Abstract
Summary
Objective:
Interictal spikes are a characteristic feature of invasive electroencephalography (EEG) recordings in children with refractory epilepsy. Spikes frequently co-occur across multiple brain regions with discernable latencies, suggesting that spikes can propagate through distributed neural networks. The purpose of this study was to examine the long-term reproducibility of spike propagation patterns over hours to days of interictal recording.
Methods:
Twelve children (mean age = 13.1 years) were retrospectively studied. An average of 47.2 ± 40.1 hours of interictal EEG were examined per patient (range = 17.5 – 166.5 hours). Interictal recordings were divided into 30-minute segments. Networks were extracted based on the frequency of spike co-activation between pairs of electrodes. For each 30-minute segment, electrodes were assigned a ‘Degree Preference (DP)’ based on the tendency to appear upstream or downstream within propagation sequences. The consistency of DPs across segments (‘DP-Stability’) was quantified using the Spearman rank correlation.
Results:
Regions exhibited highly stable preferences to appear upstream, intermediate, or downstream in spike propagation sequences. Across networks, the average DP-Stability was 0.88 ± 0.07, indicating that propagation patterns observed in 30-minute segments were representative of the patterns observed in the full interictal window. At the group level, regions involved in seizure generation appeared more upstream in spike propagation sequences.
Keywords: Interictal spike, propagation, network, epilepsy surgery, invasive EEG
1. Introduction
Interictal spikes are electrographic paroxysms arising when local populations of cortical neurons become abnormally synchronized.1 In addition to local spike activity, researchers have long recognized the tendency for regions to co-activate during spike discharges with millisecond-scale latencies,2,3 suggesting that spikes can propagate across the cortex.4–16 Spike propagation is thought to reflect the transient activation of neural networks involved in the generation and transmission of epileptiform discharges.17 Mapping spike propagation appears to be clinically relevant, as studies find that regions appearing early (i.e., ‘upstream’) in propagation sequences preferentially localize to the SOZ6,7,9 and must be excised to achieve seizure-freedom.18 Repeated co-activation of regions during spike discharges has also been theorized as a mechanism of pathological network formation and epileptogenesis.1,19,20 Given that the interictal state accounts for the vast majority of EEG recordings, improved understanding of spike propagation and network dynamics is key to advancing disease models of epilepsy and increasing the yield of the pre-surgical evaluation.
An important issue when characterizing spike propagation is establishing the reproducibility of network co-activation patterns over time.5,21 In this context, ‘co-activation patterns’ refer to the millisecond-scale latency differences observed between spikes at distinct brain regions, and ‘reproducibility’ refers to the spatiotemporal consistency of co-activation patterns over many propagation discharges. Most studies rest on the assumption that co-activation patterns observed in relatively brief EEG segments (e.g., <60 minutes) are representative of the long-term behavior of spike networks, but the well-documented spatial and temporal variability of spikes22–25 demands a skeptical evaluation of this assumption. To advance our understanding of network-level pathophysiology, we investigated the long-term reproducibility of interictal spike propagation in 12 children who underwent invasive monitoring with subdural electrodes. We tested the hypothesis that spike co-activation patterns are reproducible over hours to days of interictal recording.
2. Materials and Methods
2.1. Patient selection and intracranial EEG recording.
This study was approved by the Children’s Hospital of Philadelphia (CHOP) Institutional Review Board. Twelve patients who underwent long-term invasive EEG monitoring were retrospectively studied. Patients were selected from a larger institutional database on the basis of: (1) availability of seizure markings; (2) schematic electrode maps; (3) frequent interictal spiking documented in the EEG report; (4) at least two years of post-surgical follow-up; and (5) at least 16 hours of interictal EEG data for analysis (see Section 2.3: ‘Interictal segment selection’). Subdural electrodes manufactured by Adtech Medical Instruments Corporations (Racine, WI) were implanted (inter-electrode distance = 10 mm, exposure = 2.3 mm). Recordings were acquired at 200 Hz using a Telefactor Beehive 32–128 channel Cable Telemetry Encoder (CTE) digital synchronized video-EEG recording system (Astro Med Corp, West Warwick, RI). Online voltages were referenced to an electrode distant from the suspected epileptogenic zone. EEG data were subjected to an online 60 Hz notch filter. No offline filters or post-processing steps were applied.
All clinical decisions were made without influence from this study. Review of medical charts provided clinical information including seizure history, histopathology, and implant location. Post-surgical seizure outcome was assessed at least 24 months after surgery using the modified Engel scale.26 Study procedures were conducted in accordance with the Declaration of Helsinki.
2.2. EEG inspection.
All uncorrupted EEG files were independently inspected by two experienced pediatric epileptologists. Periods of excessive EEG artifact (e.g., malfunctioning grid) were marked, and electrodes with persistent artifact were excluded to reduce data contamination. The earliest electrographic change (EEC) and onset electrodes were marked for each seizure. Disagreement between reviewers was resolved by discussion. Ten minutes of peri-ictal activity flanking each seizure were marked for rejection. The set of electrodes constituting the seizure network (‘SZN’), which encompassed the SOZ and regions of consistent early ictal spread, was determined by expert review before conducting the interictal analyses.
2.3. Interictal segment selection.
For each patient, the longest window of interictal EEG activity without an interruption ≥2 hours in duration (e.g., peri-ictal epochs, file breaks, and artifact) was selected for analysis (Fig. 1A). Factors such as time-of-day, sleep-wake state, and anti-epileptic drug regimen were not considered when selecting the interictal window. The window was divided into non-overlapping 30-minute segments (Fig. 1A), and segments containing ≥3 minutes of rejected activity were discarded. The final dataset for each patient thus constituted a set of 30-minute segments spanning the longest available near-continuous window of interictal activity. The 30-minute segment duration was chosen to provide a sizeable number of spikes in each segment while remaining sensitive to changes unfolding over tens of minutes to hours.
Figure 1. Interictal segment selection and spike co-activation.

A) For each patient, the longest window of interictal activity without an interruption ≥2 hours in duration was selected and divided into non-overlapping 30-minute segments. Segments overlapping with file breaks, peri-ictal intervals, and artifact were excluded. In this example (Patient #10), 83/102 possible interictal segments (41.5 hours) were preserved. B) Left: Intraoperative photograph and schematic of subdural electrode implant. Right: Representative 10-second epoch containing spike discharges that co-occur across multiple electrodes in the frontal and temporal regions. C) Co-activation matrix (C) quantifying the tendency for regions to co-activate during spike discharges (i.e., spike peaks separated by <150 ms). Dotted boxes indicate clustering of matrix C to reveal two co-activation networks, which are represented in the electrode schematic in (D).
2.4. Interictal spike detection.
Each 30-minute segment was submitted to an automated spike detection algorithm, which has been previously validated against human markings.27 The mimetic-based detector identifies spikes based on a combination of amplitude and spectral characteristics. Detection parameters (e.g., amplitude threshold) were “tuned” for each patient as described in the original publication27 using representative 5-minute interictal clips. Each spike was assigned an event time corresponding to the peak amplitude. In order to focus analyses on electrodes capturing frequent spikes, a non-parametric spike frequency threshold was applied (Supplemental Fig. S1). Electrodes with an observed spike frequency (spikes/min) exceeding 1.5 standard deviations above the mean of a surrogate distribution (iterations = 1,000) were analyzed.
2.5. Spike co-activation matrix.
Spike co-activation referred to the detection of spikes at distinct electrodes with brief inter-spike latencies (Fig. 1B).28,29 Consistent with previous studies, co-activation was defined as peak times separated by <150 ms.5,20,21 Spike co-activation rates were encoded in an electrode-by-electrode matrix (C). For each electrode i, the co-activation rate between electrodes i and j (Ci,j) was given as: Ci,j = (si | sj) / si, where (si | sj) was the number of spikes at electrode i with a co-activation spike at electrode j, and si was the total number of spikes recorded at i. Thus, Ci,j represented the fraction of co-activations between electrodes i and j, normalized by the total spikes at electrode i (ranging from 0 to 1). Performing this calculation over all electrode pairs yielded the co-activation matrix, C (Fig. 1C). Note that C was non-symmetric, in that Ci,j and Cj,i need not be equal.
2.6. Co-activation networks.
Co-activation networks were defined as clusters of electrodes in matrix C with a high spike co-activation rate (Fig. 1C-D). The Louvain greedy optimization algorithm was used to partition C into electrode clusters that maximized the within-cluster co-activation rate and minimized the between-cluster co-activation rate. Clustering was performed using the Brain Connectivity Toolbox30 according to the procedures of Khambhati et al.31 To confirm that electrode clustering was appropriate, the within-cluster and between-cluster co-activation rates were compared using the Wilcoxon rank-sum test (significance threshold, α <0.001).
2.7. Spatiotemporal activation sequences.
Spike co-activation typically exhibited millisecond-scale differences in peak latencies across electrodes. Each co-activation discharge could thus be represented as a spatiotemporal activation sequence, beginning with the earliest spike in the discharge (i.e., ‘leader’) and proceeding through downstream spikes in order of peak latency. For each network, activation sequences were extracted using a previously-described algorithm.11,12 Briefly, the first detected spike in the network was initialized as a the ‘leader’ of a candidate activation sequence, and all subsequent spikes with a latency <50 ms from the leader (or <15 ms from the preceding spike) were appended to the sequence. Sequences involving ≥5 electrodes were preserved. To focus on prominent spike patterns, networks with an activation rate of <2 sequences/minute were ignored.
2.8. Quantification of regional propagation tendencies (‘Degree Preference’)
Activation sequences were represented using raster plots, with each sequence defined by the ordering of spike times in relation to the leader spike (Fig. 2A-B). To quantify electrode-level tendencies to appear upstream or downstream within activation sequences, the Degree Preference (DP) was defined:
| (Equation 1): |
Figure 2. Examination of regional propagation tendencies over time.
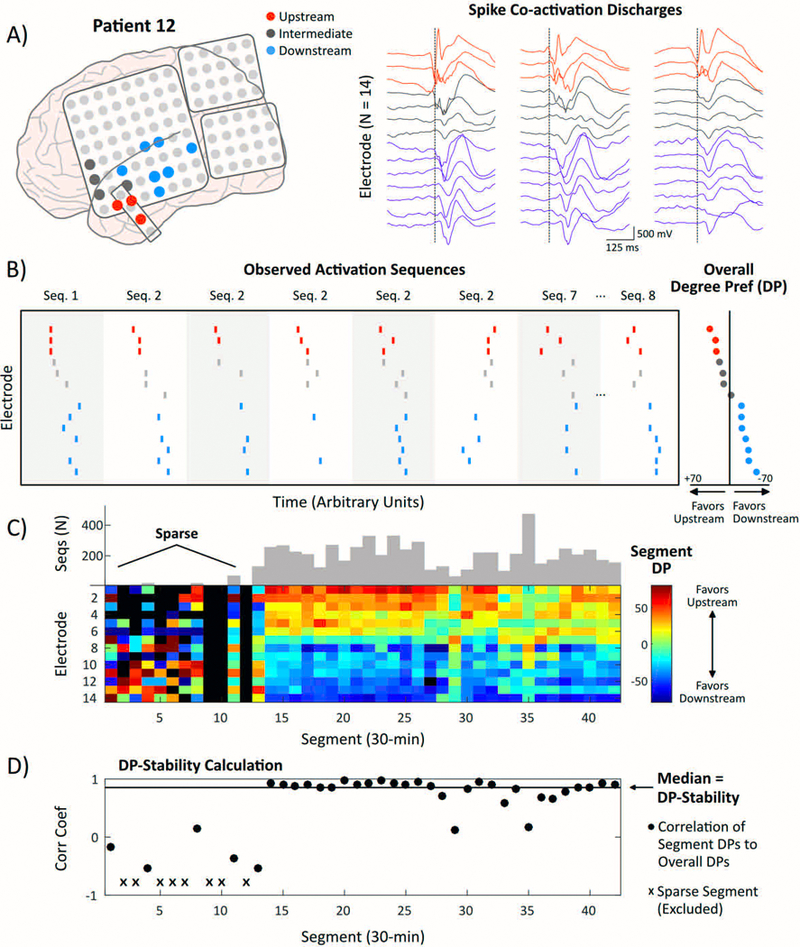
A) Left: Co-activation network encompassing 14 electrodes over the left temporal/parasylvian regions (Patient #12). Colors denote three Upstream electrodes (Overall DP ≥+20; red), four Intermediate electrodes (gray), and seven Downstream electrodes (Overall DP <−20; blue). Right: Three representative spike co-activation discharges (500-ms waveforms) revealing millisecond-scale latencies between Upstream electrodes (anterior temporal lobe) and Downstream electrodes (posterior temporal/parasylvian). Dotted lines represent the peak time of the earliest spike in the sequence. B) Rasters from 8 representative co-activation discharges marking peak times at each electrode. The Overall DP for each electrode (right) reflects the tendency to appear upstream or downstream within sequences. C) Heat map encoding Segment DPs within each 30-minute segment. Warmer colors reflect a preference to appear more upstream. The heat map is sorted top to bottom according to rank order of Overall DPs. The number of sequences captured per segment is shown in gray. The first 13 segments (6.5 hours) are sparse compared to the rest of the analysis window. D) The rank correlation between each Segment DP vector and the Overall DP vector is shown. The median of correlation values is the ‘DP-Stability.’
where upstream spikes were those recorded earlier in sequences, and downstream spikes were those recorded later in sequences (Fig. 2B). Thus, an electrode capturing the ‘leader’ spike for all sequences would be assigned the maximal DP of +100, whereas an electrode capturing the terminal spike for all sequences would have the minimal DP of −100.
2.9. Consistency of Degree Preferences (DPs) over time
To examine the stability of regional propagation tendencies over time, DPs were first calculated from the complete set of activation sequences collapsed across segments (‘Overall DPs’), and then separately for each 30-minute segment (‘Segment DPs’). The Spearman rank correlation quantified the similarity between each Segment DP vector and the Overall DP vector (Fig. 2C-D). The median correlation across segments defined the ‘DP-Stability’ (Fig. 2D), and the interquartile range (IQR) quantified the variability across segments. To account for data sparsity, segments in which <75% of electrodes had a calculable Segment DP were excluded (for representative network featuring a window of data sparsity, see Fig. 2C-D).
For grouping across networks, electrodes were classified based on Overall DPs: Upstream (Overall DP ≥+20), Intermediate (−20 ≤ Overall DP < +20), or Downstream (Overall DP <−20). For each Upstream electrode, we calculated the percentage of 30-minute segments in which an upstream Segment DP (i.e., ≥+20) was observed. This calculation was then repeated for Downstream electrodes. This analysis was performed to describe how reliably an individual segment captured the overall behavior of Upstream and Downstream electrodes.
2.10. Comparison of electrodes within and outside the seizure network (SZN).
To investigate the tendency for upstream spike regions to overlap with the neurologist-defined SZN, we compared Overall DPs for electrodes within the SZN (‘In-SZN’) to electrodes outside the SZN (‘Out-SZN’). We hypothesized that In-SZN electrodes would have a higher Overall DP than Out-SZN electrodes, indicating a relative upstream activation preference. This comparison was performed at the individual and group levels using the Wilcoxon rank-sum test.
2.11. Reproducibility of individual activation sequences.
The DP analysis captured general tendencies for regions to appear at various positions within activation sequences. A complementary approach is to examine the sequence-by-sequence reproducibility of individual co-activation sequences (Fig. 3). For each network, the ‘expected’ sequence of network activation was defined based on the rank order of Overall DPs (Fig. 3A). Then, for each individual sequence i, the Sequence Similarity (Si) was calculated as the fraction of steps in the ‘observed’ sequence that followed the order of the ‘expected’ sequence, ranging from −1 (order of the observed sequence was exactly opposite of expected order) to +1 (order of the observed sequence perfectly matched the expected order) (Fig. 3B-C).32 Performing this calculation for all sequences (collapsed across segments) yielded a distribution of S values (Fig. 3D). The network’s Reproducibility Index (R) was then computed by taking the median of S values, and variability across sequences was quantified by the IQR of S values.
Figure 3. Sequence-by-sequence reproducibility of network activation.

A) The rank order of Overall DPs constituted the ‘expected’ sequence of network activation. B) Three hypothetical sequences with peak times marked by dotted lines. From left to right, observed sequences exhibit decreasing concordance with the ‘expected’ sequence order. C) Calculation of the Sequence Similarity (S) value for each observed sequence. A point is awarded for each pairwise interaction that adheres to the expected order (e.g., for Observed (1), +1 point is awarded for the Electrode 1 → Electrode 2 interaction because Electrode 1 appears more upstream, consistent with the expected order. Zero points are awarded for ties, and −1 point is awarded when the interaction violates the expected order (e.g., Electrode 8 → 9). For each sequence (i), Si is the sum of points divided by the maximum possible score, which is given by: N*(N-1)/2, where N is the number of sequence spikes. D) Histogram of Si values for all sequences (collapsed across segments). The Reproducibility Index (Rorig) is the median Si value.
Non-parametric permutation testing was used to evaluate the statistical significance of the observed R (Rorig) for each network. To do so, the spikes times for each sequence were shuffled between electrodes to randomize the sequence order. R was then calculated on the permuted dataset, and this procedure was repeated 1,000 times to yield a distribution of surrogate R values (Rperm) (see Supplemental Figure S2 for example). To account for multiple comparisons (n = 16 networks), the significance threshold for Rorig was p <0.003.
2.12. Statistical reporting.
Unless otherwise noted, summary statistics are given as mean ± standard deviation (SD), and range is presented across networks. P-values correspond to two-tailed parametric tests or non-parametric permutation tests. All procedures were executed using Matlab 2016a (Natick, MA).
3. Results
3.1. Clinical characteristics.
Twelve children (9 males, mean age = 13.1 years, range = 7 – 18 years) with medically-refractory epilepsy were retrospectively studied. A variety of clinical histories, non-invasive findings, histopathologies, and electrode coverages (including both temporal and extra-temporal) were represented (Table 1). Patients were implanted with an average of 105.9 ± 16.2 subdural electrodes (range = 68 – 128 electrodes). Across patients, a mean of 8.1 ± 5.2 electrodes (range = 0 – 21 electrodes) were discarded due to artifact.
Table 1.
Clinical characteristics.
| Pt | Gender / Age* |
Dataset Segments / Hrs |
Recording Electrodes |
Rejected Electrodes |
Grid Coverage |
Histopathology | Engel Outcome† |
|---|---|---|---|---|---|---|---|
| 01 | F / 16 | 49 / 24.5 | 92 | 0 | R FT | Tuberous sclerosis | II |
| 02 | M / 18 | 35 / 17.5 | 100 | 8 | L FTP | FCD | IV |
| 03 | M / 11 | 113 / 56.5 | 128 | 5 | L F | Neurofibromatosis | III |
| 04 | F / 15 | 59 / 29.5 | 68 | 5 | R FP | FCD | III |
| 05 | M / 12 | 80 / 40.0 | 123 | 12 | BL O | No resection | N/A |
| 06 | M / 17 | 77 / 38.5 | 100 | 6 | L TP | Cerebral infarction | I |
| 07 | F / 11 | 134 / 67.0 | 116 | 7 | R FTP | Cerebral infarction | I |
| 08 | M / 12 | 59 / 29.5 | 110 | 21 | L Hemi | FCD Type IIA | I |
| 09 | M / 14 | 333 / 166.5 | 96 | 9 | R F | FCD Type IIA | I |
| 10 | M / 7 | 83 / 41.5 | 111 | 10 | R FTP | FCD Type IB | III |
| 11 | M / 16 | 70 / 35.0 | 118 | 4 | R FTP | FCD Type IIA | I |
| 12 | M / 8 | 42 / 21.0 | 109 | 10 | L FTP | FCD Type IIA | I |
Years;
Minimum 24-month follow-up; L: Left, R: Right, F: Frontal, P: Parietal, T: Temporal, O: Occipital, BL: Bilateral; Hemi: Hemisphere; FCD: Focal cortical dysplasia
3.2. Interictal spike detection.
The mean interictal spike frequency observed across patients was 5.3 ± 2.4 spikes/electrode/minute (range = 1.9 – 9.0 spikes/electrode/minute). Following application of the non-parametric spike frequency threshold (Supplemental Table S1), the mean spike frequency for preserved electrodes was 9.2 ± 5.6 spikes/electrode/minute (range = 2.5 – 23.6 spikes/electrode/minute).
3.3. Co-activation network extraction.
Across patients, clustering of spike co-activation matrices (C) yielded a total of 16 co-activation networks for analysis (range = 1–2 per patient). Abstractly, networks were represented as nodes (electrodes) linked by edges (co-activation rate). Networks contained an average of 14.8 ± 5.1 nodes (range = 7 – 25). The within-cluster co-activation rate significantly exceeded the between-cluster co-activation rate for all networks (Supplemental Table S2), indicating appropriate electrode clustering.
3.4. Stability of Degree Preferences over segments (DP-Stability)
The DP-Stability analysis for two representative patients is shown in Fig. 4. For Patient #2 (Fig. 4A-C, left), two co-activation networks involving the left frontal and parietal lobes exhibited high DP-Stability over time. The median (IQR) rank correlation between Segment DPs and Overall DPs (i.e., DP-Stability) for Network 1 was 0.92 (0.85–0.94). Similarly, Network 2 exhibited a DP-Stability of 0.92 (0.81–0.95).
Figure 4. Temporal stability of Degree Preferences.

Upper Left Panel: A) Two co-activation networks for Patient #2 involving the left frontal and parietal regions. B) Heat map encoding Segment DPs within each segment. Electrodes exhibit stable tendencies to appear at upstream (red), intermediate (green), or downstream (blue) positions in sequences. C) Each dot represents the correlation between the Segment DP vector and the Overall DP vector for each segment. The solid black line represents the median correlation value across segments (‘DP-Stability’). In both networks, propagation tendencies were highly stable over time (DP-Stability, IQR): Network #1: 0.92 (0.85–0.94), Network #2: 0.92 (0.81–0.95). Upper Right Panel: The same figure components are presented for Patient #10. Network #2 exhibited the lowest DP-Stability in the study: 0.77 (0.16–0.94). D) Upstream (n = 59) and Downstream (n = 75) electrodes were defined based on Overall DPs. On average, Upstream electrodes exhibited an upstream Segment DP in 77.6 ± 19.8% of segments and almost never exhibited a downstream Segment DP (2.1 ± 3.5% of segments). The same pattern was seen for Downstream electrodes. Two circled Downstream electrodes are from Patient #10, Network 2 (see asterisks in B’ showing these two electrodes, with inversion of upstream and downstream tendencies).
Patient #10 (Fig. 4A’-C’) exhibited one very stable and one dynamic co-activation network. For the right frontal network (Network 1), DP-Stability was high (0.92, 0.86–0.96). In contrast, the right temporal network (Network 2) exhibited a pronounced shift in Segment DPs after ~24 hours, with complete inversion of upstream and downstream preferences observed at several electrodes. The resultant DP-Stability was the lowest in the study: 0.77 (0.16–0.94).
Cohort-level results for the DP-Stability analysis are presented in Table 2 and Supplemental Figure S3. Across networks, the average DP-Stability was 0.88 ± 0.07 (range = 0.77–0.99). There was a significant negative correlation between DP-Stability and number of network nodes (r = −0.51, p = 0.041) (Supplemental Fig. S4). There was no significant correlation between DP-Stability and overall sequence frequency (r = −0.28, p = 0.29).
Table 2.
Reproducibility of spike co-activation patterns.
| Patient | Network | Electrodes (N) |
DP-Stability Median (IQR*) |
Rorig†
Median (IQR**) |
|---|---|---|---|---|
| 01 | 1 | 9 | 0.98 (0.97–1.00) | 0.80 (0.73–0.90) |
| 02 | 1 | 17 | 0.92 (0.85–0.94) | 0.38 (0.10–0.60) |
| 2 | 13 | 0.92 (0.81–0.95) | 0.50 (0.33–0.64) | |
| 03 | 1 | 10 | 0.99 (0.95–0.99) | 0.76 (0.60–0.87) |
| 04 | 1 | 11 | 0.86 (0.77–0.92) | 0.29 (0.00–0.50) |
| 05 | 1 | 22 | 0.81 (0.72–0.87) | 0.20 (−0.05–0.43) |
| 2 | 25 | 0.84 (0.66–0.91) | 0.23 (−0.06–0.44) | |
| 06 | 1 | 17 | 0.81 (0.64–0.91) | 0.39 (0.04–0.61) |
| 07 | 1 | 21 | 0.82 (0.70–0.88) | 0.29 (0.00–0.50) |
| 08 | 1 | 7 | 0.96 (0.89–0.96) | 0.57 (0.30–0.73) |
| 09 | 1 | 15 | 0.83 (0.53–0.94) | 0.50 (0.13–0.70) |
| 10 | 1 | 19 | 0.92 (0.86–0.96) | 0.47 (0.20–0.67) |
| 2 | 15 | 0.77 (0.16–0.94) | 0.40 (0.07–0.62) | |
| 11 | 1 | 11 | 0.79 (0.58–0.86) | 0.52 (0.18–0.76) |
| 2 | 11 | 0.92 (0.75–0.97) | 0.50 (0.24–0.67) | |
| 12 | 1 | 14 | 0.85 (0.65–0.91) | 0.38 (0.00–0.62) |
| Group (μ ±SD) | 16 | 237 | 0.88 ±0.07 | 0.45 ±0.17 |
IQR quantifies variability of correlation between Segment DPs and Overall DPs over segments
IQR quantifies variability of Sequence Similarity (Si) values across sequences
Rorig was deemed significant (p <0.003) for 16/16 networks via permutation testing
Across networks, a total of 59 Upstream electrodes (Overall DP ≥+20) and 75 Downstream electrodes (Overall DP <−20) were identified. On average, Upstream electrodes exhibited an upstream Segment DP (i.e., Segment DP ≥+20) in 77.6 ± 19.8% of segments (Fig. 4D). Upstream electrodes exhibited a downstream Segment DP in 2.1 ± 3.5% of segments. Similarly, Downstream electrodes exhibited downstream Segment DPs in 74.4 ± 17.7% of segments, and upstream Segment DPs in 4.4 ± 7.8% of segments. These results indicate that individual segments reliably captured the overall behavior of Upstream and Downstream electrodes.
At the node level, there was no significant correlation between the frequency of mono-electrode spikes (spikes/min) and Overall DP (r = 0.10, p = 0.13), nor were there any differences in spike frequency based on Overall DP category (Upstream: 10.1 ± 7.1 spikes/min; Downstream: 9.3 ± 4.8 spikes/min; Intermediate: 10.6 ± 6.1 spikes/min; one-way ANOVA, F(2, 234) = 0.89, p = 0.41).
3.5. Comparison between propagation tendencies and SZN localization.
The relationship between propagation tendencies and the neurologist-defined SZN was examined (Supplemental Table S3). In total, 86 electrodes were identified within the SZN (‘In-SZN’), whereas 151 electrodes were located outside the SZN (‘Out-SZN’). At the group level, there was a significant difference in Overall DPs for In-SZN versus Out-SZN electrodes, with a preference for In-SZN nodes to appear more upstream (Overall DP: +6.4 ± 36.1) compared to Out-SZN nodes (Overall DP: −6.3 ± 35.8; Wilcoxon rank sum, p = 0.015). At the network-level, differences in Overall DP between In-SZN and Out-SZN nodes were not significant. However, this analysis could only be performed in 9/16 networks (56.3%) due to sparse representation of Out-SZN or In-SZN electrodes in individual networks (see Supplemental Table S3).
3.6. Sequence-by-sequence reproducibility of activation patterns
The mean sequence frequency across networks was 8.9 ± 7.4 sequences/min (range = 2.2 – 29.3 sequences/min). The Reproducibility Index (R) quantified the concordance between the observed and expected activation sequences, collapsing sequences across all segments. The average Rorig across networks was 0.45 ± 0.17 (Table 2). Non-parametric permutation testing revealed that Rorig was statistically significant (p <0.003) for 16/16 networks. There was a significant negative correlation between Rorig and number of network nodes (r = −0.75, p <0.001) (Supplemental Fig. S4). The correlation between Rorig and DP-Stability across networks was high (r = 0.68, p = 0.004), reflecting the fact that these metrics are closely related (Supplemental Fig. S4).
4. Discussion
Regions frequently co-activate during interictal spike discharges with millisecond-scale latencies, suggesting that spikes can propagate across the cortex. Although the distinction between primary and propagated spikes has been recognized for decades,3 relatively little is known about the mechanisms and spatiotemporal characteristics of spike propagation. In this study, we mapped the sequential activation of cortical networks during spike propagation as recorded by subdural grid and strip electrodes. Our main findings indicate that spike propagation is a spatiotemporally reproducible network phenomenon over hours to days of interictal recording.
4.1. Spike propagation as a biomarker of pathological network activity.
The concept of network-level dysfunction has been widely influential in modern epilepsy research.33,34 Whereas earlier disease models focused on discrete “zones” of suspected epileptogenicity,35 newer models incorporate the time-varying interactions between brain regions as a fundamental driver of seizure activity.36,37 Propagation of epileptiform discharges has been raised as an intuitive measure of functional connectivity, offering a unique perspective of both the strength and directionality of inter-regional relationships.17,38 Previous studies suggest that spike propagation is a meaningful interictal biomarker of epileptic network activity, with notable work by Alarcon et al.18 showing that excision of regions appearing upstream in activation sequences correlated with higher rates of post-surgical seizure freedom. This study and others have identified a potential role of spike propagation analysis in the surgical work-up, but the clinical value of this approach is limited by uncertainties about the reproducibility of propagation patterns over time.
4.2. Spike propagation is a highly reproducible phenomenon.
The main purpose of this study was to characterize the long-term reproducibility of spike propagation sequences. In routine visual inspection of brief EEG segments, we have appreciated the tendency for regions to co-activate during spike discharges and for certain electrodes to reliably peak earlier or later in these discharges. Similar qualitative observations were reported in an influential study by Badier & Chauvel,2 who examined 13 patients with temporal lobe epilepsy (TLE) undergoing stereo-EEG investigation. The need to support these visual observations using computational techniques is obvious considering the abundance of interictal spikes, long recording durations, and the large number of electrodes implanted per patient. The road towards clinical implementation of spike propagation analysis clearly demands an efficient and intuitive signal processing method.
Our approach considers each propagation event in terms of a spatiotemporal activation sequence, in which networks of cortical regions are sequentially recruited into propagating spike discharges. The method was adapted from an algorithm introduced by Kreuz et al.,32 which was originally purposed for quantifying the consistency of propagation patterns in spike train data but is generalizable to any spatiotemporal process. In our analysis, we found that spike co-activation networks were characterized by highly reproducible propagation patterns, using both a measure of sequence-by-sequence reproducibility (Rorig) and temporal stability of regional preferences to appear at varying latencies within sequences (DP-Stability). Our results indicate that spike propagation patterns observed in 30-minutes epochs correlate strongly with the ‘overall’ patterns (i.e., from the complete interictal dataset), suggesting that these overall patterns can be assayed from brief epochs.
Demonstrating the reproducibility of activation sequences in this manner is novel, as no studies to our knowledge have leveraged the individual variation of spike activation sequences to generate continuous measures of propagation reproducibility. Our approach should be distinguished from previous studies by Bourien and Wendling,5,21 who developed an automated method to identify subsets of regions frequently co-activated during spikes. Similarly, Baud et al.10 developed a machine learning technique for unsupervised analysis of spike activity, but their approach focused on extracting archetypal propagation patterns rather than variations in individual sequences. Khoo et al.39 used a waveform-averaging approach to quantify regional tendencies to appear early or late in spike propagation, but in doing so averaged away the variation between sequences that constituted the basis of the Rorig calculation. We suspect our methods will be particularly valuable when comparing spike patterns across states, such as interictal and pre-ictal time windows. In this setting, the variability of activation sequences may offer insights into the dynamic re-organization of the epileptogenic zone, potentially revealing changes that herald an upcoming seizure.
Interestingly, 1/16 networks (Patient #10, Network 2) exhibited an impressive shift in Segment DPs over time, with several electrodes exhibiting inversion of upstream and downstream preferences. The neurobiology underlying this transition is unclear. We speculate that time-varying factors such as sleep-wake state, medication withdraw, and temporal proximity to seizures can influence propagation patterns. The shift in Segment DPs also corresponded to an apparent state-change in the overall frequency of spike sequences (see Fig. 4A’-B’). Future studies incorporating scalp-based EEG for sleep staging, as well as detailed analysis of medication regimen and seizure proximity, will help identify factors contributing to these transitions.
The negative correlation observed between both reproducibility measures (DP-Stability and Rorig) and number of network nodes also warrants exploration. One hypothesis is that smaller, more spatially-contiguous networks generate more reproducible spike activation sequences, which could be tested by implementing precise methods to localize electrodes in space. Relatedly, networks that span large cortical territories may participate in multiple, partially-independent spike networks, thereby generating more heterogeneous activation sequences. Refinement of the methodology may allow us to detect networks with multiple independent (but themselves highly reproducible) propagation patterns.
4.3. Propagation tendencies relate to SZN localization, but only at the group level.
Studies comparing interictal spike propagation with seizure onset have identified a tendency for upstream regions to overlap with the SOZ.6,7,9 However, similar to the recent study by Maharathi et al.,17 our study provides only weak support for this observation. Specifically, differences in DPs between In-SZN and Out-SZN were only detected at the group level, and even there, the within-group variability was very high. The relationship between spike and seizure networks has been controversial for decades.40 Previous studies have shown that complete excision of regions generating spike discharges (i.e., irritative zone) is correlated with seizure-free outcomes,41,42 whereas others find that complete irritative zone resection is neither necessary nor sufficient to ensure favorable outcomes.43 Network-based analysis of spike propagation is relatively understudied compared to the standard clinical approach, which emphasizes the spatial localization of spike discharges rather than spike co-activation between brain regions.44 Mapping directional relationships using inter-spike latencies enables clinicians to distinguish between upstream and downstream regions, which may differentially correlate with the seizure onset zone.6 However, poor concordance between the upstream spike regions and the SZN in many patients (see Supplemental Table S3) suggests that this correlation is variable at the patient-level.
4.4. Study limitations and future directions.
Several weaknesses of the current study should be considered. Although small sample sizes are typical of pediatric intracranial EEG studies, validating our findings in a larger, preferably multi-institutional sample with a more representative gender distribution is warranted. The use of subdural electrodes prevented us from examining interactions between superficial and deep brain regions, which may reveal nuances in the propagation patterns captured at the surface.13 The somewhat limited acquisition frequency (200 Hz) is another source of error in detecting subtle peak-time differences between brain regions. Future work extending this paradigm to SEEG recordings with higher sampling rate is underway in our lab. All invasive EEG studies share the limitation of incomplete sampling of the epileptic brain. We note that our reproducibility analyses would translate to a host of non-invasive electrophysiology modalities, including scalp EEG and MEG, as the methodology is based generically on event times and locations. Finally, due to non-uniform availability of post-resection imaging, we did not attempt to correlate spike propagation patterns with the extent of surgical resection, which has been examined elsewhere.12,18
4.5. Conclusions.
We investigated the reproducibility of interictal spike propagation in a retrospective cohort of twelve children implanted with subdural electrodes. We found that the sequential activation of network regions during spike propagation was reproducible over thousands of spike discharges across hours to days of interictal recording. These findings shed new light on long-term spatiotemporal patterns that likely constrain the network mechanisms of refractory epilepsy.
Supplementary Material
Left: Histogram of Sequence Similarity (Si) values (S1,2,3,…N) for N = 6,320 sequences from Patient #12. The darker shade represents the observed data, whereas the lighter shade represents one surrogate iteration in which spike times were randomly permuted. Solid vertical lines represent the distribution median, or ‘Reproducibility Index,’ R (Rorig for observed data, Rperm(1) for one iteration of surrogate data). Right: The observed Reproducibility Index (Rorig) was compared to a distribution of Rperm values formed by 1,000 permutation iterations. In this example, observed activation sequences were significantly more reproducible than would be expected by chance (i.e., Rorig vs. Rperm, p <0.003).
A) Subdural electrode implantation and representative 10-second EEG clip for Patient #2. B) In order to focus on electrodes capturing frequent spike discharges, a non-parametric threshold was applied. The observed spike frequency (spikes/min) was calculated for each recording electrode (Left), and then each spike in the detector output was randomly re-assigned to an electrode. Repeating this process (n = 1,000) yielded a surrogate distribution corresponding to a random allocation of spike activity across the recording field (Middle). Electrodes with an observed spike frequency exceeding 1.5 standard deviations above the mean of the surrogate distribution were analyzed (Right).
For each network, the schematic electrode map and network electrodes (black) are shown. Heat maps show the Segment DPs (color limits: DP −100, blue to DP +100, red). The rank correlation of Segment DPs and Overall DPs is shown for each segment, and the solid black line is the median across segments (‘DP-Stability’).
A) Correlation between DP-Stability and number of network nodes across networks. B) Correlation between Rorig and number of network nodes across networks. C) Correlation between DP-Stability and Rorig.
5. Key Points.
The long-term reproducibility of interictal spike propagation was studied in a retrospective cohort of 12 children implanted with subdural electrodes.
The sequential activation of network regions during spike propagation was highly reproducible over thousands of spike discharges and many hours of recording.
These findings shed new light on spatiotemporal dynamics that may constrain the network mechanisms of refractory epilepsy.
Significance:
Interictal spike propagation is a highly reproducible output of epileptic networks. These findings shed new light on spatiotemporal dynamics that may constrain the network mechanisms of refractory epilepsy.
6. Acknowledgements
SBT was supported by the University of Rochester Medical Center (URMC) Clinical & Translational Science Institute (CTSI) TL1 Academic Research Track Award (5 TL1 TR 2000–3) and the Neurosurgery Research and Education Foundation (NREF) Aaron-Cohen Gadol Medical Student Summer Research Fellowship (MSSRF). EDM was supported by NINDS R01NS099348. The authors thank neurosurgeons Gregory G. Heuer, MD PhD (CHOP), and Dr. Phillip B. Storm, MD (CHOP), for their contributions to the study dataset. We thank Brenda E. Porter, MD PhD (Stanford University) for her role in EEG inspection and seizure annotation. We acknowledge the following individuals for their conceptual contributions: Camilo Bermudez, Chiara Conley, and Merritt W. Brown, MD. The authors are indebted to the patients and families who participated in this research.
Footnotes
We confirm that we have read the Journal’s position on issues involved in ethical publication and affirm that this report is consistent with those guidelines.
Competing interests and ethical publication statement
None of the authors has any conflict of interest to disclose.
References
- 1.Staley KJ, Dudek FE. Interictal spikes and epileptogenesis. Epilepsy currents 2006; 6: 199–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Badier JM, Chauvel P. Spatio-temporal characteristics of paroxysmal interictal events in human temporal lobe epilepsy. J Physiol Paris 1995; 89: 255–264. [DOI] [PubMed] [Google Scholar]
- 3.Penfield W, Jasper H. Epilepsy and the Functional Anatomy of the Human Brain. J. & A. Churchill: London; 1954. [Google Scholar]
- 4.Baumgartner C, Lindinger G, Ebner A, et al. Propagation of interictal epileptic activity in temporal lobe epilepsy. Neurology 1995; 45: 118–122. [DOI] [PubMed] [Google Scholar]
- 5.Bourien J, Bartolomei F, Bellanger JJ, et al. A method to identify reproducible subsets of co-activated structures during interictal spikes. Application to intracerebral EEG in temporal lobe epilepsy. Clin Neurophysiol 2005; 116: 443–455. [DOI] [PubMed] [Google Scholar]
- 6.Hufnagel A, Dumpelmann M, Zentner J, et al. Clinical relevance of quantified intracranial interictal spike activity in presurgical evaluation of epilepsy. Epilepsia 2000; 41: 467–478. [DOI] [PubMed] [Google Scholar]
- 7.Lai Y, van Drongelen W, Hecox K, et al. Cortical activation mapping of epileptiform activity derived from interictal ECoG spikes. Epilepsia 2007; 48: 305–314. [DOI] [PubMed] [Google Scholar]
- 8.Tanaka N, Hamalainen MS, Ahlfors SP, et al. Propagation of epileptic spikes reconstructed from spatiotemporal magnetoencephalographic and electroencephalographic source analysis. Neuroimage 2010; 50: 217–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Asano E, Muzik O, Shah A, et al. Quantitative interictal subdural EEG analyses in children with neocortical epilepsy. Epilepsia 2003; 44: 425–434. [DOI] [PubMed] [Google Scholar]
- 10.Baud MO, Kleen JK, Anumanchipalli GK, et al. Unsupervised Learning of Spatiotemporal Interictal Discharges in Focal Epilepsy. Neurosurgery 2018; 83: 683–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tomlinson SB, Bermudez C, Conley C, et al. Spatiotemporal Mapping of Interictal Spike Propagation: A Novel Methodology Applied to Pediatric Intracranial EEG Recordings. Front Neurol 2016; 7: 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tamilia E, Park EH, Percivati S, et al. Surgical resection of ripple onset predicts outcome in pediatric epilepsy. Ann Neurol 2018; 84: 331–346. [DOI] [PubMed] [Google Scholar]
- 13.Alarcon G, Guy CN, Binnie CD, et al. Intracerebral propagation of interictal activity in partial epilepsy: implications for source localisation. J Neurol Neurosurg Psychiatry 1994; 57: 435–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zumsteg D, Friedman A, Wieser HG, et al. Propagation of interictal discharges in temporal lobe epilepsy: correlation of spatiotemporal mapping with intracranial foramen ovale electrode recordings. Clin Neurophysiol 2006; 117: 2615–2626. [DOI] [PubMed] [Google Scholar]
- 15.Tanaka N, Stufflebeam SM. Clinical application of spatiotemporal distributed source analysis in presurgical evaluation of epilepsy. Front Hum Neurosci 2014; 8: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin FH, Hara K, Solo V, et al. Dynamic Granger-Geweke causality modeling with application to interictal spike propagation. Hum Brain Mapp 2009; 30: 1877–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maharathi B, Wlodarski R, Bagla S, et al. Interictal spike connectivity in human epileptic neocortex. Clinical Neurophysiology 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alarcon G, Garcia Seoane JJ, Binnie CD, et al. Origin and propagation of interictal discharges in the acute electrocorticogram. Implications for pathophysiology and surgical treatment of temporal lobe epilepsy. Brain 1997; 120 ( Pt 12): 2259–2282. [DOI] [PubMed] [Google Scholar]
- 19.Staley K, Hellier JL, Dudek FE. Do interictal spikes drive epileptogenesis? Neuroscientist 2005; 11: 272–276. [DOI] [PubMed] [Google Scholar]
- 20.Karunakaran S, Rollo MJ, Kim K, et al. The interictal mesial temporal lobe epilepsy network. Epilepsia 2018; 59: 244–258. [DOI] [PubMed] [Google Scholar]
- 21.Wendling F, Bartolomei F, Senhadji L. Spatial analysis of intracerebral electroencephalographic signals in the time and frequency domain: identification of epileptogenic networks in partial epilepsy. Philos Trans A Math Phys Eng Sci 2009; 367: 297–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karoly PJ, Freestone DR, Boston R, et al. Interictal spikes and epileptic seizures: their relationship and underlying rhythmicity. Brain 2016; 139: 1066–1078. [DOI] [PubMed] [Google Scholar]
- 23.Marsh ED, Peltzer B, Brown MW, et al. Interictal EEG spikes identify the region of seizure onset in some, but not all pediatric epilepsy patients. Epilepsia 2010; 51: 592–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spencer SS, Goncharova II, Duckrow RB, et al. Interictal spikes on intracranial recording: behavior, physiology, and implications. Epilepsia 2008; 49: 1881–1892. [DOI] [PubMed] [Google Scholar]
- 25.Sammaritano M, Gigli GL, Gotman J. Interictal spiking during wakefulness and sleep and the localization of foci in temporal lobe epilepsy. Neurology 1991; 41: 290–297. [DOI] [PubMed] [Google Scholar]
- 26.Wieser HG, Blume WT, Fish D, et al. ILAE Commission Report. Proposal for a new classification of outcome with respect to epileptic seizures following epilepsy surgery. Epilepsia 2001; 42: 282–286. [PubMed] [Google Scholar]
- 27.Brown MW 3rd, Porter BE, Dlugos DJ, et al. Comparison of novel computer detectors and human performance for spike detection in intracranial EEG. Clin Neurophysiol 2007; 118: 1744–1752. [DOI] [PubMed] [Google Scholar]
- 28.Lambert I, Roehri N, Giusiano B, et al. Brain regions and epileptogenicity influence epileptic interictal spike production and propagation during NREM sleep in comparison with wakefulness. Epilepsia 2018; 59: 235–243. [DOI] [PubMed] [Google Scholar]
- 29.Malinowska U, Badier JM, Gavaret M, et al. Interictal networks in magnetoencephalography. Hum Brain Mapp 2014; 35: 2789–2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rubinov M, Sporns O. Complex network measures of brain connectivity: uses and interpretations. Neuroimage 2010; 52: 1059–1069. [DOI] [PubMed] [Google Scholar]
- 31.Khambhati AN, Davis KA, Oommen BS, et al. Dynamic Network Drivers of Seizure Generation, Propagation and Termination in Human Neocortical Epilepsy. PLoS Comput Biol 2015; 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomas K, Eero S, Martin P, et al. Leaders and followers: quantifying consistency in spatio-temporal propagation patterns. New Journal of Physics 2017; 19: 043028. [Google Scholar]
- 33.Kramer MA, Cash SS. Epilepsy as a disorder of cortical network organization. Neuroscientist 2012; 18: 360–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yaffe RB, Borger P, Megevand P, et al. Physiology of functional and effective networks in epilepsy. Clin Neurophysiol 2015; 126: 227–236. [DOI] [PubMed] [Google Scholar]
- 35.Luders HO, Najm I, Nair D, et al. The epileptogenic zone: general principles. Epileptic Disord 2006; 8 Suppl 2: S1–9. [PubMed] [Google Scholar]
- 36.Panzica F, Varotto G, Rotondi F, et al. Identification of the Epileptogenic Zone from Stereo-EEG Signals: A Connectivity-Graph Theory Approach. Front Neurol 2013; 4: 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Mierlo P, Papadopoulou M, Carrette E, et al. Functional brain connectivity from EEG in epilepsy: seizure prediction and epileptogenic focus localization. Prog Neurobiol 2014; 121: 19–35. [DOI] [PubMed] [Google Scholar]
- 38.Lemieux L, Daunizeau J, Walker MC. Concepts of connectivity and human epileptic activity. Front Syst Neurosci 2011; 5: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khoo HM, von Ellenrieder N, Zazubovits N, et al. The spike onset zone: The region where epileptic spikes start and from where they propagate. Neurology 2018; 91: e666–e674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Staley KJ, White A, Dudek FE. Interictal spikes: harbingers or causes of epilepsy? Neurosci Lett 2011; 497: 247–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coutin-Churchman PE, Wu JY, Chen LL, et al. Quantification and localization of EEG interictal spike activity in patients with surgically removed epileptogenic foci. Clin Neurophysiol 2012; 123: 471–485. [DOI] [PubMed] [Google Scholar]
- 42.Greiner HM, Horn PS, Tenney JR, et al. Preresection intraoperative electrocorticography (ECoG) abnormalities predict seizure-onset zone and outcome in pediatric epilepsy surgery. Epilepsia 2016; 57: 582–589. [DOI] [PubMed] [Google Scholar]
- 43.Huang C, Marsh ED, Ziskind DM, et al. Leaving tissue associated with infrequent intracranial EEG seizure onsets is compatible with post-operative seizure freedom. J Pediatr Epilepsy 2012; 1: 211–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Janca R, Krsek P, Jezdik P, et al. The Sub-Regional Functional Organization of Neocortical Irritative Epileptic Networks in Pediatric Epilepsy. Front Neurol 2018; 9: 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Left: Histogram of Sequence Similarity (Si) values (S1,2,3,…N) for N = 6,320 sequences from Patient #12. The darker shade represents the observed data, whereas the lighter shade represents one surrogate iteration in which spike times were randomly permuted. Solid vertical lines represent the distribution median, or ‘Reproducibility Index,’ R (Rorig for observed data, Rperm(1) for one iteration of surrogate data). Right: The observed Reproducibility Index (Rorig) was compared to a distribution of Rperm values formed by 1,000 permutation iterations. In this example, observed activation sequences were significantly more reproducible than would be expected by chance (i.e., Rorig vs. Rperm, p <0.003).
A) Subdural electrode implantation and representative 10-second EEG clip for Patient #2. B) In order to focus on electrodes capturing frequent spike discharges, a non-parametric threshold was applied. The observed spike frequency (spikes/min) was calculated for each recording electrode (Left), and then each spike in the detector output was randomly re-assigned to an electrode. Repeating this process (n = 1,000) yielded a surrogate distribution corresponding to a random allocation of spike activity across the recording field (Middle). Electrodes with an observed spike frequency exceeding 1.5 standard deviations above the mean of the surrogate distribution were analyzed (Right).
For each network, the schematic electrode map and network electrodes (black) are shown. Heat maps show the Segment DPs (color limits: DP −100, blue to DP +100, red). The rank correlation of Segment DPs and Overall DPs is shown for each segment, and the solid black line is the median across segments (‘DP-Stability’).
A) Correlation between DP-Stability and number of network nodes across networks. B) Correlation between Rorig and number of network nodes across networks. C) Correlation between DP-Stability and Rorig.


