Abstract
Considerable progress has been made in understanding the contribution of E3 ubiquitin ligases to health and disease, including the pathogenesis of immunological disorders. Ubiquitin ligases exert exquisite spatial and temporal control over protein stability and function and are thus critical for regulation of both innate and adaptive immunity. Given that immune responses can be both detrimental (autoimmunity) and beneficial (antitumor immunity), it is crucial to understand how ubiquitin ligases maintain immunological homeostasis. Such knowledge could reveal novel mechanisms underlying immune regulation and identify new therapeutic approaches to enhance antitumor immunity and safeguard against autoimmunity.
Keywords: E3 ubiquitin ligase, Autoimmunity, Antitumor immunity, Immune checkpoint
Emerging Challenges in Cancer immunotherapy
FDA approval of immune checkpoint blockers (ICBs) for melanoma and non-small cell lung cancer and recently for other malignancies, is a major breakthrough in cancer treatment [1, 2]. ICBs are monoclonal antibodies or antibody-based molecules that block inhibitory receptors or their ligands, most notably cytotoxic T lymphocyte antigen 4 (CTLA-4), programmed cell death 1 (PD-1), and its ligand, PD-ligand 1 (PD-L1). The physiological functions of these molecules are in part T cell development and maintenance of tolerance to self-antigens, processes dysregulated in the pathogenesis of autoimmune disorders [3, 4]. While blocking these proteins disinhibits T cells, stimulating their expansion and effector function in the tumor microenvironment (TME), activation of auto-reactive T cells, which could promote immune-related adverse events (IRAEs) is often seen with these therapies (Box 1) and remains a major complication [5]. The main challenge, however, remains on how to increase clinical efficacy across tumor (sub-)types in more patients. Currently, combined ICB treatments or regimens that engage neoadjuvant, low dose chemotherapies or radiation have been designed to address this limitation; yet, in most cases the risk of IRAEs remains a concern [2, 5–7]. Paradoxically, as IRAEs also serve as indicators for immune activation [5] but can often be treated without impairing immune checkpoint therapy (ICT) efficiency, a mechanistic link between IRAE and therapeutic efficacy remains elusive [2, 5, 8]. The link between autoimmunity, cancer immunotherapy and IRAEs underscores the need to define how the immune system balances responses to self- and non-self-antigens to improve the efficacy and safety of immunotherapies.
Box 1: Autoimmune phenotypes associated with CTLA-4 and PD-1 blockade.
Despite remarkable success of immune checkpoint blockers (ICBs), autoimmune adverse events have been reported in clinical trials of anti-CTLA-4 and anti-PD-L1/PD-1 antibodies as anti-cancer therapies [5]. Severity varies with the ICB, but the most commonly affected organs are skin, gastrointestinal tract, liver, and endocrine organs, with thyroid events being more common in patients treated with anti-PD-1 [6]. Side effects affecting the central nervous, cardiovascular, pulmonary, musculoskeletal, and hematological systems are less frequently reported [5, 6]. Interestingly, in patients, anti-CTLA-4 treatment generally induces autoimmune effects more frequently than do PD-1/PD-L1-targeting therapies [5, 6], and similar observations are reported in mouse models [99, 100]. In contrast, older PD-1−/− mice develop a mild lupus-like proliferative glomerulonephritis, arthritis [101], or cardiomyopathy [102], whereas PD-L1−/− mice show no evidence of spontaneous autoimmunity [103]. It is important to note that organs affected in animal models and in humans treated with ICBs are different, highlighting the need to develop more appropriate models to investigate ICB-induced IRAEs.
It remains unknown how ICB elicit IRAEs, but elucidating these mechanisms is crucial to prevent life-threatening events and to guide development of ICBs [5]. Aberrant T and B cell activation seen following ICB treatment can elicit IRAEs, since both antitumor and self-reactive cells are reinvigorated (reviewed in [5]). Accordingly, non-specific expansion of T cell clonotypes following anti-CTLA-4 [104] and combined CTLA-4/PD1 therapy [105] and altered effector function of circulating T cells following PD-1 blockade [105] are correlated with IRAE. T cells recognizing tumor cells may also cross-react with self-antigens following ICB or other immunotherapies [106, 107]. Similarly, alterations in B cells following combination ICB may predict IRAE risk in melanoma patients [108] and baseline antibody levels (suggestive of a preexisting autoimmune condition) may predict IRAEs in melanoma [109] or non-small cell lung cancer [110] patients. CTLA-4 and to a lesser extent PD-1 inhibition affects homeoastsis of regulatory T cells (Treg) (reviewed in [111]). Resulting decrease in Treg number or suppressor function can shift the balance between anti-inflammatory responses of Treg and pro-inflammatory responses of T helper subsets, which could contribute to IRAE in tissues where Tregs are abundant, including the gastrointestinal tract, skin, and lungs [111–113]. In support of this, anti-CTLA-4-treated patients exhibit high serum IL-17 levels correlating with IRAE onset [114]. Interestingly, a panel of 11 proinflammatory cytokines has recently been demonstrated to predict risk of IRAE in melanoma patients treated with PD-1 targeted therapy [115].
By controlling protein abundance and activity, ubiquitination serves as a key regulatory mechanism in innate and adaptive immunity and E3 ubiquitin ligases (E3) have been found to perturb both autoimmune and antitumor immune responses [10–14]. E3s are a family of approximately 600 enzymes, many of which are expressed in immune cells, that serve as the specific recognition module of the ubiquitination machinery; E3s bind and ubiquitinate substrate proteins at their lysine (K) residues (Box 2). The topology of ubiquitin chains attached dictates the fate of ubiquitinated proteins, wherein for example K48-linked polyubiquitin chains are usually implicated in proteasome-dependent degradation, K63-linkages function in signaling-complex assembly [9]. This review focuses on E3 function in the context of T cell tolerance, autoimmunity, and antitumor immunity and concludes with an outlook on how the ubiquitin system may be therapeutically harnessed to improve efficacy and safety of cancer immunotherapies.
Box 2: E3 ubiquitin ligases regulate protein stability and function by ubiquitination.
Ubiquitination is a posttranslational modification through which a covalent bond is formed between an ubiquitin protein and either a lysine (K) or a methionine (M) residue in the substrate protein. The process is mediated by the concerted action of three classes of proteins: ubiquitin-activating enzymes (E1; 2 members), ubiquitin-conjugating enzymes (E2; ~35 members), and the ubiquitin ligases (E3; ~600 members). E3 ligases have defined substrate specificities depending on their subcellular localization and posttranslational modifications. The E3s are subcategorized into three major classes based on the domains required for substrate recognition and the mode of Ub transfer: (i) RING (really interesting new gene) domain-containing and U-box-containing E3s, which function as scaffolds for E2 enzymes; (ii) HECT (homologous to the E6-associated protein C-terminus) domain-containing E3s, which form a thiol ester bond with ubiquitin prior to conjugation to the substrate; and (iii) RBR (RING1-between-RING2) E3s, which function as hybrids of RING and HECT E3s [116]. Substrate proteins can be modified by transfer of a single Ub molecule to K (monoubiquitination) or by attachment of multiple ubiquitin moieties to K or M residues to generate homotypic or heterotypic, branched or linear ubiquitin chains (polyubiquitination). This complex Ub code can be read and translated by Ub-binding domains to mediate the distinct outcomes of ubiquitination. For example, monoubiquitination commonly serves as a signal to alter protein function, whereas polyubiquitination regulates proteasomal/lysosomal degradation, protein trafficking, protein complex formation, and protein activity [9].
Control of protein ubiquitination occurs at multiple levels. The activity of E3s is regulated at the transcriptional and posttranslational level, and similarly, posttranslational modification (e.g., phosphorylation, acetylation) of the substrate can modify recognition by E3s. These modifications are often affected by the microenvironment (oxygen, nutrients, stress), enabling E3 ubiquitin ligases to recognize their substrates in a spatial and temporal manner. Conversely, ubiquitin can be removed from proteins by a family of ~100 deubiquitinating enzymes (DUBs). Thus, the complementary activities of E1, E2, and E3 proteins and DUBs regulate protein stability, activity, and localization by controlling the ubiquitin code [117]).
E3 Ubiquitin ligases regulate central and peripheral T cell tolerance
During the random process of somatic recombination, T cell receptors (TCRs) that recognize self-antigens are commonly generated. Notably, meticulously precise filtering mechanisms, termed tolerance, are necessary to eliminate or inactivate these self-reactive T cells in order to prevent autoimmunity. Establishment of T cell tolerance starts during T cell development in the thymus (central tolerance) and is maintained in the periphery (peripheral tolerance).
During T cell development, T cells are first positively selected by their ability to bind to major histocompatibility complex (MHC)-peptides presented by cortical thymic epithelial cells (cTECs), T cells that cannot bind to peptide/MHC complexes undergo death by neglect, while those that can bind survive and progress towards the next developmental stage where tolerance is established by negative selection [15]. Medullary thymic epithelial cells (m)TECs and dendritic cells (DCs) (see Glossary) present an array of self-peptides associated with major MHC class I or II molecules and T cells that express TCRs with high affinity binding to self-peptide/MHC complexes undergo apoptosis (clonal deletion), while those with intermediate TCR affinity differentiate into natural regulatory T cells (nTreg; clonal diversion), which together with CD4+ and CD8+ T cells with low affinity to the self-peptide/MHC complexes exit the thymus and seed the periphery (Figure 1A).
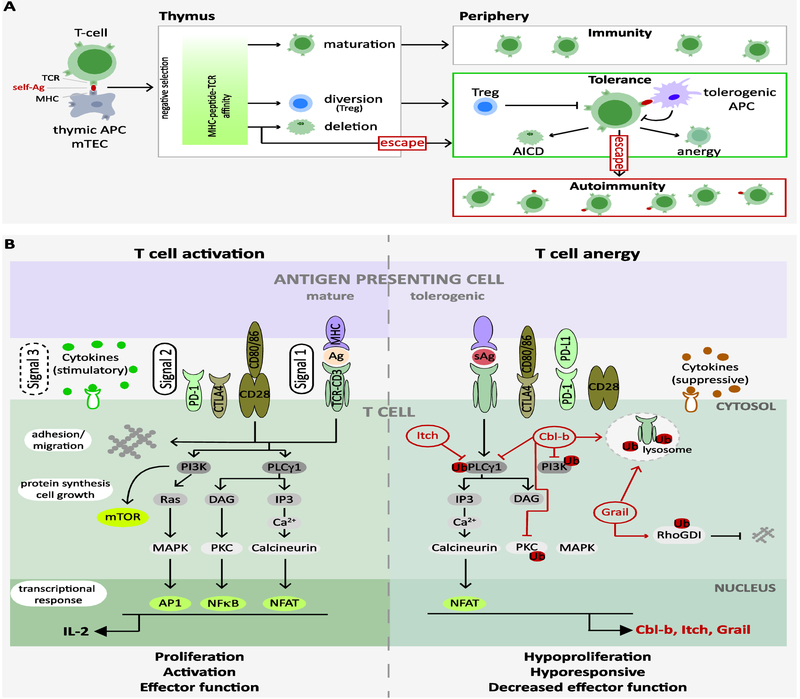
Figure 1: Mechanism of T cell tolerance and activation.
A) Mechanism of central and peripheral tolerance. Binding strength of the TCR to the MHC-self-peptide complex determines T cell fate during negative selection. T cells that bind with high affinity to self-antigens presented by mTECs or DCs are eliminated by a process termed clonal deletion. T cells that bind with lower affinity differentiate/mature and are released into the periphery. Peripheral tolerance is maintained by concerted activity of Tregs, APCs and T cell intrinsic regulatory cues that regulate T cell effector function, survival (Activation induced cell death, AICD) and responsiveness (anergy).
B) T cell activation and anergy. TCR engagement in the context of CD28 costimulation (Signals 1 and 2) leads to reorganization of cell surface molecules into the “immune synapse”, which is critical for recruitment of proximal signaling proteins. A cascade of signaling events is initiated to cooperatively regulate T cell adhesion/migration, growth and proliferation, survival, differentiation and effector function. Production of the effector cytokine IL-2, for example, requires concerted activity of calcineurin, Ras, and PKC-θ, which, in turn, activate the transcription factors NFAT, AP-1, and NF-κB. In contrast, under anergy-inducing conditions, a distinct, largely NFAT-dependent transcriptional program is initiated that fails to upregulate IL-2 production but increases expression of several anergy-regulating genes, including the E3 Ubiquitin ligases Cbl-b, Itch, and Grail (read). These in turn limit expression/activity of major signaling proteins to reprogram T cells into a long-lasting, hyporesponsive state termed anergy. The cytokine milieu (Signal 3) also affects signaling proteins downstream of the TCR to modulate T cell activation and differentiation.
APC: antigen presenting cell; MHC: major histocompatibility complex; mTEC: medullary thymic epithelial cell; Treg: regulatory T cell; AICD: activation-induced cell death; TCR: T cell receptor; Ag: Antigen; sAg: self-Ag;
The E3 ligase tumor necrosis factor (TNF) receptor-associated factor 6 (TRAF6) functions in development and distribution of mTECs in the thymic stroma. TRAF6 inactivation in mice promotes multiorgan autoimmunity [16], while TEC-specific deletion of TRAF6 results in a narrow spectrum of autoimmunity, only affecting the liver [17]. Although the TEC-specific deletion clearly illustrates a role for TRAF6 in negative selection, the more limited autoimmune manifestation as compared to systemic TRAF6 loss also highlights that additional tolerance mechanisms in the periphery limit autoimmunity when central tolerance fails.
Peripheral tolerance is mediated by one of three mechanism, namely activation induced cell death (AICD), promotion of anergy, and suppression by Treg (Figure 1A). As outlined below, E3s function in all of these activities, and many serve to prevent autoimmunity (Table 1).
Table 1.
(Key Table): Mouse models with altered E3 ligase activity and autoimmune phenotypes
| E3 ligase | Mouse model (genetic background) |
Autoimmune phenotypes | Immune cell phenotype | Ref |
|---|---|---|---|---|
| T cells | ||||
| GRAIL |
Rnf128−/− (C57BL/6 × 129 or C57BL/6) |
- Spontaneous autoimmunity in aged mice; splenomegaly, increased mesenteric lymph nodes, lymphocyte infiltration in multiple organs; autoantibody production; - Enhanced experimental autoimmunity upon transfer of Rnf128−/−CD4+ T cells in Rag1−/− mice |
- T cell activation (independent of costimulation) - Treg show impaired suppressor function |
[24] |
| Cbl-b |
CBLB−/− (C57BL/6) |
Spontaneous autoimmunity in aged mice (6 months); autoantibody production, infiltration of activated T and B lymphocytes into multiple organs; parenchymal damage |
- B cells show enhanced proliferation upon stimulation; - T cell activation (independent of costimulation) |
[32] |
|
CBLB−/− (C57BL/6 – 129) |
- No spontaneous autoimmunity (8 month); - High susceptibility to EAE |
- T cell activation independent of costimulation |
[31] | |
|
CBLBC373A (KI of E3 ligase defective mutant) (C57BL/6) |
Spontaneous autoimmunity; autoantibody production; infiltration of mononuclear cells into multiple organs |
- T cell activation independent of costimulation |
[74] | |
| ITCH | Itchy mice (aH18 agouti mice) (C57BL/6J or JU/Ct-C,A) |
Itchy JU/Ct-C,A (age 3–4 months): - intestinal infiltration of inflammatory and mononuclear cells; rare scarring of skin due to scratching Itchy C57BL/6J (lethality between 4–6 months): - no intestinal lesions but airway inflammation; enlarged lymph nodes, spleen and thymus; inflamed ulcerated skin; increased serum IgE and IgG1 |
C57BL/6J background: - enhanced TH2 differentiation; - TH2 resistance to tolerance - diminished generation of inducible Treg |
[26, 33, 126–128] |
| Itchf/lFoxp3Cre | - Spontaneous autoimmunity and lymphoproliferative disease (6 weeks): enlargement and higher cellularity of peripheral lymphoid organs; massive infiltration of multiple organs; - exacerbated airway inflammation upon experimental challenge |
- Treg acquire TH2 properties - enhanced TH2 differentiation from naive T cells |
[49] | |
|
Cbl-b + ITCH |
CBLB−/−ITCH−/− DKO (C57BL/6) |
Spontaneous autoimmunity (6– 12 weeks): Enlarged spleen, lymph nodes; massive lymphocyte infiltration in multiple organs; high concentration of autoantibodies |
- CD4+ T cells (independent of costimulation) |
[129] |
|
WWP2 + ITCH |
Wwp2−/− :Itchf/f:Cd4-cre (C57BL/6) |
Spontaneous autoimmunity: autoantibody production, inflammatory phenotypes, myeloid infiltration of lungs; elevated serum IL6 |
- enhanced TH2 differentiation; - elevated IL4 production |
[130] |
| Roquin1 |
sanroque mice (Rc3h1M199R) (C57BL/6 and CBA) |
Spontaneous autoimmunity: SLE-like glomerulonephritis and deposition of immune complexes; necrotizing hematitis; anemia; enlarged spleen and lymph nodes; spontaneous GC formation in spleen; plasma cell infiltration in multiple organs; autoantibodies |
- CD4+ T cells prone to differentiate into TFH cells |
[131] |
|
Roquin1 + Roquin2 |
Rc3h1f/f: Rc3h2f/f:Lck-cre (RING-domain deleted cDKO; T cell) |
elevated TFH formation, and increase in GC B cells upon immunization from 8 to 12 weeks |
[132] | |
|
Roquin1 + Roquin2 |
Rc3h1f/f:Rc3h2f/f: Cd4-cre (T cell cKO) |
- lymphadenopathy; splenomegaly; - no spontaneous GC formation; no autoantibodies; - perturbed splenic architecture |
- Activation of CD4 and CD8 T cells - Increase in Treg numbers - increase in TFH cells - increase in GC B cells |
[133] |
| TRAF6 |
TRAF6f/f:Cd4-cre mixed background (C57BL/6 – 129) |
spontaneous autoimmunity: multi-organ infiltration with B and CD4 T cells, increased autoantibodies |
- CD4 T cells resistant to suppression by Treg - proliferation independent of CD28 costimulation |
[134] |
| Peli1 | Peli1−/− (C57BL/6) | - Spontaneous autoimmunity (6 months): T and B cell infiltration of kidney, liver, lung; autoantibodies; immune complexes in kidney glomeruli; - Increased EAE upon T cell transfer in Rag1−/− mice |
- T cell activation independent of costimulation - resistant to Tregmediated suppression |
[135] |
| B cells | ||||
| Peli1 |
Peli1−/− (C57BL/6) |
increased severity of lupus-like disease in experimental lupus (BM12) model |
- B cell proliferation in response to CD40, BAFF - T cell-dependent antibody response |
[124] |
| TRAF3 |
Traf3f/f:Cd19-cre (C57BL/6 – 129) |
spontaneous autoimmunity (12 months): autoantibodies (from 10wks on); lymphocyte infiltration of kidney, liver; glomerular immune complex deposits |
- B cell survival - T cell-dependent antibody response |
[136] |
| A20 |
Tnfaip3f/f:Cd19- cre |
Spontaneous autoimmunity: - SLE-like: increase in GC B cell numbers, autoantibodies, glomerular immune complex deposits OR - inflammatory autoimmune syndrome, splenomegaly, plasma cell hyperplasia, IgG autoantibodies |
B cell proliferation and survival |
Reviewed in [137] |
| Innate immune cells | ||||
| A20 |
Tnfaip3f/f:Cd11ccre (C57BL/6) |
Spontaneous autoimmunity: - SLE-like autoimmunity - IBD-like autoimmunity: lymphocyte-dependent colitis, seronegative ankylosing arthritis, enthesitis |
increased survival and DC hyper-responsiveness |
Reviewed in [137] |
|
Tnfaip3f/f:LysMcre (C57BL/6) |
Spontaneous destructive arthritis | increased cytokine production by macrophages |
Reviewed in [137] |
|
| Peli1 |
Peli1−/− (C57BL/6 – 129/Sv) |
Reduced severity of EAE | Microglia activation | [138] |
EAE: experimental autoimmune encephalomyelitis; KO: knockout; DKO: double KO; cKO: conditional KO; TFH: T follicular helper cell; GC: germinal center; Teff: effector T cells; Treg: regulatory T cells; cDKO: conditional DKO; NA: not applicable; IBD: inflammatory bowel disease; SLE: systemic lupus erythematosus
Anergy Anergy is a state of hyporesponsiveness in which T cells do not fully differentiate or acquire full effector function. Anergy can be induced in the absence of a costimulatory (e.g. B7, ICOSL, OX40L, CD40) secondary signal that is in addition to TCR stimulation required for T cell activation [18], or by the presence of a co-inhibitory (PD-L1, HVEM, or Galectin-9) signal. These signals are presented by APCs and bind respective stimulating (among them CD28, ICOS, OX40, CD40L) or inhibitory (PD-1, BTLA, or TIM3) receptors on T cells (reviewed in [19]). For example TCR/MHC-peptide engagement paired with CD28/B7 costimulation initiates calcineurin, Ras, and PKC-θ signaling that activates NFAT, AP1, and NF-κB transcription factors required to induce IL-2 production, T cell proliferation and differentiation (Figure 1B). In contrast, T cells with TCR/MHC-peptide engagement in the absence of CD28/B7 costimulatory signals do not upregulate IL-2 and enter anergy [12] (Figure 1B). Several E3s are transcriptionally upregulated under anergic conditions, among them Cbl-b, Itch, and Grail (also known as RNF128) [20–22], which function to limit TCR signaling output. This is achieved through proteolysis-dependent mechanisms, such as ubiquitin-dependent TCR/CD3 receptor downmodulation by Cbl-b [23] and Grail [24] or downregulation of downstream TCR signaling components including PLC-γ and PKC-θ by Itch and Cbl-b [20, 25]. Itch further regulates AP1 by proteasomal degradation of junB, a mechanism involved in CTLA-4-mediated T cell inhibition [26, 27]. Cbl-b and Grail also exhibit proteolysis-independent functions that promote anergy: Cbl-b inhibits recruitment of the phosphoinositide 3-kinase subunit p85 to CD28 and TCRζ [28], preventing TCR clustering [29], and Grail ubiquitination of Rho GDP-dissociation inhibitor stabilizes it, preventing IL-2 expression [30]. Thus, by modulating the abundance/activity of critical TCR signaling molecules, E3s serve as T cell intrinsic checkpoints to limit T cell activation. These molecular mechanisms partially explain spontaneous autoimmunity or enhanced experimentally-induced autoimmunity observed in mice deficient for Cbl-b, Itch, or Grail [24, 31–34] (Table 1) and may serve to develop targeted therapeutics for patients suffering multisystem autoimmune disease, mediated e.g. by polymorphisms in the human homolog of Itch [35].
Activation induced cell death Interestingly, overstimulated of T cells by the effector cytokine IL-2 results in co-expression of the apoptosis-related factors Fas (CD95) and Fas ligand (FasL). Engagement of Fas by FasL induces T cell apoptosis, thereby providing an additional safety mechanism to prevent T cell overactivation and mice with defects in Fas, FasL or IL-2R develop autoimmunity due to failure of T cells to undergo AICD [15, 36]. Among E3s that either stimulate or inhibit AICD are WWP2 [37], c-Cbl [38], and A20 [38]. WWP2 limits AICD in vitro in T cells by ubiquitinating and destabilizing the transcription factor EGR2, thereby limiting EGR2-mediated FasL upregulation to promotes T cell survival [37]. As A20 has been implicated in the control of RIPK and NF-κB signaling, the role of A20 in AICD [38] and its contribution to the phenotypes observed in A20 knockout mice, which exhibit multi-organ inflammation [39] and in men with A20 polymorphisms that are associated with systemic lupus erythematous, Crohn’s disease or psoriasis [40] remain to be determined.
Suppression by Treg In addition to nTregs that are selected in thymus [15], immunosuppressive Treg can also emerge from naïve CD4+ T cells following TCR activation in the presence of the suppressor cytokine TGF-β [41–43] secreted by tolerogenic DCs [44]; this Treg subset is called induced (i)Treg. The immunosuppressive function of Treg is crucial for tolerance induction, and decreased Treg number or function is associated with a variety of autoimmune pathologies [45]. Interestingly, E3s are implicated in both Treg development and function. For example, Stub1 and Cbl-b activity destabilizes Foxp3, which is required for Treg identity, by ubiquitin-dependent degradation [46, 47]. The E3s von Hippel-Lindau (VHL) and Itch support maintenance of functional Treg [48, 49]. Treg-specific loss of VHL results in HIF1α-dependent conversion of Treg into TH1-like, IFNγ-producing effector T cells, which culminates in multi-organ lymphocyte infiltration and early mouse mortality [48]. Additionally, in an adoptive transfer murine colitis model, VHL-deficient Tregs were unable to prevents colitis, further exemplifying their role in autoimmunity [48]. Similar, Treg-specific Itch deficiency in mice results in severe airway inflammation, mediated by increased TH2 cytokine production by Itch-deficient Tregs [49]. Furthermore, Grail is critical for Treg function, as Grail−/− Treg are less immunosuppressive, and express TH17 cell-related genes [24]. Given the crucial role of E3s in maintaining Treg homeostasis, along with their ability to induce anergy in self-reactive T cells, they serve as critical T cell checkpoints to maintain T cell tolerance, thereby preventing autoimmunity. We note that B cells play an equally important role in autoimmunity (recently reviewed [50]) and point to the fact that E3s are crucial regulators of B cell reponses (Table 1; Box 3).
Box 3: E3 ubiquitin ligases in B cell tolerance.
While this review focuses on E3 functions in T cell-mediated immunity, B cells also play a pivotal role in autoimmunity (recently reviewed in [50]), and E3s regulate B cell responses (Table 1). For example, B cell-specific ablation of Cbl and Cbl-b induces lupus-like disease in mice [118]. Cbl acts as a B cell-intrinsic tolerance checkpoint, possibly by modulating BCR-proximal signaling pathways to induce anergy [119, 120]. In addition, E3s have emerged as crucial regulators of the noncanonical NF-κB pathway, which is activated downstream of CD40 and BAFFR (B cell activating factor receptor) in B cells [121, 122]. Overexpression of the cytokine BAFF is associated with autoimmunity in SLE or multiple sclerosis patients [123], and targeting BAFF with the monoclonal antibody belimumab has been exploited therapeutically to treat SLE patients [50], highlighting the importance of this signaling pathway in autoimmunity. Activation of noncanonical NF-κB signaling requires degradation of TRAF3, which binds NIK and recruits the E3 ligases cellular inhibitor of apoptosis 1 and 2 (cIAP) into a cytosolic complex to facilitate proteasomal degradation of NIK. Engagement of CD40 or BAFFR induces recruitment of TRAF2, TRAF3 and cIAP to the receptor, where cIAP mediates K48-linked ubiquitination and degradation of TRAF3, stabilizing NIK [121, 122]. Active NIK phosphorylates and activates IKKα, which then phosphorylates the NF-κB subunit p100, a precursor of p52. Phosphorylation-induced ubiquitination of p100 leads to proteasomal processing and release of the mature p52 subunit, which associates with Rel-B to initiate transcription. In addition to cIAP, the E3 ligase Peli1 also affects NIK degradation in B cells, and Peli1 levels are inversely correlated with disease severity in SLE patients [124].
Thus, E3-dependent signaling via CD40 and BAFFR promotes expression of genes required for B cell survival, maturation, and activation. Dysregulation of these pathways is implicated in autoimmunity [125].
E3 ubiquitin ligase function in innate immune cells affects T cell responses
In addition to T-cell intrinsic effects, E3 ligases are also important regulators of APCs, which affect T cell homeostasis and autoimmunity on several levels, e.g. by altering antigen presentation or the cytokine milieu.
Major histocompatibility complex in autoimmunity MHC loci were the first genetic associations identified to correlate with autoimmune diseases, exemplifying the importance of antigen presentation in the etiology of autoimmune diseases [51]. Ubiquitination regulates expression of cell surface MHC proteins and costimulatory receptors by multiple mechanisms. The E3 ligase membrane-associated RING-CH-1 (MARCH1), for example, ubiquitinates MHC class II [52] and the costimulatory molecule CD86 [53, 54] in DCs, promoting their endocytosis and lysosomal degradation. Second, mono-ubiquitination of the transcriptional coactivator CIITA increases its association with transcription factors and the MHC class II promoter, leading to increased MHCII mRNA levels [55]. Additionally, ubiquitination and degradation of the MHCII transcriptional repressor B lymphocyte-induced maturation protein 1 (BLIMP1) by the E3 Hrd1 promotes MHCII transcription [56]. Hrd1 deletion in DCs decreases MHCII expression, which impairs DC-mediated priming of CD4+ T cells and protects mice from experimental encephalomyelitis, a T cell-dependent autoimmune disease [56].
E3 Ubiquitin ligases in immune sensing Autoimmune diseases can develop by altered activity of innate immune cells that culminate in aberrant B or T cell regulation. T cell responses are strongly influenced by the cytokine milieu, which is considered a third signal for T cell activation. For example, Type I interferons (IFN I) modulate clonal T cell expansion and CD8+ T cell effector function [57], and prolonged overactivation of IFN production is associated with systemic autoinflammation and autoimmunity [58]. Interestingly, mice expressing a mutant form of IFN receptor (IFNAR1S526A) resistant to SCF-ß-Trcp-dependent ubiquitination and degradation, display exacerbated experimental autoimmune hepatitis relative to control mice [59], exemplifying the role of E3 ligases in mediating IFN-related immune pathologies. Among mechanism that lead to abberrant IFN I production is cytosolic accumulation of endogenous DNA, which is also linked to the production of antinuclear antibodies, a hallmark of several autoimmune diseases [57, 60].
Sensing of cytosolic nucleic acids via pattern recognition receptors (PRR, such as Rig-I-like helicases, cGAS, and DDX41) activates the adaptor protein STING, which recruits and activates the downstream kinase TBK1. TBK1 then activates the transcriptional regulator IRF3, driving IFN I production. STING activity is controlled by several ubiquitin ligases, among them TRIM56 and TRIM32. Both induce K63-linked STING polyubiquitination, which facilitates STING dimerization and association with TBK1, enhancing cGAS/STING signaling [61, 62]. K27-linked STING polyubiquitination by the E3 ligase AMFR and the adaptor protein INSIG1 also promotes TBK1 recruitment [63]. Conversely, TRIM29 regulates antiviral immune responses by targeting STING for degradation [64]. Ubiquitination also regulates other DNA sensors: for example, TRIM21 promotes K48-linked ubiquitination and degradation of DDX41 and negatively regulates the innate immune response to intracellular dsDNA [65]. Thus, cytosolic sensors which control cytokine stimulation and innate immune cell activation are controlled by E3s to assure temporal control of type I IFN signaling to prevent autoimmunity.
Autoimmune phenotypes seen in some E3-deficient mice may also depend on immune cell-extrinsic mechanisms. RNF5-deficient mice for example display enhanced dextran sodium sulfate (DSS)-induced colitis compared with wild-type mice [66]. Naïve RNF5−/− mice also display higher basal levels of mature DC infiltrates and inflammation in the intestine, outcomes attributable to increased secretion of the RNF5 substrate S100A8 from intestinal epithelial cells. S100A8 is part of the calprotectin complex, a damage-associated molecular pattern, which binds to PRR on innate immune cells to elicit inflammatory responses. DSS treatment of RNF5−/− mice not only enhances these phenotypes but increases activation of mucosal CD4+ T cells, leading to a TH1-type proinflammatory response [66]. In all, E3 ligase function in non-immune, innate immune and adaptive immune cells emerges as a central regulatory node to maintain immune homeostasis and protect from the development of immunopathologies, including inflammatory and autoimmune diseases.
E3 Ubiquitin Ligases in tumor immunosurveillance and escape
By analogy to autoimmunity, the contribution of E3s to tumor immunosurveillance extends beyond their role in immune cells and includes important regulatory roles in the tumor stroma and the tumor itself. These mechanisms collectively impact T cell dysfunction within or exclusion from the TME – the two major mechanisms of tumor immune escape [67] (Figure 2).
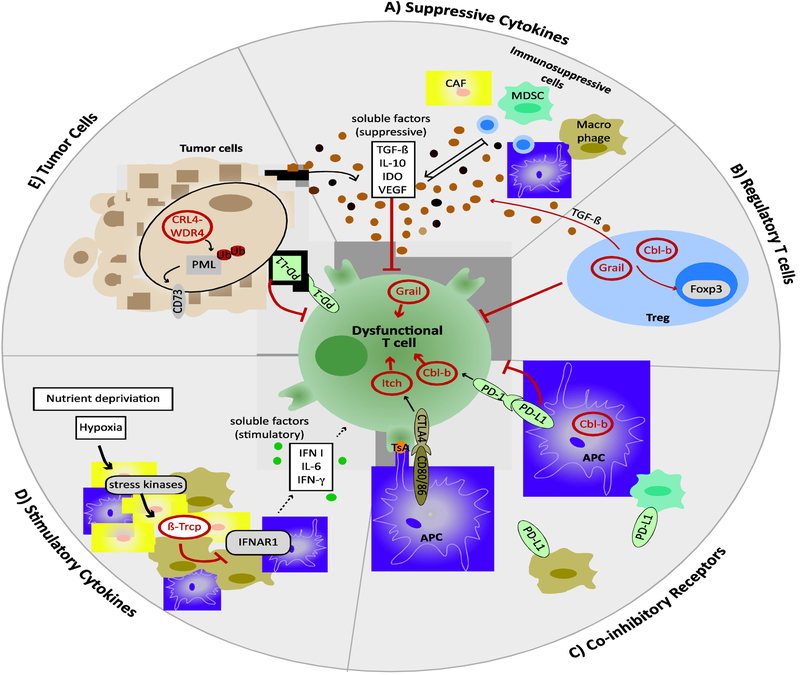
Figure 2: E3 ubiquitin ligases regulate T cell dysfunction in cancer.
E3 Ubiquitin ligases (red) can serve as T cell intrinsic checkpoints to inhibit T cell function. A) T cell activity is modulated by immunosuppressive cytokines that are present in the tumor microenvironment and E3s are crucial regulators of cytokine production. B) E3s also function in Tregs where they increase immunosuppressive cytokine production and limit Treg suppressor function. C) In innate immune cells E3s can inhibit immune-stimulatory function and/or upregulate co-inhibitory surface receptors (e.g. PD-1, CTLA-4). D) Dyregulation of ubiquitination also affects the production immunostimulatory cytokines such as type I Interferons (IFN), an effect that is influenced by stresses commonly present in the tumor microenvironment. E) Finally, E3 ubiquitin ligase function in cancer cells contributes to generate an iummunosuppressive microenvironment.
CAF: cancer associated fibroblasts; MDSC: myeloid-derived suppressor cell; APC: antigen-presenting cell; PML: promyelocytic leukemia protein; TsA: tumor-specific antigen;
E3 Ubiquitin ligases as T cell intrinsic regulators of T cell dysfunction in cancer The identification of genes that, similarly to PD-1 or CTLA4, negatively regulate T cell proliferation and function are of therapeutic interest as they have the potential to boost antitumor immune responses. To identify these novel target genes, large-scale in vivo shRNA [68] and CRISPR knockout [69] screens have been performed in murine or primary human T cells. Interestingly, these studies identified a number of E3 ligases, most notably Cbl-b, whose loss boosts T cell activation, in accordance with its role in T cell anergy. The importance of Cbl-b and Grail in tumor immunosurveillance has previously been recognized, as both Cbl-b−/− and Grail−/− mice reject lymphomas or TC-1 tumors when xenotransplanted into mice, and in Cbl-b−/− mice, spontaneous UVB-induced tumor formation is inhibited [70–72]. Moreover, adoptive transfer of either Cbl-b- or Grail-deficient CD8+ T cells into tumor-bearing mice is sufficient to promote tumor rejection [70–73], indicating that CD8+ T cell-intrinsic effects mediate tumor rejection. Mice harboring a mutation that specifically disrupts Cbl-b ligase function phenocopy Cbl-b−/− mice in terms of T cell hyperactivation, spontaneous autoimmunity and tumor rejection, demonstrating that the catalytic activity of Cbl-b is required to mediate these effects [74]. Mechanistically, Cbl-b−/− and Grail−/− CD8+ T cells were better effectors than wildtype CD8+ T cells as they could be fully activated without need of costimulation in vitro and were also less susceptible to Treg-mediated inhibition [70–72]. Cbl-b also functions downstream of TGF-ß receptor signaling, which mediates both T cell tolerance [43] and immunosuppression in the TME [75–77]. Naïve T cells from Cbl-b-deficient mice cannot undergo TGF-β-induced upregulation of Foxp3 or Treg differentiation [78]. These studies suggest that Cbl-b and Grail may represent therapeutic targets to render T cells less susceptible to immunosuppression mediated by the TME.
Furthermore, Cbl-b functions downstream of the T cell co-inhibitory receptor PD-1, since Cbl-b deficiency renders T and natural killer (NK) cells refractory to PD-1-mediated regulation [79, 80]. This raises the possibility that E3 ligase activity could serve as a biomarker for a favorable response to PD-1/PD-L1-targeted therapies. Conversely, downregulation of the E3 SCF-FBXO38 may be an indicator of responsiveness to PD-1/PD-L1-targeted therapies, as this E3 was recently found to regulate PD-1 protein levels by ubiquitin-dependent degradation [81]. FBXO38 is downregulated in the TME and correlates with increased PD-1 expression in CD8+ T cells of colorectal and hepatocellular carcinoma patients [81]. In all, E3s serve as T cell intrinsic checkpoints that limit antitumor immunity, which may in part be mediated by the modulation of the PD-1 signaling pathway, a topic that deserves further study.
E3 activity in innate immune or tumor cells contributes to cancer immune escape. Cbl-b expressed in non-T cells also regulates tumor progression, as illustrated by the finding that double knockout Cblb−/−Rag2−/− T cell-deficient mice show delayed rejection of xenotransplanted tumors as compared with Cblb+/+ Rag2−/− mice [82]. Cbl-b activity also limits NK cell function by mediating ligand-induced downregulation of TAM (TYRO, AXL, MER) receptor tyrosine kinases, which function in various innate immune cells [82]. Furthermore, adoptive transfer of Cbl-b-deficient NK cells or pharmacological inhibition of TAM receptors enhances rejection of metastatic melanoma or breast cancer tumors in wild-type mice [82]. Thus, Cbl-b activity in both innate and adaptive immune cells contributes to antitumor immunity (Figure 2).
Dysregulation of E3s is a common event in cancer cells [9] and contributes to tumor immune escape. For example, CRL4-WDR4 ubiquitinates and targets the tumor suppressor promyelocytic leukemia protein (PML) for proteasomal degradation in lung cancer cells [83] (Figure 2). Downregulation of PML promotes metastatic progression by increasing invasion and migration and by inducing an immunosuppressive microenvironment via upregulation of CD73, an ecto-5′-nucleotidase that converts AMP into adenosine. CD73 upregulation in tumor cells increases the abundance of intratumoral Treg and M2-like macrophages while decreasing that of CD8+ T cells, all of which lead to defective tumor control [83].
Several stress signals that are commonly associated with the TME, such as hypoxia, nutrient deprivation, inflammatory cytokines, and activation of the unfolded protein response can also contribute to immune escape. These stresses can induce downregulation of the IFN receptor IFNAR1 in stromal cells, mediated by ubiquitin-dependent degradation of the IFNAR1 receptor by the E3 SCF-ß-Trcp, which binds to IFNAR1 phosphorylated at Ser526/Ser535 [84, 85] (Figure 2). This mechanism has recently been demonstrated to diminish viability of cytotoxic T lymphocytes (CTLs) in the TME of murine colorectal cancer models, allowing tumors to flourish [86]. The resulting decrease in type I IFN production can limit T cell survival [86, 87] and T cell effector function. The latter is in accordance with the important role of DC-derived type I IFN for T cell priming [88] and the fact that IFN I serves as a “third” signal for T cell activation [57].
Collectively, these studies indicate that E3s contribute to antitumor immunity by serving as T cell-intrinsic checkpoints; controlling APC maturation; modulating CD4+ T cell differentiation, Treg generation, and CTL effector function; and reprogramming the TME. Thus, E3s constitutes novel targets that may improve the efficacy of cancer immunotherapy and serve as biomarkers to predict responses to approved ICBs (e.g. high Cbl-b or low FBXO38 [79–81].
E3 Ubiquitin ligases in balancing autoimmunity and antitumor immunity – therapeutic implications
Many factors that restrain autoimmunity (such as PD-1/PD-L1, CTLA-4, TGF-β) are equally relevant to cancer therapy, as they may be targeted to promote tumor eradication. As several E3s function as negative regulators of immune responses (Table 1), and some have been demonstrated to also regulate immunosurveillance in the TME (Figure 2), targeting E3s may represent a promising approach to boost antitumor immune responses. As multiple inhibitory pathways present in the TME (e.g. PD-L1 [79, 80], TGF-β [78], Treg [70, 71]) use the same E3 ligase (e.g. Cbl-b) as downstream signaling effector to inhibit T cell effector function, one may speculate that targeting T cell intrinsic checkpoints (e.g. Cbl-b) may be more efficient in activating antitumor immune responses than targeting individual cell surface inhibitory receptors.
Although targeting E3s remains a more challenging task, technological advances (e.g. CRISPR gene editing, or harnessing synthethic gene circuits [89]), a better biological understanding of their mode of action (e.g. systematic mapping of degrons, the specific motifs in substrates that are recognized by the E3 [90, 91]) along with advancements in compound screening now allow to envision an efficient way to target E3 ligases in the near future: such approaches may include genetic engineering of T cells for adoptive cell transfer [92] or chimeric antigen receptor (CAR) T cell therapy and efforts to identify small molecule inhibitors, e.g. for Cbl-b are ongoing [93, 94]. E3 themselves or their substrates could also be targeted by a protein targeting complex approach (PROTAC), which directs a susbtrate to a E3 ligase by chemical linkers [95]. In addition to Cbl-b and Grail, other ubiquitin ligases that function in autoimmunity (such as Itch, Peli1, Roquin, and RNF5) may regulate stroma-mediated antitumor immunity and may thus serve as putative therapeutic targets to promote antitumor immunity. With that noted, we recognize that a better understanding of E3 function in distinct tissues will be necessary and further improvements in the therapeutic targeting of protein-protein interactions will be required to move E3-targeting therapies into the clinic.
Given the link between E3s, autoimmunity and antitumor immunity, it is reasonable to assume that, similar other ICBs, IRAEs may limit the clinical use of E3-targeting therapies – further stressing the need to better understand the underlying mechanisms of these unwanted effects. Despite clinical experience, biomarkers that predict IRAEs associated with CTLA-4-, PD-1-, and PD-L1-targeted therapies deserve further studies (Box 1). Notably, IRAEs occur in only a subset of ICB-treated patients. The selectivity may be attributed to inter-individual variation in germline-encoded genetic factors, although analysis of HLA-A*0201 genotypes among advanced melanoma patients treated with CTLA4-blocking antibody did not identify a significant association with IRAE occurrence [96]. The gut microbiome may help in predicting IRAE emergence after ICB treatment: select commensal Bacteroidetes strains were shown to elicit protective effects against CTLA-4-blockade-induced colitis [97], and transfer of bifidobacteria to anti-CTLA-4-treated mice mitigates colitis independently of antitumor immunity [98]. Given that changes in E3 activities impact autoimmune responses and sometimes promote IRAE-like phenotypes, understanding their function in this context may suggest therapeutic options to mitigate IRAE after ICB.
Concluding Remarks & Future Perspectives
Ubiquitin ligases serve at critical regulatory nodes of signaling pathways and elicit selective functions as they may be i) mutant/dysfunctional in a tumor [9], ii) expressed in select tissues, and iii) target specific substrates in a spatial and temporal manner. Thus, they may allow to distinguish regulatory modules in immune, tumor or stroma compartments, thus become favorable targets for select manipulation of a given pathway in a tissue / cell type dependent manner. The review focused on a relative small fraction of ubiquitin ligases that are expected to play a role in antitumor immunity and autoimmunity. Some were only partially studied, for example Roquin, which has been implicated in autoimmunity, yet, the catalytic activity of its E3 ligase domain has not been studied in this context; and others are yet to be identified and characterized. Yet, the emerging theme reflect on the important role E3s play in immune homeostasis and the topic will become more translationally relevant when better mechanistic and clinical data become available (see outstanding questions).
Outstanding questions.
Can the expression of selective E3s and/or their substrates be used to stratify patients to predict immunotherapy responses and immune-related adverse events associated with these therapies?
Would targeting E3 or their substrates allow the fine-tuning of a given immune response? Would such fine tuning allow clinicians to selectively harness antitumor immune response while preventing autoimmunity?
Can modulation of E3 activity improve tumor recognition, presentation, and activation of the innate immune response?
Can tumor intrinsic or immune cell specific expression of E3 (and their substrates) guide the development of novel cancer immunotherapies?
Can a comprehensive map of tissue-specific E3 ubiquitin ligase expression and function be generated to reveal targets for anticancer immunotherapies and increase safety of existing therapies?
Better appreciation of the greater landscape of ubiquitin ligases in immune system’s regulation and function in health and disease will allow to establish a better resolution map, and correspondingly, identify the most prevalent targets to exploit therapeutically for their use in monotherapies or combination therapies.
Highlights.
E3 ubiquitin ligases recognize their substrates in a spatial and temporal manner, requiring cell type specific post translational modifications that are often affected by the microenvironment (oxygen, nutrients, stress).
E3 ubiquitin ligases are important negative regulators of T cell responses that restrain autoimmunity but also antitumor immunity.
Targeting E3 ubiquitin ligases may be of therapeutic benefit in cancer immunotherapy, highlighting the need to comprehensively characterize E3 ligase expression and function in immune cells.
Clinician’s Corner.
As regulators of innate and adaptive immune responses, E3 ubiquitin ligases have been identified as crucial factors relevant to T cell function. Accordingly, E3s contribute to the pathogenesis of autoimmune disorders and have emerged as important regulators of antitumor immunity. Mutation and epigenetic changes in E3 ligases or their substrates are commonly found in cancer.
Immune-related adverse events (IRAEs) limit widespread use of (combination) immunotherapies, and biomarkers to identify patients at increased risk for IRAEs are lacking.
Understanding E3 function in innate and adaptive immunity may allow us to identify i) biomarkers and ii) therapeutic targets for both classical and immunotherapy-induced autoimmune diseases.
Level of E3 ligases can be monitored by assessing the expression of their surrogate substrates, and related signaling components in either immune cells, tumor, stroma or bodily fluids.
Targeting E3 (or their regulated substrates) using novel technologies (i.e. PROTAC based) offers innovative means for dampening autoimmunity while enhancing antitumor immunity.
ACKNOWLEDGEMENTS
The authors gratefully acknowledge funding by NCI OIA grant R35CA197465, R01CA216187 and DOD grant CA171013 (to Z.A.R.), MRA grant 571135 and DOD grant CA171198 (to R.T.), and a JSPS Postdoctoral Fellowship for Research Abroad (to Y.F.),.
Glossary
- Autocrine motility factor receptor (AMFR)
an ER membrane-anchored RING-type E3 that functions in ER-associated degradation (ERAD).
- Casitas B-lineage lymphoma proto-oncogene-b (Cbl-b)
A RING-type E3 that belongs together with c-Cbl and Cbl-3 to the Cbl-E3-ligase family that are expressed in a variety of immune cells.
- cGMP-AMP (cGAMP) synthase (cGAS)
cGAS is a cytosolic, sequence-independent DNA sensor protein which binds double-stranded DNA and catalyzes the synthesis of 20,30-cyclic GMP-AMP (cGAMP). In turn, cGAMP engages STING.
- Cullin-RING ligase 4 (CRL4)
The Cullin4 E3 ligase complex uses one of several WD40-containing proteins such as WD repeat domain 4 (WDR4) as substrate recognition modules.
- Dendritic cells (DCs)
bone marrow-derived cells with the ability to present antigens to T cells and orchestrate adaptive immunity. DCs also trigger inhibitory circuits that ensure immunological tolerance.
- Gene related to anergy in lymphocytes (GRAIL)
Also known as RNF128, this transmembrane RING-type E3 is localized at endosomes and catalyzes K-48 or K-63-linked ubiqutination.
- Inducible T cell costimulator (ICOS)
an immune checkpoint protein expressed on activated T cells and which is triggered by ICOS ligand, ICOS-L, a B7-related transmembrane glycoprotein.
- Itchy homolog (ITCH)
Member of the NEDD4-family of HECT-type E3 ligases. Itch-deficiency in humans is associated with multisystem autoimmune disease and developmental abnormalities.
- Membrane-associated RING-CH-1 (MARCH1)
a membrane-bound E3 ligase that downregulates MHC class II molecules and other glycoproteins by directing them to the late endosomal/lysosomal compartment.
- NK cells (NK)
natural killer cells are antigen-independent cytotoxic lymphocytes of the innate immune system, responding to viral infection and tumors.
- Pathogen recognition receptor (PRR)
host-derived surface and intracellular sensors that detect pathogen- or danger-associated molecular patterns initiating an innate immune response.
- Skp1-cullin1-F-box protein (SCF)
multisubunit E3 complex whose specificity is mediated by one of ~68 F-box proteins such as β-transducin repeat-containing protein (ß-Trcp) or FBXO38, which function as substrate recognition subunits. SCF-complexes regulate the degradation of proteins involved in cell signaling and cell cycle control.
- stimulator of interferon genes (STING)
an endoplasmic reticulum-membrane adaptor protein involved in innate immune signalling. Upon activation, STING recruits and activates several signaling proteins culminating in activation of IFNs.
- (Tyro-3, Axl, and Mertk) TAM
TAM receptors are homologous type I receptor tyrosine kinases that have important roles in cellular homeostasis and inflammation.
- T cell receptors (TCRs)
a heterodimeric transmembrane protein which recognizes antigen presented on MHC molecules. Variable TCRs are generated by somatic recombination allowing recognition of a wide range of pathogens by the T cell population. CD4 or CD8 are coreceptors defining the T cell subtype.
- Tumor necrosis factor (TNF) receptor-associated factor 6 (TRAF6)
a signaling adaptor and RING-type E3 ligase that mainly forms K63-linked polyubiquitin chains to regulate NF-κB and MAPK pathways downstream of multiple immune receptors.
- von Hippel-Lindau (VHL)
substrate-recognition protein required for the Cullin2-RING E3 ligase complex to regulate HIF1α stability. pVHL recognizes and binds to HIF1α under normoxia due to hydroxylation of 2 proline residues within the HIF1α protein.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement: The authors declare no competing interests.
REFERENCES
- 1.Couzin-Frankel J (2013) Breakthrough of the year 2013. Cancer immunotherapy. Science 342 (6165), 1432–3. [DOI] [PubMed] [Google Scholar]
- 2.Wei SC et al. (2018) Fundamental Mechanisms of Immune Checkpoint Blockade Therapy. Cancer Discov 8 (9), 1069–1086. [DOI] [PubMed] [Google Scholar]
- 3.Lo B and Abdel-Motal UM (2017) Lessons from CTLA-4 deficiency and checkpoint inhibition. Curr Opin Immunol 49, 14–19. [DOI] [PubMed] [Google Scholar]
- 4.Zamani MR et al. (2016) PD-1/PD-L and autoimmunity: A growing relationship. Cell Immunol 310, 27–41. [DOI] [PubMed] [Google Scholar]
- 5.Postow MA et al. (2018) Immune-Related Adverse Events Associated with Immune Checkpoint Blockade. N Engl J Med 378 (2), 158–168. [DOI] [PubMed] [Google Scholar]
- 6.Boutros C et al. (2016) Safety profiles of anti-CTLA-4 and anti-PD-1 antibodies alone and in combination. Nat Rev Clin Oncol 13 (8), 473–86. [DOI] [PubMed] [Google Scholar]
- 7.Hassel JC et al. (2017) Combined immune checkpoint blockade (anti-PD-1/anti-CTLA-4): Evaluation and management of adverse drug reactions. Cancer Treat Rev 57, 36–49. [DOI] [PubMed] [Google Scholar]
- 8.Horvat TZ et al. (2015) Immune-Related Adverse Events, Need for Systemic Immunosuppression, and Effects on Survival and Time to Treatment Failure in Patients With Melanoma Treated With Ipilimumab at Memorial Sloan Kettering Cancer Center. J Clin Oncol 33 (28), 3193–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Senft D et al. (2018) Ubiquitin ligases in oncogenic transformation and cancer therapy. Nat Rev Cancer 18 (2), 69–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhoj VG and Chen ZJ (2009) Ubiquitylation in innate and adaptive immunity. Nature 458 (7237), 430–7. [DOI] [PubMed] [Google Scholar]
- 11.Malynn BA and Ma A (2010) Ubiquitin makes its mark on immune regulation. Immunity 33 (6), 843–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nurieva RI et al. (2011) Molecular mechanisms of T-cell tolerance. Immunol Rev 241 (1), 133–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zinngrebe J et al. (2014) Ubiquitin in the immune system. EMBO Rep 15 (1), 28–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nurieva R et al. (2013) T-cell tolerance in cancer. Immunotherapy 5 (5), 513–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodnow CC et al. (2005) Cellular and genetic mechanisms of self tolerance and autoimmunity. Nature 435 (7042), 590–7. [DOI] [PubMed] [Google Scholar]
- 16.Akiyama T et al. (2005) Dependence of self-tolerance on TRAF6-directed development of thymic stroma. Science 308 (5719), 248–51. [DOI] [PubMed] [Google Scholar]
- 17.Bonito AJ et al. (2013) Medullary thymic epithelial cell depletion leads to autoimmune hepatitis. J Clin Invest 123 (8), 3510–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lafferty KJ and Cunningham AJ (1975) A new analysis of allogeneic interactions. Aust J Exp Biol Med Sci 53 (1), 27–42. [DOI] [PubMed] [Google Scholar]
- 19.Chen L and Flies DB (2013) Molecular mechanisms of T cell co-stimulation and co-inhibition. Nat Rev Immunol 13 (4), 227–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heissmeyer V et al. (2004) Calcineurin imposes T cell unresponsiveness through targeted proteolysis of signaling proteins. Nat Immunol 5 (3), 255–65. [DOI] [PubMed] [Google Scholar]
- 21.Anandasabapathy N et al. (2003) GRAIL: an E3 ubiquitin ligase that inhibits cytokine gene transcription is expressed in anergic CD4+ T cells. Immunity 18 (4), 535–47. [DOI] [PubMed] [Google Scholar]
- 22.Nurieva R et al. (2006) T-cell tolerance or function is determined by combinatorial costimulatory signals. EMBO J 25 (11), 2623–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Naramura M et al. (2002) c-Cbl and Cbl-b regulate T cell responsiveness by promoting ligand-induced TCR down-modulation. Nat Immunol 3 (12), 1192–9. [DOI] [PubMed] [Google Scholar]
- 24.Nurieva RI et al. (2010) The E3 ubiquitin ligase GRAIL regulates T cell tolerance and regulatory T cell function by mediating T cell receptor-CD3 degradation. Immunity 32 (5), 670–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeon MS et al. (2004) Essential role of the E3 ubiquitin ligase Cbl-b in T cell anergy induction. Immunity 21 (2), 167–77. [DOI] [PubMed] [Google Scholar]
- 26.Fang D et al. (2002) Dysregulation of T lymphocyte function in itchy mice: a role for Itch in TH2 differentiation. Nat Immunol 3 (3), 281–7. [DOI] [PubMed] [Google Scholar]
- 27.Hoff H et al. (2010) CTLA-4 (CD152) inhibits T cell function by activating the ubiquitin ligase Itch. Mol Immunol 47 (10), 1875–81. [DOI] [PubMed] [Google Scholar]
- 28.Fang D and Liu YC (2001) Proteolysis-independent regulation of PI3K by Cbl-b-mediated ubiquitination in T cells. Nat Immunol 2 (9), 870–5. [DOI] [PubMed] [Google Scholar]
- 29.Krawczyk C et al. (2000) Cbl-b is a negative regulator of receptor clustering and raft aggregation in T cells. Immunity 13 (4), 463–73. [DOI] [PubMed] [Google Scholar]
- 30.Su L et al. (2006) A novel E3 ubiquitin ligase substrate screen identifies Rho guanine dissociation inhibitor as a substrate of gene related to anergy in lymphocytes. J Immunol 177 (11), 7559–66. [DOI] [PubMed] [Google Scholar]
- 31.Chiang YJ et al. (2000) Cbl-b regulates the CD28 dependence of T-cell activation. Nature 403 (6766), 216–20. [DOI] [PubMed] [Google Scholar]
- 32.Bachmaier K et al. (2000) Negative regulation of lymphocyte activation and autoimmunity by the molecular adaptor Cbl-b. Nature 403 (6766), 211–6. [DOI] [PubMed] [Google Scholar]
- 33.Perry WL et al. (1998) The itchy locus encodes a novel ubiquitin protein ligase that is disrupted in a18H mice. Nat Genet 18 (2), 143–6. [DOI] [PubMed] [Google Scholar]
- 34.Kriegel MA et al. (2009) E3 ubiquitin ligase GRAIL controls primary T cell activation and oral tolerance. Proc Natl Acad Sci U S A 106 (39), 16770–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lohr NJ et al. (2010) Human ITCH E3 ubiquitin ligase deficiency causes syndromic multisystem autoimmune disease. Am J Hum Genet 86 (3), 447–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang J et al. (2004) Activation-induced cell death in T cells and autoimmunity. Cell Mol Immunol 1 (3), 186–92. [PubMed] [Google Scholar]
- 37.Chen A et al. (2009) The HECT-type E3 ubiquitin ligase AIP2 inhibits activation-induced T-cell death by catalyzing EGR2 ubiquitination. Mol Cell Biol 29 (19), 5348–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li M et al. (2016) Ubiquitin A20 maintains activation-induced CD4+ T cell death. Int J Clin Exp Pathol 9 (8), 8247–8253. [Google Scholar]
- 39.Lee EG et al. (2000) Failure to regulate TNF-induced NF-kappaB and cell death responses in A20-deficient mice. Science 289 (5488), 2350–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vereecke L et al. (2011) Genetic relationships between A20/TNFAIP3, chronic inflammation and autoimmune disease. Biochem Soc Trans 39 (4), 1086–91. [DOI] [PubMed] [Google Scholar]
- 41.Hong J et al. (2005) Induction of CD4+CD25+ regulatory T cells by copolymer-I through activation of transcription factor Foxp3. Proc Natl Acad Sci U S A 102 (18), 6449–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen W et al. (2003) Conversion of peripheral CD4+CD25- naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J Exp Med 198 (12), 1875–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rubtsov YP and Rudensky AY (2007) TGFbeta signalling in control of T-cell-mediated self-reactivity. Nat Rev Immunol 7 (6), 443–53. [DOI] [PubMed] [Google Scholar]
- 44.Kanamori M et al. (2016) Induced Regulatory T Cells: Their Development, Stability, and Applications. Trends Immunol 37 (11), 803–811. [DOI] [PubMed] [Google Scholar]
- 45.Kasper IR et al. (2016) Empowering Regulatory T Cells in Autoimmunity. Trends Mol Med 22 (9), 784–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen Z et al. (2013) The ubiquitin ligase Stub1 negatively modulates regulatory T cell suppressive activity by promoting degradation of the transcription factor Foxp3. Immunity 39 (2), 272–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao Y et al. (2015) E3 Ubiquitin Ligase Cbl-b Regulates Thymic-Derived CD4+CD25+ Regulatory T Cell Development by Targeting Foxp3 for Ubiquitination. J Immunol 194 (4), 1639–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee JH et al. (2015) E3 Ubiquitin Ligase VHL Regulates Hypoxia-Inducible Factor-1alpha to Maintain Regulatory T Cell Stability and Suppressive Capacity. Immunity 42 (6), 1062–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jin HS et al. (2013) Itch expression by Treg cells controls Th2 inflammatory responses. J Clin Invest 123 (11), 4923–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rawlings DJ et al. (2017) Altered B cell signalling in autoimmunity. Nat Rev Immunol 17 (7), 421–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sollid LM et al. (2014) Molecular mechanisms for contribution of MHC molecules to autoimmune diseases. Curr Opin Immunol 31, 24–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.De Gassart A et al. (2008) MHC class II stabilization at the surface of human dendritic cells is the result of maturation-dependent MARCH I down-regulation. Proceedings of the National Academy of Sciences 105 (9), 3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baravalle G et al. (2011) Ubiquitination of CD86 is a key mechanism in regulating antigen presentation by dendritic cells. J Immunol 187 (6), 2966–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Corcoran K et al. (2011) Ubiquitin-mediated regulation of CD86 protein expression by the ubiquitin ligase membrane-associated RING-CH-1 (MARCH1). J Biol Chem 286 (43), 37168–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Greer SF et al. (2003) Enhancement of CIITA transcriptional function by ubiquitin. Nat Immunol 4 (11), 1074–82. [DOI] [PubMed] [Google Scholar]
- 56.Yang H et al. (2014) Hrd1-mediated BLIMP-1 ubiquitination promotes dendritic cell MHCII expression for CD4 T cell priming during inflammation. J Exp Med 211 (12), 2467–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Curtsinger JM et al. (2005) Type I IFNs provide a third signal to CD8 T cells to stimulate clonal expansion and differentiation. J Immunol 174 (8), 4465–9. [DOI] [PubMed] [Google Scholar]
- 58.Kretschmer S and Lee-Kirsch MA (2017) Type I interferon-mediated autoinflammation and autoimmunity. Curr Opin Immunol 49, 96–102. [DOI] [PubMed] [Google Scholar]
- 59.Bhattacharya S et al. (2014) Triggering ubiquitination of IFNAR1 protects tissues from inflammatory injury. EMBO Mol Med 6 (3), 384–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Crowl JT et al. (2017) Intracellular Nucleic Acid Detection in Autoimmunity. Annual Review of Immunology 35 (1), 313–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tsuchida T et al. (2010) The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity 33 (5), 765–76. [DOI] [PubMed] [Google Scholar]
- 62.Zhang J et al. (2012) TRIM32 protein modulates type I interferon induction and cellular antiviral response by targeting MITA/STING protein for K63-linked ubiquitination. J Biol Chem 287 (34), 28646–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang Q et al. (2014) The E3 ubiquitin ligase AMFR and INSIG1 bridge the activation of TBK1 kinase by modifying the adaptor STING. Immunity 41 (6), 919–33. [DOI] [PubMed] [Google Scholar]
- 64.Xing J et al. (2016) Identification of a role for TRIM29 in the control of innate immunity in the respiratory tract. Nat Immunol 17 (12), 1373–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Z et al. (2013) The E3 ubiquitin ligase TRIM21 negatively regulates the innate immune response to intracellular double-stranded DNA. Nat Immunol 14 (2), 172–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fujita Y et al. (2018) Regulation of S100A8 Stability by RNF5 in Intestinal Epithelial Cells Determines Intestinal Inflammation and Severity of Colitis. Cell Rep 24 (12), 3296–3311 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Joyce JA and Fearon DT (2015) T cell exclusion, immune privilege, and the tumor microenvironment. Science 348 (6230), 74–80. [DOI] [PubMed] [Google Scholar]
- 68.Zhou P et al. (2014) In vivo discovery of immunotherapy targets in the tumour microenvironment. Nature 506 (7486), 52–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shifrut E et al. (2018) Genome-wide CRISPR Screens in Primary Human T Cells Reveal Key Regulators of Immune Function. Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chiang JY et al. (2007) Ablation of Cbl-b provides protection against transplanted and spontaneous tumors. J Clin Invest 117 (4), 1029–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Loeser S et al. (2007) Spontaneous tumor rejection by cbl-b-deficient CD8+ T cells. J Exp Med 204 (4), 879–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Haymaker C et al. (2017) Absence of Grail promotes CD8(+) T cell anti-tumour activity. Nat Commun 8 (1), 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stromnes IM et al. (2010) Abrogating Cbl-b in effector CD8(+) T cells improves the efficacy of adoptive therapy of leukemia in mice. J Clin Invest 120 (10), 3722–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Paolino M et al. (2011) Essential role of E3 ubiquitin ligase activity in Cbl-b-regulated T cell functions. J Immunol 186 (4), 2138–47. [DOI] [PubMed] [Google Scholar]
- 75.Gorelik L and Flavell RA (2001) Immune-mediated eradication of tumors through the blockade of transforming growth factor-beta signaling in T cells. Nat Med 7 (10), 1118–22. [DOI] [PubMed] [Google Scholar]
- 76.Thomas DA and Massague J (2005) TGF-beta directly targets cytotoxic T cell functions during tumor evasion of immune surveillance. Cancer Cell 8 (5), 369–80. [DOI] [PubMed] [Google Scholar]
- 77.Tauriello DVF et al. (2018) TGFbeta drives immune evasion in genetically reconstituted colon cancer metastasis. Nature 554 (7693), 538–543. [DOI] [PubMed] [Google Scholar]
- 78.Wohlfert EA et al. (2006) Cutting edge: deficiency in the E3 ubiquitin ligase Cbl-b results in a multifunctional defect in T cell TGF-beta sensitivity in vitro and in vivo. J Immunol 176 (3), 1316–20. [DOI] [PubMed] [Google Scholar]
- 79.Fujiwara M et al. (2017) Cbl-b Deficiency Mediates Resistance to Programmed Death-Ligand 1/Programmed Death-1 Regulation. Front Immunol 8, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Peer S et al. (2017) Cblb-deficient T cells are less susceptible to PD-L1-mediated inhibition. Oncotarget 8 (26), 41841–41853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Meng X et al. (2018) FBXO38 mediates PD-1 ubiquitination and regulates anti-tumour immunity of T cells. Nature 564 (7734), 130–135. [DOI] [PubMed] [Google Scholar]
- 82.Paolino M et al. (2014) The E3 ligase Cbl-b and TAM receptors regulate cancer metastasis via natural killer cells. Nature 507 (7493), 508–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang YT et al. (2017) Ubiquitination of tumor suppressor PML regulates prometastatic and immunosuppressive tumor microenvironment. J Clin Invest 127 (8), 2982–2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kumar KG et al. (2003) SCF(HOS) ubiquitin ligase mediates the ligand-induced down-regulation of the interferon-alpha receptor. EMBO J 22 (20), 5480–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bhattacharya S et al. (2013) Anti-tumorigenic effects of Type 1 interferon are subdued by integrated stress responses. Oncogene 32 (36), 4214–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Katlinski KV et al. (2017) Inactivation of Interferon Receptor Promotes the Establishment of Immune Privileged Tumor Microenvironment. Cancer Cell 31 (2), 194–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hiroishi K et al. (2000) IFN-alpha-expressing tumor cells enhance generation and promote survival of tumor-specific CTLs. J Immunol 164 (2), 567–72. [DOI] [PubMed] [Google Scholar]
- 88.Fuertes MB et al. (2011) Host type I IFN signals are required for antitumor CD8+ T cell responses through CD8{alpha}+ dendritic cells. J Exp Med 208 (10), 2005–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nissim L et al. (2017) Synthetic RNA-Based Immunomodulatory Gene Circuits for Cancer Immunotherapy. Cell 171 (5), 1138–1150 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mészáros B et al. (2017) Degrons in cancer. Science Signaling 10 (470), eaak9982. [DOI] [PubMed] [Google Scholar]
- 91.Koren I et al. (2018) The Eukaryotic Proteome Is Shaped by E3 Ubiquitin Ligases Targeting C-Terminal Degrons. Cell 173 (7), 1622–1635.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kunert A and Debets R (2018) Engineering T cells for adoptive therapy: outsmarting the tumor. Curr Opin Immunol 51, 133–139. [DOI] [PubMed] [Google Scholar]
- 93.Riling C et al. (2018) Abstract A206: Small-molecule Cbl-b inhibitors as novel intracellular checkpoint inhibitors for cancer immunotherapy. Molecular Cancer Therapeutics 17 (1 Supplement), A206. [Google Scholar]
- 94.Gabrielsen M et al. (2017) A General Strategy for Discovery of Inhibitors and Activators of RING and U-box E3 Ligases with Ubiquitin Variants. Mol Cell 68 (2), 456–470 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Maniaci C et al. (2017) Homo-PROTACs: bivalent small-molecule dimerizers of the VHL E3 ubiquitin ligase to induce self-degradation. Nat Commun 8 (1), 830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wolchok JD et al. (2010) Ipilimumab efficacy and safety in patients with advanced melanoma: a retrospective analysis of HLA subtype from four trials. Cancer Immun 10, 9. [PMC free article] [PubMed] [Google Scholar]
- 97.Dubin K et al. (2016) Intestinal microbiome analyses identify melanoma patients at risk for checkpoint-blockade-induced colitis. Nat Commun 7, 10391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang F et al. (2018) Bifidobacterium can mitigate intestinal immunopathology in the context of CTLA-4 blockade. Proc Natl Acad Sci U S A 115 (1), 157–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tivol EA et al. (1995) Loss of CTLA-4 leads to massive lymphoproliferation and fatal multiorgan tissue destruction, revealing a critical negative regulatory role of CTLA-4. Immunity 3 (5), 541–7. [DOI] [PubMed] [Google Scholar]
- 100.Waterhouse P et al. (1995) Lymphoproliferative disorders with early lethality in mice deficient in Ctla-4. Science 270 (5238), 985–8. [DOI] [PubMed] [Google Scholar]
- 101.Nishimura H et al. (1999) Development of lupus-like autoimmune diseases by disruption of the PD-1 gene encoding an ITIM motif-carrying immunoreceptor. Immunity 11 (2), 141–51. [DOI] [PubMed] [Google Scholar]
- 102.Nishimura H et al. (2001) Autoimmune dilated cardiomyopathy in PD-1 receptor-deficient mice. Science 291 (5502), 319–22. [DOI] [PubMed] [Google Scholar]
- 103.Latchman YE et al. (2004) PD-L1-deficient mice show that PD-L1 on T cells, antigen-presenting cells, and host tissues negatively regulates T cells. Proc Natl Acad Sci U S A 101 (29), 10691–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Oh DY et al. (2017) Immune Toxicities Elicted by CTLA-4 Blockade in Cancer Patients Are Associated with Early Diversification of the T-cell Repertoire. Cancer Res 77 (6), 1322–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Das R et al. (2015) Combination therapy with anti-CTLA-4 and anti-PD-1 leads to distinct immunologic changes in vivo. J Immunol 194 (3), 950–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Johnson DB et al. (2016) Fulminant Myocarditis with Combination Immune Checkpoint Blockade. N Engl J Med 375 (18), 1749–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dudley ME et al. (2002) Cancer regression and autoimmunity in patients after clonal repopulation with antitumor lymphocytes. Science 298 (5594), 850–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Das R et al. (2018) Early B cell changes predict autoimmunity following combination immune checkpoint blockade. J Clin Invest 128 (2), 715–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gowen MF et al. (2018) Baseline antibody profiles predict toxicity in melanoma patients treated with immune checkpoint inhibitors. J Transl Med 16 (1), 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Osorio JC et al. (2017) Antibody-mediated thyroid dysfunction during T-cell checkpoint blockade in patients with non-small-cell lung cancer. Ann Oncol 28 (3), 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kumar P et al. (2018) A comprehensive review on the role of co-signaling receptors and Treg homeostasis in autoimmunity and tumor immunity. J Autoimmun. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Suarez-Almazor ME et al. (2017) Review: Immune-Related Adverse Events With Use of Checkpoint Inhibitors for Immunotherapy of Cancer. Arthritis & Rheumatology 69 (4), 687–699. [DOI] [PubMed] [Google Scholar]
- 113.Nancey S et al. (2012) Blockade of cytotoxic T-lymphocyte antigen-4 by ipilimumab is associated with a profound long-lasting depletion of Foxp3+ regulatory T cells: A mechanistic explanation for ipilimumab-induced severe enterocolitis? Inflammatory Bowel Diseases 18 (8), E1598–E1600. [DOI] [PubMed] [Google Scholar]
- 114.Callahan MK et al. (2011) Evaluation of serum IL-17 levels during ipilimumab therapy: Correlation with colitis. Journal of Clinical Oncology 29 (15). [Google Scholar]
- 115.Lim SY et al. (2018) Circulating cytokines predict immune-related toxicity in melanoma patients receiving anti-PD-1-based immunotherapy. Clinical Cancer Research, clincanres.2795.2018. [DOI] [PubMed] [Google Scholar]
- 116.Berndsen CE and Wolberger C (2014) New insights into ubiquitin E3 ligase mechanism. Nat Struct Mol Biol 21 (4), 301–7. [DOI] [PubMed] [Google Scholar]
- 117.Sahtoe DD and Sixma TK (2015) Layers of DUB regulation. Trends Biochem Sci 40 (8), 456–67. [DOI] [PubMed] [Google Scholar]
- 118.Kitaura Y et al. (2007) Control of the B cell-intrinsic tolerance programs by ubiquitin ligases Cbl and Cbl-b. Immunity 26 (5), 567–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sohn HW et al. (2003) Cbl-b negatively regulates B cell antigen receptor signaling in mature B cells through ubiquitination of the tyrosine kinase Syk. J Exp Med 197 (11), 1511–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yasuda T et al. (2000) Cbl suppresses B cell receptor-mediated phospholipase C (PLC)-gamma2 activation by regulating B cell linker protein-PLC-gamma2 binding. J Exp Med 191 (4), 641–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Matsuzawa A et al. (2008) Essential cytoplasmic translocation of a cytokine receptor-assembled signaling complex. Science 321 (5889), 663–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Vallabhapurapu S et al. (2008) Nonredundant and complementary functions of TRAF2 and TRAF3 in a ubiquitination cascade that activates NIK-dependent alternative NF-kappaB signaling. Nat Immunol 9 (12), 1364–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Steri M et al. (2017) Overexpression of the Cytokine BAFF and Autoimmunity Risk. N Engl J Med 376 (17), 1615–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Liu J et al. (2018) Peli1 negatively regulates noncanonical NF-kappaB signaling to restrain systemic lupus erythematosus. Nat Commun 9 (1), 1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Neumann M and Naumann M (2007) Beyond IkappaBs: alternative regulation of NF-kappaB activity. FASEB J 21 (11), 2642–54. [DOI] [PubMed] [Google Scholar]
- 126.Hustad CM et al. (1995) Molecular genetic characterization of six recessive viable alleles of the mouse agouti locus. Genetics 140 (1), 255–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Venuprasad K et al. (2008) The E3 ubiquitin ligase Itch regulates expression of transcription factor Foxp3 and airway inflammation by enhancing the function of transcription factor TIEG1. Nat Immunol 9 (3), 245–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Venuprasad K et al. (2006) Convergence of Itch-induced ubiquitination with MEKK1-JNK signaling in Th2 tolerance and airway inflammation. J Clin Invest 116 (4), 1117–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Huang H et al. (2010) K33-linked polyubiquitination of T cell receptor-zeta regulates proteolysis-independent T cell signaling. Immunity 33 (1), 60–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Aki D et al. (2018) The E3 ligases Itch and WWP2 cooperate to limit TH2 differentiation by enhancing signaling through the TCR. Nat Immunol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Vinuesa CG et al. (2005) A RING-type ubiquitin ligase family member required to repress follicular helper T cells and autoimmunity. Nature 435 (7041), 452–8. [DOI] [PubMed] [Google Scholar]
- 132.Pratama A et al. (2013) Roquin-2 shares functions with its paralog Roquin-1 in the repression of mRNAs controlling T follicular helper cells and systemic inflammation. Immunity 38 (4), 669–80. [DOI] [PubMed] [Google Scholar]
- 133.Vogel KU et al. (2013) Roquin paralogs 1 and 2 redundantly repress the Icos and Ox40 costimulator mRNAs and control follicular helper T cell differentiation. Immunity 38 (4), 655–68. [DOI] [PubMed] [Google Scholar]
- 134.King CG et al. (2006) TRAF6 is a T cell-intrinsic negative regulator required for the maintenance of immune homeostasis. Nat Med 12 (9), 1088–92. [DOI] [PubMed] [Google Scholar]
- 135.Chang M et al. (2011) The ubiquitin ligase Peli1 negatively regulates T cell activation and prevents autoimmunity. Nat Immunol 12 (10), 1002–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Xie P et al. (2007) Tumor necrosis factor receptor-associated factor 3 is a critical regulator of B cell homeostasis in secondary lymphoid organs. Immunity 27 (2), 253–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Catrysse L et al. (2014) A20 in inflammation and autoimmunity. Trends Immunol 35 (1), 22–31. [DOI] [PubMed] [Google Scholar]
- 138.Xiao Y et al. (2013) Peli1 promotes microglia-mediated CNS inflammation by regulating Traf3 degradation. Nat Med 19 (5), 595–602. [DOI] [PMC free article] [PubMed] [Google Scholar]