Abstract
Background
In rice (Oryza sativa) and other grains, weak growth (dwarfism, short panicle length, and low seed-setting rate) and early senescence lead to reduced yield. The molecular mechanisms behind these processes have been widely studied; however, the complex genetic regulatory networks controlling growth and senescence require further elucidation.
Results
We isolated a mutant exhibiting weak growth throughout development and early senescence of leaf tips, and designated this mutant weakness and leaf senescence5 (wls5). Histological analysis showed that the poor growth of wls5 plants involved a reduction in cell length and number. Physiological analysis and transmission electron microscopy revealed that the wls5 cells had abnormal chloroplasts, and the mutants underwent chlorophyll degradation triggered by accumulation of reactive oxygen species. Consistent with this, RNA sequencing revealed changes in senescence-related gene expression in wls5 plants. The wls5 mutants also exhibited significantly higher stomatal density and altered phytohormone contents compared with wild-type plants. Fine mapping delimited WLS5 to a 29-kb region on chromosome 5. DNA sequencing of wls5 identified a 3-bp deletion in the first exon of LOC_Os05g04900, resulting in a deletion of a lysine in the predicted protein. Knockout of LOC_Os05g04900 in Nipponbare plants caused leaf senescence, confirming this locus as the causal gene for WLS5.
Conclusions
We identified a novel mutant (wls5) that affects plant development and leaf senescence in rice. LOC_Os05g04900, encoding a protein of unknown function, is the causal gene for wls5. Further molecular study of WLS5 will uncover the roles of this gene in plant growth and leaf senescence.
Electronic supplementary material
The online version of this article (10.1186/s12284-019-0288-8) contains supplementary material, which is available to authorized users.
Keywords: wls5, Weak growth, Leaf senescence, Rice
Background
Abnormal development, such as weak growth (dwarf height and small tissues and organs) and early senescence, has major effects on rice yield (Sakamoto and Matsuoka 2008; Pan et al. 2013; Liu et al. 2016; Hong et al. 2018). Additionally, premature leaf senescence leads to low photosynthetic efficiency and decreased accumulation of photosynthetic assimilates in reproductive organs, thus decreasing yield (Mitchell and Sheehy 2006; Yang et al. 2016a; Mao et al. 2017). These two factors are regulated by genetics and often triggered by environmental stresses. Therefore, understanding the molecular mechanisms underlying growth and leaf senescence will benefit rice breeding and production.
Plant senescence is controlled by complex regulatory networks involving coordinated action at the cellular, tissue, organ and organism levels (Lim et al. 2007). Typical senescence symptoms include physiological alterations such as chlorophyll degradation, reactive oxygen species (ROS) scavenging, carbon and nitrogen imbalances, and hormone responses (Lim et al. 2007; Hong et al. 2018). Genes associated with leaf senescence in rice have been isolated and characterized (Leng et al. 2017a), including transcription factors, receptors and signaling components for hormones and stress responses, and metabolic regulators (Mitchell and Sheehy 2006; Yamada et al. 2014; Wu et al. 2016; Yang et al. 2016a; Yang et al. 2016b; Zhao et al. 2016; Deng et al. 2017; Leng et al. 2017b; Mao et al. 2017; Hong et al. 2018; Ke et al. 2019). Senescence of plants can be induced by many factors, including reproductive growth, phytohormones, and environmental cues (Lim et al. 2007); moreover, the senescence-associated genes regulated during early leaf senescence remain largely unknown.
Plant growth and development are driven by cell division and expansion, fundamental and dynamic cellular processes that enable plants and various organs to develop to suitable sizes (Duan et al. 2012). Orderly growth and development involve many genes and pathways that affect plant organ size by altering cell number, cell size, or both (Krizek 2009). Plant development and leaf senescence involve some of the same regulatory factors. Indeed, several early senescence mutants show accompanying defects in plant development (Wu et al. 2016; Xie et al. 2016; El et al. 2017; Leng et al. 2017b; He et al. 2018; Hong et al. 2018). For instance, a point mutation in O. sativa nicotinate phosphoribosyltransferase (OsNaPRT1) leads to dwarfism and a withered leaf tip phenotype (Wu et al. 2016); dwarf and early-senescence leaf1 (del1) mutants exhibit dwarfism and an early leaf senescence phenotype, with abnormal growth caused by a reduction in cell number (Leng et al. 2017b). Moreover, premature leaf senescence 3 (pls3) mutants show a semi-dwarf phenotype, early leaf senescence, and an early heading date compared to wild type (Hong et al. 2018), and the premature senescence leaf 85 (psl85) mutant shows a distinct dwarfism and premature leaf senescence phenotype (He et al. 2018). Despite in-depth study of these mutants, the linkage between development and senescence is still poorly understood in rice.
In this study, we identified a mutant displaying abnormal plant growth and novel premature leaf senescence, designated weakness and leaf senescence5 (wls5). Mutation of WLS5 caused decreases in cell length, perturbed chloroplast development, and disturbed hormonal balance, resulting plants with fewer cells, increased ROS activity, and altered expression of senescence-associated genes. Our findings thus suggest that WLS5 is a critical gene for plant growth and leaf senescence in rice.
Main text
Results
wls5 mutants exhibit weak growth in the whole plant
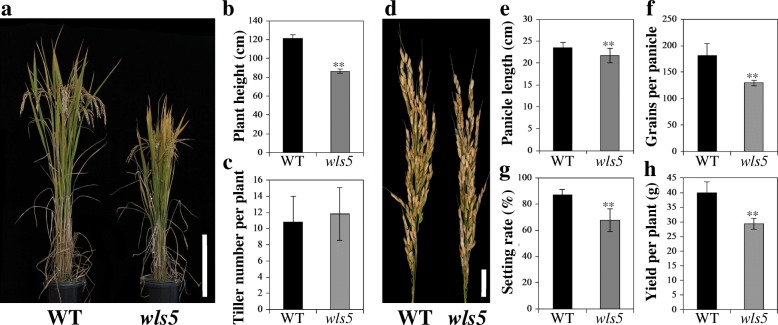
The wls5 mutant was obtained from an ethyl methane sulfonate (EMS) mutant bank of the indica rice cultivar ‘93–11’. Under normal growth conditions, wls5 plants exhibited weak growth (Fig. 1a). Compared with the wild type, tiller number was not altered, but plant height was only about 71.1% that of the wild type at the mature stage (Fig. 1b, c). The wls5 mutant also had shorter panicles, fewer grains per panicle, and lower seed-setting rate than the wild type (Fig. 1d-g). These reductions in major agronomic traits led to significant yield reduction in wls5, to just 73.2% of that in the wild type (P < 0.01, Student’s t-test; Fig. 1h).
Fig. 1.
Phenotypic comparison between wild-type (WT) and wls5 plants. a WT (‘93–11’) and wls5 plants at maturity. Bar = 40 cm. b, c Statistical analysis of plant height and tiller number between WT and wls5 plants. Twenty plants were measured. Error bars indicate SD; **P < 0.01 (Student’s t-test). d Panicle phenotype of WT and wls5 plants. Bar = 3 cm. e-h Statistical analysis of panicle length, grains per panicle, setting rate and yield per plant between WT and wls5 plants. Twenty panicles were measured. Error bars indicate SD; **P < 0.01 (Student’s t-test)
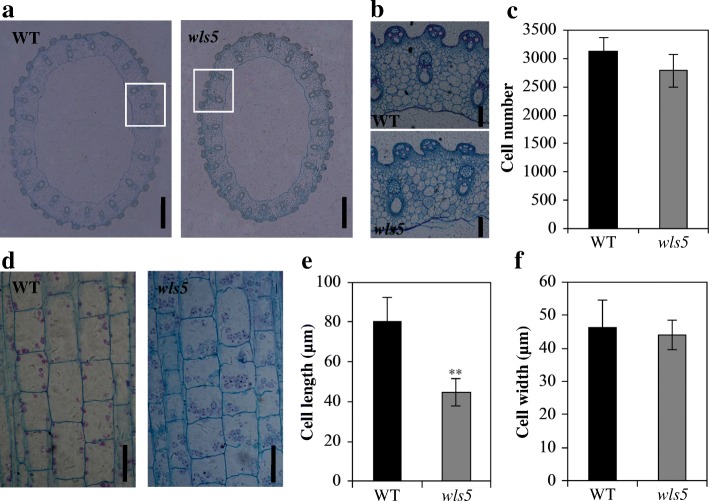
Plant organ size is determined by the number and size of cells, which are regulated by cell division and cell expansion, respectively (Krizek 2009). To investigate the causes of organ size reduction in wls5 plants, we compared paraffin sections of the second culms of wild-type and wls5 plants. Cross sections revealed that culm size in wls5 was smaller than that in the wild type (Fig. 2a, b). Statistical analysis showed that the cell number in wls5 was only 89.2% of that in the wild type (Fig. 2c). Longitudinal sections of culms revealed a dramatic change in cell size of wls5 compared with the wild type (Fig. 2d). Cell length in wls5 was 55.9% of that in the wild type, while cell width was similar in both (Fig. 2e, f). In addition, longitudinal sections of leaves indicated that the development and arrangement of mesophyll cells in wls5 were also abnormal (Additional file 1: Figure S1).
Fig. 2.
Histological characterization of culms in wild-type (WT) and wls5 plants. a Cross sections of internode II of WT (‘93–11’) and wls5 plants at heading stage. b Magnification of a. Bars = 500 μm (a), 50 μm (b). c Statistical analysis of cell number between WT and wls5 plants; means ± SD of five independent replicates. d Longitudinal sections of internode II of WT and wls5 plants. Bars = 50 μm. e-f Statistical analysis of cell length and cell width between WT and wls5 plants; means ± SD of 30 cells. **P < 0.01 (Student’s t-test)
wls5 undergoes early leaf senescence
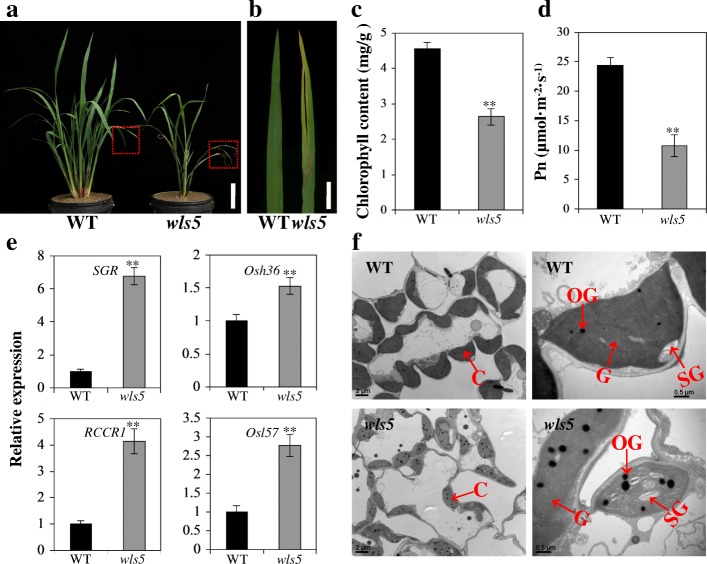
In addition to developmental weakness, wls5 also exhibited an early senescence phenotype, displaying yellow spots at the tip of each leaf at the tillering stage (Fig. 3a, b). The chlorophyll content in wls5 plants was significantly lower, only 57.9%, of that in the wild type (P < 0.01, Student’s t-test; Fig. 3c). In addition, the photosynthetic rate (Pn) in wls5 plants was only 44.3% of that in the wild type (Fig. 3d). To confirm senescence in wls5 plants, we determined the expression levels of two chlorophyll degradation related genes (CDGs), STAY-GREEN (SGR) and RED CHLOROPHYLL CATABOLITE REDUCTASE 1 (RCCR1) (Jiang et al. 2007; Tang et al. 2011), and two other senescence-associated genes (SAGs), Osh36 and OsI57 (Lee et al. 2001), by reverse-transcription quantitative PCR (RT-qPCR). The wls5 plants had higher expression levels of these senescence-related genes than wild-type plants (Fig. 3e).
Fig. 3.
Identification of leaf senescence in wls5. a Wild-type (WT; ‘93–11’) and wls5 plants at the tillering stage. Bar = 10 cm. b Leaf phenotype of WT and wls5 plants. Bar = 2 cm. c Chlorophyll content of leaves in wild-type and wls5 plants. Error bars indicate SD, n = 10. **P < 0.01 (Student’s t-test). d Photosynthesis rate of leaves in wild-type and wls5 plants. Error bars indicate SD, n = 15. **P < 0.01 (Student’s t-test). e Expression of CDGs (SGR and RCCR1) (left) and other SAGs (Osh36 and Osl57) (right) in WT and wls5 plants. Error bars indicate SD, n = 3. **P < 0.01 (Student’s t-test). f Transmission electron microscopy of senescing leaves of WT and wls5 plants at seedling stage. C, chloroplast; OG, osmiophilic granule; SG, starch granule; G, grana thylakoid
To explore the cause of low chlorophyll levels in wls5 during senescence, we compared the chloroplast ultrastructure of mesophyll cells in fully expanded leaves of wls5 and wild-type plants by transmission electron microscopy (TEM). In comparison with the wild type, the number and size of chloroplasts were dramatically lower in leaves of wls5 plants showing yellow spots (Fig. 3f, left). Furthermore, degenerated thylakoid membrane and a decreased number of grana thylakoid were observed in wls5 (Fig. 3f, right). The wls5 mutant cells also contained many more osmiophilic granules than wild-type cells (Fig. 3f, right). These observations suggest that the wls5 mutant may undergo abnormal chloroplast development.
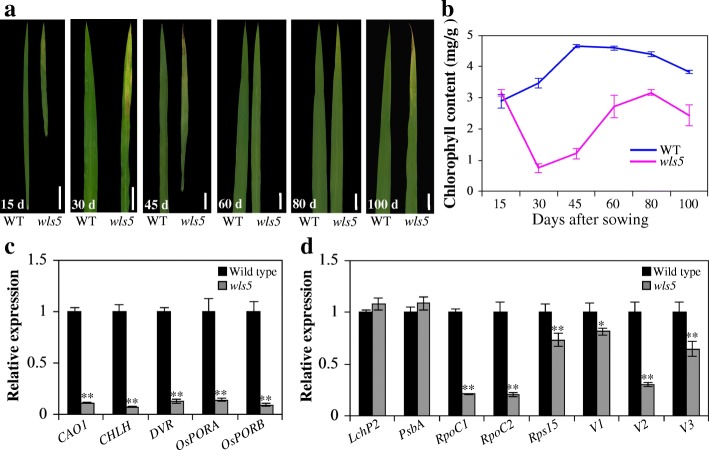
We monitored the leaf senescence phenotype of wls5 plants throughout plant growth, observing that wls5 showed early senescence to varying degrees at different growth stages (Fig. 4a). No leaf senescence was visible in wls5 mutants or wild-type plants during early germination. Around 15 days after germination, the mutant leaf apex exhibited a faint yellow color. With continued development, the yellow spots on wls5 leaves (30 and 45 days) expanded and became more yellow. When wls5 plants (60 and 80 days) progressed to the jointing stage, however, the young leaves (100 days) turned green at heading time then exhibited apparent senescence again at the grain-filling stage (Fig. 4a). Consistent with the leaf senescence phenotype, the chlorophyll content of wls5 plants decreased dramatically at the tillering stage, then increased until heading stage, and subsequently decreased rapidly again at maturation (Fig. 4b). These results indicate that internal or external factors influenced the leaf senescence process in wls5 mutants in different growth stages.
Fig. 4.
Comparison of leaf senescence over time between wild-type (WT) and wls5 plants. a Leaf phenotype of WT (‘93–11’) and wls5 plants at different stages (The youngest fully expanded leaf). Bars = 2 cm. b Chlorophyll content of leaves in wild-type and wls5 plants. Error bars indicate SD, n = 10. c, d Expression of chlorophyll synthesis (c) and chloroplast development (d) related genes in WT and wls5 plants at tillering stage. Error bars indicate SD, n = 3. **P < 0.01 (Student’s t-test)
Leaf senescence is usually accompanied by a change in expression of numerous genes, including those for chlorophyll synthesis and chloroplast development (Wang et al. 2015a, b). Therefore, we detected the expression levels of chlorophyll synthesis related genes (CAO1, CHLH, DVR, OsPORA, and OsPORB) and chloroplast development related genes (LchP2, PsbA, RpoC1, RproC2, Rps15, V1, V2, and V3) in wls5 and wild-type plants. Expression levels of CAO1, CHLH, DVR, OsPORA, OsPORB, RpoC1, RproC2, Rps15, V1, V2, and V3 were significantly lower in wls5 plants compared to those in the wild type (P < 0.01, Student’s t-test; Fig. 4c, d), further supporting the notion that early leaf senescence and abnormal chloroplast development occurred in wls5 plants.
ROS accumulation is enhanced in wls5 plants
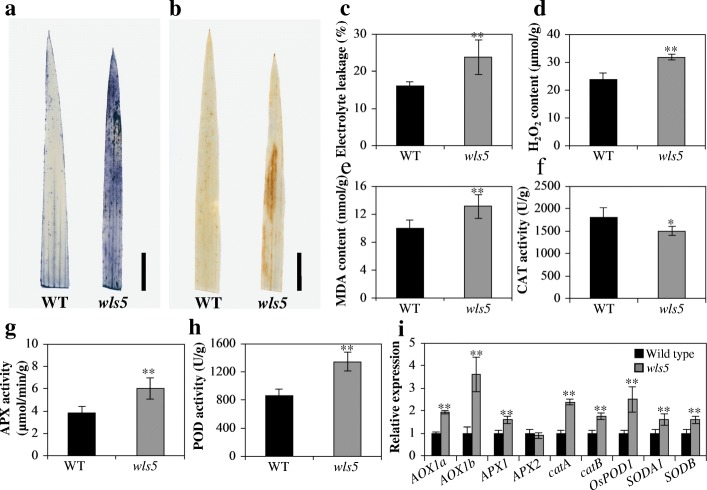
ROS accumulation promotes premature senescence (Roy et al. 2012). We therefore used the same part of the top leaves from wls5 and wild-type plants at the seedling stage to perform nitro blue tetrazolium (NBT) staining and 3,3′-diaminobenzidine (DAB) staining tests. As shown in Fig. 5a, extensive NBT staining was observed in wls5 plants, whereas staining was minimal in wild-type leaves. Similarly, there was little sign of DAB staining in wild-type leaves, but a brown color was visible in wls5 mutant leaves, consistent with the area of leaf senescence (Fig. 5b). These results indicated that ROS accumulated in wls5 plants. We then measured electrolyte leakage, an indicator of plant cell membrane damage (Blum and Ebercon 1981), and concentrations of senescence-related substances, including the ROS hydrogen peroxide (H2O2) and the lipid oxidation byproduct malondialdehyde (MDA). Electrolyte leakage in wls5 plants was 48.6% higher than that in the wild type (Fig. 5c). Similarly, higher concentrations of H2O2 and MDA were observed in wls5 leaves than in wild-type leaves (Fig. 5d, e), indicating that ROS accumulation did occur during premature leaf senescence in wls5 mutants.
Fig. 5.
ROS accumulation in wild-type (WT) and wls5 plants. a, b NBT (a) and DAB (b) staining of leaves from WT (‘93–11’) and wls5 plants at seedling stage. Bars = 1 cm. c-h Statistical analysis of electrolyte leakage (c), H2O2 content (d), MDA content (e), CAT activity (f), APX activity (g) and POD activity (h) in leaves of WT and wls5 plants. Mean ± SD of five independent replicates, **P < 0.01, *P < 0.05 (Student’s t-test). i Relative expression levels of ROS detoxification related genes in WT and wls5 plants; mean ± SD of three independent replicates. **P < 0.01 (Student’s t-test)
ROS scavenging enzymes, such as catalase (CAT), ascorbate peroxidase (APX), superoxide dismutase (SOD) and peroxidase (POD), play significant regulatory roles in plant senescence (Miller et al. 2010). We therefore quantified the activity of these enzymes. In comparison with the wild type, CAT activity was significantly lower in wls5 (P < 0.05, Student’s t-test; Fig. 5f). By contrast, the activities of APX and POD were much higher in wls5 leaves (6.0 μmol/min/g and 1344 U/mg fresh weight, respectively) than in wild-type leaves (3.8 μmol/min/g and 864 U/mg fresh weight, respectively) (Fig. 5g, h). The wls5 and wild-type plants showed no significant difference in SOD activity (318.6 and 316.6 U/mg fresh weight, respectively). As ROS-scavenging systems have an important role in ROS detoxification (Tan et al. 2014), we detected expression levels of ROS scavenging related genes. Expression levels of AOX1a, AOX1b, APX1, catA, catB, OsPOD1, SODA, and SODB were significantly higher in wls5 plants than in the wild type (P < 0.01, Student’s t-test; Fig. 5i).
wls5 affects the density of leaf stomata
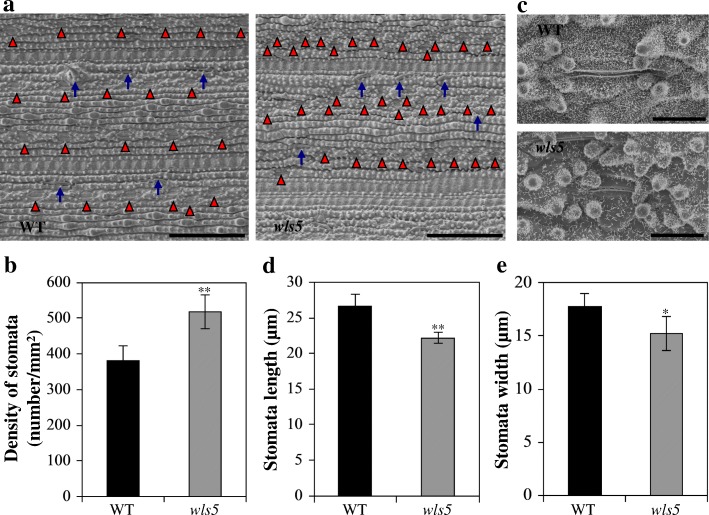
The rice early senescence mutant early senescence 1 has greater stomatal density than wild-type rice (Rao et al. 2015). To examine whether wls5 has the same phenotype, we compared leaves of wls5 and wild-type plants by scanning electron microscopy. Stomatal density was 36.4% higher in wls5 plants (518.3/mm2) than in wild-type leaves (380.0/mm2) (Fig. 6a, b). Stomatal length and width were 22.2 and 15.2 μm, respectively, in wls5 plants, much smaller than the 26.6 and 17.8 μm observed in the wild type (Fig. 6c-e). These results indicate that wls5 affects the density of leaf stomata in rice. In addition, the tips of trichomes and the setae were severely degraded on the leaf surface of wls5 mutants (Fig. 6a, c).
Fig. 6.
Observation of stomata in wild-type (WT) and wls5 plants. a Stomatal density of WT (‘93–11’) and wls5 leaves at tillering stage. Red triangles and blue arrows indicate the positions of stomata and setae, respectively. Bars = 100 μm. b Statistical analysis of stomatal density. c Morphological characteristics of stomata of WT and wls5 plants at tillering stage. Bars = 10 μm. d, e Statistical analysis of stomata length (d) and stomata width (e). Three independent replicates for stomatal density, and 20 independent replicates for stomata size. **P < 0.01 (Student’s t-test)
Hormone levels are disturbed in wls5 mutants
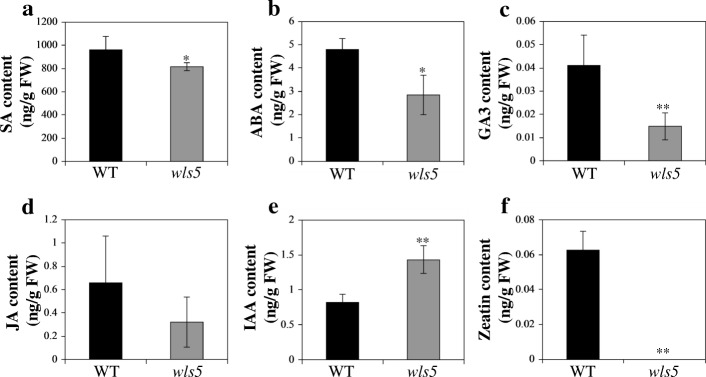
Phytohormone levels regulate leaf senescence (Jan et al. 2019). For example, gibberellic acid (GA) inhibits senescence (Yu et al. 2009), and auxins decrease the expression of SAG12, thereby delaying senescence (Mueller-Roeber and Balazadeh 2014). We therefore measured endogenous hormone contents in leaves at the tillering stage. Compared with wild type, the salicylic acid (SA; P < 0.05), abscisic acid (ABA; P < 0.05) and GA (P < 0.01) contents of wls5 mutants were significantly lower, being only about 84.9%, 59.1% and 35.9% of those in wild-type plants (Student’s t-test; Fig. 7a-c). The jasmonic acid (JA) content in wls5 was also lower than that in the wild type (Fig. 7d). By contrast, wls5 plants had a higher indole-3-acetic acid (IAA) content than wild-type plants: 1.43 and 0.82 ng/g fresh weight, respectively (Fig. 7e). The zeatin content was 0.06 ng/g fresh weight in the wild type, but zeatin levels were too low to measure in wls5 mutants (Fig. 7f). These results indicate that endogenous hormone levels were disturbed in wls5 mutants.
Fig. 7.
Comparison of endogenous hormone contents between wild-type (WT) and wls5 plants. SA (a), ABA (b), GA3 (c), JA (d), IAA (e) and zeatin (f) content in leaves of WT (‘93–11’) and wls5 plants at tillering stage. Means ± SD of three independent replicates. *P < 0.05; **P < 0.01 (Student’s t-test)
RNA sequencing analysis of wls5 mutants
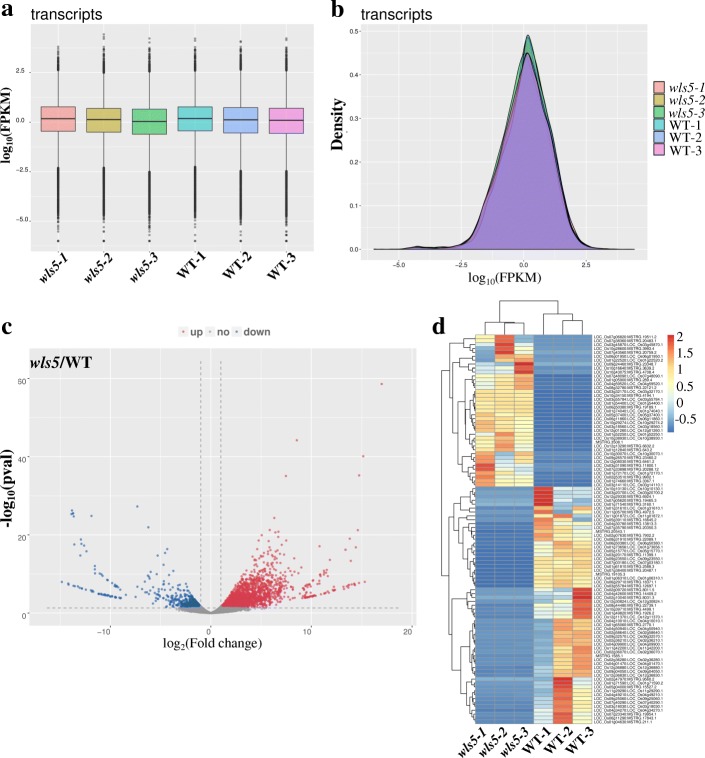
Regulation of leaf senescence is highly complex, with many specific genes involved (Lin et al. 2015). We performed RNA sequencing (RNA-seq) to analyze the effect of the wls5 mutation on gene expression. Leaf tip samples of wls5 and wild-type plants at tillering stage were selected for sequencing analysis with three replicates. Boxplots and density profiles of transcript levels in wild-type and wls5 plants indicated good reproducibility among biological replicates (Fig. 8a, b). More than 50 million reliable, clean reads were obtained from wls5 and wild-type samples, representing 2606 up-regulated genes and 840 repressed genes in wls5 compared to the wild type (Fig. 8c, d). Gene ontology and Kyoto Encyclopedia of Genes and Genomes pathway enrichment analyses indicated that genes involved in oxidoreductase activity, the oxidation-reduction process, response to ABA, multicellular organism development and ABA-activated signaling pathways were altered in wls5 plants (Additional file 1: Figure S2-S4).
Fig. 8.
RNA-seq analysis of wild-type (WT) and wls5 plants. mRNA was purified from total RNA isolated from tillering-stage plants of WT (‘93–11’) and wls5. a Transcript expression boxplots of WT and wls5 plants. b Transcript expression density of WT and wls5. c Volcano plot showing the overall alterations in gene expression between WT and wls5. d Cluster analysis of differentially expressed genes in WT and wls5. Red represents highly expressed genes. Blue represents poorly expressed genes
Candidate gene analysis of WLS5
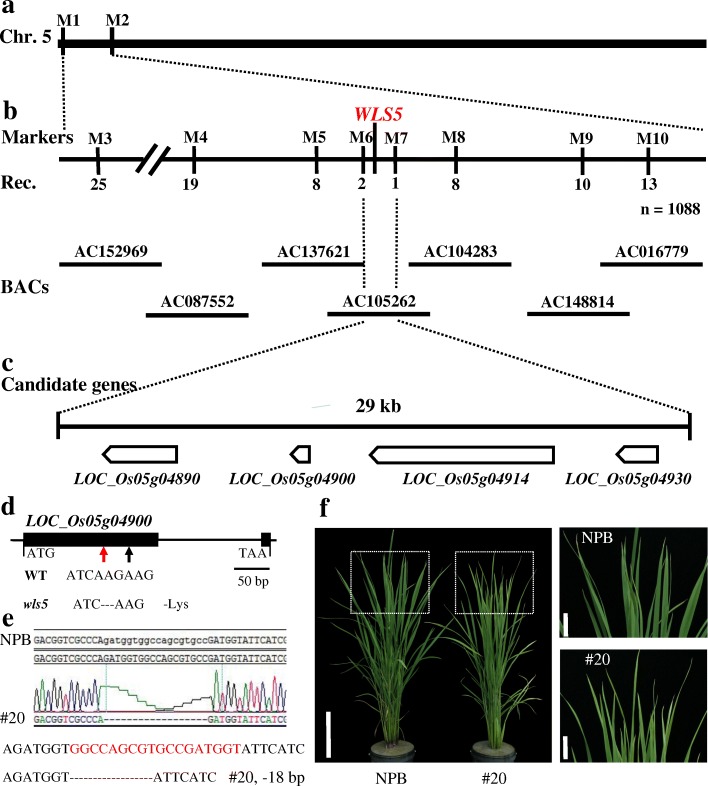
We employed a map-based cloning approach to isolate the gene responsible for the wls5 phenotype. A mapping population was generated by crossing wls5 with the japonica cultivar ‘Nipponbare’. All F1 plants showed a normal phenotype matching that of the wild type. A segregating population of 1088 F2 plants was obtained, among which 821 plants displayed the normal wild-type phenotype and 267 plants exhibited the early leaf senescence phenotype similar to wls5, fitting a typical segregation ratio of 3:1 (χ2 = 0.12 < χ20.05 = 3.84). This suggested that wls5 was controlled by a single recessive nuclear gene.
Twenty-one F2 plants with the wls5 phenotype were used for primary mapping, which located WLS5 on the short arm of chromosome 5 between markers M1 and M2 (Fig. 9a). To further narrow the WLS5 region, we developed a number of additional insertion/deletion (InDel) markers between M1 and M2, as shown in Additional file 2: Table S1. These allowed mapping of WLS5 to a region of 29 kb between markers M6 and M7, with two and one recombinants, respectively (Fig. 9b). This region does not contain any known senescence-related loci. Four genes were predicted within this 29-kb region, based on the Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/) (Fig. 9c), LOC_Os05g04890, LOC_Os05g04900, LOC_Os05g04914, and LOC_Os05g04930. Sequence alignment of these four genes from wild-type and wls5 plants identified only a 3-bp deletion in the coding sequence of LOC_Os05g04900 in wls5, which led to a lysine (Lys) deletion in the protein (Fig. 9d). LOC_Os05g04900 encodes a predicted protein of 87 amino acids with no predicted function; however, the protein has two potential transmembrane domains.
Fig. 9.
Candidate gene analysis for WLS5. a Location of WLS5 on rice chromosome 5. b Coarse linkage map of WLS5. Markers used for mapping are indicated. Numbers below lines indicate number of recombinants. c Candidate genes for WLS5. d LOC_Os05g04900 structure and sequence variations between wild type and wls5. Black and red arrows indicate mutation site and target knockout position, respectively. e, f LOC_Os05g04900 knockout in ‘Nipponbare’ (NPB). CRISPR/Cas9-induced 18-bp deletion in LOC_Os05g04900 (e). Phenotype of NPB and line #20. Bars = 25 cm (left) and 12.5 cm (right) (f)
To test whether LOC_Os05g04900 is responsible for the wls5 phenotype, we used genome editing to generate an additional mutant allele. Using CRISPR/Cas9, we obtained a homozygous mutant, line #20, with an 18-bp deletion in LOC_Os05g04900, in the ‘Nipponbare’ genetic background (Fig. 9e). This genetically edited line showed similar leaf senescence phenotype to the wls5 mutant (Fig. 9f), indicating that LOC_Os05g04900 might be the candidate gene for WLS5.
Discussion
As the final phase of plant development, senescence is under the control of a highly regulated genetic program (Lim et al. 2007). A growing number of leaf senescence-associated genes have been cloned and functionally characterized in rice (Mitchell and Sheehy 2006; Yamada et al. 2014; Wu et al. 2016; Yang et al. 2016a; Yang et al. 2016b; Zhao et al. 2016; Deng et al. 2017; Leng et al. 2017b; Mao et al. 2017; Hong et al. 2018). However, the molecular mechanism underlying leaf senescence remains poorly understood. In this study, we characterized a novel premature senescence mutant, wls5, which also showed weak plant growth.
Previous studies have identified and characterized several mutants involved in both premature leaf senescence and weak plant growth in rice (Wu et al. 2016; Xie et al. 2016; El Mannai et al. 2017; Leng et al. 2017b; He et al. 2018; Hong et al. 2018). In Arabidopsis, mutations in some genes also lead to similar phenotype. For instance, the mod1 (mosaic death1) mutant displayed multiple morphological phenotypes, including chlorotic and semidwarfism (Mou et al. 2000). The T-DNA knockout plants of RPI2 gene (At2g01290) were smaller than its wild-type and exhibited chlorosis phenotype (Xiong et al. 2009), and PIC1 (permease in chloroplasts 1) mutants showed severely impaired plastid development and plant growth (Duy et al. 2011). These results suggest that leaf senescence and plant growth processes might be regulated by the same genetic factors in plants. Here, wls5 mutant plants not only showed leaf senescence throughout their growth (Fig. 3a, b, Fig. 4a), but also exhibited dwarfism and small organs (Fig. 1, Fig. 3a). Similar to previously characterized mutants, leaf senescence in wls5 was directly related to low chlorophyll levels and high ROS accumulation (Fig. 3c, Fig. 4b, Fig. 5), and abnormal growth of wls5 was caused by a reduction in cell number and small cell size compared to the wild type (Fig. 2). Thus, one genetic factor affects these two processes, although the underlying mechanism remains largely unclear and requires further study. The wls5 mutant provides ideal material for this research.
Leaf senescence is generally accompanied by a decrease in chlorophyll levels and an increase in various ROS (He et al. 2018; Hong et al. 2018). In the present study, chlorophyll was degraded in wls5 (Fig. 3c, Fig. 4b), and chloroplast structure was also abnormal (Fig. 3f). Meanwhile, expression levels of genes involved in chlorophyll synthesis and chloroplast development were significantly down-regulated in wls5 plants compared to those in the wild type (Fig. 4c, d). Therefore, we speculate that degradation of chlorophyll and chloroplasts plays an important role in leaf senescence in wls5 plants. Various metabolic processes generate ROS in plants, causing oxidative damage to thylakoid membranes and other cellular components (Apel and Hirt 2004); (Wang et al. 2015a, b). Here, we found greater accumulation of superoxide anions (O2−) and H2O2 in wls5 plants than wild-type plants (Fig. 5a, b, d). Consistent with this, more electrolyte leakage was observed in wls5 than in the wild type (Fig. 5c). In addition, APX and POD activities were higher in wls5 than in wild-type plants (Fig. 5g, h). RNA-seq indicated that expression of genes involved in oxidoreductase activity and oxidation-reduction processes was remarkably altered in wls5 plants (Additional file 1: Figure S2-S4). Taking these finding together, it is clear that leaf senescence in wls5 plants involves ROS.
Phytohormones play important regulatory roles in promoting or delaying leaf senescence (Jan et al. 2019). In our study, wls5 mutants had lower SA, GA3, ABA, JA, and zeatin contents, and higher IAA content compared to the wild type (Fig. 7). These altered levels of endogenous hormones in wls5 mutants may contribute to its leaf senescence phenotype. Furthermore, genes responsive to ABA and involved in ABA-activated signaling pathways were dramatically up-regulated or down-regulated in wls5 plants compared to the wild type (Additional file 1: Figure S4). However, ABA is an important endogenous factor that induces leaf senescence in plants (Lin et al. 2015). This is not consistent ith the low ABA content in wls5 plants, indicating that leaf senescence in the wls5 mutant may not involve ABA signaling pathways.
Using a map-based cloning approach, we fine mapped WLS5 to a 29-kb region containing four predicted genes on the short arm of chromosome 5 (Fig. 9a-c). No evidence currently suggests that these four genes are associated with premature leaf senescence or weakness in plant growth; thus, wls5 is most likely a novel mutant for weakness and leaf senescence in rice. Sequence comparison of these four genes between the wild type and wls5 identified only a 3-bp deletion in the coding sequence region of LOC_Os05g04900, a gene encoding an expressed protein of unknown function, in wls5 (Fig. 9d). This mutation caused a lysine (Lys) deletion in the predicted protein. Given the leaf senescence phenotype of the LOC_Os05g04900 knockout line (Fig. 9e, f), we inferred that LOC_Os05g04900 is the candidate gene for WLS5. By multiple sequences alignment and phylogeny evolution analysis of LOC_Os05g04900 sequence with the other species, we found no homologous genes to WLS5/wls5 except for the GRMZM2G111850 in maize, whose gene function is also unidentified. Therefore, WLS5/wls5 is a new genetic factor for growth and senescence in plants, but its molecular function needs further study.
Conclusions
A novel mutant involved in plant development and leaf senescence was identified in rice. LOC_Os05g04900, encoding a protein of unknown function, is the candidate gene for WLS5. Further molecular study of wls5 will uncover the functional roles of this gene in plant growth and leaf senescence.
Methods
Plant materials and growth conditions
The rice (Oryza sativa) wls5 mutant was isolated from the ethyl methane sulfonate treated indica cultivar ‘93–11’. An F2 mapping population was generated from a cross between wls5 and the japonica cultivar ‘Nipponbare’. All plants were grown in paddy fields in either Zhejiang or Hainan provinces in China (2015–2018).
Paraffin sectioning and microscopic analysis
Internodes and leaves of wild-type and wls5 plants were collected at heading stage and fixed in 50% ethanol, 0.9 M glacial acetic acid, and 3.7% formaldehyde overnight at 4 °C. These samples were then dehydrated with a graded series of ethanol, infiltrated with xylene and embedded in paraffin (Sigma). The specimens were sectioned (8 mm thickness) with a Leica RM2245 microtome, then transferred onto poly-L-Lys-coated glass slides, deparaffinized in xylene and dehydrated through an ethanol series. After staining with 1% safranin (Amresco) and 1% Fast Green (Amresco), sections were dehydrated through an ethanol series, infiltrated with xylene and finally mounted beneath a coverslip (Ren et al. 2016). Light microscopy was performed using a Nikon SMZ1500 microscope.
Measurement of pigment content and photosynthetic rate
Fresh leaf samples (0.05 g) of wild-type and wls5 plants were cut into small segments and incubated with 5 ml 80% acetone in the dark for 24 h. Absorbance of the supernatant was then measured with an ultraviolet spectrophotometer (DU800, BECKMAN, Fullerton, USA) at 470, 645, and 663 nm (Chen et al. 2018; Wang et al. 2018). Total chlorophyll was determined according to the methods of Arnon (1949) and Wellburn (1994). Ten biological replicates were analyzed for each condition.
Photosynthetic rates of wild-type and wls5 plants grown in Zhejiang were measured at 65 days post-sowing at 10:30 am on a sunny day using a LI-6400 portable photosynthesis device (LICOR, USA). Fifteen biological replicates were included.
TEM analysis
TEM assays were performed according to the methods described by Leng et al. (2017a). Fresh leaves of wild-type and wls5 plants were fixed in 2.5% glutaraldehyde in phosphate-buffered saline (PBS, 137 mM NaCl, 2.7 mM KCl, 8 mM Na2HPO4, and 2 mM KH2PO4, pH 7.4) for at least 4 h and washed in PBS three times. The samples were then postfixed with 1% (w/v) OsO4 for 2 h after extensive washing in PBS, dehydrated with a graded ethanol series and infiltrated with Spurr Kit (Sigma). The specimens were sectioned (70 nm ultrathin) with a Leica EM UC7 ultramicrotome, and sections were stained with uranyl acetate and alkaline lead citrate for 10 min. TEM was performed using a Hitachi model H-7650.
Histochemical staining and ROS-scavenging enzyme assays
DAB and NBT staining were performed on leaves from wild-type and wls5 plants to detect H2O2 and superoxide anions, as described by Blum and Ebercon (1981). Electrolyte leakage was determined in accordance with a previous study (Zhou and Guo, 2009). H2O2, MDA content and CAT, APX, and POD activity of wild-type and wls5 mutant leaves were measured using an Assay Kit (Suzhou Keming Biotechnology Co., Ltd.).
RT-qPCR analysis
Total RNA was extracted from fresh leaf samples of wild-type and wls5 plants using a Micro RNA Extraction kit (Axygen) and reverse transcribed into cDNA using a Rever Tra Ace qPCR-RT kit (TOYOBA, Japan). qPCR was conducted on an ABI PRISM 7900HT Sequence Detector (Applied Biosystems) according to the manufacturer’s instructions. Primers for RT-qPCR are listed in Additional file 2: Table S1. The rice UBQ5 gene was used as an internal control. Three biological replicates of each sample were prepared.
Hormone extraction and determination of hormone levels
Levels of SA, ABA, GA3, JA, IAA, and zeatin were determined according to a previously described method with little modification (You et al. 2016). Fresh leaf samples (1.0 g) of wild-type and wls5 plants were ground in pre-cooled extraction buffer (10 ml) and shaken at 4 °C for 30 min. Dichloromethane (20 mL) was added, and the sample was shaken at 4 °C for 30 min and then centrifuged at 16,020 g for 5 min. The organic phase was extracted and dried under liquid nitrogen. Pellets were dissolved in 150 mL methanol (0.1% methane acid) and filtered with a 0.22-mm filter membrane. The purified product was then subjected to high-performance liquid chromatography tandem mass spectrometry (HPLC-MS/MS) analysis. The injection volume was 2 mL. Mass spectrometry conditions were as follows: spray voltage was 4500 V; pressure of the air curtain, nebulizer and aux gas was 15, 65 and 70 psi, respectively; and atomizing temperature was 400 °C.
RNA-seq analysis
Total RNA was extracted using a TRK1001 Total RNA Purification Kit (LC Science, Houston, TX, USA) following the manufacturer’s procedure, then quantified using a Bioanalyzer 2100 and RNA 6000 Nano LabChip Kit (Agilent, CA, USA) with RIN number > 7.0. Poly(A) mRNA was isolated from approximately 5 μg total RNA by using magnetic beads with poly-T oligo attached (Invitrogen). cDNA was synthesized using random hexamer primers. A library was constructed and sequenced using an Illumina Hiseq 2000/2500 (LC Sciences, USA). More than 50 million reads from wild-type and wls5 plants were obtained. Gene ontology analysis was performed using GOseq (Young et al. 2010). Pathway enrichment analysis was performed using the Kyoto Encyclopedia of Genes and Genomes database (Kanehisa et al. 2008).
Map-based cloning
A fine-mapping population was generated from a cross between a wls5 mutant (in the indica background) and the japonica rice cultivar ‘Nipponbare’. A total of 267 individual plants from a segregating population of 1088 F2 plants showed the same phenotype as the wls5 mutant, and were sampled for mapping. The initial location of wls5 was determined using 163 simple sequence repeat (SSR) markers scattered across the 12 rice chromosomes (http://www.gramene.org). For fine mapping, eight InDel markers between the M1 and M2 markers were designed based on genomic DNA sequences from ‘Nipponbare’ and the indica rice variety ‘93–11’, to narrow down the wls5 region. All markers used are detailed in Additional file 2: Table S1.
CRISPR/Cas9 vector construction and rice transformation
CRISPR/Cas9 genome editing vector construction was performed as described by (Wang et al. 2015a, b). One specific target guide RNA sequence, GATGGTGGCCAGCGTGCCGATGG, located in the first exon was selected to generate mutants of LOC_Os05g04900. Primers for vector construction are listed in Additional file 2: Table S1. The fragment was inserted into a pC1300-UBI:Cas9 vector and introduced into ‘Nipponbare’ by Agrobacterium tumefaciens (EHA105) mediated transformation. Twenty independent transgenic plants (T0) were obtained and sequenced. One homozygous mutant, line #20, and its wild type (‘Nipponbare’) were grown in the field for phenotype determination.
Additional files
Figure S1. Histological characterization of leaves in wild-type and wls5 plants. a, b Cross sections of leaves of wild-type (‘93-11’) and wls5. c-f Longitudinal sections of leaves of wild-type and wls5 plants. Scale bar = 50 μm. Figure S2. Statistics of GO enrichment analysis of differential expression genes between wild-type (WT) and wls5 plants. mRNA was purified from total RNA isolated from tillering-stage plants of WT (‘93-11’) and wls5. Figure S3. Statistics of pathway enrichment analysis of differential expression genes between wild-type (WT) and wls5 plants. mRNA was purified from total RNA isolated from tillering-stage plants of WT (‘93-11’) and wls5. Figure S4. The number of differential expression genes involved in different biological processes, cellular components and molecular functions. mRNA was purified from total RNA isolated from tillering-stage plants of WT (‘93-11’) and wls5. (PDF 2035 kb)
Table S1. The list of primers used in this study (PDF 77 kb)
Acknowledgements
Not applicable.
Funding
This study was supported by the Natural Science Foundation of China (Grant Nos. 31461143014), the National Science-technology Support Projects (Grant No.2015BAD01B02–02), the National Key Research and Development Program of China (Grant No.2016YFD0100902–07).
Availability of data and materials
All data generated or analyzed during this study are available in this published article (and its additional files).
Abbreviations
- CRISPR/Cas9
Clustered regularly interspaced short palindromic repeats and CRISPR-associated protein 9
- DAB
3,3′-diaminobenzidine
- NBT
Nitro blue tetrazolium
- qPCR
Quantitative PCR
- TEM
Transmission electron microscopy
Authors’ contributions
C-YZ, C-LL and L-BG designed the experiments; C-YZ, C-LL, YZ, Y-TC, H-TH, NJ and YL performed the experiments; C-YZ and C-LL analyzed the data; C-LL and C-YZ wrote the manuscript. L-BG and QQ revised the manuscript. All authors read and approved the manuscript.
Ethics approval and consent to participate
There are no ethics issues associated with this article.
Consent for publication
These co-authors involved in the paper all consent to publish this article in Rice.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Chunyan Zhao, Email: zhaochunyan525@163.com.
Chaolei Liu, Email: liuchaolei505@163.com.
Yu Zhang, Email: zhangyu881005@163.com.
Yongtao Cui, Email: cuiyongtao20@163.com.
Haitao Hu, Email: haitao-hu@zjnu.cn.
Noushin Jahan, Email: noushincnrri@yahoo.com.
Yang Lv, Email: 342771473@qq.com.
Qian Qian, Email: qianqian188@hotmail.com.
Longbiao Guo, Email: guolongbiao@caas.cn.
References
- Apel K, Hirt H. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol. 2004;55(1):373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- Arnon DI. Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 1949;24:1. doi: 10.1104/pp.24.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum A, Ebercon A. Cell membrane stability as a measure of drought and heat tolerance in wheat 1. Crop Sci. 1981;21(1):43–47. [Google Scholar]
- Chen P, Hu HT, Zhang Y, Wang ZW, Dong GJ, Cui YT, Qian Q, Ren DY, Guo LB. Genetic analysis and fine-mapping of a new rice mutant, white and lesion mimic leaf1. Plant Growth Regul. 2018;85:425–435. [Google Scholar]
- Deng L, Qin P, Liu Z, Wang G, Chen W, Tong J, Xiao L, Tu B, Sun Y, Yan W, He H, Tan J, Chen X, Wang Y, Li S, Ma B. Characterization and fine-mapping of anovel premature leaf senescence mutant yellow leaf and dwarf 1 in rice. Plant Physiol Biochem. 2017;111:50–58. doi: 10.1016/j.plaphy.2016.11.012. [DOI] [PubMed] [Google Scholar]
- Duan Y, Li S, Chen Z, Zheng L, Diao Z, Zhou Y, Lan T, Guan H, Pan R, Xue Y, Wu W. Dwarf and deformed flower 1, encoding an F-box protein, is critical forvegetative and floral development in rice (Oryza sativa L.) Plant J. 2012;72(5):829–842. doi: 10.1111/j.1365-313X.2012.05126.x. [DOI] [PubMed] [Google Scholar]
- Duy D, Stübe R, Wanner G, Philippar K. The chloroplast permease PIC1 regulates plant growth and development by directing homeostasis and transport of iron. Plant Physiol. 2011;155(4):1709–1722. doi: 10.1104/pp.110.170233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El MY, Akabane K, Hiratsu K, Satoh-Nagasawa N, Wabiko H (2017) The NAC transcription factor gene OsY37 (ONAC011) promotes leaf senescence and accelerates heading time in rice. Int J Mol Sci 18(10) [DOI] [PMC free article] [PubMed]
- He Yan, Zhang Zhihong, Li Liangjian, Tang Shaoqing, Wu Jian-Li. Genetic and Physio-Biochemical Characterization of a Novel Premature Senescence Leaf Mutant in Rice (Oryza sativa L.) International Journal of Molecular Sciences. 2018;19(8):2339. doi: 10.3390/ijms19082339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y, Zhang Y, Sinumporn S, Yu N, Zhan X, Shen X, Chen D, Yu P, Wu W, Liu Q, Cao Z, Zhao C, Cheng S, Cao L. Premature leaf senescence 3, encoding a methyltransferase, is required for melatonin biosynthesis in rice. Plant J. 2018;95(5):887–891. doi: 10.1111/tpj.13995. [DOI] [PubMed] [Google Scholar]
- Jan S, Abbas N, Ashraf M, Ahmad P. Roles of potential plant hormones and transcription factors in controlling leaf senescence and drought tolerance. Protoplasma. 2019;256(2):313–329. doi: 10.1007/s00709-018-1310-5. [DOI] [PubMed] [Google Scholar]
- Jiang H, Li M, Liang N, Yan H, Wei Y, Xu X, Liu J, Xu Z, Chen F, Wu G. Molecular cloning and function analysis of the stay green gene in rice. Plant J. 2007;52(2):197–209. doi: 10.1111/j.1365-313X.2007.03221.x. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36(Database issue):D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke S, Liu S, Luan X, Xie XM, Hsieh TF, Zhang XQ. Mutation in a putative glycosyltransferase-like gene causes programmed cell death and early leaf senescence in rice. Rice. 2019;12(1):7. doi: 10.1186/s12284-019-0266-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek BA. Making bigger plants: key regulators of final organ size. Curr Opin Plant Biol. 2009;12(1):17–22. doi: 10.1016/j.pbi.2008.09.006. [DOI] [PubMed] [Google Scholar]
- Lee RH, Wang CH, Huang LT, Chen SC. Leaf senescence in rice plants: cloning and characterization of senescence up-regulated genes. J Exp Bot. 2001;52(358):1117–1121. doi: 10.1093/jexbot/52.358.1117. [DOI] [PubMed] [Google Scholar]
- Leng Y, Yang Y, Ren D, Huang L, Dai L, Wang Y, Chen L, Tu Z, Gao Y, Li X, Zhu L, Hu J, Zhang G, Gao Z, Guo L, Kong Z, Lin Y, Qian Q, Zeng D. A rice PECTATELYASE-LIKE gene is required for plant growth and leaf senescence. Plant Physiol. 2017;174(2):1151–1166. doi: 10.1104/pp.16.01625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng Yujia, Ye Guoyou, Zeng Dali. Genetic Dissection of Leaf Senescence in Rice. International Journal of Molecular Sciences. 2017;18(12):2686. doi: 10.3390/ijms18122686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim PO, Kim HJ, Nam HG. Leaf senescence. Annu Rev Plant Bio. 2007;58:115–136. doi: 10.1146/annurev.arplant.57.032905.105316. [DOI] [PubMed] [Google Scholar]
- Lin M, Pang C, Fan S, Song M, Wei H, Yu S. Global analysis of the Gossypium hirsutum L. transcriptome during leaf senescence by RNA-Seq. BMC Plant Biol. 2015;15(1):43. doi: 10.1186/s12870-015-0433-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Wei X, Sheng Z, Jiao G, Tang S, Luo J, Hu P. Polycomb protein OsFIE2 affects plant height and grain yield in rice. PLoS One. 2016;11(10):e0164748. doi: 10.1371/journal.pone.0164748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao C, Lu S, Lv B, Zhang B, Shen J, He J, Luo L, Xi D, Chen X, Ming F. A rice NAC transcription factor promotes leaf senescence via ABA biosynthesis. Plant Physiol. 2017;174(3):1747–1763. doi: 10.1104/pp.17.00542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G, Suzuki N, Ciftci-Yilmaz S, Mittler R. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 2010;33(4):453–467. doi: 10.1111/j.1365-3040.2009.02041.x. [DOI] [PubMed] [Google Scholar]
- Mitchell PL, Sheehy JE. Supercharging rice photosynthesis to increase yield. New Phytol. 2006;171(4):688–693. doi: 10.1111/j.1469-8137.2006.01855.x. [DOI] [PubMed] [Google Scholar]
- Mou Z, He Y, Dai Y, Liu X, Li J. Deficiency in fatty acid synthase leads to premature cell death and dramatic alterations in plant morphology. Plant Cell. 2000;12(3):405–418. doi: 10.1105/tpc.12.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller-Roeber B, Balazadeh S. Auxin and its role in plant senescence. J Plant Growth Regul. 2014;33(1):21–33. [Google Scholar]
- Pan S, Rasul F, Li W, Tian H, Mo Z, Duan M, Tang X. Roles of plant growth regulators on yield, grain qualities and antioxidant enzyme activities in super hybrid rice (Oryza sativa L.) Rice. 2013;6(1):9. doi: 10.1186/1939-8433-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao Y, Yang Y, Xu J, Li X, Leng Y, Dai L, Huang L, Shao G, Ren D, Hu J, Guo L, Pan J, Zeng D. EARLY SENESCENCE1 encodes a SCAR-LIKE PROTEIN2 that affects water loss in rice. Plant Physiol. 2015;169(2):1225–1239. doi: 10.1104/pp.15.00991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D, Rao Y, Huang L, Leng Y, Hu J, Lu M, Zhang G, Zhu L, Gao Z, Dong G, Guo L, Qian Q, Zeng D. Fine mapping identifies a new QTL for brown rice rate in rice (Oryza sativa L.) Rice. 2016;9(1):4. doi: 10.1186/s12284-016-0076-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy N, Bagchi S, Raychaudhuri P. Damaged DNA binding protein 2 in reactive oxygen species (ROS) regulation and premature senescence. Int J Mol Sci. 2012;13(9):11012–11026. doi: 10.3390/ijms130911012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T, Matsuoka M. Identifying and exploiting grain yield genes in rice. Curr Opin Plant Biol. 2008;11(2):209–214. doi: 10.1016/j.pbi.2008.01.009. [DOI] [PubMed] [Google Scholar]
- Tan J, Tan Z, Wu F, Sheng P, Heng Y, Wang X, Ren Y, Wang J, Guo X, Zhang X, Cheng Z, Jiang L, Liu X, Wang H, Wan J. A novel chloroplast-localized pentatricopeptide repeat protein involved in splicing affects chloroplast development and abiotic stress response in rice. Mol Plant. 2014;7(8):1329–1349. doi: 10.1093/mp/ssu054. [DOI] [PubMed] [Google Scholar]
- Tang Y, Li M, Chen Y, Wu P, Wu G, Jiang H. Knockdown of OsPAO and OsRCCR1 cause different plant death phenotypes in rice. J Plant Physiol. 2011;168(16):1952–1959. doi: 10.1016/j.jplph.2011.05.026. [DOI] [PubMed] [Google Scholar]
- Wang C, Shen L, Fu Y, Yan C, Wang K. A simple CRISPR/Cas9 system for multiplex genome editing in rice. J Genet Genomics. 2015;42(12):703–706. doi: 10.1016/j.jgg.2015.09.011. [DOI] [PubMed] [Google Scholar]
- Wang Z, Wang Y, Hong X, Hu D, Liu C, Yang J, Li Y, Huang Y, Feng Y, Gong H, Li Y, Fang G, Tang H, Li Y. Functional inactivation of UDP-N-acetylglucosamine pyrophosphorylase 1 (UAP1) induces early leaf senescence and defence responses in rice. J Exp Bot. 2015;66(3):973–987. doi: 10.1093/jxb/eru456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZW, Lv J, Xie SZ, Zhang Y, Qiu ZN, Chen P, Cui YT, Niu YF, Hu SK, Jiang HZ, Sheng-zhen Ge SZ, Trinh HP, Lei KR, Bai WQ, Zhang Y, Guo LB, Ren DY. OsSLA4 encodes a pentatricopeptide repeat protein essential for early chloroplast development and seedling growth in rice. Plant Growth Regul. 2018;84:249–260. [Google Scholar]
- Wellburn AR. The spectral determination of chlorophylls a and b, as well as total carotenoids, using various solvents with spectrophotometers of different resolution. J Plant Physiol. 1994;144:307–313. [Google Scholar]
- Wu L, Ren D, Hu S, Li G, Dong G, Jiang L, Hu X, Ye W, Cui Y, Zhu L, Hu J, Zhang G, Gao Z, Zeng D, Qian Q, Guo L. Down-regulation of a nicotinate phosphoribosyltransferase gene, OsNaPRT1, leads to withered leaf tips. Plant Physiol. 2016;171(2):1085–1098. doi: 10.1104/pp.15.01898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q, Liang Y, Zhang J, Zheng H, Dong G, Qian Q, Zuo J. Involvement of a putative bipartite transit peptide in targeting rice pheophorbide a oxygenase into chloroplasts for chlorophyll degradation during leaf senescence. J Genet Genomics. 2016;43(3):145–154. doi: 10.1016/j.jgg.2015.09.012. [DOI] [PubMed] [Google Scholar]
- Xiong Y, DeFraia C, Williams D, Zhang X, Mou Z. Deficiency in a cytosolic ribose-5-phosphate isomerase causes chloroplast dysfunction, late flowering and premature cell death in Arabidopsis. Physiol Plant. 2009;137(3):249–263. doi: 10.1111/j.1399-3054.2009.01276.x. [DOI] [PubMed] [Google Scholar]
- Yamada Y, Furusawa S, Nagasaka S, Shimomura K, Yamaguchi S, Umehara M. Strigolactone signaling regulates rice leaf senescence in response to a phosphate deficiency. Planta. 2014;240(2):399–408. doi: 10.1007/s00425-014-2096-0. [DOI] [PubMed] [Google Scholar]
- Yang X, Nian J, Xie Q, Feng J, Zhang F, Jing H, Zhang J, Dong G, Liang Y, Peng J, Wang G, Qian Q, Zuo J. Rice ferredoxin-dependent glutamate synthase regulates nitrogen-carbon metabolomes and is genetically differentiated between japonica and indica subspecies. Mol Plant. 2016;9(11):1520–1534. doi: 10.1016/j.molp.2016.09.004. [DOI] [PubMed] [Google Scholar]
- Yang Y, Xu J, Huang L, Leng Y, Dai L, Rao Y, Chen L, Wang Y, Tu Z, Hu J, Ren D, Zhang G, Zhu L, Guo L, Qian Q, Zeng D. PGL, encoding chlorophyllide aoxygenase 1, impacts leaf senescence and indirectly affects grain yield and quality in rice. J Exp Bot. 2016;67(5):1297–1310. doi: 10.1093/jxb/erv529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You C, Zhu H, Xu B, Huang W, Wang S, Ding Y, Liu Z, Li G, Chen L, Ding C, Tang S. Effect of removing superior spikelets on grain filling of inferior spikelets in rice. Frontiers Plant Sci. 2016;7:1161. doi: 10.3389/fpls.2016.01161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11(2):R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, Wei J, Ma Q, Yu D, Li J. Senescence of aerial parts is impeded by exogenous gibberellic acid in herbaceous perennial Paris polyphylla. J Plant Physiol. 2009;166(8):819–830. doi: 10.1016/j.jplph.2008.11.002. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Chan Z, Gao J, Xing L, Cao M, Yu C, Hu Y, You J, Shi H, Zhu Y, Gong Y, Mu Z, Wang H, Deng X, Wang P, Bressan RA, Zhu JK. ABA receptor PYL9 promotes drought resistance and leaf senescence. Proc Natl Acad Sci U S A. 2016;113(7):1949–1954. doi: 10.1073/pnas.1522840113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Guo Z. Calcium is involved in the abscisic acid-induced ascorbate peroxidase, superoxide dismutase and chilling resistance in Stylosanthes guianensis. Biol Plant. 2009;53(1):63–68. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Histological characterization of leaves in wild-type and wls5 plants. a, b Cross sections of leaves of wild-type (‘93-11’) and wls5. c-f Longitudinal sections of leaves of wild-type and wls5 plants. Scale bar = 50 μm. Figure S2. Statistics of GO enrichment analysis of differential expression genes between wild-type (WT) and wls5 plants. mRNA was purified from total RNA isolated from tillering-stage plants of WT (‘93-11’) and wls5. Figure S3. Statistics of pathway enrichment analysis of differential expression genes between wild-type (WT) and wls5 plants. mRNA was purified from total RNA isolated from tillering-stage plants of WT (‘93-11’) and wls5. Figure S4. The number of differential expression genes involved in different biological processes, cellular components and molecular functions. mRNA was purified from total RNA isolated from tillering-stage plants of WT (‘93-11’) and wls5. (PDF 2035 kb)
Table S1. The list of primers used in this study (PDF 77 kb)
Data Availability Statement
All data generated or analyzed during this study are available in this published article (and its additional files).