Abstract
Syntrophy is essential for the efficient conversion of organic carbon to methane in natural and constructed environments, but little is known about the enzymes involved in syntrophic carbon and electron flow. Syntrophus aciditrophicus strain SB syntrophically degrades benzoate and cyclohexane-1-carboxylate and catalyzes the novel synthesis of benzoate and cyclohexane-1-carboxylate from crotonate. We used proteomic, biochemical, and metabolomic approaches to determine what enzymes are used for fatty, aromatic, and alicyclic acid degradation versus for benzoate and cyclohexane-1-carboxylate synthesis. Enzymes involved in the metabolism of cyclohex-1,5-diene carboxyl-CoA to acetyl-CoA were in high abundance in S. aciditrophicus cells grown in pure culture on crotonate and in coculture with Methanospirillum hungatei on crotonate, benzoate or cyclohexane-1-carboxylate. Incorporation of 13C-atoms from 1-[13C]-acetate into crotonate, benzoate, and cyclohexane-1-carboxylate during growth on these different substrates showed that the pathways are reversible. A protein conduit for syntrophic reverse electron transfer from acyl-CoA intermediates to formate was detected. Ligases and membrane-bound pyrophosphatases make pyrophosphate needed for the synthesis of ATP by an acetyl-CoA synthetase. S. aciditrophicus, thus, uses a core set of enzymes that operates close to thermodynamic equilibrium to conserve energy in a novel and highly efficient manner.
Introduction
Syntrophy is a dynamic interaction among two or more microorganisms in which the syntrophic partnership is the only means to sustain energy production for all members involved. It is essential for carbon cycling in natural and constructed anaerobic ecosystems and is a viable source for biogenetic methane production (McInerney, 1999; McInerney et al., 2009). In methanogenic environments, syntrophy often involves the transfer of hydrogen and formate between a fermentative, syntrophic metabolizer and a methanogenic archaeon (Schink, 1997; McInerney et al., 2008; Sieber et al., 2012). The syntrophic partner produces hydrogen and formate that must be kept at very low concentrations (ca. 10 Pa or 10 µM, respectively) by the methanogen to allow the degradative reaction to be energetically favorable. Alternatively, syntrophic metabolism can be achieved by direct electron transfer via conductive pili (Gorby et al., 2006; El-Naggar et al., 2010; Leang et al., 2010; Summers et al., 2010; Qian et al., 2011) or conductive minerals (Kato et al., 2012). Recently, it has been shown that Syntrophus aciditrophicus has electrically conductive pili that allow syntrophic growth by direct electron transfer (Walker et al., 2018).
S. aciditrophicus serves as the model organism for syntrophic fatty, aromatic, and alicyclic acid (cyclohexane-1-carboxylate) degradation, producing acetate, CO2, formate and/or H2 in coculture with H2- and/or formate-using microorganisms (Hopkins et al., 1995; Jackson et al., 1999; Elshahed et al., 2001; Mouttaki et al., 2009). Although S. aciditrophicus is considered a syntrophic specialist, it also ferments crotonate and benzoate (Jackson et al., 1999; Elshahed and McInerney, 2001; Mouttaki et al., 2007) and respires benzoate in pure culture (Mouttaki et al., 2008). Interestingly, S. aciditrophicus catalyzes the novel synthesis of benzoate and cyclohexane-1-carboxylate from crotonate (Mouttaki et al., 2007) (Fig. S1). The detection of similar metabolites during both benzoate and cyclohexane-1-carboxylate degradation and benzoate and cyclohexane-1-carboxylate formation (Elshahed et al., 2001; Elshahed and McInerney, 2001; Mouttaki et al., 2007) suggests that the same enzymes may be used for both benzoate and cyclohexane-1-carboxylate degradation and synthesis. An earlier study indicated that benzoate and cyclohexane-1-carboxylate degradation proceed by a pathway involving 2-hydroxycyclohexane-1-carboxyl-CoA (Elshahed et al., 2001) as found in Rhodopseudomonas palustris (Harwood et al., 1998; Pelletier and Harwood, 2000) (Fig. S1). However, later genomic and enzymatic studies indicated that benzoate and cyclohexane-1-carboxylate degradation proceeds via 6-hydroxycyclohex-1-ene-1-carboxyl-CoA (Peters et al., 2007; Kuntze et al., 2008; Löffler et al., 2011) as found in Geobacter metallireducens (Wischgoll et al., 2005; Fuchs, 2008) (Fig. S1). The proposed S. aciditrophicus substrate degradative pathways are depicted in Figure 1. Remarkably, S. aciditrophicus uses a novel approach for ATP synthesis from acetyl-CoA involving an acetyl-CoA synthetase (Acs1), which uses AMP, pyrophosphate and acetyl-CoA to make ATP and acetate, rather than the classical bacterial approach involving phosphate acetyl transferase and acetate kinase (James et al., 2016). However, it is still unclear how net ATP synthesis needed for growth occurs during syntrophic aromatic acid metabolism as aromatic ring reduction and reverse electron transfer needed in hydrogen and formate production are energy requiring processes (Sieber et al., 2012).
Figure 1. Summary of substrate utilization pathways operative in S. aciditrophicus.
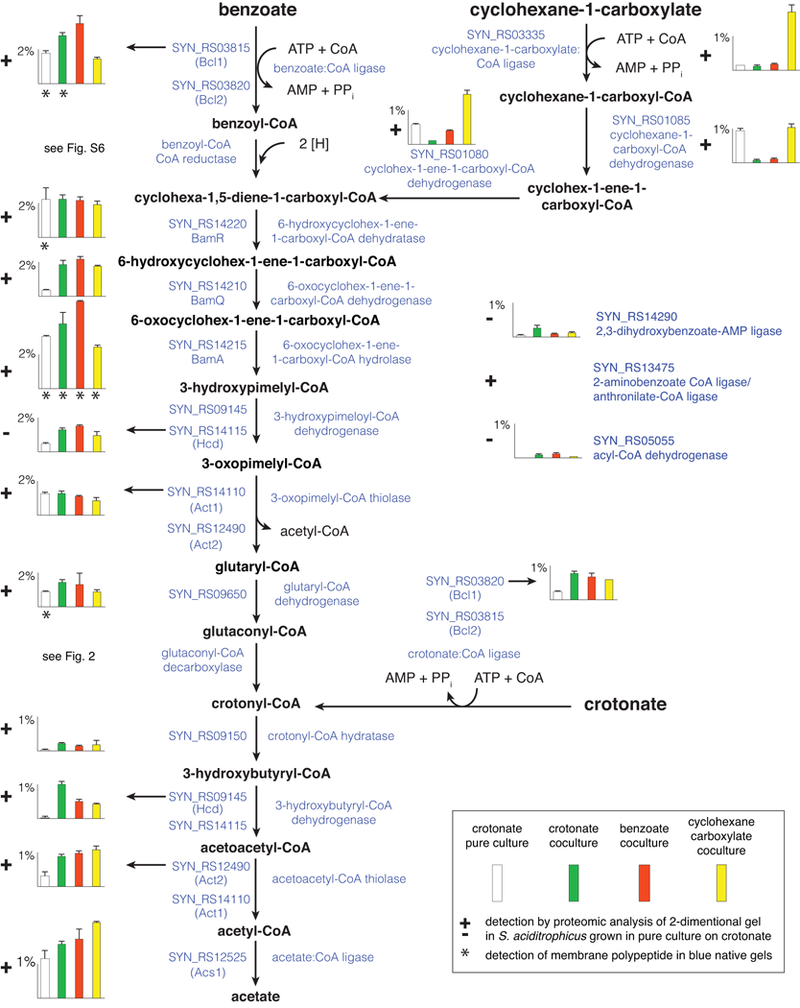
Peptide abundance of proteins is indicated in histograms as the percentage of the total peptides detected in S. aciditrophicus grown: in pure culture on crotonate (white) (duplicate cultures), in coculture on crotonate (green) (triplicate cultures), in coculture on benzoate (red) (duplicate cultures), and in coculture on cyclohexane-1-carboxylate (yellow) (duplicate cultures) (see box in figure). Error bars show the standard deviation when it was larger than the width of the horizontal capping bar. A plus sign indicates that the polypeptide was detected by proteomic analysis of 2-dimensional gel spots with S. aciditrophicus cells grown in pure culture on crotonate (duplicates). Asterisk underneath the histogram bars indicates that the polypeptide was detected in blue native gels with membranes from S. aciditrophicus cells grown under that growth condition (single culture for each condition). Gene locus tags of the RefSeq version of the genome (accession number, NC_007759) are listed under each gene product. For reactions with multiple locus tags, the arrow indicates to which locus tag the histogram corresponds.
Here, we use several proteomic approaches and stable isotope metabolomics to determine whether the same or different enzymes are involved in syntrophic crotonate, benzoate and cyclohexane-1-carboxylate degradation and in benzoate and cyclohexane-1-carboxylate synthesis. We used recombinant DNA and classical biochemical approaches to characterize the ligases involved in substrate activation and to determine whether they make pyrophosphate. In this comprehensive study, we show that a) S. aciditrophicus uses a reversible set of core enzymes for syntrophic substrate degradation and benzoate and cyclohexane-1-carboxylate synthesis; b) substrate activation occurs by pyrophosphate-producing ligases; and c) the thermodynamically difficult redox reaction involving the formation of hydrogen or formate from electrons generated in the oxidation of acyl-CoA intermediates involves a protein conduit comprised of an electron transfer flavoprotein and an iron-sulfur oxidoreductase that interacts with the quinone pool.
Results
Common central metabolic pathway.
Shotgun whole-cell proteomics combined with proteomic analysis of 2D-PAGE of cell extracts and of BN-PAGE of membrane proteins revealed a core set of enzymes that S. aciditrophicus uses to degrade crotonate, benzoate, and cyclohexane-1-carboxylate and to synthesize cyclohexane-1-carboxylate and benzoate (Fig. 1; Table S1; Datasets 1, 2, 3, and 4). BamR, BamQ, and BamA, which convert cyclohex-1,5-diene-1-carboxyl-CoA to 3-hydroxypimelyl-CoA, were among the most abundant proteins detected in the proteome, ranging from 0.36 to 5.1% of the total peptides detected under the four different growth conditions examined (Fig. 1). The activities of BamR (SYN_RS14220 gene product) and BamA (SYN_RS14215 gene product) have been previously demonstrated in S. aciditrophicus (Peters et al., 2007; Kuntze et al., 2008; Löffler et al., 2011). To confirm the proposed BamQ function, SYN_RS14210 was cloned and expressed in E. coli BL21. The purified recombinant SYN_RS14210 enzyme had a Vmax of 14 U • mg−1, and KM of 72 ± 10 µM for 6-hydroxycyclohex-1-ene-1-carboxyl-CoA, confirming that it is a 6-hydroxycyclohex-1-ene-1-carboxyl-CoA dehydrogenase (BamQ activity). These values are similar to those determined for the related enzymes from Thauera aromatica (Vmax of 12 U • mg−1, KM of 60 ± 20 µM) and G. metallireducens (Vmax of 35 U • mg−1) (Laempe et al., 1999). We did not detect the formation of 2-hydroxycyclohexane-1-carboxyl-CoA from cyclohex-1-ene-1-carbonyl-CoA, or NAD+ reduction or NADH oxidation when 2-hydroxycyclohexane-1-carboxyl-CoA or 2-oxocyclohexane-1-carboxyl-CoA, respectively, were added to cell-free extracts of S. aciditrophicus grown in pure culture on crotonate (data not shown), arguing against the pathway found in R. palustris (Harwood et al., 1998) (Fig. S1).
Next, 3-hydroxypimelyl-CoA is converted to glutaryl-CoA and acetyl-CoA by two enzymes: a 3-hydroxyacyl-CoA dehydrogenase (Hcd) (encoded by SYN_RS09145 (hcd) and an acetyl-CoA acetyltransferase (Act) (encoded by SYN_RS14110 (act1) or SYN_RS12490 (act2)) (Fig. 1). Act1 and Act2 were abundant in the proteome under all tested conditions (0.3 to 1.5% of the total peptides detected) (Fig. 1). Hcd was detected in low amounts in cells grown in pure culture on crotonate (0.02% of total peptides detected) but was abundant in cells grown in coculture on crotonate, benzoate, and cyclohexane-1-carboxylate (0.4 to 1% of the total peptides detected) (Fig. 1). The SYN_RS14115 gene product annotates as a 3-oxoacyl-CoA reductase and might function as a 3-hydroxyacyl-CoA dehydrogenase. Its peptides were abundant under all tested conditions (0.47 to 1.5% of the total peptides detected).
Glutaryl-CoA is converted to glutaconyl-CoA by non-decarboxylating glutaryl-CoA dehydrogenase (Ghd) (SYN_RS09650 gene product) (Djurdjevic, 2010), which was abundant in the proteome (0.9 to 1.4% of the total peptides detected) (Fig. 1). Another putative acyl-CoA dehydrogenase was present in coculture-grown S. aciditrophicus cells but at lower levels than Ghd; its function is not clear (Fig. 1). The decarboxylation of glutaconyl-CoA to crotonyl-CoA and CO2 is accomplished by a Na+-translocating, glutaconyl-CoA decarboxylase (GcdABC)) (Beatrix et al., 1990; Schöcke and Schink, 1999) and the carboxytransferase subunit (GcdA) was abundant in the proteome under all growth conditions (0.4 to 1.3% of the total peptides detected) (Fig. 2).
Figure 2.
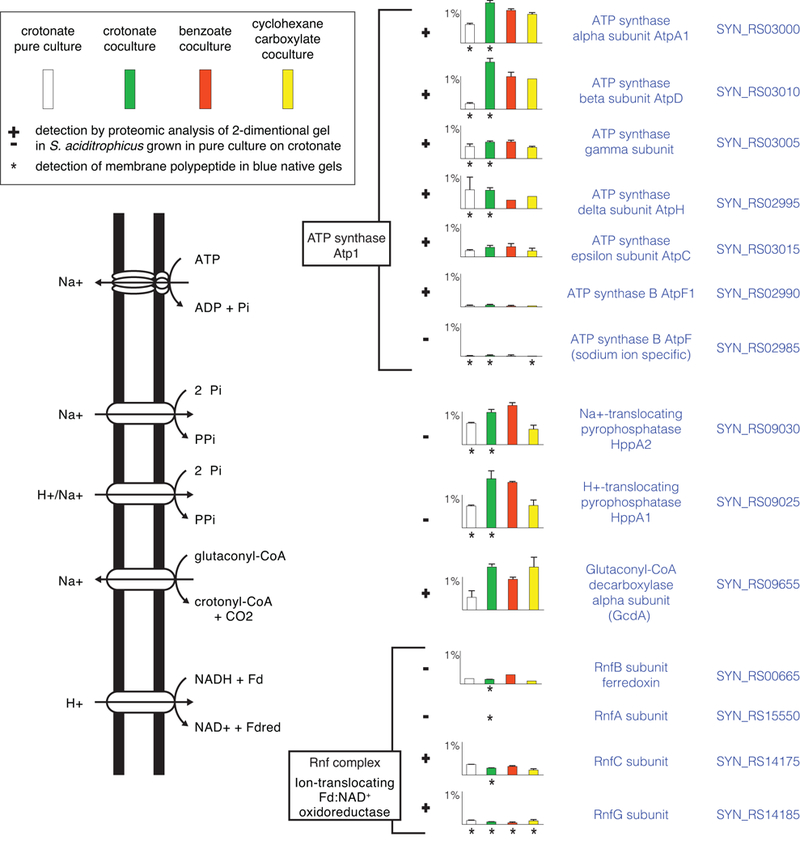
Membrane proteins involved in formation and use of ion gradients. Peptide abundance of proteins is indicated in histograms as the percentage of the total peptides detected in S. aciditrophicus grown: in pure culture on crotonate (white) (duplicates), in coculture on crotonate (green) (triplicates), in coculture on benzoate (red) (duplicates), and in coculture on cyclohexane-1-carboxylate (yellow) (duplicates) (see box in figure). Error bars show the standard deviation when it was larger than the width of the horizontal capping bar. A plus sign indicates that the polypeptide was detected by proteomic analysis of 2-dimensional gel spots with S. aciditrophicus cells grown in pure culture on crotonate (duplicates). Asterisk underneath the histogram bars indicates that the polypeptide was detected in blue native gels with membranes from S. aciditrophicus cells grown under that growth condition (single culture for each condition). Gene locus tags of the RefSeq version of the genome (accession number, NC_007759) are listed by each gene product.
The conversion of crotonyl-CoA to 3-hydroxybutyryl-CoA is most likely catalyzed by SYN_RS09150 gene product, as it was the only enoyl-CoA hydratase detected in the S. aciditrophicus proteome (Fig. 1). Hcd, the SYN_RS14115 gene product (possible 3-hydroxyacyl-CoA dehydrogenase), Act1 and Act2 discussed above for 3-hydroxypimelyl-CoA metabolism could also be involved in the conversion of 3-hydroxybutyryl-CoA to acetyl-CoA (Fig. 2). Enoyl-CoA hydratase, 3-hydroxyacyl-CoA dehydrogenase, and acetyl-CoA acetyltransferase activities have been demonstrated in cell-free extracts of S. aciditrophicus grown in pure culture on crotonate and in coculture on benzoate (Elshahed et al., 2001).
All of the central core enzymes except the SYN_RS14115 gene product were also detected by proteomic analysis of 2D gel slices of S. aciditrophicus cells grown in pure culture on crotonate (Fig. 1; Dataset 3).
Aromatic and fatty acid activation.
The bioenergetic model proposed for S. aciditrophicus involves substrate activation by AMP- and pyrophosphate-forming ligases and the subsequent use of the pyrophosphate and AMP to make ATP by an AMP-forming acetyl-CoA synthetase (Acs1) (James et al., 2016) (Fig. 1). The S. aciditrophicus genome contains four genes that annotate as aromatic acid ligases and four genes that annotate as fatty acid ligases (McInerney et al., 2007). To determine which gene products are involved in substrate activation, crotonate:CoA ligase and benzoate:CoA ligase activities were purified from cell-free extracts of S. aciditrophicus. The purification of the crotonate:CoA ligase activity recovered 8% of the initial activity (Table S2) and yielded a homogenous protein that migrated as a single band with an apparent mass of 60 kDa after denaturing gel electrophoresis (Fig. S2). Mass spectrometric analysis assigned 52 unique polypeptides to one protein encoded by SYN_RS03815 (Table S3). The purification of benzoate:CoA ligase activity recovered 36% of the initial activity (Table S4) and resulted in a final preparation that contained two proteins with masses of approximately 60 kDa on denaturing gels (Fig. S3). Eleven unique polypeptides were assigned to the SYN_RS03820 gene product, which has a mass of 58951 daltons; and 8 unique polypeptides were assigned to the SYN_RS03815 gene product, which has a mass of 59049 daltons (Table S4). The SYN_RS03815 and SYN_RS03820 gene products were abundant under all growth conditions (McInerney et al., 2007). A ligase encoded by SYN_RS14290 was present in low abundance and a ligase encoded by SYN_RS13475 was only detected in 2D gel slices with crotonate-grown pure culture cells (Fig. 1).
The following analyses show that the SYN_RS03815 and SYN_RS03820 gene products (Bcl1 and Bcl2, respectively) function as benzoate:CoA and crotonate:CoA ligases. The purified Bcl1 had high activity with benzoate and crotonate, very low activity with cyclohexane-1-carboxylate, and undetectable activity with cyclohex-1-ene-1-carboxylate, and hydroxylated benzoates (Table S5). The nearly pure fraction with Bcl1 and Bcl2 had high benzoate:CoA ligase activity, much less activity with crotonate or cyclohexane-1-carboxylate, and very low activity with 3-hydroxybenzoate and 4-hydroxybenzoate (Table S5). To verify the substrate specificities, each gene was heterologously expressed in Escherichia coli. The purified, recombinant Bcl1 had a Vmax and Km for benzoate of 43 U • mg−1 and 0.16 mM, respectively, and a Vmax and Km for crotonate of 23 U • mg−1 and 4.2 mM, respectively (Table 1). These values are similar to the respective Vmax and Km of the crotonate:CoA ligase activity purified from cell-free extracts of S. aciditrophicus (Table 1). The purified, recombinant Bcl2 had a Vmax of 77 U • mg−1 and Km value of 0.11 mM for benzoate and a Vmax of 2.3 U • mg−1 and Km value of 2.3 mM for crotonate, both of which are in agreement with the values obtained from the preparation purified from S. aciditrophicus cell-free extracts (Table 1). The purified, recombinant enzyme had high cyclohexane-1-carboxyl-CoA ligase activity (Table S5), but its involvement in cyclohex-1-ene-1-carboxylate activation is unlikely as the Km value for cyclohexane-1-carboxylate was 8 mM (Table 1).
Table 1.
Kinetic properties of purified ligase activities and recombinantly produced ligases from S. aciditrophicus.
| Substrate | Purified crotonate:CoA ligase activitya | Purified benzoate:CoA ligase activity a | Recombinant Bcl1 | Recombinant Bcl2 | Recombinant SYN_RS03335 gene product | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Km
(mM) |
Vmax
(U•mg−1) |
Km
(mM) |
Vmax
(U•mg−1) |
Km
(mM) |
Vmax
(U•mg−1) |
Km
(mM) |
Vmax
(U•mg−1) |
Km
(mM) |
Vmax
(U•mg−1) |
|
| Benzoate | 0.3 ± 0.04 | 54 ± 2 | 0.3 ± 0.2 | 35 ± 0.6 | 0.16 | 43 | 0.11 | 77 | ND | ND |
| Crotonate | 4.7 ± 1.9 | 14 ± 2 | 10 | 26 | 4.2 | 23 | 2.4 | 2.3 | 38 ± 6 | 0.7 ± 0.04 |
| Cyclohexane-1-carboxylate | 12 | 15 | 19 | 13 | 0.04 ± 0.007 | 15 ± 0.5 | ||||
| Cyclohex-1-ene-1-carboxylate | 8 | 7.6 | 0.006 ± 0.002 | 0.6 ± 0.04 | ||||||
| CoA | 0.5 ± 0.2 | 17 ± 4 | 0.3 | 27 | 0.3 ± 0.05 | |||||
The purified crotonate:CoA ligase activity contained Bcl and the benzoate:CoA ligase activity contained Bcl1 and Bcl2.
Although the Km values for crotonate are high for the purified recombinant Bcl1 and Bcl2 (4.2 and 2.4 mM, respectively), the Km values are consistent with the growth behavior of S. aciditrophicus on crotonate. Very little growth of S. aciditrophicus occurs at crotonate concentrations of 2.5 mM and optimal growth occurs when the crotonate concentration is >10 mM (Fig. S4).
Cyclohexane-1-carboxylate activation and metabolism.
The SYN_RS03335 gene product was differentially abundant when S. aciditrophicus was grown in coculture on cyclohexane-1-carboxylate (Fig. 1). SYN_RS03335 annotates as a long chain fatty acid ligase (McInerney et al., 2007). The purified recombinant SYN_RS03335 gene product (Fig. S5; Table S5) had a high Vmax (15 ± 0.5 U • mg−1) and a low Km (40 ± 7 µM) for cyclohexane-1-carboxylate, and a low Vmax (0.6 ± 0.002 U • mg−1) and a low Km (6 ± 2 µM) for cyclohex-1-ene-1-carboxylate (Table 1). The enzyme had low activity for benzoate, crotonate, acetate and succinate (Table S5). Thus, the SYN_RS03335 gene product is a cyclohexane-1-carboxylate:CoA ligase.
Peptides of cyclohex-1-ene-1-carboxyl-CoA dehydrogenase (SYN_RS01080 gene product) and cyclohexane-1-carboxyl-CoA dehydrogenase (SYN_RS01085 genes product) (Kung et al., 2013; Kung et al., 2014) were also differentially abundant in the proteome when S. aciditrophicus was grown in coculture on cyclohexane-1-carboxylate and in pure culture on crotonate where cyclohexane-1-carboxylate is formed as an end product (Elshahed et al., 2001; Mouttaki et al., 2007) (Fig. 2), supporting their role in cyclohexane-1-carboxylate degradation and synthesis. SYN_RS03335, SYN_RS01080 and SYN_RS01085 gene products were detected by proteomic analysis of 2-D gel slices in S. aciditrophicus grown in pure culture on crotonate (Fig. 2).
Benzoyl-CoA reduction.
The S. aciditrophicus genome has two gene loci with the same synteny as a gene locus found in G. metallireducens that codes for the ATP-independent, benzoyl-CoA reductase (BamBCDEF) and a NADH dehydrogenase (BamGHI) (Wischgoll et al., 2005; McInerney et al., 2007; Kung et al., 2009; Kung et al., 2014). Additionally, S. aciditrophicus has two other gene clusters with paralogs of bamBCDEF, but lacking bamGHI genes, one set of two genes homologous to bamBC and one set of two genes homologous to bamEF (Fig. S6). While all of the five benzoyl-CoA reductase subunits were detected in the proteome, they were not encoded by a single gene cluster, but by genes from multiple clusters (Fig. S6). BamB4, BamC5, BamD4, BamE5, BamF5 were the most abundant of the BamBCDEF subunits detected. Two NADH dehydrogenase subunits (SYN_RS00430 and SYN_RS00435 gene products) were detected under all growth conditions (Fig. 3), but are encoded by genes not linked to any of the bamBCDE genes (Fig. S6).
Figure 3.
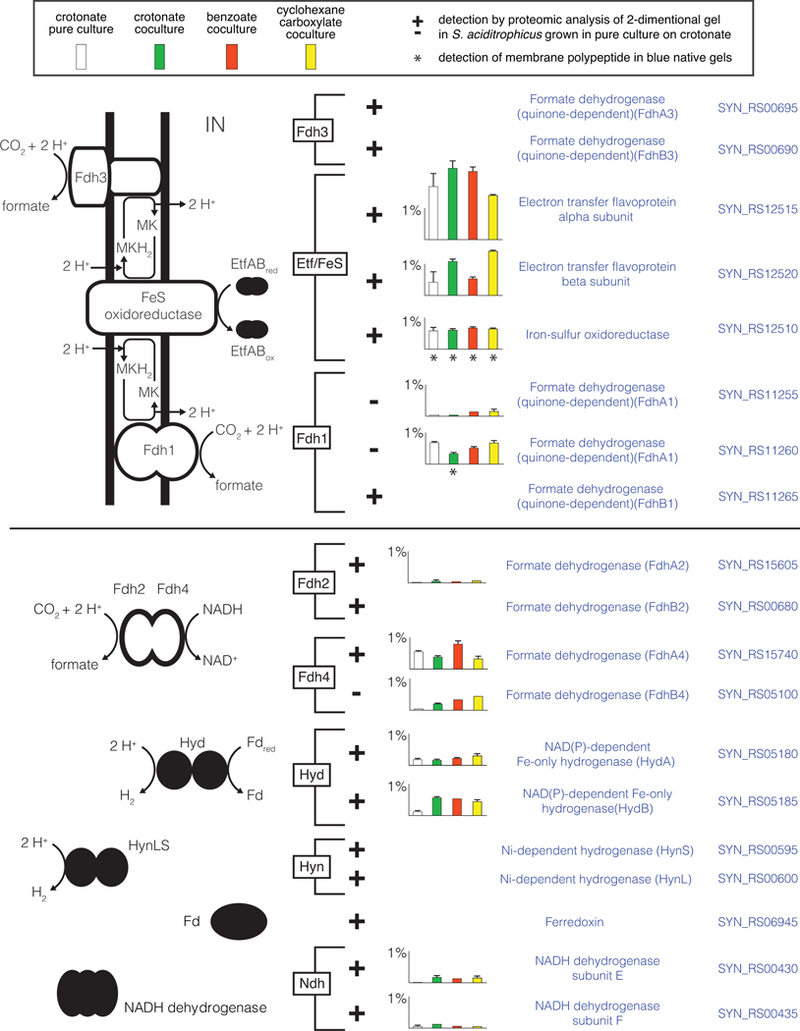
Proteins involved in electron transfer and formate and hydrogen production in S. aciditrophicus. Peptide abundance of proteins is indicated in histograms as the percentage of the total peptides detected in S. aciditrophicus grown: in pure culture on crotonate (white) (duplicates), in coculture on crotonate (green) (triplicates), in coculture on benzoate (red) (duplicates), and in coculture on cyclohexane-1-carboxylate (yellow) (duplicates) (see box in figure). Error bars show the standard deviation when it was larger than the width of the horizontal capping bar. A plus sign indicates that the polypeptide was detected by proteomic analysis of 2-dimensional gel spots with S. aciditrophicus cells grown in pure culture on crotonate (duplicates). Asterisk underneath the histogram bars indicates that the polypeptide was detected in blue native gels with membranes from S. aciditrophicus cells grown under that growth condition (single culture for each condition). Gene locus tags of the RefSeq version of the genome (accession number, NC_007759) are listed by each gene product.
ATP synthesis from acetyl-CoA.
S. aciditrophicus uses a novel approach to make ATP by substrate-level phosphorylation involving acetyl-CoA synthetase (Acs1) (James et al., 2016). However, two genes are present that annotated as butyrate kinases (SYN_RS09865, buk1 and SYN_RS11570, buk2) (McInerney et al., 2007) that could be used to make ATP from acetyl-phosphate as in other bacteria. To test this possibility, both genes were cloned into an acetate kinase mutant of Escherichia coli. After induction, cell-free extracts of the mutant strains with plasmid buk1or plasmid buk2 each had high butyrate kinase activities (Table S5). Acetate kinase activity was detected at levels less than the background levels present in the mutant without the plasmid (Table S6). Thus, Buk1 and Buk2 are butyrate kinases not acetate kinases.
Electron flow and bioenergetics.
Electron transfer flavoprotein EtfAB (SYN_RS12515 and SYN_RS12520 gene products) and a membrane-bound iron sulfur oxidoreductase (Fes) (SYN_RS12510 gene product) were highly abundant in the proteome under all growth conditions and probably serve as the membrane input module for electrons derived from the oxidation of acyl-CoA intermediates (Fig. 3; Datasets 1, 2, 3 and 4). Two membrane-bound formate dehydrogenases (FdhA1B1 and FdhA3B3) were also detected (Fig. 3; Table S7; Dataset 4).
Reduced ferredoxin required for benzoyl-CoA reduction (Kung et al., 2009) may be generated by a membrane-bound NADH:ferredoxin oxidoreductase called Rnf (Biegel and Müller, 2010). Shotgun proteomic analyses detected two Rnf subunits (RnfC and RnfG) under all growth conditions (Fig. 2). RnfA, RnfC and RnfG were also detected in 2-D gels from crotonate-grown cells as well as in BN gels (Fig. 2; Datasets 3 and 4). NADH reoxidation can occur by a NADH-dependent formate dehydrogenase (FdhA4B4) and a NADH-dependent hydrogenase (HydAB), whose peptides were abundant in the proteome under all growth conditions (Fig. 3). Another NADH-linked formate dehydrogenase (FdhA2) was detected under all growth conditions, but at low levels (Fig. 3).
Two membrane-bound pyrophosphates, HppA1 and HppA2 were detected in the proteome under all growth conditions (Fig. 2). Their membrane location was confirmed by detection in blue native gels from cells grown in pure culture and in coculture on crotonate. The chemiosmotic gradient needed for pyrophosphate synthesis and reverse electron transfer can be formed by the activity of glutaconyl-CoA decarboxylase discussed above (Fig. 2). In addition, S. aciditrophicus has genes for two ATP synthases (atp1 and atp2), which could generate a chemiosmotic potential by ATP hydrolysis (McInerney et al., 2007). Shotgun proteomic and 2-D gel analyses detected seven subunits of Atp1 whose peptides were abundant under all growth conditions while peptides for six subunits were detected in BN gels. Peptides from subunits of the Atp2 complex were not detected (Fig. 2).
Reversibility of the pathway.
Isotopomer analysis showed that 13C-atoms from 1-[13C]-acetate were detected in crotonate, benzoate, and cyclohexane-1-carboxylate during growth of S. aciditrophicus on these individual substrates (Table 2). The incorporation of [13C]-carbon from acetate into substrates during their degradation shows that the enzyme reactions involved in these pathways are reversible.
Table 2.
Evidence for the reversibility of the pathways for crotonate, benzoate, and cyclohexane-1-carboxylate metabolism.a
| Metabolite detected | 1-[13C]-Acetate + Crotonate | 1-[13C]-Acetate + Benzoate | 1-[13C]-Acetate + Cyclohexane-1-carboxylate | Unlabeled authentic standard | Number of 13C atoms present in the substrate |
|---|---|---|---|---|---|
| Crotonate | NDb | BDLb | 144, 145, 146 | 144 | 1 or 2 |
| Benzoate | 179, 180, 181,182 | ND | 179, 180, 181,182 | 179 | 1, 2, or 3 |
| Cyclohexane-1-carboxylate | 185, 186, 187,188 | 185, 186, 187,188 | ND | 185 | 1, 2, or 3 |
The mass of m/z-15 ions of trimethylsilane-derivatized substrates detected by GC-MS in culture fluids of S. aciditrophicus when amended with [13C1]-acetate.
Abbreviations: ND, not determined; BDL, below detection limit.
Discussion
S. aciditrophicus has an interesting physiology in that it degrades benzoate and cyclohexane-1-carboxylate to acetate, and synthesizes benzoate and cyclohexane-1-carboxylate from crotonate (Elshahed et al., 2001; Elshahed and McInerney, 2001; Mouttaki et al., 2007, 2008). The synthesis of benzoate and cyclohexane-1-carboxylate from crotonate and/or acetate (Mouttaki et al., 2007) is unconventional, as cyclohexane-1-carboxylate formation was thought to occur only by the dehydration and reduction of shikimate, the classic precursor used to form aromatic amino acids (McInerney et al., 2009). However, proteomic analyses showed that the same enzymes (Fig. 1 and 2) are used to convert cyclohex-1,5-diene-1-carboxyl-CoA to acetyl-CoA under all tested growth conditions. 13C-Labeling studies show that 1, 2, or 3 13C-atoms from 1-[13C]-acetate were found in TMS derivatives of benzoate and cyclohexane-1-carboxylate during growth on crotonate and 1-[13C]-acetate (Table 2), consistent with the reversibility of the pathway. While the protein levels varied somewhat, there was no appreciable modulation of protein levels with exception of the three branch pathway reactions leading from cyclohexane-1-carboxylate. BamR, BamQ, and BamA, which catalyze the conversion of cyclohex-1,5-diene-1-carboxyl-CoA to 3-hydroxypimelyl-CoA via a 6-substituted cyclohex-1-ene-1-carboxyl-CoA intermediate, were among the most abundant proteins in the proteome (Fig. 1). Based on its high abundance in the proteome, BamA is the likely candidate to catalyze ring formation when making benzoate and cyclohexane-1-carboxylate from crotonate and acetate (Fig. 1). However, purified recombinant BamA from S. aciditrophicus did not form 6-oxocyclohex-1-ene-1-carboxyl-CoA from pimelyl-CoA or 3-hydroxypimeplyl-CoA (Kuntze et al., 2008). It may be that the in vivo conditions needed for 6-oxocyclohex-1-ene-1-carboxyl-CoA formation were not replicated in vitro or that another enzyme is involved in ring formation. Our proteomic and enzymatic analyses show that benzoate and cyclohexane-1-carboxylate degradation and synthesis likely proceed via a 6-substituted cyclohex-1-ene-1-carboxyl-CoA intermediate by using the same enzymes in a reversible manner.
The current bioenergetic model for energy conservation in S. aciditrophicus involves the unique synthesis of ATP by an AMP-forming, acetyl-CoA synthetase (Acs1) (James et al., 2016) (Fig. 4). It was thought that Buk1 and Buk2 might function as acetate kinases to make ATP (McInerney et al., 2007) but, Buk1 and Buk2 had butyrate kinase activity not acetate kinase activity (Table S6). Substrate activation in S. aciditrophicus occurs by pyrophosphate-forming ligases (Table 1). As more acetyl-CoA is formed during substrate degradation than the amount of pyrophosphate formed in substrate activation, another source of pyrophosphate is needed. Membrane-bound pyrophosphatases (Schöcke and Schink, 1998; Biegel and Müller, 2011) were detected in the proteome (Fig. 3), which could synthesize the additional pyrophosphate. The chemiosmotic gradient needed for pyrophosphate synthesis can be formed by a sodium-dependent ATP synthase (Atp1) or glutaconyl-CoA decarboxylase (Fig. 2 and 3). Syntrophus gentianae has membrane-bound pyrophosphatases that catalyze ATP-dependent synthesis of pyrophosphate (Schöcke and Schink, 1998). It should be noted that S. aciditrophicus lacks soluble pyrophosphatases (McInerney et al., 2007) (James et al., 2016). Thus, the pyrophosphate formed during substrate activation or by membrane-bound pyrophosphatases is available for use by Acs1 rather than being rapidly cleaved if soluble pyrophosphatases were present.
Figure 4.
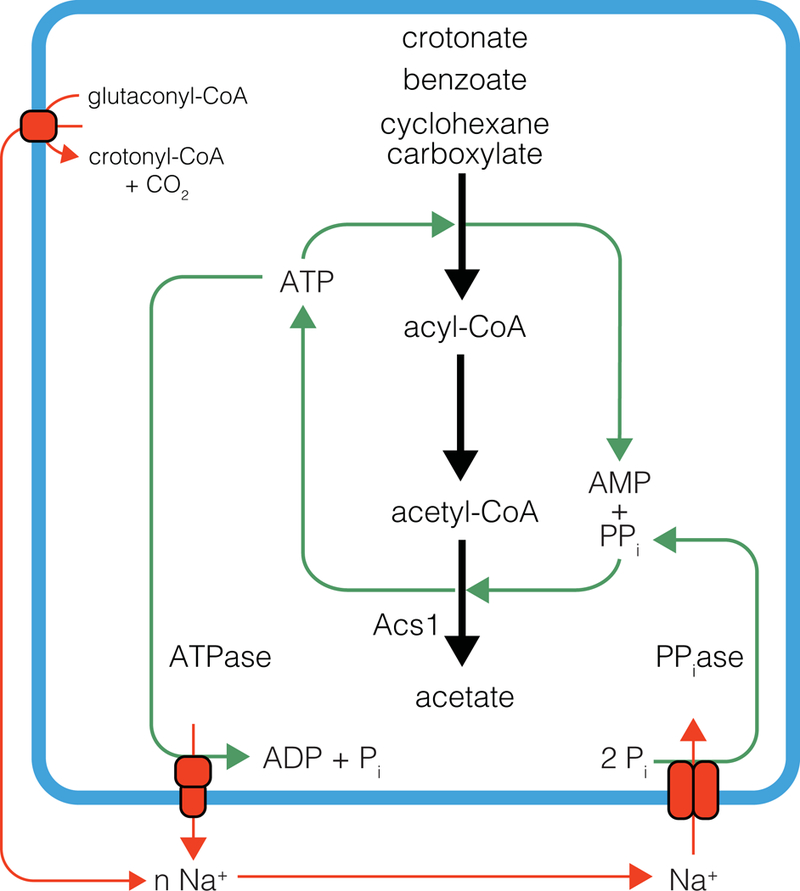
Bioenergetic model for pyrophosphate cycling and ATP synthesis by Acs1 in S. aciditrophicus.
The formation of hydrogen or formate from electrons released in the oxidation of acyl-CoA intermediates generated during syntrophic fatty and aromatic acid metabolism is a defining feature of syntrophic metabolism (Sieber et al., 2012; Sieber et al., 2014; Sieber et al., 2015). Proteomic analysis identified a protein conduit of electron flow from butyryl-CoA to hydrogen or formate in the syntrophic butyrate-degrading Syntrophomonas wolfei involving acyl-CoA dehydrogenase, EtfA2B2, Fes, and a membrane-bound hydrogenase or formate dehydrogenase (Schmidt et al., 2013; Sieber et al., 2014; Sieber et al., 2015). We found a similar protein conduit of electron flow in S. aciditrophicus (Fig. 3). Thus, there is an emerging, unifying model for syntrophic reverse electron transfer that involves a quinone loop where Fes acts to receive electrons from acyl-CoA dehydrogenases via EtfAB and subsequently reduces menaquinone to menaquinol (Schmidt et al., 2013; Sieber et al., 2014; Sieber et al., 2015; Crable et al., 2016) (Fig. 3). The reliance on formate production for reverse electron transfer in S. aciditrophicus is supported by the fact that formate dehydrogenase inhibitors blocked syntrophic benzoate and cyclohexane-1-carboxylate degradation by cocultures of S. aciditrophicus and M. hungatei (Sieber et al., 2014).
Multiple approaches show that S. aciditrophicus uses a single reversible core set of enzymes for the degradation of its substrates and for benzoate and cyclohexane-1-carboxylate synthesis (Fig. 1 and 2) (Table 2), as was suggested for two syntrophic acetate oxidizers, strain AOR and Thermacetogenium phaeum (Lee and Zinder, 1988b, a; Hattori et al., 2005). Reversibility suggests that the enzyme reactions operate at near-equilibrium conditions as expected from catabolic reactions that operate close to thermodynamic equilibrium (∆G ~ −10 to −30 kJ/mol) (Schink and Friedrich, 1994; Jackson and McInerney, 2002; Hoehler et al., 2018). The low free energy changes associated with syntrophic metabolism suggest that their pathways lack strongly irreversible reactions, i.e., those that are associated with large negative free energy changes. In support of this concept, S. aciditrophicus uses a) a reversible reaction for ATP synthesis from acetyl-CoA (acetyl-CoA synthetase rather than phosphate acetyl transferase and acetate kinase) (James et al., 2016); b) an ATP-independent, benzoyl-CoA reductase that catalyzes both the rearomatization of cyclohex-1,5-diene-1-carboxyl-CoA and dearomatization of benzoyl-CoA (Löffler et al., 2011; Kung et al., 2014); and c) a reversible glutaconyl-CoA decarboxylase that catalyzes both the decarboxylation of glutaconyl-CoA and carboxylation of crotonyl-CoA (Dimroth and Hilpert, 1984; Beatrix et al., 1990; Schöcke and Schink, 1999) (Fig. 1, 2 and 4). Thus, syntrophic metabolizers are highly dependent on their environment, as their metabolic pools would be very susceptible to environmental perturbations. The ability of S. aciditrophicus to reverse its biochemical pathway from substrate to product and vice versa using the same enzyme machinery is a remarkable strategy for energy conservation and provides a model to understand how microorganisms evolve and persist under thermodynamic constraints.
Materials and Methods
Cultivation of Syntrophus aciditrophicus.
S. aciditrophicus strain SB (DSM 26646) was grown anaerobically in pure culture on 20 mM crotonate and in coculture with M. hungatei JF1 (ATCC 27890) on 20 mM crotonate, 10 mM benzoate, or 10 mM cyclohexane-1-carboxylate in basal medium without rumen fluid (Elshahed et al., 2001). The Wolin’s metal solution was modified to include Na2MoO4 • 2H2O (0.01g/L), Na2SeO4 (0.01g/L) and Na2WO4 • 2H2O (0.01g/L) (Sieber et al., 2014; Sieber et al., 2015). Stock solutions of the modified Wolin’s trace metals and Balch vitamins were added at a volume of 5 ml•l−1 and 10 ml•l−1, respectively (Sieber et al., 2014). The headspace was pressurized to 27.5 kPa with N2/CO2 (80:20 v/v) (Balch and Wolfe, 1976). Cultivation was conducted at 37°C without shaking. All cultures were transferred at least three times in the same medium prior to use in proteomic analyses. All cultures were examined microscopically and checked for contamination using thioglycollate medium. Culture manipulations were performed in the anaerobic chamber and all centrifuge steps were done with sealed, anoxic, centrifuge tubes (Balch and Wolfe, 1976). Escherichia coli strains BL21, TOP10, and JW2293 were grown in Luria-Bertani (LB) medium with 100 µg/ml of ampicillin, 50 µg/ml of carbenicillin, and 50 µg/ml of kanamycin.
Cell harvesting, separation, and extract preparation.
Cultures were harvested by centrifugation, washed in anoxic phosphate buffer, and the pellets were stored at −80 °C. S. aciditrophicus grown in coculture was separated from M. hungatei cells using a Percoll gradient as previously described (Sieber et al., 2014). Cell-free extracts and membrane and soluble fractions were prepared as previously described (Sieber et al., 2014).
Proteomic analyses.
S. aciditrophicus pure cultures and cocultures grown in duplicate 500-ml Schott bottles (triplicate for crotonate cocultures) with 250 ml of medium were used for whole-cell proteomic analysis. Washed cell pellets were digested with trypsin in solution with deoxycholate present, in accordance with the enhanced Filter Assisted Sample Prepartion (eFASP) method (Erde et al., 2014). Tryptic digests were analyzed by shotgun LC-MSE analysis or fractionated by hydrophilic interaction chromatography (HILIC) prior to LC-MSE analysis (James et al., 2016). Data were searched with ProteinLynx Global Server v3.0.2 (PLGS, Waters) embedded inside Progenesis QI software v2 (Nonlinear Dynamics, Waters). Proteins were quantified based on the top 3 most abundant unique peptides. The Waters Expression Informatics component of the PLGS software was utilized to quantify data using glycogen phosphorylase B (Uniprot accession P00489) as the quantification reference (Silva et al., 2005; Hughes et al., 2006; Silva et al., 2006).
Alternatively, peptides from eFASP processing were separated on an EASY-Spray column (25 cm × 75 µm ID, PepMap RSLC C18, 2 µm, Thermo Scientific) connected to an EASY-nLC 1000 nUPLC (Thermo Scientific) using a gradient of 5 – 35% acetonitrile in 0.1% formic acid, and a flow rate of 300 nl • min−1 (total time 90 minutes). Tandem mass spectra were acquired in a data-dependent manner with an Orbitrap Q Exactive mass spectrometer (Thermo Fisher Scientific) interfaced to a nanoelectrospray ionization source. The raw MS/MS data were converted into MGF format by Thermo Proteome Discoverer 1.4 (Thermo Scientific) and searched with Mascot version 2.5 (Matrix Science, Boston, MA). Carbamidomethylation on cysteine and methionine oxidation were specified as variable modifications. The search allowed for up to two missed cleavages and employed a precursor mass tolerance of ±5 ppm and a product mass tolerance of ±0.01 Da. Quantification was based on the three most intense peptides.
Duplicate pure cultures of S. aciditrophicus grown in 250 ml of 20 mM crotonate medium in 500-ml Schott bottles were used for proteomic analysis after 2-dimensional polyacrylamide gel electrophoresis (2-D PAGE) (see Supplemental Information). For blue-native polyacrylamide gel electrophoresis (BN-PAGE), one-liter of sterile medium in a 2-L Schott bottle was inoculated with 200 ml of the respective culture. BN-PAGE, tryptic digestion of protein bands, and peptide sequencing and analysis were conducted as previously described (Crable et al., 2016).
13C-acetate labeling.
S. aciditrophicus was grown in pure culture on crotonate and in coculture with M. hungatei on benzoate or cyclohexane-1-carboxylate in 500-ml Schott bottles with 300-ml of medium. At mid-log phase, three cultures were amended with 1 mM sodium acetate-1-13C (Cambridge Isotope Laboratories, Inc., Andover, MA) and one culture was amended with unlabeled sodium acetate. Controls included uninoculated medium amended with each labeled substrate. Fifty-ml samples were withdrawn with time after the addition of acetate. The pH of each sample was brought to 12 N with 1 N NaOH, and then acidified to a pH less than 2 with 12 N HCl (Elshahed et al., 2001). Each culture was extracted with 20 ml of ethyl acetate three times; the ethyl acetate extracts were pooled, filtered through anhydrous sodium sulfate, and concentrated by rotary evaporation at 65°C to 2 ml (Elshahed et al., 2001). The extracted samples were then dried under a stream of nitrogen gas and resuspended in 250 µl of ethyl acetate (Elshahed et al., 2001). The ethyl-extracted samples were derivatized by the addition of 50 µl N,O-bis-(trimethylsilyl)trifluoroacetamide (Pierce Chemicals, Rockford, IL) at 70°C for 10 minutes and analyzed by gas chromatography-mass spectrometry (Elshahed et al., 2001; Mouttaki et al., 2007).
Enzyme assays.
6-hydroxycyclohex-1-ene-1-carboxyl-CoA dehydrogenase activity was measured with a continuous assay at 30°C in a 1-cm cuvette containing 200 mM 3-N-morpholino-propanesulfonic acid (MOPS) (pH 7.5), 15 mM MgCl2, 1 mM nicotinamide adenine dinucleotide (NAD+), and 1–5 µg per ml enzyme. The reaction was started by the addition of 200 μM 6-hydroxycyclohex-1-ene-1-carboxyl-CoA. NADH formation was monitored at 365 nm (Δε365=3,400 M−1 cm−1). Enzyme assays for 2-hydroxycyclohexanecarboxyl-CoA oxidation and 2-oxo-cyclohexanecarboxyl-CoA reduction were carried out as described previously (Pelletier and Harwood, 2000).
Ligase activities were measured by tracking AMP formation with its conversion to ADP via myokinase and the subsequent oxidation of NADH with the coupling enzymes pyruvate kinase and lactate dehydrogenase (Auburger and Winter, 1992; Schuhle et al., 2003; James et al., 2016). The oxidation of NADH was measured spectrophotometrically at 340 nm (ε = 6220 M−1 cm−1) (McComb et al., 1976). The reaction was started by the addition of the acid substrate.
Kinase activity was determined by measuring the formation of the hydroxamate (Bowman et al., 1976). The assay mixture contained: 50 mM Tris (pH 8.3), 10 mM MgCl2, 500 mM hydroxylamine (pH 7), 10 mM nucleotide triphosphate (ATP, GTP, or CTP), and 20 mM of either potassium acetate or sodium butyrate. The reaction was stopped after 20 minutes of incubation at 37°C by the addition of ferric reagent (10% iron (III) chloride, 3% trichloroacetic acid, in 0.7 N hydrochloric acid) and set at room temperature for 15 minutes. The reaction mixture was centrifuged for 5 min at 13,000 x g and the absorbance was measured at 535 nm. The molar extinction coefficient for acetyl hydroxamate under the assay conditions was 594 M−1 cm-1.
Enzyme activities were linear with time and proportional to protein concentration. Controls included the deletion of each substrate and the cell-free extract, and the use of heat-treated extracts. All assays were performed at 37°C. Assay buffers were transferred to 1-cm cuvettes and warmed to 37°C in water bath prior to addition of reagents.
Purification of ligase activity from extracts.
Benzoate:CoA-ligase activity was purified from cell-free extracts obtained from crotonate-grown S. aciditrophicus cells using a combination of 45% ammonium sulfate fractionation, anion chromatography, and hydroxyapatite chromatography (see Supplemental Information). Crotonate:CoA ligase activity was partially purified using the methods for purification of benzoate:CoA ligase with the exception of the hydroxyapatite fractionation and following activity with crotonate (see Supplemental Information). Kinetic parameters were determined and coenzyme A substrates were synthesized as described in Supplemental Information.
Expression of S. aciditrophicus genes.
The genes SYN_RS14210 (putative 6-oxocyclohex-1-ene-1-carboxyl-CoA dehydrogenase) (bamQ) and SYN_RS03335 (putative long chain fatty acid CoA ligase), and SYN_RS03815 (bcl1) and SYN_RS03820 (bcl2) (both putative 4-hydroxybenzoate-CoA ligase/benzoate CoA ligases) were cloned and overexpressed in E. coli BL21 using the Champion™ pET101 Directional TOPO® Expression Kit (Invitrogen). The recombinantly produced proteins were purified and characterized as described in Supplemental Information. Genes SYN_RS09865 (buk1) and SYN_RS11570 (buk2), annotated as butyrate kinases, were cloned into an acetate kinase mutant of Escherichia coli as described in Supplemental Information.
Analytical procedures.
Crotonate, benzoate and cyclohexane-1-carboxylate were measured by high performance liquid chromatography as previously described (Sieber et al., 2014). Except for 2-D PAGE analyses, the protein concentration was determined by the Bradford assay using Coomassie Plus Protein Assay Reagent (ThermoFisher Scientific) with bovine serum albumin as the standard. For 2-D PAGE analyses, the protein concentration was estimated by using a commercial Non-interfering Protein Assay (Geno Technology, Inc., St. Louis, MO, USA) and bovine serum albumin as standard.
Data availability.
S. aciditrophicus whole-cell quantitative proteomic data are available at the PRIDE proteomics data repository (http://www.ebi.ac.uk/pride/archive/) with the dataset identifier PXD004638. 2-D PAGE proteomic data are available at the Japan ProteOme Standard Repository (jPOSTrepo, https://repository.jpostdb.org/) with the identifiers JPST000497 and ProteomicXChangeidentifier PXD011100. BN-PAGE proteomics data have been deposited in jPOSTrepo with the identifiers JPST000498 and ProteomicXChangeidentifier PXD011101. Protein identifications were made using the S. aciditrophicus genome in GenBank (accession number CP000252) and a RefSeq version of this genome (accession number NC_007759). Dataset 5 provides equivalencies between our original gene identifications (GeneBank accession number CP000252) (McInerney et al., 2007) and a more recent RefSeq version of the genome (accession number of NC_007759).
Supplementary Material
Originality-Significance Statement.
How microorganisms grow using catabolic reactions that operate close to thermodynamic equilibrium is not well understood. The model syntrophic metabolizer, Syntrophus aciditrophicus, uses the same enzymes in a reversible manner to degrade but also to make aromatic and alicyclic compounds. Reversibility means that the core reactions operate at near-equilibrium conditions as expected from catabolic reactions that are close to thermodynamic equilibrium. Reversibility also means that the metabolic pools of syntrophic metabolizers are susceptible to environmental perturbations, which is consistent with the observed inhibition of syntrophic metabolism by the accumulation of their end products of metabolism, acetate, hydrogen and formate. Energy is conserved by the unique synthesis of ATP by an AMP-forming, acetyl-CoA synthetase using pyrophosphate made from substrate activation and by membrane-bound pyrophosphatases. Proteomic analyses identified the protein machinery needed to catalyze the energetically difficult step of making formate from high potential electrons generated from acyl-CoA oxidation. The ability of S. aciditrophicus to reverse its biochemical pathway from substrate to product and vice versa using the same enzyme machinery is a remarkable strategy for energy conservation and provides a model to understand how microorganisms evolve and persist under thermodynamic constraints.
Acknowledgements
Cultivation, enzymology, metabolomics, and gene expression were supported by Department of Energy contract DE-FG02–96ER20214 from the Physical Biosciences Division, Office of Basic Energy Sciences to M.J.M. Proteomic analyses were supported by the National Institutes of Health grants R01GM085402 and R01GM104610 to J.A.L. and R.R.O.L. and Department of Energy Office of Science (BER) contract DE-FC-02–02ER63421 to J.A.L., R.R.O.L. and R. P. G. at the UCLA-DOE Institute.
Footnotes
Authors declare that there are no conflicts of interest.
References
- Auburger G, and Winter J (1992) Purification and characterization of benzoyl-CoA ligase from a syntrophic, benzoate-degrading, anaerobic mixed culture. Appl Microbiol Biotechnol 37: 789–795. [DOI] [PubMed] [Google Scholar]
- Balch WE, and Wolfe RS (1976) New approach to the cultivation of methanogenic bacteria: 2-mercaptoethanesulfonic acid (HS-CoM)-dependent growth of Methanobacterium ruminantium in a pressureized atmosphere. Appl Environ Microbiol 32: 781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatrix B, Bendrat K, Rospert S, and Buckel W (1990) The biotin-dependent sodium ion pump glutaconyl-CoA decarboxylase from Fusobacterium nucleatum (subsp. nucleatum). Comparison with the glutaconyl-CoA decarboxylases from gram-positive bacteria. Arch Microbiol 154: 362–369. [DOI] [PubMed] [Google Scholar]
- Biegel E, and Müller V (2010) Bacterial Na+-translocating ferredoxin: NAD+ oxidoreductase. Proc Natl Acad Sci U S A 107: 18138–18142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biegel E, and Müller V (2011) A Na+-translocating pyrophosphatase in the acetogenic bacterium Acetobacterium woodii. J Biol Chem 286: 6080–6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman CM, Valdez RO, and Nishimura JS (1976) Acetate kinase from Veillonella alcalescens. Regulation of enzyme activity by succinate and substrates. J Biol Chem 251: 3117–3121. [PubMed] [Google Scholar]
- Crable BR, Sieber JR, Mao X, Alvarez-Cohen L, Gunsalus RP, Ogorzalek Loo RR et al. (2016) Membrane complexes of Syntrophomonas wolfei involved in syntrophic butyrate degradation and hydrogen formation. Front Microbiol 7: 1795. doi 1710.3389/fmicb.2016.01795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimroth P, and Hilpert W (1984) Carboxylation of pyruvate and acetyl-coenzyme A by reversal of sodium pumps oxaloacetate decarboxylase and methylmalonyl-CoA decarboxylase. Biochem 23: 5360–5366. [Google Scholar]
- Djurdjevic I (2010) Production of glutaconic acid in recombinant Escherichia coli Thesis, Philipps-Universität Marburg, Germany. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Naggar MY, Wanger G, Leung KM, Yuzvinsky TD, Southam G, Yang J et al. (2010) Electrical transport along bacterial nanowires from Shewanella oneidensis MR-1. Proc Natl Acad Sci U S A 107: 18127–18131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshahed MS, and McInerney MJ (2001) Benzoate fermentation by the anaerobic bacterium Syntrophus aciditrophicus in the absence of hydrogen-using microorganisms. Appl Environ Microbiol 67: 5520–5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshahed MS, Bhupathiraju VK, Wofford NQ, Nanny MA, and McInerney MJ (2001) Metabolism of benzoate, cyclohex-1-ene carboxylate, and cyclohexane carboxylate by “Syntrophus aciditrophicus” strain SB in syntrophic association with H2-using microorganisms. Appl Environ Microbiol 67: 1728–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erde J, Loo RR, and Loo JA (2014) Enhanced FASP (eFASP) to increase proteome coverage and sample recovery for quantitative proteomic experiments. J Proteome Res 13: 1885–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs G (2008) Anaerobic metabolism of aromatic compounds. Ann N Y Acad Sci 1125: 82–99. [DOI] [PubMed] [Google Scholar]
- Gorby YA, Yanina S, McLean JS, Rosso KM, Moyles D, Dohnalkova A et al. (2006) Electrically conductive bacterial nanowires produced by Shewanella oneidensis strain MR-1 and other microorganisms. Proc Natl Acad Sci USA 103: 11358–11363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood CS, Burchdardt G, Herrmann H, and Fuchs G (1998) Anaerobic metabolism of aromatic compounds via the benzoyl-CoA pathway. FEMS Microbiol Rev 22: 439–458. [Google Scholar]
- Hattori S, Galushko AS, Kamagata Y, and Schink B (2005) Operation of the CO dehydrogenase/acetyl coenzyme A pathway in both acetate oxidation and acetate formation by the syntrophically acetate-oxidizing bacterium Thermacetogenium phaeum. J Bacteriol 187: 3471–3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoehler T, Losey NA, Gunsalus RP, and J., M.M. (2018) Environmental constraints that limit methanogenesis. In Biogenesis of Hydrocarbons Handbook of Hydrocarbon and Lipid Microbiology Stams A, and Sousa D (eds): Springer, Cham, pp. 1–26. [Google Scholar]
- Hopkins BT, McInerney MJ, and Warikoo V (1995) Anaerobic syntrophic benzoate degradation: evidence for a threshold and isolation of a syntrophic benzoate degrader. Appl Environ Microbiol 61: 526–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes MA, Silva JC, Geromanos SJ, and Townsend CA (2006) Quantitative proteomic analysis of drug-induced changes in mycobacteria. J Proteome Res 5: 54–63. [DOI] [PubMed] [Google Scholar]
- Jackson BE, and McInerney MJ (2002) Anaerobic microbial metabolism can proceed close to thermodynamic limits. Nature 415: 454–456. [DOI] [PubMed] [Google Scholar]
- Jackson BE, Bhupathiraju VK, Tanner RS, Woese CR, and McInerney MJ (1999) Syntrophus aciditrophicus sp. nov., a new anaerobic bacterium that degrades fatty acids and benzoate in syntrophic association with hydrogen-using microorganisms. Arch Microbiol 171: 107–114. [DOI] [PubMed] [Google Scholar]
- James KL, Ríos-Hernández LA, Wofford NQ, Mouttaki H, Sieber JR, Sheik CS et al. (2016) Pyrophosphate-dependent ATP formation from acetyl coenzyme A in Syntrophus aciditrophicus, a new twist on ATP formation. mBio 7: e01208–01216. doi: 01210.01128/mBio.01208–01216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato S, Hashimoto K, and Watanabe K (2012) Microbial interspecies electron transfer via electric currents through conductive minerals. Proc Natl Acad Sci U S A 109: 10042–10046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung JW, Seifert J, von Bergen M, and Boll M (2013) Cyclohexanecarboxyl-coenzyme A (CoA) and cyclohex-1-ene-1-carboxyl-CoA dehydrogenases, two enzymes involved in the fermentation of benzoate and crotonate in Syntrophus aciditrophicus. J Bacteriol 195: 3193–3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung JW, Meier A-K, Mergelsberg M, and Boll M (2014) Enzymes involved in a novel anaerobic cyclohexane carboxylic acid degradation pathway. J Bacteriol 196: 3667–3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung JW, Löffler C, Dörner K, Heintz D, Gallien S, Dorsselaer AV et al. (2009) Identification and characterization of the tungsten-containing class of benzoyl-coenzyme A reductases. Proc Natl Acad Sci USA 106: 17687–17692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuntze K, Shinoda Y, Mouttaki H, McInerney MJ, Vogt C, Richnow H-H, and Boll M (2008) 6-Oxocyclohexene-1-carbonyl-CoA hydrolases from obligately anaerobic bacteria: characterization and identification of its gene and development of a functional marker for aromatic compounds-degrading anaerobes. Environ Microbiol 10: 1547–1556. [DOI] [PubMed] [Google Scholar]
- Laempe D, Jahn M, and Fuchs G (1999) 6-Hydroxycyclohex-1-ene-1-carbonyl-CoA cehydrogenase and 6-oxocyclohex-1-ene-1-carbonyl-CoA hydrolase, enzymes of the benzoyl-CoA pathway of anaerobic aromatic metabolism in the denitrifying bacterium Thauera aromatica. Eur J Biochem 263: 420–429. [DOI] [PubMed] [Google Scholar]
- Leang C, Qian X, Mester T, and Lovley DR (2010) Alignment of the c-type cytochrome OmcS along pili of Geobacter sulfurreducens. Appl Environ Microbiol 76: 4080–4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MJ, and Zinder SH (1988a) Carbon monoxide pathway enzyme activities in a thermophilic anaerobic bacterium grown acetogenically and in asyntrophic acetate-oxidizing coculture. Arch Microbiol 150: 513–518. [Google Scholar]
- Lee MJ, and Zinder SH (1988b) Isolation and characterization of a thermophilic bacterium which oxidizes acetate in syntrophic association with a methanogen and which grows acetogenically on H2-CO2. Appl Environ Microbiol 54: 124–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löffler C, Kuntze K, Vazquez JR, Rugor A, Kung JW, Böttcher A, and Boll M (2011) Occurrence, genes and expression of the W/Se-containing class II benzoyl-coenzyme A reductases in anaerobic bacteria. Environ Microbiol 13: 696–709. [DOI] [PubMed] [Google Scholar]
- McComb RB, Bond LW, Burnett RW, Keech RC, and Bowers GN (1976) Determination of the molar absorptivity of NADH. Clin Chem 22: 141–150. [PubMed] [Google Scholar]
- McInerney MJ (1999) Anaerobic metabolism and its regulation. In Biotechnology, Second, Completely Revised Edition Rehm H-J, and Reed G (eds): Wiley-VCH, Weinheim, Germany, pp. 445–478. [Google Scholar]
- McInerney MJ, Sieber JR, and Gunsalus RP (2009) Syntrophy in anaerobic global carbon cycles. Curr Opin Biotechnol 20: 623–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerney MJ, Struchtemeyer CG, Sieber J, Mouttaki H, Stams AJM, Schink B et al. (2008) Physiology, ecology, phylogeny and genomics of microorganisms capable of syntrophic metabolism. Ann NY Acad Sci 1125: 58–72. [DOI] [PubMed] [Google Scholar]
- McInerney MJ, Rohlin L, Mouttaki H, Kim U, Krupp RS, Rios-Hernandez L et al. (2007) The genome of Syntrophus aciditrophicus: life at the thermodynamic limit of microbial growth. Proc Natl Acad Sci U S A 104: 7600–7605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouttaki H, Nanny MA, and McInerney MJ (2007) Cyclohexane carboxylate and benzoate formation from crotonate in Syntrophus aciditrophicus. Appl Environ Microbiol 73: 930–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouttaki H, Nanny MA, and McInerney MJ (2008) Use of benzoate as an electron acceptor by Syntrophus aciditrophicus grown in pure culture with crotonate. Environ Microbiol 10: 3265–3274. [DOI] [PubMed] [Google Scholar]
- Mouttaki H, Nanny MA, and McInerney MJ (2009) Metabolism of hydroxylated and fluorinated benzoates by Syntrophus aciditrophicus and detection of a fluorodiene metabolite. Appl Environ Microbiol 75: 998–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier DA, and Harwood CS (2000) 2-Hydroxycyclohexanecarboxyl coenzyme A dehydrogenase, an enzyme characteristic of the anaerobic benzoate degradation pathway used by Rhodopseudomonas palustris. J Bacteriol 182: 2753–2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters F, Shinoda Y, McInerney MJ, and Boll M (2007) Cyclohex-1,5-diene-1-carbonyl-coenzyme A hydratases of Geobacter metallireducens and Syntrophus aciditrophicus: evidence for a common benzoyl-CoA pathway in facultative and obligate anaerobes. J Bacteriol 189: 1055–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X, Mester T, Morgado L, Arakawa T, Sharma ML, Inoue K et al. (2011) Biochemical characterization of purified OmcS, a c-type cytochrome required for insoluble Fe(III) reduction in Geobacter sulfurreducens. Biochim Biophys Acta 1807: 404–412. [DOI] [PubMed] [Google Scholar]
- Schink B (1997) Energetics of syntrophic cooperation in methanogenic degradation. Microbiol Mol Biol Rev 61: 262–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schink B, and Friedrich M (1994) Energetics of syntrophic fatty acid oxidation. FEMS Microbiol Rev 15: 85–94. [Google Scholar]
- Schmidt A, Müller N, Schink B, and Schleheck D (2013) A proteomic view at the biochemistry of syntrophic butyrate oxidation in Syntrophomonas wolfei. PloS ONE 8: e56905. doi: 56910.51371/journal.pone.0056905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöcke L, and Schink B (1998) Membrane-bound proton-translocating pyrophosphatase of Syntrophus gentianae, a syntrophically benzoate-degrading fermenting bacterium. Eur J Biochem 256: 589–594. [DOI] [PubMed] [Google Scholar]
- Schöcke L, and Schink B (1999) Energetics and biochemistry of fermentative benzoate degradation by Syntrophus gentianae. Arch Microbiol 171: 331–337. [DOI] [PubMed] [Google Scholar]
- Schuhle K, Gescher J, Feil U, Paul M, Jahn M, and Schagger H (2003) Benzoate-coenzyme A ligase from Thauer aromatica: an enzyme acting in anaerobic and aerobic pathways. J Bacteriol 185: 4920–4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber JR, McInerney MJ, and Gunsalus RP (2012) Genomic insights into syntrophy: the paradigm for anaerobic metabolic cooperation. Ann Rev Microbiol 66: 429–452. [DOI] [PubMed] [Google Scholar]
- Sieber JR, Le HM, and McInerney MJ (2014) The importance of hydrogen and formate transfer for syntrophic fatty, aromatic and alicyclic metabolism. Environ Microbiol 16: 177–188. [DOI] [PubMed] [Google Scholar]
- Sieber JR, Crable BR, Sheik CS, Hurst GB, Rohlin L, Gunsalus RP, and McInerney MJ (2015) Proteomic analysis reveals metabolic and regulatory systems involved the syntrophic and axenic lifestyle of Syntrophomonas wolfei. Front Microbiol 6: 115. doi: 110.3389/fmicb.2015.00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva JC, Gorenstein MV, Li G-Z, Vissers JPC, and Geromanos SJ (2006) Absolute quantification of proteins by LCMS. A virtue of parallel MS acquisition. Mol Cell Proteomics 5: 144–156. [DOI] [PubMed] [Google Scholar]
- Silva JC, Denny R, Dorschel CA, Gorenstein M, Kass IJ, Li GZ et al. (2005) Quantitative proteomic analysis by accurate mass retention time pairs. Anal Chem 77: 2187–2200. [DOI] [PubMed] [Google Scholar]
- Summers ZM, Fogarty HE, Leang C, Franks AE, Malvankar NS, and Lovley DR (2010) Direct exchange of electrons within aggregates of an evolved syntrophic coculture of anaerobic bacteria. Science 330: 1413–1415. [DOI] [PubMed] [Google Scholar]
- Walker DJF, Nevin KP, Holmes DE, Rotaru A-E, Ward JE, Woodard TL et al. (2018) Syntrophus conductive pili demonstrate that common hydrogen-donating syntrophs can have a direct electron transfer option. bioRxiv: 10.1101/479683. [DOI] [PMC free article] [PubMed]
- Wischgoll S, Heintz D, Peters F, Erxleben A, Sarnighausen E, Reski R et al. (2005) Gene clusters involved in anaerobic benzoate degradation of Geobacter metallireducens. Mol Microbiol 58: 1238–1252. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
S. aciditrophicus whole-cell quantitative proteomic data are available at the PRIDE proteomics data repository (http://www.ebi.ac.uk/pride/archive/) with the dataset identifier PXD004638. 2-D PAGE proteomic data are available at the Japan ProteOme Standard Repository (jPOSTrepo, https://repository.jpostdb.org/) with the identifiers JPST000497 and ProteomicXChangeidentifier PXD011100. BN-PAGE proteomics data have been deposited in jPOSTrepo with the identifiers JPST000498 and ProteomicXChangeidentifier PXD011101. Protein identifications were made using the S. aciditrophicus genome in GenBank (accession number CP000252) and a RefSeq version of this genome (accession number NC_007759). Dataset 5 provides equivalencies between our original gene identifications (GeneBank accession number CP000252) (McInerney et al., 2007) and a more recent RefSeq version of the genome (accession number of NC_007759).


