Abstract
Background:
B cells play a critical role in the development and maintenance of food allergy by producing allergen-specific IgE. Despite the importance of B cells in IgE-mediated food allergy, the identity of sIgE-producing human B cells and how IgE is regulated are poorly understood.
Objective:
To identify the immunophenotypes of circulating B cells associated with the production of galactose-alpha-1,3-galactose specific IgE production in patients with red meat allergy.
Methods:
B cells in PBMC samples obtained from 19 adults with physician-diagnosed red meat allergy and 20 non-meat allergic healthy controls were assessed by mass cytometry along with a bioinformatics analysis pipeline to identify discrete B cell phenotypes that associated with serum sIgE. Fluorescent flow cytometry was then applied to sort purify discrete B cell subsets, and B cells were functionally evaluated on an individual cell level for the production of sIgE by ELISPOT.
Results:
Discrete B cell phenotypes abundant in meat allergic subjects compared to non-meat allergic controls were found in peripheral blood that do not share typical characteristics of classical isotype-switched memory B cells that express high levels of CD27. These B cell subsets shared higher IgD and lower IgM expression levels coupled with CXCR4, CCR6 and CD25 expression. In vitro polyclonal stimulation of purified B cell subsets from meat allergic subjects demonstrated that these subsets were enriched for cells induced to secrete sIgE.
Conclusions and Clinical Relevance:
Circulating B cells display increased abundance of discrete B cell subsets in meat allergic subjects. This observation, coupled with the capacity of individual B cell subsets to produce sIgE following activation, implicates these novel B cell phenotypes in promoting IgE in meat allergy.
INTRODUCTION
Red meat allergy, also known as alpha-gal syndrome, is among a minority of food allergies that pose a serious acute health risk through induction of IgE-mediated anaphylactic reactions. This novel form of food allergy develops in adults worldwide and is thought to result from tick bites through mechanisms that remain unknown [1–9]. Allergic reactions in patients following consumption of red meat are driven by allergen-specific IgE (sIgE) against the oligosaccharide galactose-α−1,3-galactose (alpha-gal) [1], which is present in the tissues of all non-primate mammals [10, 11]. Despite the importance of IgE in the pathogenesis of allergic diseases, the identity of sIgE-producing human B cells and their frequency are poorly understood. The reason for this is because B cells that express IgE are found at very low frequencies and that serum IgE binds to Fc receptors for IgE on the surface of B cells [12–16]. Moreover, there has been a lack of robust assays that allow for comprehensive immunophenotyping of B cells within complex biological samples. Although some studies have described IgE-expressing B cells in the blood of allergic and healthy individuals [17–19], the contribution of such cells to IgE responses is unclear. These observations underscore a need to evaluate IgE-producing B cells with greater resolution to determine their clinical relevance in allergic diseases.
Here, we sought to interrogate the phenotypes of circulating B cells in patients with food allergy to red meat. The study was designed to sample B cells in peripheral blood of patients actively avoiding meat who had positive alpha-gal sIgE titers and histories of delayed urticaria after eating mammalian meat. Using mass cytometry with a bioinformatics analysis pipeline and traditional fluorescent-based flow cytometric cell sorting approaches, we aimed to determine whether discrete B cell subsets could be identified in meat allergic subjects that associated with alpha-gal sIgE production. Mass cytometry by time-of-flight (CyTOF) combines antibodies labeled with metal isotopes with mass spectrometry, which allows for single-cell analysis of more than 40 parameters simultaneously with minimal interference from signal overlap between channels that are encountered with highly-multiparametric flow cytometry [20–22]. We describe the use of viSNE, an algorithm for single-cell visualization based on t-SNE embedding [23, 24], SPADE, a density-based algorithm for identifying subpopulations of distinct cell types [25] and flowType, an algorithm that defines all possible cell subsets that correlate with a clinical parameter [26, 27]. Application of these computational tools to CyTOF datasets led to identification of discrete B cell subsets whose abundance were enriched in blood of meat allergic patients. Our analytical approach also facilitated the transition from CyTOF to fluorescence-based cell sorting, enabling functional examination of cultured B cell subsets that cannot be achieved with mass cytometry since cells are vaporized. Testing the capacity of these rare B cell subsets to secrete antibody following in vitro stimulation demonstrated that such cells produced alpha-gal sIgE in patients with red meat allergy. Our findings support a novel B cell signature in meat allergic subjects that associates with alpha-gal sIgE production, which may play a role in the pathogenesis of this food allergy.
MATERIALS AND METHODS
Human subjects
All participants were adults (ages 23–77) and included patients with physician-diagnosed red meat allergy with alpha-gal sIgE positive titers > 0.35 IU/mL, and non-meat allergic healthy controls with no alpha-gal sIgE antibodies (Supplementary Table 1). Inclusion criteria for meat allergic subjects included histories of delayed urticaria after meat consumption. All meat allergic subjects were actively avoiding meat. Non-meat allergic controls had no history of reactions to mammalian meat by self-report, no history of other food allergies or anaphylaxis, and had normal total IgE levels for age. Subjects were non-pregnant without a history of chronic illness. All subjects were recruited through the UVA Allergic Diseases Clinic, or else by advertisement. All studies were approved by the University of Virginia Institutional Review Board under protocols #13166 and #13298, and all study participants gave written informed consent.
Total and alpha-gal specific IgE
Total and alpha-gal IgE antibodies in blood serum were measured using the ImmunoCAP system (Phadia US, Portage, Mich) with the ImmunoCAP 250 instrument, according to the manufacturer’s instructions. The streptavidin CAP technique was used to measure IgE levels to alpha-gal as described previously [28, 29], in which 5 μg of biotinylated cetuximab, a chimeric mouse-human IgG1 mAb against epidermal growth factor receptor that contains alpha-gal moieties on the Fab portion of the cetuximab heavy chain that are recognized by IgE specific for alpha-gal, was added to each streptavidin-coated CAP before serum was added. Results were calculated as kUA/L. Alpha-gal sIgE antibody-secreting cells were quantified by ELISPOT. Briefly, ELISPOT plates were coated overnight at 4°C with 10 μg/mL unlabeled cetuximab mAb. Plates were blocked with 10% FBS in RPMI, washed and samples were added in appropriate dilutions for 18 h. After washing, biotin-conjugated mouse anti-human IgE antibody (BioLegend, San Diego, Calif) was added overnight. Plates were washed and HRP-avidin (SouthernBiotech) was added for 24 h. AEC substrate solution was used to obtain coloration, and washing plates with water stopped the reaction. The number of antibody-secreting spots was quantified using a dual-axis light-dissecting microscope and confirmed using an Immunospot Microanalyzer (Cellular Technology Limited, Shaker Heights, OH). The numbers of IgE ASCs per million cells were calculated.
Cell preparation and culture
Peripheral blood mononuclear cells (PBMC) of 14 meat allergic subjects and 10 non-meat allergic controls were isolated from venous blood in collection tubes with acid citrate dextrose (ACD) by density-gradient centrifugation on Ficoll-Paque PREMIUM media (GE Healthcare, Pittsburgh, Penn), and washed twice in PBS. Cells were either frozen in FBS (ThermoFisher Scientific, Rockford, Ill) with 20% DMSO achieving > 95% cell viability, or freshly enriched for B cells by negative selection using the Human Pan B cell isolation kit (Miltenyi Biotech) and sort purified. B cells were cultured in RPMI (ThermoFisher Scientific) with 15% FBS, L-glutamine-penicillin-streptomycin (Sigma Aldrich, St. Louis, MO), beta mercaptoethanol (Sigma-Aldrich), and 20 ng/mL recombinant human IL-4 (Peprotech, Rocky Hill, NJ). In 96-well plates, cells were cultured in the presence of 12.5 ng/mL PMA (EMD Millipore Corp, Billerica, Mass) and 500 ng/mL ionomycin (EMD Chemicals, San Diego, Calif) at 37°C in an atmosphere of 5% CO2. Cells were collected at day 8 to quantify alpha-gal sIgE-secreting cells by ELISPOT.
Mass cytometry data acquisition and analysis
Thawed PBMCs from 19 meat allergic subjects and 20 non-meat allergic controls were washed in PBS with 5% FBS and incubated with 5 μM Cell-ID Cisplatin (Fluidigm, San Francisco, Calif) to determine viability, as previously described [30]. Mean PBMC viability after thawing was 94%, as determined by trypan blue exclusion. Patient samples were obtained over a period of two years, with initial samples analyzed prior to the availability of barcoding reagents. Therefore, barcoding was not used in cell preparation and analysis to maintain consistency across all samples. For cell surface marker analysis, 3 × 106 live cells were first incubated with Fc block to prevent non-specific binding, washed, and stained in 100 μL PBS with 5% FBS and Ab cocktail at room temperature for 30 min. All of the antibodies used in the panel (Supplemental Table 2) were purchased pre-conjugated to metals (Fluidigm) and concentrations optimized through the Mass Cytometry Antibody Bank services provided by the UVA Flow Cytometry Core Facility. Surface phenotypes between freshly isolated and cryopreserved PBMCs were compared and confirmed to be equivalent. Cells were washed twice in PBS with 5% FBS and then stained with Cell-ID Intercalator-Ir (Fluidigm) in Maxpar Fix and Perm Buffer (Fluidigm) at 4°C overnight. Samples were washed once in PBS with 5% FBS, once in Maxpar H2O, resuspended in Maxpar H2O, and then collected on a CyTOF 2 instrument (Fluidigm). For quality control, the acquisition rate was maintained under 400 events/s, and events were normalized using bead standards to minimize batch effects, as previously described [31]. FCS files containing the normalized single-cell data were uploaded to Cytobank for analysis. All datasets used here are publicly available at http://www.cytobank.org.
Negative staining for cisplatin and positive staining for iridium identified cells, and doublets were excluded by higher DNA content and longer event length. Human immune cell subsets were identified using standard markers as reported previously [32, 33]. Total CD4+ T cells were identified by gating on CD3+CD20−CD4+ cells. Total CD8+ T cells were identified by gating on CD3+CD20−CD8+ cells. NK cells were identified by gating on CD3−CD20−CD14−CD56+ cells. DCs were identified by gating on CD3−CD20−CD56−CD14−HLA-DR+ cells. Monocytes were identified by gating on CD3−CD20−CD14+ cells. Total B cells were identified by gating on CD20+ CD3− cells, and were analyzed using the data analysis platforms viSNE, SPADE and flowType. viSNE is a CyTOF analysis tool that uses the t-distributed stochastic neighbor embedding (t-SNE) algorithm to analyze and display high-dimensional data on a two-dimensional map, with cells colored according to frequency or expression of a marker (available at http://www.cytobank.org) [34]. SPADE uses a density-based algorithm to visualize and enable cellular hierarchy of distinct cell types on the basis of similarities across user-selected markers (available at http://www.bioconductor.org) [25]. The Bioconductor package flowType is a method that defines all possible cell subpopulations within a sample based on a certain combination of marker expression levels from all parameters measured (available at http://www.bioconductor.org). The heterogeneity of B cell populations was first measured using t-SNE inferred clustering by viSNE. SPADE was used to identify discrete B cell subsets and to quantify the frequency of cells within each population. FlowType was used on the arsinh transformed data (cofactor 5) to partition the cells into a immunophenotype hierarchy as described previously [35]. To identify immunophenotypes of B cells found to be differentially abundant between allergic and control subjects, a generalized linear model was fitted to the immunophenotyped hierarchy, with sIgE titers and age as covariates, using a quasi-likelihood framework in edgeR to estimate the dispersion of cell counts as described previously [35, 36]. A more conservative approach based on observation weights, described by Zhou and colleagues [37], was used to reduce the effect of outliers on count-based differential expression analyses. Immunophenotypes enriched in allergic subjects and having adjusted P-values better than a false discovery rate (FDR) using the Benjamini-Hochberg method of 5% were plotted in a heat map. The code to perform this set of analyses is available on github at https://github.com/bc2zb/Cox-et-al-meat-allergy, and the binary results files are available at http://www.cytobank.org.
Flow cytometry and cell sorting
For flow cytometric analysis, PBMCs were stained with fluorescent-labeled surface markers (Supplemental Table 3), followed by the live/dead stain AQUA (Invitrogen). Cells were acquired on an Invitrogen Attune NxT, or sorted with a Becton Dickinson Influx Cell Sorter, and analysis was performed using FlowJo software version 9.3.3 (TreeStar Inc., Ashland, Ore). Dead cells and doublets were excluded from the analysis and cell population gates were determined using fluorescence-minus-one controls.
Statistics
Data are presented as the mean ± SD. Statistical analyses were performed using GraphPad Prism, version 6 (GraphPad Software, La Jolla, Calif). Statistics were performed with the Fisher exact test, and data with a 2-sided P-value < 0.05 was considered significant. Pearson’s correlation coefficient was used to determine the strength of association between 2 variables. The Mann-Whitney U nonparametric test was used to compare differences between allergic and control groups for the frequencies of alpha-gal sIgE ASCs in cultures of sorted B cells.
RESULTS
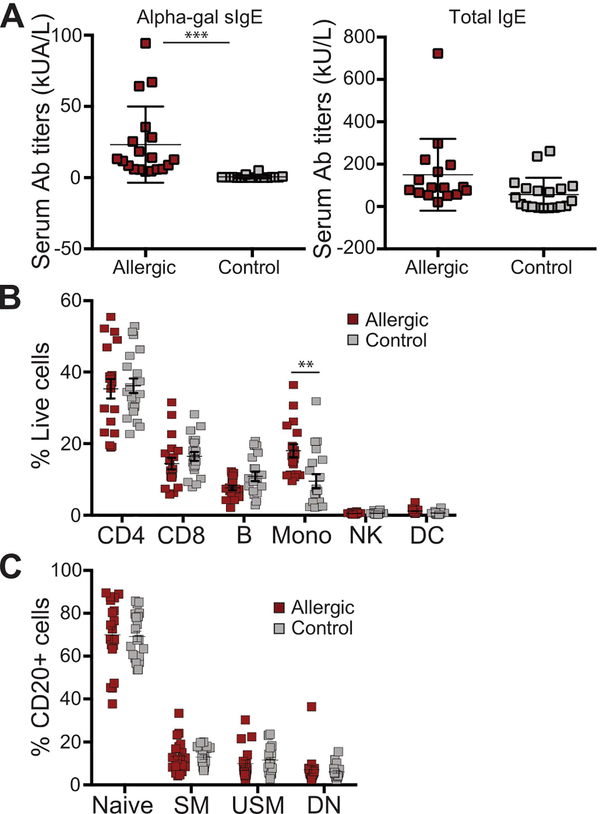
Meat allergic subjects and healthy controls show equivalent distribution of major circulating immune cell populations
We first established a CyTOF staining panel for testing the cellular heterogeneity within PBMCs isolated from venous blood. This panel contained 23 leukocyte markers that allowed us to identify all of the major immune cell populations, and to profile B cells with greater dimensionality by incorporating various markers based on prior indications of their utility in polychromatic flow cytometry curated from the literature (Supplementary Table 2). CyTOF was performed on cryopreserved PBMC samples obtained from the blood of meat allergic patients with a range of positive alpha-gal sIgE titers and healthy controls (Figure 1A). Characteristics of subject groups are summarized in Supplementary Table 1. After data normalization, we initially assessed the frequencies of major immune cell populations (CD4+ and CD8+ T cells, CD56+ NK cells, HLA-DR+ DCs, CD14+ monocytes, and total CD20+ B cells), identified by manual gating on standard individual markers. No significant differences in the frequencies of T cells, B cells, NK cells, and DCs between allergic and control samples were observed (Figure 1B). Monocytes from allergic subjects were found at higher frequencies compared with controls. We further compared the frequencies of the major mature B cell populations found in peripheral blood when manually gated on the standard phenotypic markers CD3, CD20, IgD, and CD27. No significant differences in the frequencies of naïve B cells (CD3−CD20+IgD+CD27−), unswitched memory B cells (CD3−CD20+IgD+CD27+) and switched memory B cells (CD3−CD20+IgD−CD27+) were observed in meat allergic subjects compared with healthy controls (Figure 1C). There was also no significant difference in the frequency of the double negative B cell population (IgD−CD27−), which includes switched memory B cells [38, 39]. Interestingly, this B cell population has been shown to contain IgE+CD27− memory B cells that are enriched in the blood of children with food allergy [40]. Flow cytometric analysis using fluorescent-labeled CD3, CD20, IgD, and CD27 confirmed these results (Supplemental Figure 1).
Figure 1. Frequencies of circulating immune cell populations in meat allergic and control subjects.
(A) Serum alpha-gal sIgE and total IgE titers in meat allergic (n = 19) and non-meat allergic controls (n = 20), ***P < 0.001. (B) CD4+ and CD8+ T cells, natural killer cells (NK), dendritic cells (DC), monocytes (Mono), and total B cells expressed as proportion of live CD45+ leukocytes, as well as (C) naïve B cells, immunoglobulin unswitched (USM) and switched (SM) memory B cells, and double negative (DN) B cells expressed as proportion of total CD20+ B cells of subjects were quantified by manual gating.
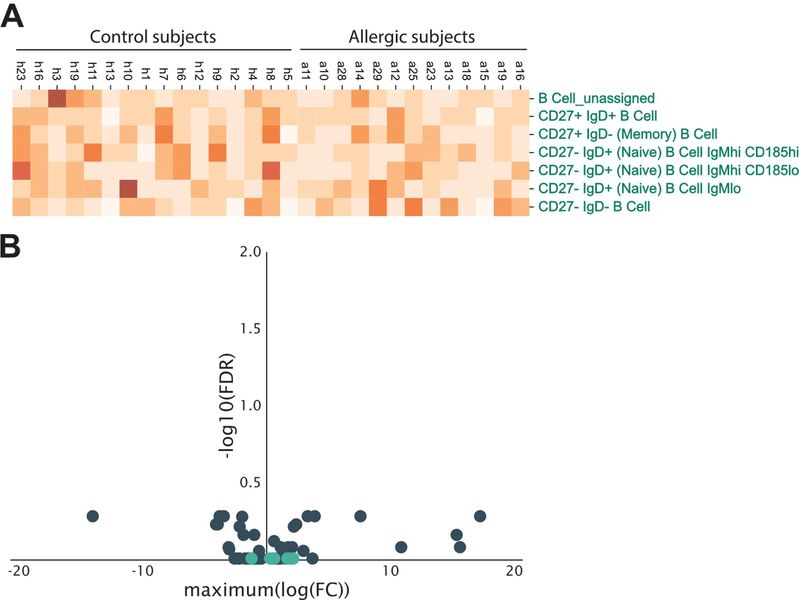
Comparable results were also observed when B cells were automatically gated using the cloud-based Astrolabe platform designed for mass cytometry data (astrolabediagnostics.com). Using machine learning, Astrolabe identifies cell subsets based on curation of phenotypic signatures reported in the literature and performs differential abundance analysis. Moreover, this platform allows for the stratification of cellular subsets based on expression of markers that correlate with clinical parameters, including disease states. No significant differences in the frequencies of 7 curated B cell subsets, representing naïve, memory B cells and double negative B cells based on IgD and CD27 expression, were found between subject groups (Figure 2A and2B). Taken together, these findings support the view that defining major immune non-B cell and B cell populations based solely on conventional immune parameters shows no difference in cell frequencies of meat allergic subjects, and does not account for potential cellular heterogeneity within these populations.
Figure 2. Comparable frequencies of curated B cell populations defined by automated gating in meat allergic and control subjects.
(A) Heatmap of the relative frequency of 7 different B cell populations in meat allergic (n = 19) and non-meat allergic control (n = 20) subjects. B cell populations were assigned by the Astrolabe Diagnostics analysis platform. (B) Differential abundance analysis of the frequencies of B cell populations (light blue dots) compared to non-B cell immune cell populations (dark blue dots) shows no difference between allergic and control subjects.
Distinct B cell phenotypes are observed in meat allergic subjects
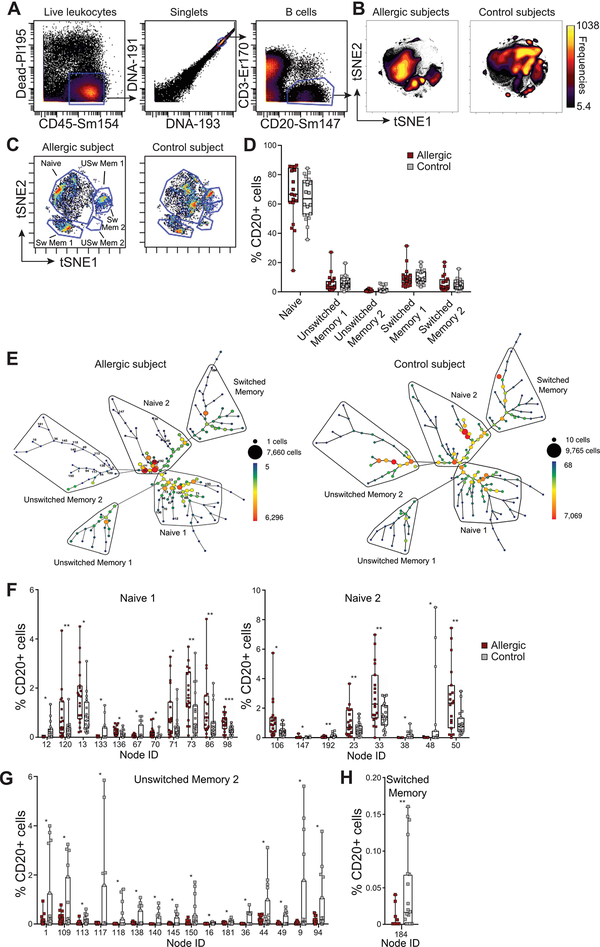
Having established that the standard B cell markers used to delineate total, naïve and memory B cells did not reveal changes in cell numbers from meat allergic and healthy subjects, we applied the t-SNE dimensionality reduction approach, viSNE [23, 24] to the CyTOF datasets. The datasets from each subject group were concatenated to visualize in the form of a scatter plot cohort-level phenotypes. Analysis of total CD20+ B cells using viSNE revealed heterogeneity both within the major mature B cell populations and between meat allergic and control subjects (Figure 3A and3B). By manually gating on IgD and CD27 to annotate naïve, switched and unswitched memory B cells (Figure 3C, Supplemental Figure 2), we identified multiple islands that were spatially separated, indicating phenotypic heterogeneity within these B cell populations that has not been appreciated previously. Although the location of each annotated B cell population within the viSNE plots were similar between allergic and control subjects, there were marked differences in the clustering patterns of cells within each island. The frequencies of B cells within each manual gate were quantified for each subject to determine whether there were differences between meat allergic and control cohorts. No significant differences in the frequencies of B cells within the annotated naïve, switched and unswitched B cells based on IgD and CD27 expression were found between subject groups (Figure 3D). These results confirmed the analyses performed above, despite phenotypic heterogeneity of B cells in particular within the naïve B cell population between subject groups.
Figure 3. viSNE and SPADE analysis of CyTOF data shows phenotypic changes of B cells in meat allergic subjects.
(A) Gating strategy for viSNE analysis on CD20+ B cells. (B) viSNE plots of concatenated datasets from meat allergic (n = 19) and non-meat allergic (n = 20) control subjects. Plots are colored by cell frequency. (C) Data from 1 meat allergic and 1 control subject showing heterogeneous clusters of naïve B cells, unswitched memory B cells (USw Mem 1, USw Mem 2) and switched memory B cells (Sw Mem 1, Sw Mem 2) based on IgD and CD27 expression. (D) Frequencies of naïve, unswitched memory and switched memory B cells expressed as proportion of total CD20+ B cells of subjects were quantified by manual gating. (E) A SPADE tree derived from a meat allergic patient and a control subject annotated with naïve, unswitched memory and switched memory B cell types on the basis of IgD and CD27 expression. The size and color of each node denotes the number of cells. (F-H) Frequencies of B cells of each node statistically enriched in naïve, unswitched memory and switched memory branches expressed as the percentage of total CD20+ B cells. *P < 0.05, **P < 0.01, ***P < 0.001.
Since viSNE helped visualize the data but does not explicitly identify and partition cells into subpopulations, we evaluated the CyTOF datasets using SPADE [25], a computational tool that enables cellular hierarchy inference among subpopulations of similar cells, to quantify discrete B cell subpopulations. Differences in the distribution and frequency of B cells between meat allergic and control subjects were found, as indicated by differences in the size and color of nodes within the SPADE tree (Figure 3E, Supplemental Figure 3). Fourteen nodes within the naïve B cell branches (CD27− IgD+) were statistically enriched in meat allergic subjects compared to controls (Figure 3F). In contrast, 16 nodes within unswitched (CD27+ IgD+) memory B cell branches and one node within switched (CD27+ IgD−) memory B cell branches were found enriched in non-allergic controls compared to meat allergic subjects (Figure 3G and3H). However, the enrichment of these memory B cell nodes was largely driven by a small subset of control subjects. These findings suggest that variation in the combined expression levels of all 23 markers within our CyTOF panel accounts for the differences of B cell nodes within the larger populations of naïve and memory B cells.
Markers within the CyTOF panel are identified that define B cell clusters associated with alpha-gal sIgE in meat allergic subjects
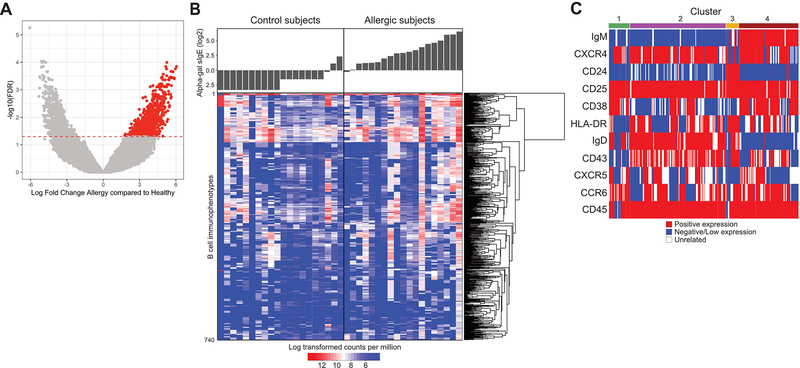
To determine the identity of markers that distinguished B cell phenotypes associated with red meat allergy subjects, we analyzed the CyTOF data using the flowType algorithm [26, 27]. This computational approach first uses automated gating to define all possible cell subsets within a dataset based on integrated marker expression levels, without manual gating on user-specified markers, and then assesses the relationship between each subset and the clinical outcome. Results demonstrated that approximately 500,000 immunophenotypes of B cells were identified within the CyTOF panel by flowType (Supplemental Figure 4A). These immunophenotypes result from further division of traditional B cell populations into related subpopulations. To identify the immunophenotypes of B cells found to be differentially abundant between meat allergic and control subjects, a generalized linear model was fitted to the data, with age as a covariate, using a quasi-likelihood framework in edgeR to estimate the dispersion of cell counts [35, 36]. The frequencies of 710 B cell immunophenotypes with adjusted P-values better than a false discovery rate (FDR) of 5% were found significantly enriched across meat allergic subjects compared to control subjects (Supplemental Figure 4A). Using a more conservative edgeR approach to reduce the effect of outliers on differential abundance analysis [37] yielded a total of 740 B cell immunophenotypes enriched across the allergic patient cohort (Figure 4A) and are plotted in a heatmap (Figure 4B). An average of 143 B cell immunophenotypes (ranging between 5 and 480) were enriched in allergic patient samples at an FDR of 5%, whereas an average of 61 of these B cell immunophenotypes (ranging between 6 and 181) were enriched in control samples. While these B cell immunophenotypes were associated with allergic status, no B cell immunophenotype signature was found to be specifically associated with age or the range of alpha-gal sIgE titer, although patients with higher sIgE titers generally had more B cell immunophenotypes. Not surprisingly, subject-to-subject variability was observed, with some phenotypes enriched in control subjects. These observations suggest that other factors such as history of tick bites or additional environmental cues may contribute to increased frequencies of certain B cell phenotypes unrelated to sIgE antibody production.
Figure 4. FlowType analysis identifies discrete B cell immunophenotypes that associate with alpha-gal sIgE in meat allergic subjects.
(A) Volcano plot of differentially abundant B cell immunophenotypes between meat allergic subjects and non-allergic controls, with negative log10-scaled FDR-corrected P-values on the Y-axis, and the log2 fold change on the X-axis. B cell immunophenotypes to the right and colored in red were comparatively found at higher frequencies in allergic patients, accounting for age and sIgE titers as covariates. The horizontal red line indicates a significance of 0.05 after adjustment of FDR using the Benjamini-Hochberg method. (B) Heat map resulting from flowType analysis of CyTOF datasets. Each row represents a distinct B cell immunophenotype based on marker expression levels, with its actual abundance in each subject expressed as log transformed counts per million indicated by the color scale. Corresponding serum alpha-gal sIgE for each subject is shown above the heat map. (C) Expression of 11 markers within the CyTOF panel needed to minimally define B cell immunophenotypes that were found significantly enriched in meat allergic subjects is represented graphically. Each row indicates the expression levels of an individual marker, with each column representing B cell immunophenotypes assigned to 4 main clusters based on the degree of shared marker expression.
Expression levels of 11 of the 23 markers in the CyTOF panel were used by flowType to define these B cell immunophenotypes. These immunophenotypes were assigned into four main clusters based on similar expression patterns of the 11 markers (Figure 4C). Cluster 1 comprised B cell immunophenotypes that shared negative expression of IgM, CD24 and CD43, variable expression of HLA-DR and IgD, and shared positive expression of CXCR4, CD38 and CD25. Cluster 2 comprised phenotypically related B cell subsets that shared negative or low expression of IgM and CD24, but generally shared positive expression of IgD, CXCR4, CCR6, and CD43. Cluster 3 comprised B cells that shared negative or low expression of IgM and CXCR4 and positive expression of CD24, CD25 and IgD. Cluster 4 comprised B cell subsets that shared positive expression of IgM, CXCR4 and CD25, negative expression of CD24 and IgD but were heterogeneous in their expression levels of other markers. Interestingly, expression levels of the classical memory B cell marker CD27 did not discriminate the four clusters of B cell subsets from the meat allergic subjects using flowType, with some B cells expressing CD27 and some B cells lacking CD27 expression (Supplemental Figure 4B). Membrane IgG and IgE expression was not detectable on B cells of any cluster identified by flowType (data not shown). Together, these results demonstrated that the B cell signature associated with red meat allergy is rare compared to the overall number of B cell immunophenotypes identified by our CyTOF panel, and does not show features of conventional human memory B cells, as measured by high expression of CD27 and intermediate antibody isotypes.
B cells from clusters identified by flowType are capable of producing alpha-gal sIgE following in vitro stimulation
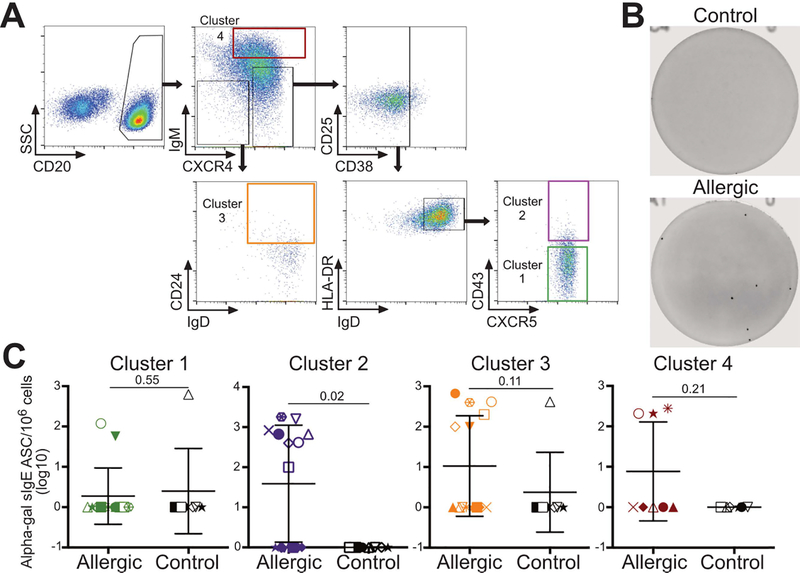
To evaluate and compare the functional properties of B cells associated with red meat allergy, we designed a fluorescent cytometry panel based on the markers used by flowType to group B cell subsets into the four main clusters (Supplementary Table 3). This approach allowed us to sort live B cells within each cluster to test their capacity to secrete alpha-gal sIgE in vitro following stimulation (Figure 5A). The frequencies of sorted B cells negatively correlated with age in Cluster 1 (Pearson’s r = −0.07), Cluster 2 (r = −0.15), Cluster 3 (r = −0.17), and Cluster 4 (r = −0.43). In addition, no significant differences in the frequencies of B cell clusters were found between males and females (Student’s t-test p > 0.5). We tested the ability for polyclonal stimulation of B cells to induce alpha-gal sIgE production to preclude suppositions of the activation requirements for these novel B cell phenotypes. The structural conformation of alpha-gal has not been determined, nor is it clear if and how the structural conformation affects IgE binding and B cell activation [41]. After stimulation with PMA, ionomycin and IL-4, we first assessed alpha-gal sIgE in culture supernatants by ImmunoCAP. However, given the small number of cells of each B cell cluster from subjects and the short half-life of soluble IgE the amount of IgE antibodies contained in supernatants was below the level of detection (data not shown). We therefore employed the ELISPOT assay as a sensitive method to enumerate alpha-gal sIgE antibody-secreting cells (ASCs) from the B cell cultures (Figure 5B and5C). Results demonstrated that alpha-gal sIgE-secreting B cells following in vitro stimulation were detected in 9 of 14 meat allergic subjects, with alpha-gal sIgE ASCs from the same individual found in Clusters 2 and 3 of four patients, Clusters 1 and 3 in one patient, Clusters 1, 2 and 3 in one patient, and in Cluster 2 only of three patients. In contrast, alpha-gal sIgE-secreting B cells following stimulation were detected in Clusters 1 and 3 from one control subject, with no ASCs found in Cluster 2. Analysis of Cluster 4 revealed that three allergic patients but no control subjects had alpha-gal sIgE ASCs following stimulation. These findings demonstrated that Clusters 2 and 3, which contain rare subsets of B cells with higher IgD and lower IgM expression levels coupled with CXCR4, CCR6 and CD25 expression, are enriched for cells that are able to differentiate into alpha-gal sIgE producing B cells following in vitro polyclonal activation.
Figure 5. Polyclonally stimulated B cells sorted from B cell clusters identified by flowType are capable of producing alpha-gal sIgE in meat allergic subjects.
(A) Gating strategy for sort purification of B cell subsets grouped into 4 main clusters, with each cluster indicated by a colored gate. (B and C) B cells of each cluster from meat allergic (n = 14) and non-meat allergic control (n = 10) subjects were cultured 8 days and analyzed for alpha-gal sIgE antibody-secreting cells (ASC) by ELISPOT. (B) Representative ELISPOT assay images from 1 allergic and 1 control subject. (C) Frequencies of alpha-gal sIgE ASCs of each B cell cluster after culture expressed as the number of ASCs per million cells. P-values are shown in each panel.
DISCUSSION
Using an integrated framework that combined mass cytometry with a bioinformatics pipeline, we have demonstrated that a novel B cell signature is associated with meat allergic subjects. We have taken advantage of the algorithms, viSNE, SPADE and flowType, to identify the minimal number of markers within our CyTOF panel that defined increased abundance of novel B cell phenotypes in PBMCs of meat allergic subjects. Eleven surface markers defined 740 phenotypically-related but discrete subsets of B cells in meat allergic subjects, which can be grouped into four main clusters based on shared expression levels. Moreover, we confirmed these findings using fluorescent flow cytometry that enabled subsequent functional analysis of purified B cells contained within each cluster to secrete alpha-gal sIgE following in vitro stimulation.
Our findings demonstrated that the B cell subsets capable of secreting sIgE following activation are found within the naïve population and do not share typical characteristics of classical isotype-switched memory B cells that express high levels CD27 [42, 43]. These cells may represent a novel population of memory B cells lacking CD27 expression found in the peripheral circulation [38, 39] that are prone to differentiate into IgE-secreting cells specific for alpha-gal in meat allergic patients. Early work in human adults identified peripheral blood memory B cells that express IgM with or without IgD [42, 44, 45], or express IgD only [46, 47]; these cells show somatically mutated IgV genes suggesting the involvement of T cell help for their development. Interestingly, given the cutaneous route of sensitization implicated in the development of meat allergy, memory B cells displaying low to negative CD27 expression have been described in human tissues near epithelial surfaces [48–50]. Further work will be required to assess the antigen experience state of the B cell subsets we have identified, including analysis of the IgV genes.
Several questions remain regarding the mechanisms of alpha-gal sIgE production from the B cell subsets in meat allergic subjects. First, although it is not feasible to functionally interrogate each B cell subset within an assigned cluster, further work is needed to explore the composition of potentially functionally distinct subsets to determine whether one exhibits a propensity for alpha-gal sIgE production following stimulation. Moreover, it will be necessary to elucidate how these B cells respond to T cell-dependent signals as well as T cell-independent signals, as activation of B cells responding to carbohydrate antigens such as alpha-gal is thought to be independent of cognate T cell help. Finally, an important question in the study of food allergy is whether sIgE-producing B cells traffic to the gut and mediate local IgE responses to digested allergens. Notably, B cell subsets within Clusters 2 and 3 of meat allergic subjects shared high expression levels of the chemokine receptors CXCR4 and CCR6, which have been implicated in regulating mucosal immunity and driving intestinal inflammation during disease pathogenesis [51, 52]. Additional work is needed to better understand the contribution of these chemokine receptors on B cells for trafficking to the gut mucosa in food allergy.
Increasing our understanding of the B cells that maintain allergic sensitization is critical for identifying new targets to treat and prevent allergies. Additionally, detection of novel B cell immunophenotypes associated with red meat allergy may serve as a biomarker to identify individuals who have, or will become sensitized to red meat, as well as identifying affected individuals who may develop severe allergic reactions such as anaphylaxis. Longitudinal studies assessing the frequency of these B cell subset in the circulation of individuals with red meat allergy, and those who are at high risk of obtaining bites from the lone star tick—and thus developing the allergy—would be useful in determining the utility of these B cell subsets as biomarkers for diagnosis.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Mark Conaway, PhD, of the Department of Public Health Sciences at the University of Virginia for assistance with statistical analyses.
FUNDING
This work was supported by the National Institutes of Health (grant no. NIAID R21AI124490 to LDE and SPC, and R01AI20565 to TPM).
Footnotes
CONFLICT OF INTEREST
The authors have declared that no conflict of interest exists.
REFERENCES
- 1.Commins SP, Satinover SM, Hosen J, Mozena J, Borish L, Lewis BD, et al. Delayed anaphylaxis, angioedema, or urticaria after consumption of red meat in patients with IgE antibodies specific for galactose-alpha-1,3-galactose. J Allergy Clin Immunol 2009;123:426–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Commins SP, James HR, Kelly LA, Pochan SL, Workman LJ, Perzanowski MS, et al. The relevance of tick bites to the production of IgE antibodies to the mammalian oligosaccharide galactose-alpha-1,3-galactose. J Allergy Clin Immunol 2011;127:1286–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Commins SP, Platts-Mills TA. Delayed anaphylaxis to red meat in patients with IgE specific for galactose alpha-1,3-galactose (alpha-gal). Curr Allergy Asthma Rep 2013;13:72–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamsten C, Starkhammar M, Tran TA, Johansson M, Bengtsson U, Ahlen G, et al. Identification of galactose-alpha-1,3-galactose in the gastrointestinal tract of the tick Ixodes ricinus; possible relationship with red meat allergy. Allergy 2013;68:549–52. [DOI] [PubMed] [Google Scholar]
- 5.Caponetto P, Fischer J, Biedermann T. Gelatin-containing sweets can elicit anaphylaxis in a patient with sensitization to galactose-alpha-1,3-galactose. J Allergy Clin Immunol 2013;1:302–3. [DOI] [PubMed] [Google Scholar]
- 6.Morisset M, Richard C, Astier C, Jacquenet S, Croizier A, Beaudouin E, et al. Anaphylaxis to pork kidney is related to IgE antibodies specific for galactose-alpha-1,3-galactose. Allergy 2012;67:699–704. [DOI] [PubMed] [Google Scholar]
- 7.Van Nunen SA, O’Connor KS, Clarke LR, Boyle RX, Fernando SL. An association between tick bite reactions and red meat allergy in humans. Med J Aust 2009;190:2. [DOI] [PubMed] [Google Scholar]
- 8.Sekiya K, Fukutomi Y, Nakazawa T, Taniguchi M, Akiyama K. Delayed anaphylactic reaction to mammalian meat. J Investig Allergol Clin Immunol 2012;22:446–7. [PubMed] [Google Scholar]
- 9.Nunez R, Carballada F, Gonzalez-Quintela A, Gomez-Rial J, Boquete M, Vidal C. Delayed mammalian meat-induced anaphylaxis due to galactose-alpha-1,3-galactose in 5 European patients. J Allergy Clin Immunol 2011;128:1122–4. [DOI] [PubMed] [Google Scholar]
- 10.Galili U, Clark MR, Shohet SB, Buehler J, Macher BA. Evolutionary relationship between the natural anti-Gal antibody and the Gal alpha 1--−−3Gal epitope in primates. Proc Natl Acad Sci U S A 1987;84:1369–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galili U, Swanson K. Gene sequences suggest inactivation of alpha-1,3-galactosyltransferase in catarrhines after the divergence of apes from monkeys. Proc Natl Acad Sci U S A 1991;88:7401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He JS, Narayanan S, Subramaniam S, Ho WQ, Lafaille JJ, Curotto de Lafaille MA. Biology of IgE production: IgE cell differentiation and the memory of IgE responses. Curr Top Microbiol Immunol 2015;388:1–19. [DOI] [PubMed] [Google Scholar]
- 13.Irsch J, Hunzelmann N, Tesch H, Merk H, Maggi E, Ruffilli A, et al. Isolation and characterization of allergen-binding cells from normal and allergic donors. Immunotechnology 1995;1:115–25. [DOI] [PubMed] [Google Scholar]
- 14.King CL, Poindexter RW, Ragunathan J, Fleisher TA, Ottesen EA, Nutman TB. Frequency analysis of IgE-secreting B lymphocytes in persons with normal or elevated serum IgE levels. J Immunol 1991;146:1478–83. [PubMed] [Google Scholar]
- 15.Karnowski A, Achatz-Straussberger G, Klockenbusch C, Achatz G, Lamers MC. Inefficient processing of mRNA for the membrane form of IgE is a genetic mechanism to limit recruitment of IgE-secreting cells. Eur J Immunol 2006;36:1917–25. [DOI] [PubMed] [Google Scholar]
- 16.Mitre E, Norwood S, Nutman TB. Saturation of immunoglobulin E (IgE) binding sites by polyclonal IgE does not explain the protective effect of helminth infections against atopy. Infect Immun 2005;73:4106–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donohoe PJ, Heddle RJ, Sykes PJ, Fusco M, Flego LR, Zola H. IgE+ cells in the peripheral blood of atopic, nonatopic, and bee venom-hypersensitive individuals exhibit the phenotype of highly differentiated B cells. J Allergy Clin Immunol 1995;95:587–96. [DOI] [PubMed] [Google Scholar]
- 18.Stein LD, Chan MA, Hibi T, Dosch HM. Epstein-Barr virus-induced IgE production in limiting dilution cultures of normal human B cells. Eur J Immunol 1986;16:1167–70. [DOI] [PubMed] [Google Scholar]
- 19.Steinberger P, Bohle B, di Padova F, Wrann M, Liehl E, Scheiner O, et al. Allergen-specific IgE production of committed B cells from allergic patients in vitro. J Allergy Clin Immunol 1995;96:209–18. [DOI] [PubMed] [Google Scholar]
- 20.Bandura DR, Baranov VI, Ornatsky OI, Antonov A, Kinach R, Lou X, et al. Mass cytometry: technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Anal Chem 2009;81:6813–22. [DOI] [PubMed] [Google Scholar]
- 21.Simoni Y, Chng MHY, Li S, Fehlings M, Newell EW. Mass cytometry: a powerful tool for dissecting the immune landscape. Curr Opin Immunol 2018;51:187–96. [DOI] [PubMed] [Google Scholar]
- 22.Bendall SC, Nolan GP, Roederer M, Chattopadhyay PK. A deep profiler’s guide to cytometry. Trends Immunol 2012;33:323–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amir el AD, Davis KL, Tadmor MD, Simonds EF, Levine JH, Bendall SC, et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol 2013;31:545–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Diggins KE, Ferrell PB Jr., Irish JM Methods for discovery and characterization of cell subsets in high dimensional mass cytometry data. Methods 2015;82:55–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qiu P, Simonds EF, Bendall SC, Gibbs KD Jr., Bruggner RV, Linderman MD, et al. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat Biotechnol 2011;29:886–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aghaeepour N, Chattopadhyay P, Chikina M, Dhaene T, Van Gassen S, Kursa M, et al. A benchmark for evaluation of algorithms for identification of cellular correlates of clinical outcomes. Cytometry Part A 2016;89:16–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aghaeepour N, Chattopadhyay PK, Ganesan A, O’Neill K, Zare H, Jalali A, et al. Early immunologic correlates of HIV protection can be identified from computational analysis of complex multivariate T-cell flow cytometry assays. Bioinformatics 2012;28:1009–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Erwin EA, Custis NJ, Satinover SM, Perzanowski MS, Woodfolk JA, Crane J, et al. Quantitative measurement of IgE antibodies to purified allergens using streptavidin linked to a high-capacity solid phase. J Allergy Clin Immunol 2005;115:1029–35. [DOI] [PubMed] [Google Scholar]
- 29.Chung CH, Mirakhur B, Chan E, Le Q-T, Berlin J, Morse M, et al. Cetuximab-induced anaphylaxis and IgE specific for galactose-α−1,3-galactose. N Engl J Med 2008;358:1109–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fienberg HG, Simonds EF, Fantl WJ, Nolan GP, Bodenmiller B. A platinum-based covalent viability reagent for single-cell mass cytometry. Cytometry Part A 2012;81:467–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Finck R, Simonds EF, Jager A, Krishnaswamy S, Sachs K, Fantl W, et al. Normalization of mass cytometry data with bead standards. Cytometry Part A 2013;83:483–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maecker HT, McCoy JP, Nussenblatt R. Standardizing immunophenotyping for the Human Immunology Project. Nat Rev Immunol 2012;12:191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Orsini G, Legitimo A, Failli A, Massei F, Biver P, Consolini R. Enumeration of human peripheral blood dendritic cells throughout the life. Int Immunol 2012;24:347–56. [DOI] [PubMed] [Google Scholar]
- 34.Kimball AK, Oko LM, Bullock BL, Nemenoff RA, van Dyk LF, Clambey ET. A beginner’s guide to analyzing and visualizing mass cytometry data. J Immunol 2018;200:3–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lun ATL, Richard AC, Marioni JC. Testing for differential abundance in mass cytometry data. Nat Methods 2017;14:707–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou X, Lindsay H, Robinson MD. Robustly detecting differential expression in RNA sequencing data using observation weights. Nucleic Acids Res 2014;42:e91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wirths S, Lanzavecchia A. ABCB1 transporter discriminates human resting naive B cells from cycling transitional and memory B cells. Eur J Immunol 2005;35:3433–41. [DOI] [PubMed] [Google Scholar]
- 39.Wei C, Anolik J, Cappione A, Zheng B, Pugh-Bernard A, Brooks J, et al. A new population of cells lacking expression of CD27 represents a notable component of the B cell memory compartment in systemic lupus erythematosus. J Immunol 2007;178:6624–33. [DOI] [PubMed] [Google Scholar]
- 40.Heeringa JJ, Rijvers L, Arends NJ, Driessen GJ, Pasmans SG, van Dongen JJM, et al. IgE-expressing memory B cells and plasmablasts are increased in blood of children with asthma, food allergy, and atopic dermatitis. Allergy 2018;73:1331–6. [DOI] [PubMed] [Google Scholar]
- 41.Steinke JW, Platts-Mills TA, Commins SP. The alpha-gal story: lessons learned from connecting the dots. J Allergy Clin Immunol 2015;135:589–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klein U, Rajewsky K, Kuppers R. Human immunoglobulin (Ig)M+IgD+ peripheral blood B cells expressing the CD27 cell surface antigen carry somatically mutated variable region genes: CD27 as a general marker for somatically mutated (memory) B cells. J Exp Med 1998;188:1679–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Agematsu K, Nagumo H, Yang FC, Nakazawa T, Fukushima K, Ito S, et al. B cell subpopulations separated by CD27 and crucial collaboration of CD27+ B cells and helper T cells in immunoglobulin production. Eur J Immunol 1997;27:2073–9. [DOI] [PubMed] [Google Scholar]
- 44.Klein U, Kuppers R, Rajewsky K. Evidence for a large compartment of IgM-expressing memory B cells in humans. Blood 1997;89:1288–98. [PubMed] [Google Scholar]
- 45.Giesecke C, Frolich D, Reiter K, Mei HE, Wirries I, Kuhly R, et al. Tissue distribution and dependence of responsiveness of human antigen-specific memory B cells. J Immunol 2014;192:3091–100. [DOI] [PubMed] [Google Scholar]
- 46.Liu YJ, de Bouteiller O, Arpin C, Briere F, Galibert L, Ho S, et al. Normal human IgD+IgM- germinal center B cells can express up to 80 mutations in the variable region of their IgD transcripts. Immunity 1996;4:603–13. [DOI] [PubMed] [Google Scholar]
- 47.Seifert M, Steimle-Grauer SA, Goossens T, Hansmann ML, Brauninger A, Kuppers R. A model for the development of human IgD-only B cells: Genotypic analyses suggest their generation in superantigen driven immune responses. Mol Immunol 2009;46:630–9. [DOI] [PubMed] [Google Scholar]
- 48.Ehrhardt GR, Hsu JT, Gartland L, Leu CM, Zhang S, Davis RS, et al. Expression of the immunoregulatory molecule FcRH4 defines a distinctive tissue-based population of memory B cells. J Exp Med 2005;202:783–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moir S, Ho J, Malaspina A, Wang W, DiPoto AC, O’Shea MA, et al. Evidence for HIV-associated B cell exhaustion in a dysfunctional memory B cell compartment in HIV-infected viremic individuals. J Exp Med 2008;205:1797–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weiss GE, Crompton PD, Li S, Walsh LA, Moir S, Traore B, et al. Atypical memory B cells are greatly expanded in individuals living in a malaria-endemic area. J Immunol 2009;183:2176–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Uo M, Hisamatsu T, Miyoshi J, Kaito D, Yoneno K, Kitazume MT, et al. Mucosal CXCR4+ IgG plasma cells contribute to the pathogenesis of human ulcerative colitis through FcgammaR-mediated CD14 macrophage activation. Gut 2013;62:1734–44. [DOI] [PubMed] [Google Scholar]
- 52.Ito T, Carson WFt, Cavassani KA, Connett JM, Kunkel SL. CCR6 as a mediator of immunity in the lung and gut. Exp Cell Res 2011;317:613–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.