How do our individual genomes and life histories influence our well-being, risk for diseases, and responses to medical treatments? This is the fundamental question precision medicine seeks to address. Understand how the confluence of genes and environment defines pathophysiological traits, and we can in theory prescribe the most suitable treatments to each individual, better predict population health to improve policy-making, and perhaps even unlock some of the mysteries behind the circuitry of life itself.
While we have not yet achieved this goal, for the first time we possess the investigative tools that suggest how it can be accomplished. It is with recent technological advances in mind that for this Circulation Research Omics Compendium, we invited leaders in our field to discuss essential aspects of ‘omics’ technologies from genomics and transcriptomics to proteomics, metabolomics, phenomics, and beyond, and to explore what the integration of large-scale digital data means for precision health and medicine.
We begin with two essays on how evolving technologies are changing the ways health status can be assessed. Kellogg et al. (1) describe the emergence of mobile health (“m-health”) devices and sensors that have revolutionized the measurement of human “dynamic physiology,” a concept which encompasses not only genetic information, but also continuous measurements of high-dimensional phenotypes. Small devices and smartphones can now be used to collect quasi-continuous data on blood pressure, heart rhythm, oxygen saturation, brain waves, air quality, radiation, and an ever-expanding list of metrics. The resulting physiological and environmental information can be connected to other omics layers such as genomes, metabolomes and microbiomes to discover subclinical imbalances or elevated disease risk in otherwise healthy individuals.
Cranley and MacRae (2) further expand on the theme of deriving a “phenotypic repertoire” at scale. Using atherosclerosis as an example, the authors argue that the slow progress on disease mechanisms comes not from incomplete genotyping to identify associated variants, but rather from our inability to make causal connections between identified variants (e.g., 9p21) with and disease pathways. They contend that the difficulty of finding novel pathways is related to empirical science’s tendency to mostly build on known paradigms, channeling the science historian Thomas Kuhn (3). A proposed solution is to keep pace with genotyping efforts by phenotyping to establish comprehensive baseline physiology, define bona fide absence of subclinical disease, and enable better case control separation. To fully redeem the promise of precision medicine, we need data on all fronts from genomes to phenomes, including the intermediary molecular “endophenotypes” which often provide critical mechanistic information.
Indeed, emerging and rapidly progressing technologies can now measure the molecular phenotypes of genes, chromatin, transcripts, proteins, metabolites, and environmental exposure (Figure 1). Six articles in the issue introduce readers to the forefront of technologies and concepts in each respective omics domain. The omics revolution began with the sequencing of the human genome, and genomics continues to lead the way by bringing revolutionary technologies to researchers and providing an anchor upon which all other omics layers are built. Costs of gene sequencing have plummeted, enabling routine and large-scale sequencing to power association studies between genes and traits. In addition to the human genome, the genomes of our gut flora are now under the spotlight, revealing important links to health and metabolism. Beyond conventional traits such as height and binary disease status, genome-wide association studies (GWAS) can now provide insight into the pharmacokinetics and pharmacodynamics of prescribed pharmaceutical compounds as traits displaying individual variabilities. Pharmacogenomics studies, expertly discussed by Roden et al. (4), have leveraged the study designs of GWAS to unearth a plethora of rare and common variants in different populations that control individual drug responses, and in the process also connected new dots in disease mechanisms. Precision medicine also begets precision trials, because drug candidates can be tested in more targeted subpopulations, in which drug efficacy is not masked by the inclusion of predicted non-responders.
Figure 1: Omics, Big Data and Precision Medicine.
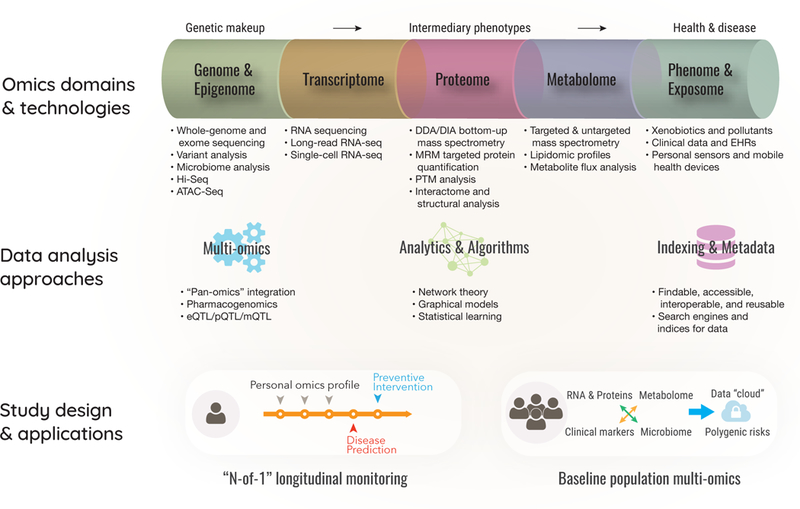
Top: Emerging omics technologies allow genomes, transcriptomes, proteomes, and other intermediary phenotypes to be measured at scale. Middle: Advances in data science, integration, and modeling connect high-dimensional big data to biomedical knowledge. Bottom: Multi-omics studies in longitudinal “personal omics profiles” and “dynamic data clouds” from healthy cohorts demonstrate the potential to generate actionable insights.
The genome continues to yield other secrets, with the structure and folding of chromatin a recent highlight. Unlike the neat and tidy picture of metaphase chromosomes described in textbooks, the chromosomes of non-dividing or interphase cells actually fold in complex three-dimensional structures with discernible domains and subdomains. Once considered linear and one-dimensional, it is now clear that the genome has a tertiary structure not unlike that of proteins, and this spatial architecture critically regulates gene expression and cell identity. Wang and Chang (5) review the field of epigenomics as a first connective layer between the constant genome found in every cell in the body and the diverse heterogeneity of cellular behaviors across tissues. Capitalizing on the genomic revolution made possible by next-generation sequencing, new epigenomics methods including Chromatin Interaction Analysis by Paired-end Tag Sequencing (ChIA-PET), Chromatin Conformation Capture with Sequencing (Hi-C), and Assay for Transposase-Accessible Chromatin with High-throughput Sequencing (ATAC-Seq) can now accurately depict DNA methylation, histone modifications, non-coding RNAs, transcription factor occupancy, chromatin accessibility, and higher-order chromatin structures. Many genomic variants implicated in GWAS occur in intervening regions with no immediate connections to known coding genes or biochemical pathways. Studies using ATAC-seq and other techniques are linking loci identified by GWAS to epigenetic changes such as enhancer-promoter interactions. Epigenetic engineering is also an exciting next step using modified Clustered Regularly-Interspaced Short Palindromic Repeats/Cas9 (CRISPR/Cas9) tools which can create chromatin contact and write DNA methylation.
The transcriptome offers further intriguing clues to the functions of genetic variants. Unlike the genome, the transcriptome is highly dynamic in response to acute and cumulative exposures. RNA-sequencing (RNA-seq) is now ubiquitously deployed to identify differential gene expression, and a large number of GWAS variants are now known to function as expression quantitative trait loci (eQTL), meaning that they regulate the expression level of transcripts, whereas splice-QTLs regulate the splice ratios of transcript isoforms. Wirka et al. (6) describe two emerging frontiers in transcriptomics. First is the emergence of long-read RNA-seq, which overcomes the difficulty of mapping short transcript reads to reference genomes, allowing the reconstruction of full-length isoform transcripts in high resolution. In parallel, advances in single-cell library preparation and amplification chemistry, coupled with the increasing depth and economy of sequencing, have allowed transcript profiles of individual cells to be sequenced from tens of thousands of cells. The advent of single-cell RNA-sequencing (scRNA-seq) has opened new windows into the cell-to-cell heterogeneity of transcription programs in development and disease, which are affected by factors such as transcriptional noise, cell cycle, as well as spatiotemporal differences in gene expression across tissue regions and cell types. The authors provide an accessible guide to the technical considerations arising from new developments in single-cell sample preparation, data normalization, and quantitative analysis.
Parallel to sequencing, advances in mass spectrometry have enabled the identity and quantity of proteins in biological samples to be queried with increasing depth, as discussed by Fert-Bober et al. (7). Because proteins effectuate the majority of biological processes, in a proteome-centric view, the raison d’etre of DNA is largely to make proteins. Given that we could profile transcripts so well and at a lower cost than proteins, why bother with proteomics? The authors explain the concept of proteoforms: one gene can create multiple isoforms, which diversify further by myriad post-translational modification (PTM) configurations, with each configuration representing a chemically distinct population of molecules that can and do carry out different functions. Thus proteomes are staggeringly more complex than transcriptomes and also require many physicochemical parameters to be fully described; perturbations in protein modifications, folding, localization, turnover, and activity also could well be key to disease development, in addition to transcript/protein expression. Mass spectrometry techniques are leading the way to characterize proteoforms, including many understudied PTMs such as citrullination and S-nitrosylation that were once neglected because the necessary reagents were not available to study them, but now are known to modulate many cardiac processes.
Metabolomes are the next step in bridging genetic information to chemical space. The availability of quicker and more powerful mass spectrometers has also propelled the measurements of metabolites, the detailed methodologies and experimental design considerations of which are examined in McGarrah et al. (8). In addition to steady-state abundance, the flux of molecules along metabolic pathways can also be estimated with stable isotopes to inform temporal changes. The tens of thousands of small molecules circulating in the blood can reflect many causal chains of events between genes, traits, and critically, the environment. As an example, the authors described how the baseline level of short-chain dicarboxylacylcarnitine species in >2,000 individuals were found to strongly predict myocardial infarction risk on top of clinical models. Subsequent genome-wide analysis further linked individual variations of these metabolites to metabolomics quantitative trait locus (mQTL) variants in genes that regulate endoplasmic reticulum stress, thus fleshing out a mechanistic loop involving genes, cellular mechanism, and clinical traits.
Circulating molecules comprise not only endogenous species indirectly encoded by the genome, but also various xenobiotics from ingested nutrients, pollutants, and other environmental exposures. It is well known that complex traits are the combination of genes and environment; in our efforts to define genetic causes it is easy to forget that environmental exposures also provide a critical layer between genome and phenome. Riggs et al. (9) analyze the challenges of profiling the “envirome” and provide a conceptual framework of the ways in which environmental factors can influence human health. Omics technologies can be used to detect an individual’s exposure over time to classes of chemicals including volatile organic compounds, heavy metals, and particulate matter. Here the parameter space of molecular phenotypes again expands exponentially, and we are no longer constrained by the “parts list” of the human genome. Nor does the complexity stop here. Embodied in the concept of the envirome are less well-defined compound exposures including diurnal and seasonal variations, as well as socioeconomic and lifestyle choices known to bias health on epidemiological scales. To tackle this challenge, the authors discuss a classification system that can order concepts and entities along ontological categories.
These large-scale techniques are generating an overwhelming amount of biomedical data. To avoid wasting acquisition efforts, the data must be harnessed to generate insights. Two excellent articles expound on what this task requires. Trachana et al. (10) provide a theoretical framework that conceptualizes molecular changes as the reorganization of network nodes and edges, and introduce the readers to a lexicon of terminologies from network analysis. Physiological phenomena such as the emergence of high glucose levels in the prediabetic state are recast in a new light as tipping points and bifurcation phenomena of a network with multiple alternative stable states. One power of the network approach is that it addresses a blind spot of the disease-oriented paradigm of clinical research and practice, which by definition precludes detailed knowledge about early presentations in subclinical populations. In this view, better baseline knowledge on organizational principles is key to combating diseases, and changes in co-variation between molecules are more instructive than the differential expression of individual markers. Network science approaches may also prove valuable for delineating complex environmental interactions among high number of variables, as shown in the environmental networks formulated by Bhatnagar and colleagues.
Ping et al. (11) explicate the practical aspects of data-mining in the burgeoning field of data science, in particular contemporary considerations for sharing data sets at-scale. The importance of metadata is introduced, as are indexing tools that lead users to data and help them extract meaningful information. While we may take for granted the ease of fetching a journal article with a keyword search on PubMed, a huge amount of work is involved behind the scene to design standardized catalogs and vocabularies, resolve synonyms, and match queries to data. This indexing and searching ability is being extended to omics data sets to help make biomedical data more “FAIR” (findable, accessible, interoperable, and reusable). Other emerging technologies include cloud computing, which allows users to access, store, and analyze data from anywhere without hefty infrastructure investment; and deep learning and graphical models that allow molecular signatures to be automatically extracted from rich datasets in an unsupervised manner, and can even draw inference on causality. We learn that deep learning is already deployed on electrocardiography data to detect arrhythmias with the accuracy of cardiologists.
Tying it all together, the capstone article by Leopold and Loscalzo (12) provides an insightful overview on the promise and realization of precision medicine. The power of precision medicine, suggest the authors, lies in the data and demands a synthesis of rapidly evolving datasets. The majority of cardiovascular disease factors are now known to involve perturbations in a large number of interlinked genetic and environmental factors, thus exposing the flawed logic behind the traditional paradigm of searching for single causative genes or gene products in heart diseases, and by extension, of the search for a single “magic bullet” to cure all patients. Instead, the authors propose that both a population-based preventive approach and individual-based plans to treat high-risk patients are needed to lower the societal burden of heart diseases. This in turns demands high-resolution, deep phenotype data, encompassing historical metrics, environmental and social exposures, wearable devices and sensors, and deep omics profiling with the technologies covered in the compendium.
What might this omics and precision medicine future look like? Several landmark studies have provided powerful proofs-of-concept on two parallel designs. On the individual level, “N-of-1” deep profiling studies involve high-dimensional longitudinal profiling in a single individual to provide constant monitoring and preventive intervention. The MyConnectome study (13) assessed brain images, functions, gene and metabolic profiles of one individual over 18 months to reveal a joint dynamics between brain and metabolic functions. The Integrative Personal Omics Profile study (14) traced the transcriptome, proteome, and metabolome of an individual over 14 months, discovering a subclinical pre-diabetic state during the longitudinal study and helping prevent disease by prompting the individual to self-correct in diet. On the population level, dense and dynamic “data clouds” are used to analyze individual differences and make actionable predictions. The “P100 Wellness” study (15) combined gene, protein, metabolite, and microbiome with clinical laboratory tests to create statistical associations across omics layers, deriving a polygenic score to predict risks for 127 traits including blood pressure and QT interval. The “Personalized Nutrition” study (16) integrated blood glucose monitoring, food intake questionnaires on smartphones, metabolome and microbiome surveys to predict inter-individual differences in postprandial glycemic responses. Machine learning algorithms then integrated the data to provide dietary recommendations, which outperformed a professional dietician in minimizing glucose spikes in the subjects.
Assisted by an abundance of molecular, physiological, and environmental data from various omics technologies, cardiovascular research increasingly resides in a massive, digital, data-driven world. Clinical research and practice will no longer be content with targeting only the hypothetical “average patient,” and will instead enter the realm of precise knowledge of individuals and populations. With the NIH Precision Medicine Initiative, All of Us study, and other global initiatives on the horizon extending this paradigm to massive populations around the world, we stand on the verge of realizing the promise of precision medicine and health.
Acknowledgments:
We thank Blake Wu and Katy Claiborn for reading the manuscript. This work was supported in part by American Heart Association 17MERIT336100009, Burroughs Wellcome Fund Innovation in Regulatory Science Award 1015009, and National Institutes of Health (NIH) R01 HL113006, R01 HL128170, R24 HL117756 (JCW), F32 HL139045 (EL).
Bibliography
- 1.Kellogg RA, Dunn J, Snyder MP 2018. Personal omics for precision health. Circulation Research xxx:xxx. [in this issue] [DOI] [PubMed] [Google Scholar]
- 2.Cranley J, MacRae CA 2018. A new approach to an old problem: one brave idea. Circulation Research xxx:xxx. [in this issue] [DOI] [PubMed] [Google Scholar]
- 3.Kuhn TS 1996. The structure of scientific revolutions. 3rd Edition. Chicago: University of Chicago Press; Print. [Google Scholar]
- 4.Roden DM, Van Driest S, Wells QS, Mosley JD, Denny JC, Peterson JF 2018. Opportunities and challenges in cardiovascular pharmacogenomics: from discovery to implementation. Circulation Research xxx:xxx. [in this issue] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang KC, Chang HY 2018. Epigenomics – technologies and applications. Circulation Research xxx:xxx. [in this issue] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wirka R, Pjanic M, Quertermous T 2018. Advances in transcriptomics: investigating cardiovascular disease at high resolution. Circulation Research xxx:xxx. [in this issue] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fert-Bober J, Murray CI, Parker S, Van Eyk JE 2018. Precision profiling of cardiovascular proteoforms: where there is a will, there is a way. Circulation Research xxx:xxx. [in this issue] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McGarrah RW, Crown SB, Zhang G, Shah SH, Newgard CB 2018. Cardiovascular metabolomics. Circulation Research xxx:xxx. [in this issue] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Riggs DW, Yeager RA, Bhatnagar A 2018. Defining the human envirome: an omics approach for assessing the environmental risk of cardiovascular disease. Circulation Research xxx:xxx. [in this issue] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trachana K, Bargaje R, Glusman G, Price ND, Huang S, Hood LE 2018. Taking systems medicine to heart. Circulation Research xxx:xxx. [in this issue] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ping P, Hermjakob H, Polson JS, Benos PV, Wang W 2018. Biomedical informatics on the cloud: a treasure hunt for advancing cardiovascular medicine. Circulation Research xxx:xxx. [in this issue] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leopold JA, Loscalzo J 2018. The emerging role of precision medicine in cardiovascular disease. Circulation Research xxx:xxx. [in this issue] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poldrack RA, Laumann TO, Koyejo O, Gregory B, Hover A, Chen M-Y, Gorgolewski KJ, Luci J, Joo SJ, Boyd RL, Hunicke-Smith S, Simpson ZB, Caven T, Sochat V, Shine JM, Gordon E, Snyder AZ, Adeyemo B, Petersen SE, Glahn DC and Mumford JA 2015. Long-term neural and physiological phenotyping of a single human. Nature Communications 6, p. 8885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen R, Mias GI, Li-Pook-Than J, Jiang L, Lam HYK, Chen R, Miriami E, Karczewski KJ, Hariharan M, Dewey FE, Cheng Y, Clark MJ, Im H, Habegger L, Balasubramanian S, O’Huallachain M, Dudley JT, Hillenmeyer S, Haraksingh R, Sharon D and Snyder M 2012. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell 148(6), pp. 1293–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Price ND, Magis AT, Earls JC, Glusman G, Levy R, Lausted C, McDonald DT, Kusebauch U, Moss CL, Zhou Y, Qin S, Moritz RL, Brogaard K, Omenn GS, Lovejoy JC and Hood L 2017. A wellness study of 108 individuals using personal, dense, dynamic data clouds. Nature Biotechnology 35(8), pp. 747–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeevi D, Korem T, Zmora N, Israeli D, Rothschild D, Weinberger A, Ben-Yacov O, Lador D, Avnit-Sagi T, Lotan-Pompan M, Suez J, Mahdi JA, Matot E, Malka G, Kosower N, Rein M, Zilberman-Schapira G, Dohnalová L, Pevsner-Fischer M, Bikovsky R and Segal E 2015. Personalized nutrition by prediction of glycemic responses. Cell 163(5), pp. 1079–1094. [DOI] [PubMed] [Google Scholar]


