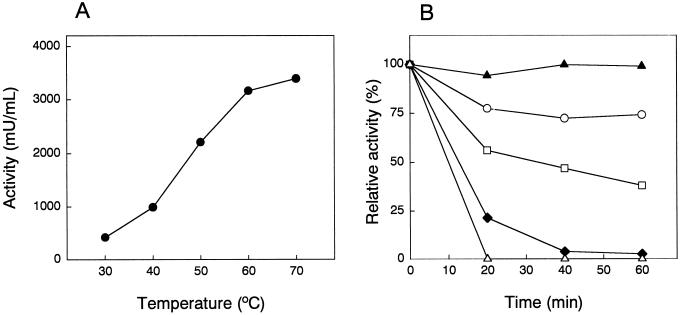
The optimal pH for ureidoglycolase activity was between 7 and 8, and the activity was fully stable between pH 6 and 9. The optimum temperature for the activity in vitro was above 70°C, and higher temperatures could not be tested due to ureidoglycolate chemical instability at such temperatures (Fig. 4A). Activation energy was 57 kJ mol−1 and the increase in rate of process produced by raising temperature by 10°C (40°C–50°C) was 2.2. The enzyme showed a remarkable stability to heat denaturation because most of the activity was retained after 60 min treatment at 80°C in the presence of manganese. The enzyme was more susceptible to temperature denaturation when Mn2+ was removed by dialysis (Fig. 4B).
Figure 4.
Effect of temperature on the activity and stability of ureidoglycolate urea-lyase from chickpea. A, Enzymatic activity was determined at the indicated temperatures. B, Dialyzed extracts were pre-incubated overnight in the presence (black symbols) or absence (white symbols) of 1 mm MnSO4. Both enzyme preparations were incubated at 60°C (circles), 70°C (squares), 80°C (triangles), and 90°C (diamonds) during 20, 40, and 60 min. After cooling down the samples, the activity was assayed under standard conditions.
DISCUSSION
We have detected and purified an ureidoglycolate-degrading enzyme from the legume chickpea. This enzyme has been characterized as an ureidoglycolate urea-lyase (EC 4.3.2.3) because it catalyzes the degradation of (−) ureidoglycolate to glyoxylate and urea. The enzyme consists of six subunits bound by disulfide bridges. This molecular structure is unique among the few ureidoglycolate-degrading enzymes studied. Ureidoglycolate lyase from fish liver consist of two subunits of 60 kD each (Takada and Noguchi, 1986), whereas the same enzyme from rat liver is a dimer of 64 kD consisting of two subunits of 33 kD each (Fujiwara and Noguchi, 1995). The only datum available from plants, the ureidoglycolate amidohydrolase from French bean, is a protein of 300 kD and no information about the subunits was reported (Wells and Lees, 1991). However, this enzyme shared some properties with other ureidoglycolate-degrading activities, such as the activation by manganese (Trijbels and Vogels, 1967; Takada and Noguchi, 1986; Takada and Tsukiji, 1987; Wells and Lees, 1991; Piedras et al., 2000) and the heat stability (Trijbels and Vogels, 1967; Wells and Lees 1991; Piedras et al., 2000).
During the last years, there has been an intense debate about which activities (ammonium or urea producers) are actually responsible for ureide degradation in plants. The presence of ureidoglycolate urea-lyase activity demonstrates the existence of a urea-producing pathway for ureide catabolism, although this does not rule out the existence of amidohydrolase activities in other plant species or even in the same one. Previous reports showed that the legume soybean degrades most ureides to ammonia, except for a minor fraction that is degraded to urea, the latter having little importance for the growth of N2-fixing soybean (Winkler et al., 1987; Stebbins and Polacco, 1995). In chickpea, a plant that is now classified as amide instead of ureide (Atkins, 1991), we saw no indication of a second ureidoglycolate-degrading activity during ureidoglycolase purification. The presence of two ureidoglycolate-degrading activities with the same role in the same plant tissue appears unlikely. Chickpea ureidoglycolase is a ureidoglycolate urea-lyase, and the enzyme shows clear differences in comparison with the enzyme from French bean, a ureide legume.
In an attempt to put together these diverse results, we hypothesize that ureide plants export ureides from the roots to the aerial organs, where there are two degradative pathways, the ammonium-producing pathway being relevant in N2-fixing conditions, whereas the urea-producing one would have a minor role in this process. In amide plants, the ammonium-releasing pathway is less important because these species export amides instead of ureides from their roots, consistent with only the ureidoglycolate urea-lyase being detected in the amide chickpea plant. On the other hand, it will be interesting to test the hypothesis that ureidoglycolate urea-lyase has a role unrelated to ureide catabolism, as suggested by Fujiwara and Noguchi (1995) for the rat ureidoglycolate urea-lyase. These authors suggest a role in creatine synthesis for the rat ureidoglycolate urea-lyase. This hypothesis is supported by the loss of the preceding enzymes in the pathway in mammals. To complete the picture of ureide catabolism in plants, it will be necessary to detect, purify, and characterize the activity responsible for the production of (-) ureidoglycolate, the substrate of ureidoglycolate urea-lyase, because there are no clear studies on ureidoglycolate-producing enzymes in plants.
MATERIALS AND METHODS
Plant Material
Developing pods of chickpea (Cicer arietinum) were obtained from nodulated plants grown in a farm near Córdoba, Spain. After harvesting, the material was immediately frozen with liquid nitrogen and stored at −80°C until use.
Preparation of Crude Extract
Plant material was homogenized with a blender (Waring, New Hartford, CT) by adding 2 mL of working buffer per gram of tissue. The working buffer was 50 mm TES [N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid]-NaOH, pH 7.8, and contained 1 mm MnSO4. The suspension was filtered through two layers of Miracloth (Calbiochem, La Jolla, CA) and the resulting homogenate was centrifuged at 22,000g for 20 min. The supernatant was considered as crude extract.
Purification
Unless otherwise stated, all purification steps were carried out at 4°C in working buffer. Crude extracts were brought to 30% saturation with ammonium sulfate by stepwise addition of the salt. After gentle stirring for 30 min, the suspension was centrifuged at 22,000g for 20 min. The supernatant was recovered and brought to 50% ammonium sulfate saturation, stirred, and centrifuged as above. The resulting pellet was resuspended in the minimal volume of working buffer, and centrifuged at 82,000g for 1 h and the supernatant was passed through an S-300 HR column (82 × 2.6 cm; Sephacryl, Uppsala) equilibrated with working buffer at a flow rate of 60 mL h−1.
Fractions (4.5 mL) with high activity were pooled and heated to 60°C for 60 min. After cooling to 4°C, the solution was centrifuged at 22,000g for 25 min. The resulting supernatant was brought to 60% ammonium sulfate and precipitated as above. The pellet was dissolved in working buffer and desalted by dialysis against 3,000 volumes.
This dialyzed solution was then subjected to an FPLC (Mono Q HR 5/5; Pharmacia, Uppsala). After loading, the column was washed with working buffer containing the following concentrations of NaCl: a gradient from 0 to 0.13 m NaCl (3 volumes), 4 volumes of 0.13 m NaCl, a final gradient (10 volumes) from 0.13 to 0.24 m NaCl, 4 volumes of 0.24 m NaCl, and finally 5 volumes of 1 m NaCl. Chromatography was carried out at a flow rate of 1 mL min−1 and the collected fractions contained 1 mL. Due to the loading capacity of the column, this chromatography was performed several times with aliquots of the original sample. The fractions with activity were pooled, dialyzed, and concentrated using Centriplus-10 and Centricon-10 (Amicon, Bedford, MA).
The dialyzed sample was subjected to native electrophoresis in a PrepCell apparatus (Bio-Rad Laboratories, Hercules, CA) with polyacrylamide concentrations of 5% (w/v) for separating (4.1 cm high and 3.6-cm2 section) and 4% (w/v) for stacking (1 cm high). The electrophoresis was run for 14 h at 6 W. Proteins were eluted with 25 mm Tris-Gly buffer (pH 8.3) at a flow rate of 45 mL h−1. Fractions of 4.6 mL were collected. Those fractions with activity were pooled, concentrated, and dialyzed against working buffer, and used as source of pure enzyme.
Enzymatic Activities
Unless otherwise stated, the standard reactions were carried out at 30°C and started by the addition of the substrate. Due to the high instability of ureidoglycolate (Pineda et al., 1994), controls to account for the nonenzymatic hydrolysis were performed in parallel.
The activity was determined by either a colorimetric or a continuous assay. The colorimetric assay was performed as described by Wells and Lees (1991). The standard reaction mixture was 50 mm TES (pH 7.8), 2.5 mm ureidoglycolate, 0.5 mm MnSO4, and 0.15% (w/v) phenylhydrazine. At different times 0.4-mL aliquots were taken and glyoxylate phenylhydrazone was determined as described by Vogels and Van der Drift (1970). The continuous assay was carried out as described by Pineda et al. (1994). The standard reaction mixture contained 50 mm TES-NaOH, 2.5 mm ureidoglycolate, 0.5 mm MnSO4, and 3 mm phenylhydrazine. The formation of glyoxylate phenylhydrazone was monitored at 324 nm. One unit of enzyme is defined as the amount that catalyzes the formation of 1 μmol of glyoxylate per minute.
Determination of Molecular Parameters
The Stokes' radius was determined according to Siegel and Monty (1966) in a Superdex 200 HR 10/30 column (Pharmacia) equilibrated with working buffer, using the following standards: carbonic anhydrase from bovine erythrocytes (2.43 nm), albumin from bovine serum (3.70 nm), alcohol dehydrogenase from yeast (4.61 nm), and apoferritin from horse spleen (7.80 nm).
The native molecular mass of the enzyme was determined by gel filtration through the same Superdex column. As markers, the following standards were used: carbonic anhydrase from bovine erythrocytes (29 kD), albumin from bovine serum (66 kD), alcohol dehydrogenase from yeast (150 kD), β-amylase from sweet potato (200 kD), and apoferritin from horse spleen (443 kD). The molecular mass was also determined by SDS-PAGE under nonreducing conditions. As markers the following standards were used: myosin from rabbit muscle (205 kD), β-galactosidase from Escherichia coli (116 kD), phosphorylase b from rabbit muscle (97.5 kD), albumin from bovine serum (66 kD), and albumin from egg (45 kD).
The molecular mass was also determined by SDS-PAGE under denaturing and reducing conditions (boiling the sample in the presence of 100 mm dithiothreitol). The following standards were used: albumin from bovine serum (66 kD), albumin from egg (45 kD), glyceraldehyde 3-phosphate dehydrogenase from rabbit muscle (36 kD), carbonic anhydrase from bovine erythrocytes (29 kD), trypsinogen from bovine pancreas (24 kD), and trypsin inhibitor from soybean (20 kD).
Analytical Determinations
Protein concentration was determined by the method of Bradford (1976) with the Bio-Rad system using bovine serum albumin as a standard. Ammonium was measured by the phenol-hypochlorite method as described by Solorzano (1969). Urea was determined either as ammonium after treatment with urease S from Canavalia ensiformis or by the diacetyl monoxime method (Kaplan, 1969). Ureidoglycolate and glyoxylate were determined as described by Vogels and Van der Drift (1970).
ACKNOWLEGMENTS
We thank the Ministerio de Educación y Cultura (Madrid) for the award of a predoctoral fellowship (A.M.) and incorporation contracts (P.P. and M.A.).
Footnotes
This work was supported by Dirección General de Enseñanza Superior e Investigación Científica (grant no. PB96–0504–C02–02) and Plan Andaluz de Investigación (grant no. CVI–0115).
LITERATURE CITED
- Atkins CA. Ammonia assimilation and export of nitrogen from the legume nodule. In: Dilworth M, Glenn A, editors. Biology and Biochemistry of Nitrogen Fixation. Amsterdam: Elsevier Science Publishers B.V.; 1991. pp. 293–319. [Google Scholar]
- Bell JA, Webb MA. Immunoaffinity purification and comparison of allantoinases from soybean root nodules and cotyledons. Plant Physiol. 1995;107:435–441. doi: 10.1104/pp.107.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Enzyme Nomenclature Recommendations of the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology. San Diego: Academic Press; 1992. [Google Scholar]
- Fujiwara S, Noguchi T. Degradation of purines: ureidoglycollate lyase out of four allantoin-degrading enzymes is present in mammals. Biochem J. 1995;312:315–318. doi: 10.1042/bj3120315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan A. The determination of urea, ammonia, and urease. Methods Biochem Anal. 1969;17:311–324. doi: 10.1002/9780470110355.ch7. [DOI] [PubMed] [Google Scholar]
- Lukaszewski KM, Blevins DG, Randall DD. Asparagine and boric acid cause allantoate accumulation in soybean leaves by inhibiting manganese-dependent allantoate amidohydrolase. Plant Physiol. 1992;99:1670–1676. doi: 10.1104/pp.99.4.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piedras P, Aguilar M, Pineda M. Uptake and metabolism of allantoin and allantoate by cells of Chlamydomonas reinhardtii (Chlorophyceae) Eur J Phycol. 1998;33:57–64. [Google Scholar]
- Piedras P, Muñoz A, Aguilar M, Pineda M. Allantoate amidinohydrolase (allantoicase) from Chlamydomonas reinhardtii: its purification and catalytic and molecular characterization. Arch Biochem Biophys. 2000;378:340–348. doi: 10.1006/abbi.2000.1833. [DOI] [PubMed] [Google Scholar]
- Pineda M, Piedras P, Cárdenas J. A continuous spectrophotometric assay for ureidoglycolase activity with lactate dehydrogenase or glyoxylate reductase as coupling enzyme. Anal Biochem. 1994;222:450–455. doi: 10.1006/abio.1994.1515. [DOI] [PubMed] [Google Scholar]
- Polacco JC, Thomas AL, Bledsoe P. A soybean seed urease-null produces urease in cell culture. Plant Physiol. 1982;69:1233–1240. doi: 10.1104/pp.69.5.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma AD, Serfozo P, Kahn K, Tipton PA. Identification and purification of hydroxyisourate hydrolase, a novel ureide-metabolizing enzyme. J Biol Chem. 1999;274:33863–33865. doi: 10.1074/jbc.274.48.33863. [DOI] [PubMed] [Google Scholar]
- Schubert KR. Products of biological nitrogen fixation in higher plants: synthesis, transport, and metabolism. Annu Rev Plant Physiol. 1986;37:539–574. [Google Scholar]
- Schubert KR, Boland MJ. The ureides. In: Miflin BJ, Lea PJ, editors. The Biochemistry of Plants. Vol. 16. San Diego: Academic Press; 1990. pp. 197–283. [Google Scholar]
- Shelp BJ, Ireland RJ. Ureide metabolism in leaves of nitrogen-fixing soybean plants. Plant Physiol. 1985;77:779–783. doi: 10.1104/pp.77.3.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel LM, Monty KJ. Determination of molecular weights and frictional ratios of proteins in impure systems by use of gel filtration and density gradient centrifugation: application to crude preparations of sulfite and hydroxylamine reductase. Biochim Biophys Acta. 1966;112:346–362. doi: 10.1016/0926-6585(66)90333-5. [DOI] [PubMed] [Google Scholar]
- Singh R. Evidence for the presence of allantoicase in germinating peanuts. Phytochemistry. 1968;7:1503–1508. [Google Scholar]
- Solorzano L. Determination of ammonia in natural waters by the phenolhypochlorite method. Limnol Oceanogr. 1969;14:799–801. [Google Scholar]
- Stahlhut RW, Widholm JM. Ureide catabolism by soybean [Glycine max (L.) Merrill] cell suspension cultures: I. Urea is not an intermediate in allantoin degradation. J Plant Physiol. 1989a;134:85–89. [Google Scholar]
- Stahlhut RW, Widholm JM. Ureide catabolism by soybean [Glycine max (L.) Merrill] cell suspension cultures: II. Assimilation of allantoin. J Plant Physiol. 1989b;134:90–97. [Google Scholar]
- Stebbins NE, Polacco JC. Urease is not essential for the ureide degradation in soybean. Plant Physiol. 1995;109:169–175. doi: 10.1104/pp.109.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada Y, Noguchi T. Ureidoglycolate lyase, a new metalloenzyme of peroxisomal urate degradation in marine fish liver. Biochem J. 1986;235:391–397. doi: 10.1042/bj2350391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada Y, Tsukiji N. Peroxisomal localization and activation by bivalent metal ions of ureidoglycolate lyase, the enzyme involved in urate degradation in Candida tropicalis. J Bacteriol. 1987;169:2284–2286. doi: 10.1128/jb.169.5.2284-2286.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trijbels F, Vogels GD. Allantoate and ureidoglycolate degradation by Pseudomonas aeruginosa. Biochim Biophys Acta. 1967;132:115–126. doi: 10.1016/0005-2744(67)90197-0. [DOI] [PubMed] [Google Scholar]
- Vogels GD, van der Drift C. Differential analysis of glyoxylate derivatives. Anal Biochem. 1970;33:143–157. doi: 10.1016/0003-2697(70)90448-3. [DOI] [PubMed] [Google Scholar]
- Webb MA, Lindell JS. Purification of allantoinase from soybean seeds and characterization of anti-allantoinase antibodies. Plant Physiol. 1993;103:1235–1241. doi: 10.1104/pp.103.4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells XE, Lees EM. Ureidoglycolate amidohydrolase from developing French bean fruits [Phaseolus vulgaris (L.)] Arch Biochem Biophys. 1991;287:151–159. doi: 10.1016/0003-9861(91)90400-d. [DOI] [PubMed] [Google Scholar]
- Wells XE, Lees EM. Properties of allantoinase from whole developing fruits of french bean, Phaseolus vulgaris L. Aust J Plant Physiol. 1992;19:201–211. [Google Scholar]
- Winkler RG, Blevins DG, Polacco JC, Randall DD. Ureide catabolism in soybeans: II. Pathway of catabolism in intact leaf tissue. Plant Physiol. 1987;83:585–591. doi: 10.1104/pp.83.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler RG, Polacco JC, Blevins DG, Randall DD. Enzymic degradation of allantoate in developing soybeans. Plant Physiol. 1985;79:787–793. doi: 10.1104/pp.79.3.787. [DOI] [PMC free article] [PubMed] [Google Scholar]