Abstract
Clinical, biochemical and molecular biology studies have identified lysosome-encapsulated cellular proteases as critical risk factors for cancer progression. Cathepsins represent a group of such proteases aimed at maintenance of cellular homeostasis. Nevertheless, recent reports suggest that Cathepsin B executes other cellular programs such as controlling tumor growth, migration, invasion, angiogenesis, and metastases development. In fact, elevated levels of Cathepsins are found under different pathological conditions including inflammation, infection, neurodegenerative disease, and cancer. Furthermore, the discovery of Cathepsin B secretion and function as an extracellular matrix protein has broadened our appreciation for the impact of Cathepsin B on cancer progression. Underneath a façade of an intracellular protease with limited therapeutic potential hides a central role of cathepsins in extracellular functions. Moreover, this role is incredibly diverse from one condition to the next – from driving caspase-dependent apoptosis to facilitating tumor neovascularization and metastasis. Here we discuss the role of Cathepsin B in the oncogenic process and perspective the use of Cathepsin B for diagnostic and therapeutic applications.
Keywords: Cathepsin B, tumor, migration, autophagy
1. Introduction
In the last decade, cancer has evolved as one of the leading causes of death worldwide. The ability of cancer cells to maintain an internal homeostasis correlates with tumor aggressiveness and represents an essential characteristic of a neoplasm. Multiple pieces of evidence highlight the importance of lysosomes in cellular homeostasis [1, 2] and in developing cellular reaction[3, 4]. The fundamental role of these membrane-bound organelles is the disposal and recycling of degraded macromolecules, along with digestion of alien structures that enter the cell via phagocytosis [5]. Nevertheless, several studies established that under conditions of cellular stress the lysosome is involved in cellular adaptation, nutrient sensing [6], drug resistance[7, 8], immune response[9] and cell death[10]. Lysosomes contain more than 60 hydrolytic enzymes which include proteases, lipases, hydrolases, nucleases, glycosidases, phospholipases, phosphatases, and sulfatases [11].
Tumor homeostasis is a multidimensional process that is regulated by cellular proteins, including cathepsin family of proteases, protein-protein interactions, alternative splicing[12] and expression of miRNAs. Among the proteases, Cathepsin B is of most interest due to its central role in pathological processes. Cathepsin B is a critical element of lysosome cascade. It is a cysteine protease that is involved in the regulation of metalloproteinases[13, 14], intracellular communications, autophagy induction, and immune resistance. Moreover, the role of Cathepsin B in cell survival and the mechanisms of its execution are vastly diverse from one condition to the next – from driving caspase-dependent apoptosis to facilitating tumor neovascularization and metastasis. Herein, we review recent studies which investigate the role of Cathepsin B in pathological processes with a focus on cancer.
2. Understanding the functions of Cathepsin B through the studies of its structure
Cathepsin B is a member of a cysteine protease family. It acts through 3 isoforms: main transcript, main transcript lacking exon 2 or main transcript lacking exon 2 and 3 (Table 1). In a common opinion, the cytosomal localization of Cathepsin B dictates its main functions such as the turnover of cellular proteins[15]. However, other functions may include regulation of angiogenesis[16, 17], invasion[16, 18], tumor proliferation[18] and immune resistance[19], neurogenesis[20] cellular differentiation[21, 22], tumor response to hypoxia [23, 24]. Furthermore, different Cathepsin B isoforms have differing subcellular localizations which may determine their distinct functions and independent regulation mechanisms[12, 25, 26].
Table 1:
Main isoforms of Cathepsin B and their function
| Wild type or splice variant | Tumor/health tissue | Function | Localization |
|---|---|---|---|
| Main transcript (13 exons), | -COS cells(green monkey adherent fibroblasts[1]), -human cultured articular chondrocytes and polyclonal T/C-28a2 chondrocyte cell line[2], -human rheumatoid synovial tissue[3]; -human colon adenocarcinoma ( tumor and mucosa[4]) -human breast adenocarcinoma[5] -human melanoma[5] |
Cellular metabolism[6] | Lysosome[8] |
| Main transcript (11 exons lacking exons 2+3), | Enzyme, Cell death[7], Eukaryotic translation[1] | Mitochondria[9] Cytoplasm[10] or nuclei[1] | |
| Main transcript(12 exons) lacking exon 2 | Extracellular space[8, 9] |
References:
S. Mehtani, Q. Gong, J. Panella, S. Subbiah, D.M. Peffley, A. Frankfater, In vivo expression of an alternatively spliced human tumor message that encodes a truncated form of cathepsin B. Subcellular distribution of the truncated enzyme in COS cells, J Biol Chem, 273 (1998) 13236–13244.
R. Zwicky, K. Muntener, M.B. Goldring, A. Baici, Cathepsin B expression and down-regulation by gene silencing and antisense DNA in human chondrocytes, Biochem J, 367 (2002) 209–217.
R. Lemaire, R.M. Flipo, H. Migaud, C. Fontaine, G. Huet, E. Dacquembronne, R. Lafyatis, Alternative splicing of the 5’ region of cathepsin B pre-messenger RNA in rheumatoid synovial tissue, Arthritis Rheum, 40 (1997) 1540–1542.
C. Hizel, M. Ferrara, H. Cure, D. Pezet, P. Dechelotte, J. Chipponi, P. Rio, Y.J. Bignon, D. Bernard-Gallon, Evaluation of the 5’ spliced form of human cathepsin B mRNA in colorectal mucosa and tumors, Oncol Rep, 5 (1998) 31–34.
Q. Gong, S.J. Chan, A.S. Bajkowski, D.F. Steiner, A. Frankfater, Characterization of the cathepsin B gene and multiple mRNAs in human tissues: evidence for alternative splicing of cathepsin B pre-mRNA, DNA Cell Biol, 12 (1993) 299–309.
S. Yan, B.F. Sloane, Molecular regulation of human cathepsin B: implication in pathologies, Biol Chem, 384 (2003) 845–854.
F. Bestvater, C. Dallner, E. Spiess, The C-terminal subunit of artificially truncated human cathepsin B mediates its nuclear targeting and contributes to cell viability, BMC Cell Biol, 6 (2005) 16.
A. Baici, K. Muntener, A. Willimann, R. Zwicky, Regulation of human cathepsin B by alternative mRNA splicing: homeostasis, fatal errors and cell death, Biol Chem, 387 (2006) 1017–1021.
E. Olivotto, R. Vitellozzi, P. Fernandez, E. Falcieri, M. Battistelli, S. Burattini, A. Facchini, F. Flamigni, S. Santi, A. Facchini, R.M. Borzi, Chondrocyte hypertrophy and apoptosis induced by GROalpha require three-dimensional interaction with the extracellular matrix and a co-receptor role of chondroitin sulfate and are associated with the mitochondrial splicing variant of cathepsin B, J Cell Physiol, 210 (2007) 417–427.
K. Muntener, R. Zwicky, G. Csucs, A. Baici, The alternative use of exons 2 and 3 in cathepsin B mRNA controls enzyme trafficking and triggers nuclear fragmentation in human cells, Histochem Cell Biol, 119 (2003) 93–101.
Musil et al. created a model of the three-dimensional structure of Cathepsin B and established that the proteasomal function of Cathepsins requires the presence of specific structural features of the enzyme. In this structure, more than a third of amino acid residues are identical to the construction of protein papain, except for the covalently closed circular region between Cys108 and Cys119 residues (“occluding loop”)[27]. Cathepsin B is first expressed as a 44kDa inactive precursor which then undergoes maturation to produce a 33 kDa lysosome enzyme later converted to a final active form composed of two (24 and 5 kDa) subunits. The earliest studies demonstrated cathepsins to be most active in acidic conditions, and also to be released in an active form at low pH of the pericellular environment [28, 29]. Other reports described the pH-dependent autoactivation of the zymogen pro-Cathepsin B, also showing that such pH-dependency could be alleviated with the introduction of glycosaminoglycans, explaining the wider window of optimal pH for Cathepsin B activity in vivo [30, 31]. Furthermore, the catalytic activity of Cathepsin B is dependent on the conformation of the “occluding loop”[32].
3. Cathepsin B in neurological disease: mechanism of action
It is thought that active Cathepsin B is a carboxypeptidase, cleaving dipeptides from the C-terminus of protein substrates [15]. Such activity of Cathepsin B may regulate the rate of cell proliferation [33]. In pathological states where neurogenesis is impaired, and the rate of cell proliferation is decreased, such as Alzheimer’s disease [34] and Huntington’s disease[35], Cathepsin B plays a protective role by degrading excessive amounts of misfolded protein inside the cell [26, 36]. In humans, the levels of Cathepsin B correlate with hippocampal-dependent memory functions and can be increased by physical exercise, while a Cathepsin B knock-down mice do not benefit from physical activity in terms of hippocampal neurogenesis and spatial memory [20]. On the other hand, the decrease in the rate of neurogenesis in AD can be secondary to the accumulation of the criticalAD proteins, which can be induced by inhibition of Cathepsin B and the consequent the lysosomal dysfunction [37].
It is also established that cells can secrete proteolytic enzymes as a means of execution of endocrine and nervous functions [38]. Specifically, the product of Cathepsin B transcript was found in the extracellular matrix (Table 1), suggesting enzyme release from active (proliferating) or passive (dead) cells, especially the cells growing under acidic pericellular conditions [11]. Extracellular release of lysosome-based enzyme Cathepsin B has been implicated in the breakdown of the connective tissue of the extracellular matrix (ECM) [39] and shedding integrins [18] and angiogenesis factors which reduce tumor progression[40]. On the other hand, experimental evidence has implicated Cathepsin B in apoptosis regulation. In fact, the mitochondria-based caspase 9 and caspase 3 activation after lysosomal destabilization and Cathepsin B release into the cytoplasm exemplify pro-apoptotic function of Cathepsin B [41]. The release of Cytochrome C (Cyt C) from mitochondria and its accumulation in the cytoplasm increases the affinity of procaspase effector Apaf-1 to ATP, which recruits procaspase-9 and initiates caspase 3 activation to induce apoptosis [42, 43]. Cathepsin B can also function to induce apoptosis independently of caspase activation [44]. Separating caspase-dependent and caspase-independent cell death made it difficult to rationalize the biological significance of Cathepsin B for therapy, especially taking into consideration the more recent publications. Specifically, Alhajala et al. reported that radioresistant pediatric glioma exhibit a high level of MMP12, MMP19 and Cathepsin B [45], conferring dependency of glioma stress response on cellular proteases. Moreover, data from our investigation[46] and study performed by Hsu et al. suggest that targeting artificial autophagy[47] with either autophagy inducer, autophagy inhibitor or their combination may argument the anti-glioma effect of oncolytic adenovirus and/or temozolomide. These therapeutic combinations represent an advantageous approach that aims to convert the aborted autophagy to apoptosis via mitochondria damage or cathepsin B release.
4. Interaction of Cathepsin B with cellular proteins: link to carcinogenesis
The expression of Cathepsin B is elevated in many, but not all, cancers. In a screen of 501 randomly collected thyroid cancer human specimens, high expression of Cathepsin B promoted patient survival (Log Rank p=5.76e-4) (www.proteinatlas.org). Furthermore, in glioblastoma patients, high expression of Cathepsin B negatively correlated with the stage of the tumor (TCGA and Rembrandt Dataset). Conversely, in 406 patients with urothelial cancer, high expression of Cathepsin B negatively impacted patient survival (Log Rank, p=9.2e-4).
Khan et al. demonstrated a negative correlation of Cathepsin B expression and laminin (ECM protein) in gastric[48] and colorectal carcinoma[49], suggesting the involvement of Cathepsin B in the remodeling of ECM. Examinations of the regulation of Cathepsin B by matrix proteins found that collagen I, through its interaction with α1β1 and α2β1 integrins, stimulated secretion of proCathepsin B by human breast fibroblasts. It was suggested that the effect may be executed at the post-transcriptional level because no change in mRNA level was found. It was also suggested that interaction of the fibroblasts with collagen I could increase translation or stabilize proCathepsin B protein [50].
Skeletal muscle differentiation was also shown to be linked to the levels of expression and excretion of Cathepsin B[51]. Small et al. demonstrated that when smooth muscle cells shift into a nonproliferative (contractile) state after termination of vascular reconstruction, expression of complement C1s, Cathepsin B, and cellular repressor of E1A-activated genes increased, as well as expression of Wilms’ tumor 1-associating protein [52]. The forementioned evidence suggests the role of Cathepsin B in cellular differentiation that may have implications in cancer progression.
The signaling cascades upstream of Cathepsin B were also worked out by previous studies. Glogowska et al. demonstrated that CTRP8 (C1q-tumor necrosis factor-related protein 8) and RLN2 (relaxin isoform) hormone induce the production and secretion of Cathepsin B in glioblastoma cells, resulting in laminin degradation [53] and glioblastoma dissemination. Notably, the same group demonstrated the role of EGFcyt (Epidermal Growth Factor cytoplasmic domain) as an inducer of lysosomal procathB expression [54]. Recent mechanistic studies revealed a few novel regulators of Cathepsin B that facilitate either the pro-oncogenic function of Cathepsin B or its pro-apoptotic function. One of the regulators is metastasis-associated protein (MTA1) which inhibits Cathepsin B expression and is negatively correlated with E-Cadherin, critical for bone metastases progression [55]. Two other reports reveal that TNF-alpha enables the Bid-dependent lysosomal permeabilization, followed by the release of Cathepsin B into the cytosol, which facilitates mitochondrial cytochrome c release and apoptosis[56].
The role of Cathepsin B-induced signaling in the promotion of tumorigenicity was also extensively studied. In fact, Yanamandra et al. and later Gupta et al. discovered that upregulation of Cathepsin B promotes angiogenesis via induction of VEGF-C and MMP-9 [17, 57]. On the contrary, in glioma, downregulation of uPAR in combination with Cathepsin B inhibits CD151 and α3β1-integrin-mediated adhesion and invasion [58]. Later, Malla et al. suggest that downregulation of uPAR and Cathepsin B in glioma facilitates apoptosis through increased translocation of calcineurin from mitochondria to cytosol, decreased phosphorylation of BAD and increased interaction of BAD with Bcl-2 [59]. It is not surprising that Cathepsin B and uPAR downregulation work synergistically as anticarcinogenic mechanisms. This combination reduces p-ERK and c-Myc, which increases levels of E2F1 and FOXO3a, upregulating p27 expression in glioma cells [60]. The discovery of the interaction between Cathepsin B, MAPK, BAD and ERK[61] helped clarify the downstream targets of Cathepsin B and opened a new venue to design pharmacological combinatorial approaches to target Cathepsin B.
The feedback loop between Cathepsin B, MMP9 (metalloproteinase 9) and VEGF (vascular endothelial growth factor) highlight the role of the CTSB gene in tumor angiogenesis. On the one hand, expression of proangiogenic VEGF-A promotes the production of cathepsins including type B[62] by glioma cells to digest basal membrane[63] and stimulate endothelial cells to release the matrix-degrading enzymes. This enables glioma cells to form capillary sprouts via MMP9-regulated migration and proliferation. On the other hand, in neuroendocrine mouse tumors, levels of Cathepsin B and MMP9 are negatively correlated [64]. An earlier study on human glioblastoma cells showed that simultaneous inhibition of Cathepsin B and MMP-9 via RNA interference reduced tumor invasion, growth, and angiogenesis [16]. Similar results were produced when MMP-9, uPAR and Cathepsin B were inhibited in prostate cancer cells [65]. Ponnala et al. showed that silencing MMP-9 in vivo in combination with either uPAR or Cathepsin B resulted in suppression of aerobic glycolysis in glioma cells and switch to oxidative phosphorylation (OXPHOS) through inhibition of Akt, ultimately leading to the production of ROS and accumulation of cytochrome C in the cytosol [66]. Although, as evidence from recent publication suggests that OXPHOS is mostly intact in cancer cells [67], the formation of OXPHOS complex may preclude metabolic transformation of the cells from oxidative to glycolytic metabolism[68]. It was shown that fibronectin degradation by Cathepsin B allows the tumor cells to invade into the blood and lymphatic vessels of bladder carcinoma [69]. A similar conclusion may be drawn from the suppression of angiogenesis in glioblastomas lacking Cathepsin B. Inhibition of Cathepsin B via RNA interference reduces VEGF release from cancer cells which prevent the development of microvessels [17]. Mai et al. and then Kong et al. suggested that Cathepsin B degradation of tenascin-C surrounding neovessels could facilitate neovascular extension resulting in the progression of gliomas[70, 71]. These findings present downregulation of Cathepsin B as a useful approach to target tumor neovascularization.
5. Role of Cathepsin B modulation in anti-cancer therapy
Stress stimulated secretion of Cathepsin B from the lysosomes, and its consequent cytoplasmic localization suggest a chain of events which may lead to toxicity. Time-dependent production of reactive oxygen species compromises the lysosomal integrity and is required for Cathepsin B and L activation and release [72]. Taking into consideration that the ROS may initiate cytoprotective and cytotoxic autophagy, it is reasonable to expect the involvement of Cathepsin B in both types of reaction depending on the stimulus. The main question is how to promote the Cathepsin-dependent cytotoxic autophagy and prevent the cytoprotective autophagy in cancer cells. For instance, Zhang et al. were able to inhibit the growth of non-small cell lung cancer in vivo via treatment with CA-5f (autophagosome-lysosome fusion inhibitor). Such treatment leads to an increase in ROS production, apoptosis, but no changes in the levels of Cathepsin B and D [73]. Another set of studies implicate Cathepsin B in the mechanisms of execution of cytotoxic effects of multiple anti-cancer drugs. Cathepsin B completely or partially prevents toxicity of drugs such as HDAC [74], ERBB1/2/4 inhibitor neratinib [75], nilotinib [76], acid ceramidasa [77], Thymoquinone [78], Tyrosine kinase inhibitors (TKI) such as sunitinib, and pazopanib[79]. Han et al. showed that SAHA promotes cytotoxic autophagy via activation of Cathepsin B. Interestingly, a block of breast cancer cells treated with SAHA in the presence of siRNA against Cathepsin B or Cystatin C reduces apoptosis and promotes cell viability, also suggesting an intrinsic role of Cathepsin B in the SAHA-induced cytotoxicity[80] and establishing a link between Cathepsin B expression and cell survival. Mechanistically, it can be explained that increasing of lysosome membrane permeability and release of Cathepsin B may trigger late stages of autophagy which can be critical for drug toxicity. Regardless, all those possibilities need to be further investigated.
On the other hand, autophagy is an essential cytoprotective system that is rapidly activated in response to various stimuli. In fact, Hong et al. revealed an increase in autophagy-related proteins Cathepsin B along with (ATG) 3, ATG7 and Rab7 during hepatic ischemia and perfusion [81]. In the same study, the necroptosis inhibitor Nec-1 attenuated those changes suggesting that Cathepsin B may be implicated in necroptotic cell death. Furthermore, Obatoclax (Bcl-2 family inhibitor) toxicity in oesophageal cancer is mediated by blockage of autophagic flux, evidenced by the concomitant accumulation of LC3-II and p62, and downregulation of lysosomal Cathepsins B, D, and L[82]. In the same study, Cathepsin knockdown induced cytotoxicity, suggesting compromised lysosomal function as a mechanism mediating the effect of Obatoclax on the progression of oesophageal cancer [82]. It is also significant to assess whether cytoprotective autophagy requires expression of Tumor Necrosis Factor Receptor-Associated Protein-1 (TRAP1), a homolog of mitochondrial-specific HSP90 [83]. This study observed decreased viability of NSCLC cells upon inhibition of autophagy, but the stimulation of autophagy above the endogenous levels had a protective effect only in TRAP-1 depleted cells.
Although functional test data suggests a prosurvival function for Cathepsin B, it remains unknown how and when cytoprotective autophagy become cytotoxic and what is the molecular mechanism of that transformation. For instance, inhibition of PERK was shown to decrease the autophagic degradation of eLF2α and promoted cell survival. Cathepsin B inhibitor CA074 also prevented IF2alpha from degradation, suggesting that Cathepsin B-mediated cytotoxic autophagy is PERK-dependent [84]. It is also unclear how malignant cells escape the cytotoxic autophagy. A recent report by Ning et al. identified a decrease in the levels of Cathepsin B along with other autophagic proteins in PTEN-knockdown Trastuzumab-resistant breast cancer cells [85].
Another possibility for Cathepsin B to regulate autophagy is an opportunity to collaborate with other Cathepsin molecules of a different type. We speculate that each type of cells death is progressing through the involvement of several Cathepsins which together act as cytoprotective or cytotoxic triggers inside cells. For example, Liu et al. noted that the release of Cathepsin B and apoptosis induction occurs in the presence of Cathepsin D [86]. Multiple reports demonstrate that release of Cathepsin B alone in combination with an increase in expression of Cathepsin D [87, 88] and Cathepsin L [89] sensitize cancer cells to chemotherapeutic drugs via caspase-dependent and caspase-independent cell death.
6. Cathepsin B is a target for therapy and diagnostics
An increasing number of studies highlight the role of autophagy and autophagy-related proteins in a broad range of physiological and pathological processes. The investigations of the molecular mechanisms of Cathepsin B regulation have attracted a lot of attention since this protein plays a pivotal role in autophagy-related events. Nowadays, Cathepsin B has become a cornerstone of novel therapeutic strategies.
Tumor progression is a sequence of choreographed actions of transcriptional regulators, heavily influenced by cellular stress. Apart from transcriptional regulators, the activity of Cathepsin B is also regulated by endogenous inhibitors. Currently, four different inhibitors have been identified in the cystatin superfamily – stefins, kininogens, thyropins, and serpins[90]. Their function is to oversee the processes of biosynthesis and trafficking of Cathepsin B to lysosomes, as well as (auto-)proteolytic cleavage of pro-peptides (Fig. 1). Despite the potential known effect of the inhibitors on Cathepsin B, several studies reported on the metastasis suppressor function of the Cathepsin B inhibitors [91] in breast cancer [92], colorectal cancer [93], pancreatic ductal adenocarcinoma [94], etc. Despite the clear anti-tumor effect, each inhibitor has different efficacy against Cathepsin B.
Figure 1.
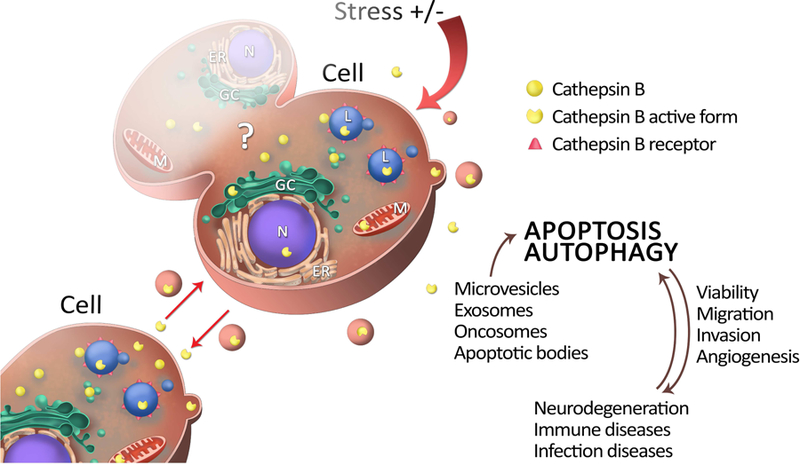
Figure 1. Lifecycle of cathepsin B. Inactive forms of enzymes, after post-translation modifications at endoplasmatic reticulum (ER) are, via Golgi complex (GC), are directed to lysosomes (L), cytosol or extracellular matrix. With location there function may differ, but main role stays to insure homeostasis and survival of the cell. Homeostasis may be corrupt due to stress cell was exposed to (infections, inflammations, chronic diseases, etc.) To insure their goal, they are living lysosomes within macrovesicles, exosomes or oncosomes. All of these cathepsin B also can be delivered by the messengers of intercellular communication -extracellular vesicles vesicles have purpose to provide optimal conditions for proteolytic activity of cathepsin B (e.g. pH) induce migration, invasion and if autophagy doesn`t succeed, cathepsin B activate apoptosis, trying to control damage.
In recent years, the efforts of pharmacological research have been focused on targeting Cathepsin B specifically. For instance, the chemically designed Cathepsin B inhibitor ankyrin repeat protein DARP demonstrated more significant benefits for anti-cancer therapy and diagnostics. As shown by Kramer et al., application of DARP in 8h6 blocked Cathepsin B activity ex vivo and was useful to monitor tumor-associated protein inhibition using non-invasive in vivo imaging [95]. Another report demonstrated that inhibition of vacuolar H+-ATPases (V-ATPases) with concanamycin A decreased Cathepsin B activity in cell lysates of metastatic breast cancer cells [96]. Liow et al. and Chow et al. recently found that Cathepsin B inhibitor benzyloxycarbonyl-phenylalanine-alanine-chloromethyl ketone (z-FA-CMK) induces apoptosis at low concentrations and necrosis at higher concentrations in leukemic T cells via oxidative stress [97]. Most recently, Tang et al. reported a new autophagy inhibitor cepharanthine (CEP) with an anti-cancer effect on non-small lung cancer cells mediated by dacomitinib (DAC) via blockage of autophagosome-lysosome fusion and inhibition of lysosomal Cathepsin B and D maturation [98]. The full list of Cathepsin B inhibitor is presented at Tab.1.
An increased level of Cathepsin B is already present in the lysosome, but it is nevertheless rapidly induced under various stimuli and leads to the production of prostaglandins, with subsequent inflammation via ATG7-dependent mechanism. Thus, it is believed that targeting of Cathepsin B via RNA interference may show a greater impact on Cathepsin B activity than chemical inhibitors due to lack of specificity. In fact, RNA interference oligonucleotide shRNA-CTSB#2 showed the most efficient inhibition of Cathepsin B at both mRNA and protein levels and resulted in suppressing endometrial cancer growth and development in vivo [33].
Another approach which experimentally provided benefits for targeting lysosome is the application of cationic liposomes. Cationic liposomes induce lysosome membrane permeabilization and inhibit late-stage autophagic flux that results in the cytoplasmic release of Cathepsin B, mitochondrial dysfunction and production of reactive oxygen species followed by cell necrosis [99].
7. Conclusions
In the last few decades, we are witnessing an exceptional stride in deciphering the perplexing biology of cathepsins in the normal and pathological environment. Owing to new techniques in genomics and proteomics, our knowledge of new functions and new substrates for each member of this group of proteins grows progressively [100]. Development of bioinformatics allows us to explore possible interactions between Cathepsin B and potential inhibitors in silico [101]. Non-invasive in vivo imaging of cathepsins has potential in becoming a novel standard in diagnostics [95]. Despite the hope of using a chemical inhibitor of Cathepsin B, the possible application of designed drugs in clinics may be limited. First, we believe that the specificity of chemical inhibitors to Cathepsin B is a severe constraint for therapeutic use. In fact, the high expression of Cathepsin B in normal cells, as well as the promiscuous expression of Cathepsin B in tumor cells raise questions related to safety and specificity. Secondly, due to the multifaceted regulation of Cathepsin B signaling, chemical inhibition may stimulate alternative routes to restore the Cathepsin B levels (Tab. 2). That possibility raises concerns about modifications in the resistant cancer cell populations, leading to the formation of cells with unknown feedback loops or activation of other cathepsins. Nevertheless, a better understanding of the cathepsins biology of humans will empower new generations of scientists to find the best treatments to a broad spectrum of diseases involving cathepsins.
Table 2.
Cathepsin B inhibitors and their appropriate mechanism of action available to inhibit cathepsin B
| Inhibitor | Chemistry | Mechanism | Source | Reference |
|---|---|---|---|---|
| Z-Phe-AlaCH2F | fluoromethyl ketone 3-(N-benzyloxycarbonylphenylalanylamido)-DL-1-fluoro-2-butanone | Irreversible covalent inhibitor | synthesized | Rasnick D. Synthesis of peptide fluoromethyl ketones and the inhibition of human Cathepsin B. Anal Biochem. 1985 Sep;149(2):461–5. |
| E-64 | C15N5H2705 | Irreversible covalent inhibitor | Aspergillus japonicus TPR-64 | Hanada K, Tamai M, Yamagishi M, Ohmura S, Sawada J, Tanaka I. Isolation and characterization of E-64, a new thiol protease inhibitor. Agric Biol Chem. 1978;42:523–528. |
| E64d | EST, Loxistatin, ethyl (2S,3S)-3-[[(2S)-4-methyl-1-(3-methylbutylamino)-1-oxopentan-2-yl]carbamoyl]oxirane-2-carboxylate | Irreversible covalent inhibitor | synthesized | Hashida S, Towatari T, Kominami E, Katunuma N. Inhibitions by E-64 derivatives of rat liver Cathepsin B and Cathepsin L in vitro and in vivo. J Biochem. 1980 Dec;88(6):1805–11. |
| iodo and diiodotyrosine E-64-c analogs | Iodo and diiodotyrosine epoxysuccinyl derivatives | Irreversible covalent inhibitor | synthesized | Giordano C, Calabretta R, Gallina C, Consalvi V, Scandurra R, Noya FC. Iodo and diiodotyrosine epoxysuccinyl derivatives as selective inhibitors of Cathepsin B. Eur J Med Chem. 1993;28:917–926. |
| CA-074 and its derivates ( CA-074 Me) | [L-3-trans-(propylcarbamoyl)oxirane-2- carbonyl]-L-isoleucyl-L-proline | Irreversible covalent inhibitor | synthesized | Montaser M, Lalmanach G, Mach L. CA-074, but not its methyl ester CA-074Me, is a selective inhibitor of Cathepsin B within living cells. Biol Chem. 2002 Jul-Aug;383(7–8):1305–8. |
| Miraziridine A | aziridinylpeptide | Irreversible covalent inhibitor | Theonella mirabilis Theonella swinhoei | Nakao Y, Fujita M, Warabi K, Matsunaga S, Fusetani N. Miraziridine A a novel cysteine protease inhibitor from the marine sponge theonella aff. mirabilis. J Am Chem Soc. 2000;122:10462– 10463. |
| Bz-Phe-Arg-CH2F | peptide derivative of arginylfluoromethane | Irreversible covalent inhibitor | synthesized | Angliker H, Wikström P, Rauber P, Stone S, Shaw E. Synthesis and properties of peptidyl derivatives of arginylfluoromethanes. Biochem J. 1988 Dec 1;256(2):481–6. |
| diazomethyl ketones | Benzyloxycarbonyl-phenylalanyl diazomethyl ketone and benzyloxy-carbonyl-phenylalanyl-phenylalanyl diazomethyl ketone | Irreversible covalent inhibitor | synthesized | Leary R, Shaw E. Inactivation of Cathepsin B1 by diazomethyl ketones. Biochem Biophys Res Commun. 1977 Dec 7;79(3):926–31. |
| peptidyl (acyloxy)methanes | peptidyl (acyloxy)methanes | Irreversible covalent inhibitor | synthesized | Krantz A. Peptidyl (acyloxy)methanes as quiescent affinity labels for cysteine proteinases. Methods Enzymol. 1994;244:656–71. |
| 1,2,4-thiadiazole derivate | thiadiazoles 3a–h | Irreversible covalent inhibitor | synthesized | Leung-Toung R, Wodzinska J, Li W, Lowrie J, Kukreja R, Desilets D, Karimian K, Tam TF. 1,2,4-thiadiazole: a novel Cathepsin B inhibitor. Bioorg Med Chem. 2003 Dec 1;11(24):5529–37. |
| Calpain inhibitor II | N-Acetyl-Leu-Leu-Methional | Reversible inhibitor | synthesized | Sasaki T, Kishi M, Saito M, Tanaka T, Higuchi N, Kominami E, Katunuma N, Murachi T. Inhibitory effect of di- and tripeptidyl aldehydes on calpains and Cathepsins. J Enzyme Inhib. 1990;3(3):195–201. |
| Peptidyl cyclopropenone 9 | Peptidyl cyclopropenone 9 (the S isomer) | Reversible inhibitor | synthesized | Cohen M, Bretler U, Albeck A. Peptidyl cyclopropenones: reversible inhibitors, irreversible inhibitors, or substrates of cysteine proteases? Protein Sci. 2013 Jun;22(6):788–99. |
| shRNA-CTSB#2 | oligonucleotide | RNA interference inhibitor | synthesized | Bao W, Fan Q, Luo X, Cheng WW, Wang YD, Li ZN, Chen XL, Wu D. Silencing of Cathepsin B suppresses the proliferation and invasion of endometrial cancer. Oncol Rep. 2013 Aug;30(2):723–30. |
| DARPin 8h6 | ankyrin repeat protein | antibody-drug conjugate like inhibitor | synthesized | Kramer L, Renko M, Završnik J, Turk D, Seeger MA, Vasiljeva O, Grütter MG, Turk V, Turk B. Non-invasive in vivo imaging of tumour-associated Cathepsin B by a highly selective inhibitory DARPin. Theranostics. 2017 Jul 8;7(11):2806–21. |
| Concanamycin A | designated folimycin | Indirect inhibitor (by specific inhibiton of V-ATPase) | synthesized | Uhlman A, Folkers K, Liston J, Pancholi H, Hinton A. Effects of Vacuolar H(+)-ATPase Inhibition on Activation of Cathepsin B and Cathepsin L Secreted from MDA-MB231 Breast Cancer Cells. Cancer Microenviron. 2017 Dec;10(1–3):49–56. |
Highlights:
Cathepsins represent a group of cysteine proteases
Cathepsins maintain homeostais in normal and pathological states of the cell
Cathepsin B role in anticancer therapy is diverse
Modulation of Cathepsin B regulates autophagy
Acknowledgments
We are thankful for Dr. Jason Miska (Northwestern University at Chicago) for his critical comments.
Funding
This research study was supported by the Russian academic excellence project “5–100”.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: Authors declare no conflict of interest
References
- [1].Li X, Qian X, Lu Z, Local histone acetylation by ACSS2 promotes gene transcription for lysosomal biogenesis and autophagy, Autophagy, 13 (2017) 1790–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Jensen SS, Aaberg-Jessen C, Christensen KG, Kristensen B, Expression of the lysosomal-associated membrane protein-1 (LAMP-1) in astrocytomas, Int J Clin Exp Pathol, 6 (2013) 1294–1305. [PMC free article] [PubMed] [Google Scholar]
- [3].Remy J, Linder B, Weirauch U, Konovalova J, Marschalek R, Aigner A, Kogel D, Inhibition of PIM1 blocks the autophagic flux to sensitize glioblastoma cells to ABT-737-induced apoptosis, Biochim Biophys Acta Mol Cell Res, 1866 (2019) 175–189. [DOI] [PubMed] [Google Scholar]
- [4].Mbah NE, Overmeyer JH, Maltese WA, Disruption of endolysosomal trafficking pathways in glioma cells by methuosis-inducing indole-based chalcones, Cell Biol Toxicol, 33 (2017) 263–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Johansson AC, Appelqvist H, Nilsson C, Kagedal K, Roberg K, Ollinger K, Regulation of apoptosis-associated lysosomal membrane permeabilization, Apoptosis : an international journal on programmed cell death, 15 (2010) 527–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Settembre C, Ballabio A, Lysosome: regulator of lipid degradation pathways, Trends Cell Biol, 24 (2014) 743–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Zhitomirsky B, Assaraf YG, Lysosomal sequestration of hydrophobic weak base chemotherapeutics triggers lysosomal biogenesis and lysosome-dependent cancer multidrug resistance, Oncotarget, 6 (2015) 1143–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zhitomirsky B, Assaraf YG, Lysosomes as mediators of drug resistance in cancer, Drug Resist Updat, 24 (2016) 23–33. [DOI] [PubMed] [Google Scholar]
- [9].Pan H, Chen L, Xu Y, Han W, Lou F, Fei W, Liu S, Jing Z, Sui X, Autophagy-associated immune responses and cancer immunotherapy, Oncotarget, 7 (2016) 21235–21246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Aits S, Jaattela M, Lysosomal cell death at a glance, J Cell Sci, 126 (2013) 1905–1912. [DOI] [PubMed] [Google Scholar]
- [11].Lim CY, Zoncu R, The lysosome as a command-and-control center for cellular metabolism, The Journal of cell biology, 214 (2016) 653–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Bestvater F, Dallner C, Spiess E, The C-terminal subunit of artificially truncated human cathepsin B mediates its nuclear targeting and contributes to cell viability, BMC Cell Biol, 6 (2005) 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Sigloch FC, Knopf JD, Weisser J, Gomez-Auli A, Biniossek ML, Petrera A, Schilling O, Proteomic analysis of silenced cathepsin B expression suggests non-proteolytic cathepsin B functionality, Biochim Biophys Acta, 1863 (2016) 2700–2709. [DOI] [PubMed] [Google Scholar]
- [14].Formolo CA, Williams R, Gordish-Dressman H, MacDonald TJ, Lee NH, Hathout Y, Secretome signature of invasive glioblastoma multiforme, J Proteome Res, 10 (2011) 3149–3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Cavallo-Medved D, Moin K, Sloane B, Cathepsin B: Basis Sequence: Mouse, AFCS Nat Mol Pages, 2011 (2011). [PMC free article] [PubMed] [Google Scholar]
- [16].Lakka SS, Gondi CS, Yanamandra N, Olivero WC, Dinh DH, Gujrati M, Rao JS, Inhibition of cathepsin B and MMP-9 gene expression in glioblastoma cell line via RNA interference reduces tumor cell invasion, tumor growth and angiogenesis, Oncogene, 23 (2004) 4681–4689. [DOI] [PubMed] [Google Scholar]
- [17].Yanamandra N, Gumidyala KV, Waldron KG, Gujrati M, Olivero WC, Dinh DH, Rao JS, Mohanam S, Blockade of cathepsin B expression in human glioblastoma cells is associated with suppression of angiogenesis, Oncogene, 23 (2004) 2224–2230. [DOI] [PubMed] [Google Scholar]
- [18].Veeravalli KK, Chetty C, Ponnala S, Gondi CS, Lakka SS, Fassett D, Klopfenstein JD, Dinh DH, Gujrati M, Rao JS, MMP-9, uPAR and cathepsin B silencing downregulate integrins in human glioma xenograft cells in vitro and in vivo in nude mice, PLoS One, 5 (2010) e11583. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [19].Fais S, Cannibalism: a way to feed on metastatic tumors, Cancer letters, 258 (2007) 155–164. [DOI] [PubMed] [Google Scholar]
- [20].Moon HY, Becke A, Berron D, Becker B, Sah N, Benoni G, Janke E, Lubejko ST, Greig NH, Mattison JA, Duzel E, van Praag H, Running-Induced Systemic Cathepsin B Secretion Is Associated with Memory Function, Cell Metab, 24 (2016) 332–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Zwicky R, Muntener K, Csucs G, Goldring MB, Baici A, Exploring the role of 5’ alternative splicing and of the 3’-untranslated region of cathepsin B mRNA, Biol Chem, 384 (2003) 1007–1018. [DOI] [PubMed] [Google Scholar]
- [22].Muntener K, Zwicky R, Csucs G, Baici A, The alternative use of exons 2 and 3 in cathepsin B mRNA controls enzyme trafficking and triggers nuclear fragmentation in human cells, Histochem Cell Biol, 119 (2003) 93–101. [DOI] [PubMed] [Google Scholar]
- [23].Xiaofei C, Yanqing L, Dongkai Z, Dong C, Feng Z, Weilin W, Identification of cathepsin B as a novel target of hypoxia-inducible factor-1-alpha in HepG2 cells, Biochem Biophys Res Commun, 503 (2018) 1057–1062. [DOI] [PubMed] [Google Scholar]
- [24].Wickramasinghe NS, Banerjee K, Nagaraj NS, Vigneswaran N, Zacharias W, Hypoxia alters cathepsin B / inhibitor profiles in oral carcinoma cell lines, Anticancer Res, 25 (2005) 2841–2849. [PubMed] [Google Scholar]
- [25].Yan S, Sloane BF, Molecular regulation of human cathepsin B: implication in pathologies, Biol Chem, 384 (2003) 845–854. [DOI] [PubMed] [Google Scholar]
- [26].Baici A, Muntener K, Willimann A, Zwicky R, Regulation of human cathepsin B by alternative mRNA splicing: homeostasis, fatal errors and cell death, Biol Chem, 387 (2006) 1017–1021. [DOI] [PubMed] [Google Scholar]
- [27].Musil D, Zucic D, Turk D, Engh RA, Mayr I, Huber R, Popovic T, Turk V, Towatari T, Katunuma N, et al. , The refined 2.15 A X-ray crystal structure of human liver cathepsin B: the structural basis for its specificity, The EMBO journal, 10 (1991) 2321–2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Rozhin J, Sameni M, Ziegler G, Sloane BF, Pericellular pH affects distribution and secretion of cathepsin B in malignant cells, Cancer Res, 54 (1994) 6517–6525. [PubMed] [Google Scholar]
- [29].Maciewicz RA, Wotton SF, Etherington DJ, Duance VC, Susceptibility of the cartilage collagens types II, IX and XI to degradation by the cysteine proteinases, cathepsins B and L, FEBS Lett, 269 (1990) 189–193. [DOI] [PubMed] [Google Scholar]
- [30].Rozman J, Stojan J, Kuhelj R, Turk V, Turk B, Autocatalytic processing of recombinant human procathepsin B is a bimolecular process, FEBS Lett, 459 (1999) 358–362. [DOI] [PubMed] [Google Scholar]
- [31].Pungercar JR, Caglic D, Sajid M, Dolinar M, Vasiljeva O, Pozgan U, Turk D, Bogyo M, Turk V, Turk B, Autocatalytic processing of procathepsin B is triggered by proenzyme activity, FEBS J, 276 (2009) 660–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Nagler DK, Storer AC, Portaro FC, Carmona E, Juliano L, Menard R, Major increase in endopeptidase activity of human cathepsin B upon removal of occluding loop contacts, Biochemistry, 36 (1997) 12608–12615. [DOI] [PubMed] [Google Scholar]
- [33].Bao W, Fan Q, Luo X, Cheng WW, Wang YD, Li ZN, Chen XL, Wu D, Silencing of Cathepsin B suppresses the proliferation and invasion of endometrial cancer, Oncol Rep, 30 (2013) 723–730. [DOI] [PubMed] [Google Scholar]
- [34].Rodriguez JJ, Jones VC, Verkhratsky A, Impaired cell proliferation in the subventricular zone in an Alzheimer’s disease model, Neuroreport, 20 (2009) 907–912. [DOI] [PubMed] [Google Scholar]
- [35].Ernst A, Alkass K, Bernard S, Salehpour M, Perl S, Tisdale J, Possnert G, Druid H, Frisen J, Neurogenesis in the striatum of the adult human brain, Cell, 156 (2014) 1072–1083. [DOI] [PubMed] [Google Scholar]
- [36].Liang Q, Ouyang X, Schneider L, Zhang J, Reduction of mutant huntingtin accumulation and toxicity by lysosomal cathepsins D and B in neurons, Molecular neurodegeneration, 6 (2011) 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Cermak S, Kosicek M, Mladenovic-Djordjevic A, Smiljanic K, Kanazir S, Hecimovic S, Loss of Cathepsin B and L Leads to Lysosomal Dysfunction, NPC-Like Cholesterol Sequestration and Accumulation of the Key Alzheimer’s Proteins, PLoS One, 11 (2016) e0167428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Murthy SRK, Dupart E, Al-Sweel N, Chen A, Cawley NX, Loh YP, Carboxypeptidase E promotes cancer cell survival, but inhibits migration and invasion, Cancer Lett, 341 (2013) 204–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Wyczalkowska-Tomasik A, Paczek L, Cathepsin B and L activity in the serum during the human aging process: cathepsin B and L in aging, Arch Gerontol Geriatr, 55 (2012) 735–738. [DOI] [PubMed] [Google Scholar]
- [40].Lakka SS, Gondi CS, Rao JS, Proteases and glioma angiogenesis, Brain Pathol, 15 (2005) 327–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Joy B, Sivadasan R, Abraham TE, John M, Sobhan PK, Seervi M, R.S. T, Lysosomal destabilization and cathepsin B contributes for cytochrome c release and caspase activation in embelin-induced apoptosis, Molecular carcinogenesis, 49 (2010) 324–336. [DOI] [PubMed] [Google Scholar]
- [42].Rodriguez J, Lazebnik Y, Caspase-9 and APAF-1 form an active holoenzyme, Genes Dev, 13 (1999) 3179–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Liu X, Kim CN, Yang J, Jemmerson R, Wang X, Induction of apoptotic program in cell-free extracts: requirement for dATP and cytochrome c, Cell, 86 (1996) 147–157. [DOI] [PubMed] [Google Scholar]
- [44].Gondi CS, Rao JS, Cathepsin B as a cancer target, Expert Opin Ther Targets, 17 (2013) 281–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Alhajala HS, Nguyen HS, Shabani S, Best B, Kaushal M, Al-Gizawiy MM, Erin Ahn EY, Knipstein JA, Mirza S, Schmainda KM, Chitambar CR, Doan NB, Irradiation of pediatric glioblastoma cells promotes radioresistance and enhances glioma malignancy via genome-wide transcriptome changes, Oncotarget, 9 (2018) 34122–34131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Kaverina NV, Kadagidze ZG, Borovjagin AV, Karseladze AI, Kim CK, Lesniak MS, Miska J, Zhang P, Baryshnikova MA, Xiao T, Ornelles D, Cobbs C, Khramtsov A, Ulasov IV, Tamoxifen overrides autophagy inhibition in Beclin-1-deficient glioma cells and their resistance to adenovirus-mediated oncolysis via upregulation of PUMA and BAX, Oncogene, (2018). [DOI] [PubMed] [Google Scholar]
- [47].Ulasov IV, Lenz G, Lesniak MS, Autophagy in glioma cells: An identity crisis with a clinical perspective, Cancer Lett, 428 (2018) 139–146. [DOI] [PubMed] [Google Scholar]
- [48].Khan A, Krishna M, Baker SP, Banner BF, Cathepsin B and tumor-associated laminin expression in the progression of colorectal adenoma to carcinoma, Mod Pathol, 11 (1998) 704–708. [PubMed] [Google Scholar]
- [49].Khan A, Krishna M, Baker SP, Malhothra R, Banner BF, Cathepsin B expression and its correlation with tumor-associated laminin and tumor progression in gastric cancer, Arch Pathol Lab Med, 122 (1998) 172–177. [PubMed] [Google Scholar]
- [50].Koblinski JE, Dosescu J, Sameni M, Moin K, Clark K, Sloane BF, Interaction of human breast fibroblasts with collagen I increases secretion of procathepsin B, J Biol Chem, 277 (2002) 32220–32227. [DOI] [PubMed] [Google Scholar]
- [51].Jane DT, DaSilva L, Koblinski J, Horwitz M, Sloane BF, Dufresne MJ, Evidence for the involvement of cathepsin B in skeletal myoblast differentiation, J Cell Biochem, 84 (2002) 520–531. [PubMed] [Google Scholar]
- [52].Small TW, Bolender Z, Bueno C, O’Neil C, Nong Z, Rushlow W, Rajakumar N, Kandel C, Strong J, Madrenas J, Pickering JG, Wilms’ tumor 1-associating protein regulates the proliferation of vascular smooth muscle cells, Circ Res, 99 (2006) 1338–1346. [DOI] [PubMed] [Google Scholar]
- [53].Glogowska A, Kunanuvat U, Stetefeld J, Patel TR, Thanasupawat T, Krcek J, Weber E, Wong GW, Del Bigio MR, Hoang-Vu C, Hombach-Klonisch S, Klonisch T, C1q-tumour necrosis factor-related protein 8 (CTRP8) is a novel interaction partner of relaxin receptor RXFP1 in human brain cancer cells, J Pathol, 231 (2013) 466–479. [DOI] [PubMed] [Google Scholar]
- [54].Glogowska A, Stetefeld J, Weber E, Ghavami S, Hoang-Vu C, Klonisch T, Epidermal growth factor cytoplasmic domain affects ErbB protein degradation by the lysosomal and ubiquitin-proteasome pathway in human cancer cells, Neoplasia, 14 (2012) 396–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Kumar A, Dhar S, Campanelli G, Butt NA, Schallheim JM, Gomez CR, Levenson AS, MTA1 drives malignant progression and bone metastasis in prostate cancer, Mol Oncol, 12 (2018) 1596–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Guicciardi ME, Bronk SF, Werneburg NW, Yin XM, Gores GJ, Bid is upstream of lysosome-mediated caspase 2 activation in tumor necrosis factor alpha-induced hepatocyte apoptosis, Gastroenterology, 129 (2005) 269–284. [DOI] [PubMed] [Google Scholar]
- [57].Gupta R, Nalla AK, Gogineni VR, Chetty C, Bhoopathi P, Klopfenstein JD, Tsung AJ, Mohanam S, Rao JS, uPAR/cathepsin B overexpression reverse angiogenesis by rescuing FAK phosphorylation in uPAR/cathepsin B down regulated meningioma, PLoS One, 6 (2011) e17123. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [58].Rao Malla R, Gopinath S, Alapati K, Gorantla B, Gondi CS, Rao JS, Knockdown of cathepsin B and uPAR inhibits CD151 and alpha3beta1 integrin-mediated cell adhesion and invasion in glioma, Mol Carcinog, 52 (2013) 777–790. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [59].Malla RR, Gopinath S, Gondi CS, Alapati K, Dinh DH, Tsung AJ, Rao JS, uPAR and cathepsin B downregulation induces apoptosis by targeting calcineurin A to BAD via Bcl-2 in glioma, J Neurooncol, 107 (2012) 69–80. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [60].Gopinath S, Alapati K, Malla RR, Gondi CS, Mohanam S, Dinh DH, Rao JS, Mechanism of p27 upregulation induced by downregulation of cathepsin B and uPAR in glioma, Mol Oncol, 5 (2011) 426–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Cheng YC, Ding YM, Hueng DY, Chen JY, Chen Y, Caffeine suppresses the progression of human glioblastoma via cathepsin B and MAPK signaling pathway, J Nutr Biochem, 33 (2016) 63–72. [DOI] [PubMed] [Google Scholar]
- [62].Chang SH, Kanasaki K, Gocheva V, Blum G, Harper J, Moses MA, Shih SC, Nagy JA, Joyce J, Bogyo M, Kalluri R, Dvorak HF, VEGF-A induces angiogenesis by perturbing the cathepsin-cysteine protease inhibitor balance in venules, causing basement membrane degradation and mother vessel formation, Cancer Res, 69 (2009) 4537–4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Mohamed MM, Sloane BF, Cysteine cathepsins: multifunctional enzymes in cancer, Nat Rev Cancer, 6 (2006) 764–775. [DOI] [PubMed] [Google Scholar]
- [64].Shchors K, Nozawa H, Xu J, Rostker F, Swigart-Brown L, Evan G, Hanahan D, Increased invasiveness of MMP-9-deficient tumors in two mouse models of neuroendocrine tumorigenesis, Oncogene, 32 (2013) 502–513. [DOI] [PubMed] [Google Scholar]
- [65].Nalla AK, Gorantla B, Gondi CS, Lakka SS, Rao JS, Targeting MMP-9, uPAR, and cathepsin B inhibits invasion, migration and activates apoptosis in prostate cancer cells, Cancer Gene Ther, 17 (2010) 599–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Ponnala S, Chetty C, Veeravalli KK, Dinh DH, Klopfenstein JD, Rao JS, Metabolic remodeling precedes mitochondrial outer membrane permeabilization in human glioma xenograft cells, Int J Oncol, 40 (2012) 509–518. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [67].Solaini G, Sgarbi G, Baracca A, Oxidative phosphorylation in cancer cells, Biochim Biophys Acta, 1807 (2011) 534–542. [DOI] [PubMed] [Google Scholar]
- [68].de Groof AJ, te Lindert MM, van Dommelen MM, Wu M, Willemse M, Smift AL, Winer M, Oerlemans F, Pluk H, Fransen JA, Wieringa B, Increased OXPHOS activity precedes rise in glycolytic rate in H-RasV12/E1A transformed fibroblasts that develop a Warburg phenotype, Mol Cancer, 8 (2009) 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Garden RJ, Liu BC, Redwood SM, Weiss RE, Droller MJ, Bacillus Calmette-Guerin abrogates in vitro invasion and motility of human bladder tumor cells via fibronectin interaction, J Urol, 148 (1992) 900–905. [DOI] [PubMed] [Google Scholar]
- [70].Kong X, Ma W, Li Y, Wang Y, Guan J, Gao J, Wei J, Yao Y, Lian W, Xu Z, Dou W, Xing B, Ren Z, Su C, Yang Y, Wang R, Does Tenascin have Clinical Implications in Pathological Grade of Glioma Patients?: A Systematic Meta-Analysis, Medicine (Baltimore), 94 (2015) e1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Mai J, Sameni M, Mikkelsen T, Sloane BF, Degradation of extracellular matrix protein tenascin-C by cathepsin B: an interaction involved in the progression of gliomas, Biol Chem, 383 (2002) 1407–1413. [DOI] [PubMed] [Google Scholar]
- [72].Radogna F, Cerella C, Gaigneaux A, Christov C, Dicato M, Diederich M, Cell type-dependent ROS and mitophagy response leads to apoptosis or necroptosis in neuroblastoma, Oncogene, 35 (2016) 3839–3853. [DOI] [PubMed] [Google Scholar]
- [73].Zhang L, Qiang P, Yu J, Miao Y, Chen Z, Qu J, Zhao Q, Chen Z, Liu Y, Yao X, Liu B, Cui L, Jing H, Sun G, Identification of compound CA-5f as a novel late-stage autophagy inhibitor with potent anti-tumor effect against non-small cell lung cancer, Autophagy, (2018) 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Booth L, Roberts JL, Rais R, Poklepovic A, Dent P, Valproate augments Niraparib killing of tumor cells, Cancer Biol Ther, (2018) 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Booth L, Roberts JL, Samuel P, Avogadri-Connors F, Cutler RE, Lalani AS, Poklepovic A, Dent P, The irreversible ERBB1/2/4 inhibitor neratinib interacts with the PARP1 inhibitor niraparib to kill ovarian cancer cells, Cancer Biol Ther, 19 (2018) 525–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Yang Q, Zhang C, Wei H, Meng Z, Li G, Xu Y, Chen Y, Caspase-Independent Pathway is Related to Nilotinib Cytotoxicity in Cultured Cardiomyocytes, Cell Physiol Biochem, 42 (2017) 2182–2193. [DOI] [PubMed] [Google Scholar]
- [77].Liu F, Li X, Lu C, Bai A, Bielawski J, Bielawska A, Marshall B, Schoenlein PV, Lebedyeva IO, Liu K, Ceramide activates lysosomal cathepsin B and cathepsin D to attenuate autophagy and induces ER stress to suppress myeloid-derived suppressor cells, Oncotarget, 7 (2016) 83907–83925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Guan JJ, Zhang XD, Sun W, Qi L, Wu JC, Qin ZH, DRAM1 regulates apoptosis through increasing protein levels and lysosomal localization of BAX, Cell Death Dis, 6 (2015) e1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Santoni M, Amantini C, Morelli MB, Liberati S, Farfariello V, Nabissi M, Bonfili L, Eleuteri AM, Mozzicafreddo M, Burattini L, Berardi R, Cascinu S, Santoni G, Pazopanib and sunitinib trigger autophagic and non-autophagic death of bladder tumour cells, Br J Cancer, 109 (2013) 1040–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Han H, Li J, Feng X, Zhou H, Guo S, Zhou W, Autophagy-related genes are induced by histone deacetylase inhibitor suberoylanilide hydroxamic acid via the activation of cathepsin B in human breast cancer cells, Oncotarget, 8 (2017) 53352–53365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Hong JM, Kim SJ, Lee SM, Role of necroptosis in autophagy signaling during hepatic ischemia and reperfusion, Toxicol Appl Pharmacol, 308 (2016) 1–10. [DOI] [PubMed] [Google Scholar]
- [82].Yu L, Wu WK, Gu C, Zhong D, Zhao X, Kong Y, Lin Q, Chan MT, Zhou Z, Liu S, Obatoclax impairs lysosomal function to block autophagy in cisplatin-sensitive and -resistant esophageal cancer cells, Oncotarget, 7 (2016) 14693–14707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Barbosa IA, Vega-Naredo I, Loureiro R, Branco AF, Garcia R, Scott PM, Oliveira PJ, TRAP1 regulates autophagy in lung cancer cells, Eur J Clin Invest, 48 (2018). [DOI] [PubMed] [Google Scholar]
- [84].Storniolo A, Alfano V, Carbotta S, Ferretti E, Di Renzo L, IRE1alpha deficiency promotes tumor cell death and eIF2alpha degradation through PERK dipendent autophagy, Cell Death Discov, 4 (2018) 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Ning L, Guo-Chun Z, Sheng-Li A, Xue-Rui L, Kun W, Jian Z, Chong-Yang R, Ling-Zhu W, Hai-Tong L, Inhibition of autophagy induced by PTEN loss promotes intrinsic breast cancer resistance to trastuzumab therapy, Tumour Biol, 37 (2016) 5445–5454. [DOI] [PubMed] [Google Scholar]
- [86].Liu L, Zhang N, Dou Y, Mao G, Bi C, Pang W, Liu X, Song D, Deng H, Lysosomal dysfunction and autophagy blockade contribute to IMB-6G-induced apoptosis in pancreatic cancer cells, Sci Rep, 7 (2017) 41862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Zhao X, Fang Y, Yang Y, Qin Y, Wu P, Wang T, Lai H, Meng L, Wang D, Zheng Z, Lu X, Zhang H, Gao Q, Zhou J, Ma D, Elaiophylin, a novel autophagy inhibitor, exerts antitumor activity as a single agent in ovarian cancer cells, Autophagy, 11 (2015) 1849–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Chen QY, Shi JG, Yao QH, Jiao DM, Wang YY, Hu HZ, Wu YQ, Song J, Yan J, Wu LJ, Lysosomal membrane permeabilization is involved in curcumin-induced apoptosis of A549 lung carcinoma cells, Mol Cell Biochem, 359 (2012) 389–398. [DOI] [PubMed] [Google Scholar]
- [89].Liu J, Peng L, Niu T, Wu Y, Li J, Wang F, Zheng Y, Liu T, PIG7 promotes leukemia cell chemosensitivity via lysosomal membrane permeabilization, Oncotarget, 7 (2016) 4841–4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Novinec M, Lenarcic B, Turk B, Cysteine cathepsin activity regulation by glycosaminoglycans, BioMed research international, 2014 (2014) 309718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Cox JL, Cystatins and cancer, Front Biosci (Landmark Ed), 14 (2009) 463–474. [DOI] [PubMed] [Google Scholar]
- [92].Zavrsnik J, Butinar M, Prebanda MT, Krajnc A, Vidmar R, Fonovic M, Grubb A, Turk V, Turk B, Vasiljeva O, Cystatin C deficiency suppresses tumor growth in a breast cancer model through decreased proliferation of tumor cells, Oncotarget, 8 (2017) 73793–73809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Oh BM, Lee SJ, Cho HJ, Park YS, Kim JT, Yoon SR, Lee SC, Lim JS, Kim BY, Choe YK, Lee HG, Cystatin SN inhibits auranofin-induced cell death by autophagic induction and ROS regulation via glutathione reductase activity in colorectal cancer, Cell death & disease, 8 (2017) e3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Komura T, Takabatake H, Harada K, Yamato M, Miyazawa M, Yoshida K, Honda M, Wada T, Kitagawa H, Ohta T, Kaneko S, Sakai Y, Clinical features of cystatin A expression in patients with pancreatic ductal adenocarcinoma, Cancer science, 108 (2017) 2122–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Kramer L, Renko M, Zavrsnik J, Turk D, Seeger MA, Vasiljeva O, Grutter MG, Turk V, Turk B, Non-invasive in vivo imaging of tumour-associated cathepsin B by a highly selective inhibitory DARPin, Theranostics, 7 (2017) 2806–2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Uhlman A, Folkers K, Liston J, Pancholi H, Hinton A, Effects of Vacuolar H(+)-ATPase Inhibition on Activation of Cathepsin B and Cathepsin L Secreted from MDA-MB231 Breast Cancer Cells, Cancer microenvironment : official journal of the International Cancer Microenvironment Society, 10 (2017) 49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Liow KY, Chow SC, The cathepsin B inhibitor z-FA-CMK induces cell death in leukemic T cells via oxidative stress, Naunyn-Schmiedeberg’s archives of pharmacology, 391 (2018) 71–82. [DOI] [PubMed] [Google Scholar]
- [98].Tang ZH, Cao WX, Guo X, Dai XY, Lu JH, Chen X, Zhu H, Lu JJ, Identification of a novel autophagic inhibitor cepharanthine to enhance the anti-cancer property of dacomitinib in non-small cell lung cancer, Cancer Lett, 412 (2018) 1–9. [DOI] [PubMed] [Google Scholar]
- [99].Yang K, Lu Y, Xie F, Zou H, Fan X, Li B, Li W, Zhang W, Mei L, Feng SS, Yin Y, Liu Y, Zhang H, Yin C, Zhong Y, Gao J, Cationic liposomes induce cell necrosis through lysosomal dysfunction and late-stage autophagic flux inhibition, Nanomedicine (Lond), 11 (2016) 3117–3137. [DOI] [PubMed] [Google Scholar]
- [100].Liu F, Zhang Y, Men T, Jiang X, Yang C, Li H, Wei X, Yan D, Feng G, Yang J, Bergquist J, Wang B, Jiang W, Mi J, Tian G, Quantitative proteomic analysis of gastric cancer tissue reveals novel proteins in platelet-derived growth factor b signaling pathway, Oncotarget, 8 (2017) 22059–22075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Law S, Panwar P, Li J, Aguda AH, Jamroz A, Guido RVC, Bromme D, A composite docking approach for the identification and characterization of ectosteric inhibitors of cathepsin K, PloS one, 12 (2017) e0186869. [DOI] [PMC free article] [PubMed] [Google Scholar]


