Abstract
Plant long non-coding RNA (lncRNA) undergoes dynamic regulation and acts in developmental and stress regulation. In this study, we surveyed the expression dynamics of lncRNAs in grapevine (Vitis vinifera L.) under cold stress using high-throughput sequencing. Two-hundred and three known lncRNAs were significantly up-regulated and 144 known lncRNAs were significantly down-regulated in cold-treated grapevine. In addition, 2 088 novel lncRNA transcripts were identified in this study, with 284 novel lncRNAs significantly up-regulated and 182 novel lncRNAs significantly down-regulated in cold-treated grapevine. Two-hundred and forty-two differentially expressed grapevine lncRNAs were predicted to target 326 protein-coding genes in a cis-regulatory relationship. Many differentially expressed grapevine lncRNAs targeted stress response-related genes, such as CBF4 transcription factor genes, late embryogenesis abundant protein genes, peroxisome biogenesis protein genes, and WRKY transcription factor genes. Sixty-two differentially expressed grapevine lncRNAs were predicted to target 100 protein-coding genes in a trans-regulatory relationship. The expression of overall target genes in both cis and trans-regulatory relationships were positively related to the expression of lncRNAs in grapevines under cold stress. We identified 31 known lncRNAs as 34 grapevine micro RNA (miRNA) precursors and some miRNAs may be derived from multiple lncRNAs. We found 212 lncRNAs acting as targets of miRNAs in grapevines, involving 150 miRNAs; additionally, 120 grapevine genes were predicted as targets of grapevine miRNAs and lncRNAs. We found one gene cluster that was up-regulated and showed the same expression trend. In this cluster, many genes may be involved in abiotic stress response such as WRKY, Hsf, and NAC transcription factor genes.
Subject terms: Genomics, Abiotic
Introduction
In eukaryotes, many transcripts are non-coding RNAs (ncRNAs)1,2. Long ncRNA (lncRNA) is a type of ncRNA that is generally >200 nt long and has no discernable coding potential3,4. Most lncRNAs can be broadly classified into three types based on their genomic positions: (1) lncRNAs transcribed from intergenic regions of lncRNAs are known as lincRNAs (long intergenic non-coding RNA); (2) lncRNAs transcribed from introgenic regions are long intronic RNAs, which can be transcribed in any orientation relative to coding genes; and (3) long non-coding nature antisense transcripts (lncNAT) that overlap with protein-coding regions or ncRNAs on the opposite strand and antisense RNA5–7. In eukaryotes, different lncRNAs have been shown to be differentially expressed in different tissues or under different stress conditions. This indicates that lncRNAs undergo dynamic regulation and act in the regulation of development and stress response8. LncRNAs have been shown to be involved in gene silencing, the control of flowering time, photomorphogenesis in seedlings, organogenesis in roots, and reproduction in plants4,9–16. Some lncRNAs can also serve as precursors to small RNAs17–22. Some lncRNAs can regulate proteins or microRNAs (miRNA) by acting as decoys that mimic target DNA or RNA. For example, the Arabidopsis microRNA target mimics the IPS1 lncRNA and the decoy ASCO-lncRNA14,23. This illustrates the competing endogenous RNA (ceRNA) theory, which is well-supported and is now widely accepted17,24. The ceRNA theory states that mRNA, lncRNAs, pseudogenes, and other miRNA sponges share common miRNA binding sites because the amount of any given miRNA is limited24.
Currently, growing evidence supports the view that non-coding RNAs, including lncRNAs, play important roles in regulating responses to a variety of abiotic and biotic stressors25–27. A previous study has identified 318 lncRNAs responsive to cold and/or drought stress in cassava28. In cotton, some lncRNAs were shown to possibly be involved in regulating plant hormone pathways in response to drought stress29. Several stress-responsive lncRNAs have been functionally characterized in plant signaling pathways such as lncRNA npc4830, At4/IPS123,31, and npc53632. In addition, miRNA, another non-coding RNA, was shown to be involved in various abiotic stress responses such as cold stress (chilling or freezing) in plants33–36. LncRNAs compete with other miRNA sponges, such as target gene mRNA, to play important roles in eukaryotes30,37–40. Therefore, lncRNA may play important roles in various abiotic stress responses via the ceRNA mechanism.
Cold stress is an important environmental factor that negatively affects grapevine productivity and quality. However, in grapevine, the function of lncRNA and the relationship between grapevine lncRNA and cold stress or cold stress tolerance are unknown. Here, cold-inducible lncRNAs in grapevine were detected using RNA-sequencing and analysis. The potential function of these lncRNAs, their target genes, and the relationship between grapevine mRNAs, lncRNAs, and miRNAs were also predicted and analyzed. Our aims were to identify the cold-responsive lncRNAs and determine if or how cold stress response in grapevine is related to lncRNA regulation.
Results
Data mining of transcriptome sequencing and identification of lncRNAs in grapevine
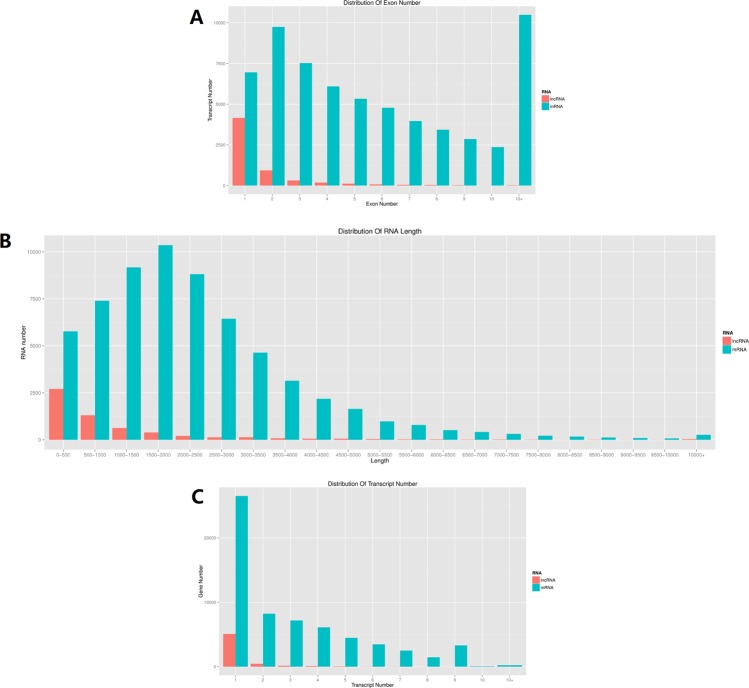
To systematically identify lncRNAs related to cold stress in grapevine, we performed whole transcriptome RNA-seq of grapevine cv. Cabernet Sauvignon that had been submitted to a cold-stress treatment of 4 °C. We generated an average of 12.65 gigabases (Gb) of raw reads per sample from the six samples used for Illumina RNA-sequencing. The total number of raw reads per control (CK) sample (plants were kept under a 16-h light/8-h dark photoperiod at 26 °C) ranged from 220842362 to 274931726, and the number of clean reads in each CK sample ranged from 216561108 to 270342092. The total number of raw reads in each cold treatment sample ranged from 191766324 to 233777742, and the number of clean reads in each cold treatment sample ranged from 186259776 to 223345340. The average mapping rate to the grapevine genome is 63.77%. In total, we identified 56732 transcripts, including 44644 known mRNA transcripts, 2031 known lncRNA transcripts, 7969 novel mRNA transcripts, and 2088 novel lncRNA transcripts. The transcripts of novel lncRNAs predicted here are listed in Table S1, and the transcript names and the related lncRNA gene IDs are listed in Table S2. The transcripts of novel mRNAs predicted here are listed in Table S3, and the transcript names and the related mRNA gene IDs are listed in Table S4. In all samples, we identified 212 novel lincRNAs, 1 933 novel long intronic RNAs, and 688 novel lncNAT. We also found 1 893 known lincRNAs, 511 known long intronic RNAs, and 803 known lncNAT in total samples. In addition, we found that it was the most common for the lncRNAs to contain only one exon; lncRNAs containing two exons were the next most common, followed by lncRNAs containing three exons and four exons (Fig. 1A). The lncRNAs less than 500 bp long were most common, followed by the 500–1000 bp long lncRNAs and 1000–1500 bp long lncRNAs (Fig. 1B). We also found that most lncRNAs were located on chromosome 1 (Fig. 1C).
Figure 1.
Characteristics of grape lncRNAs. (A) The number of exons per transcript for all mRNAs and lncRNAs. (B) Transcript size distributions for all mRNAs and lncRNAs. (C) Distribution of mRNAs and lncRNAs along each chromosome.
Variation in lncRNA expression among cold stress
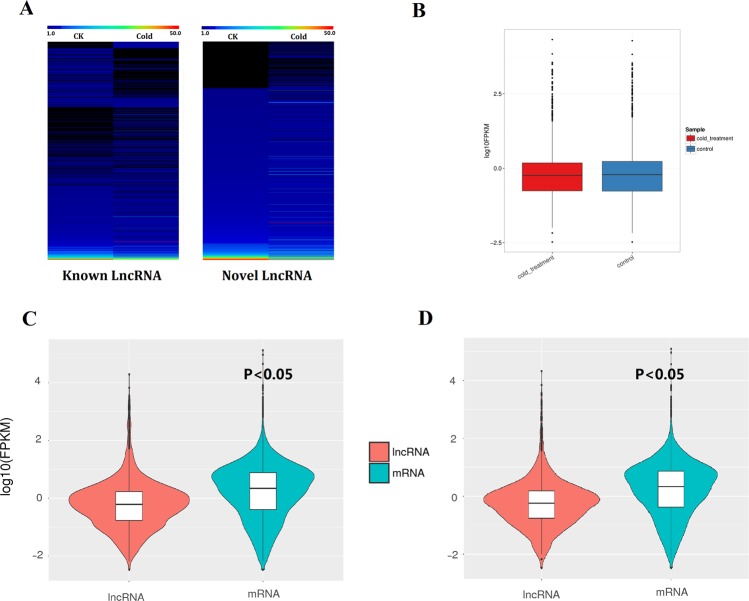
In grapevine, 17 known lncRNAs were expressed only in the CK library and 97 known lncRNAs were expressed only in the cold-treated library. The expression heatmaps of all known and novel grape lncRNAs in the CK and cold treatment based on the Fragments Per Kilobase Million (FPKM) model are shown in Fig. 2A, and the box plot of expression levels of grape lncRNAs in the CK and cold treatment are shown in Fig. 2B. In both the control and cold treatments, the average expression level of the total lncRNAs was lower than that of the mRNAs in grapevine (Fig. 2C,D).
Figure 2.
Expression models of grape lncRNAs and mRNAs. (A) The expression heatmap of all known and novel grape lncRNAs in the control and cold treatment based on the average FPKM value of each set of replicates. (B) The box plot of expression levels of grape lncRNAs under the control and cold treatment conditions. The y-axis represents the average log2 (FPKM) value of each set of replicates. (C) The violin map of expression levels of grape lncRNAs and mRNAs in the control. The y-axis represents the average log2 (FPKM) value of three replicates. T-test p-values < 0.05 are considered to be significantly different. (D) The violin map of expression levels of grape lncRNAs and mRNAs under cold treatment. The y-axis represents the average log2 (FPKM) value of three replicates. T-test p-values < 0.05 are considered to be significantly different.
Two-hundred and three known lncRNAs were significantly up-regulated (fold change > 2, P < 0.05) and 144 known lncRNAs were significantly down-regulated in cold-treated grapevine (fold change < −2, P < 0.05). In grapevine, VIT_203s0017n00360 was the lncRNA with the greatest increase of up-regulation by the cold treatment, followed by VIT_207s0031n00070 and VIT_201s0011n00530. VIT_209s0002n00340 was the lncRNA with the greatest down-regulation by cold treatment, followed by VIT_213s0158n00020 and VIT_213s0067n00110. These significantly up- and down-regulated lncRNAs were considered the differentially expressed known lncRNAs (Fig. 3A, Table S5).
Figure 3.
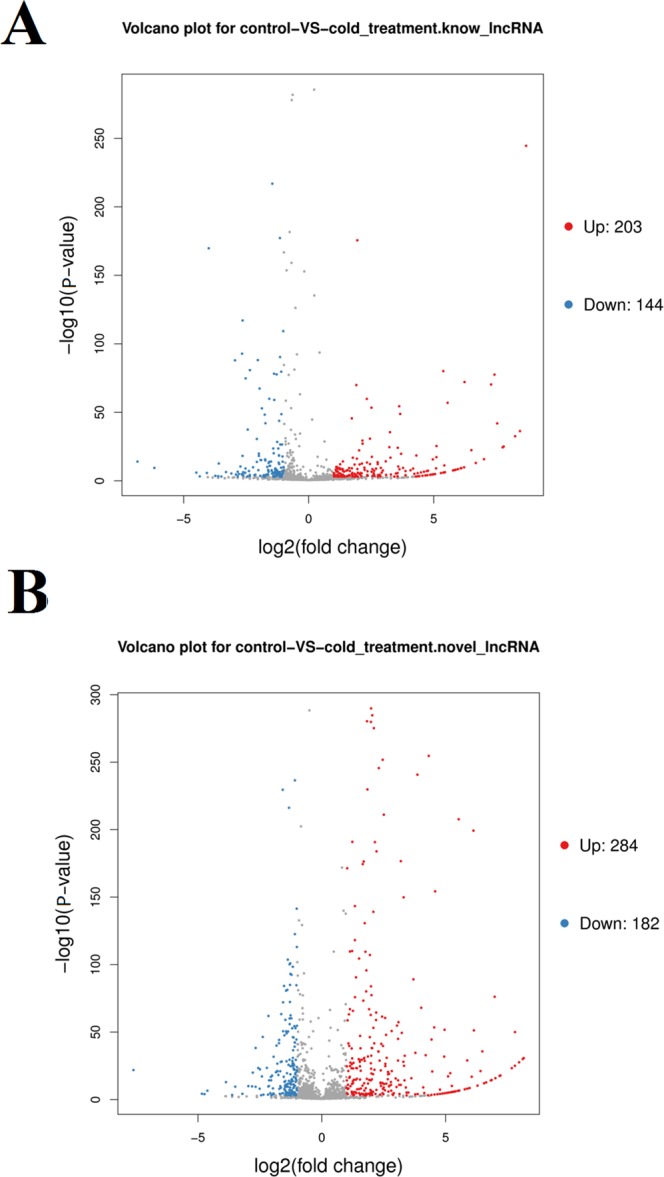
Differentially expressed lncRNAs in grapevine treated with cold stress. (A) The Volcano map of differentially expressed known lncRNAs and (B) differentially expressed novel lncRNAs. The x-axis represents the log2 (FPKM) values of the differentially expressed lncRNAs, and the y-axis represents the −log10 (P value) values of the differentially expressed lncRNAs.
In grapevine, 17 novel lncRNAs were expressed only in the untreated library and 11 novel lncRNAs were expressed only in the cold-treated library. We identified 284 novel lncRNAs as significantly up-regulated (fold change > 2, P < 0.05) and 182 novel lncRNAs were significantly down-regulated (fold change > 2, P < 0.05) in cold-treated grapevine compared with in the CK. In grapevine, LXLOC_001173 was the lncRNA with the greatest up-regulation in the cold treatment compared with the CK, followed by LXLOC_004676 and LXLOC_028762. Compared with the CK, LXLOC_003867 was the lncRNA with the greatest down-regulation in the cold treatment, followed by LXLOC_011153 and LXLOC_017876. These significantly up- and down-regulated lncRNAs were considered the differentially expressed novel lncRNAs (Fig. 3B, Table S5).
Prediction of target genes of cold-related lncRNA targets in cis-regulatory relationships
To investigate the possible functions of grape lncRNAs, we predicted the potential targets of lncRNAs in cis-regulatory relationships. We searched for known protein-coding genes located within 10 kb downstream and upstream of all the identified grape lncRNAs. These genes were thought to be the targets of lncRNAs in cis-regulatory relationships if the Pearson and Spearman correlation coefficients between the expression levels of these genes were ≥0.6 or ≤−0.6, and P < 0.0541.
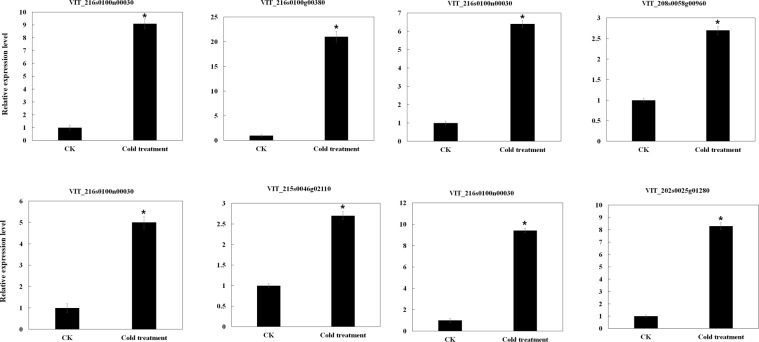
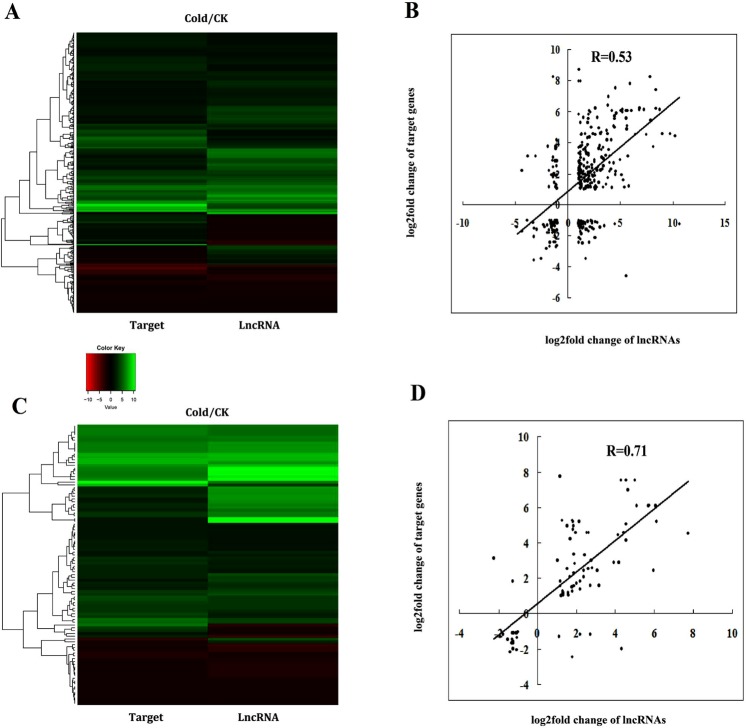
Our results predicted a total of 2 527 target genes in cis-regulatory relationships of 1 650 lncRNAs in grapevine. In our study, significantly up-regulated or down-regulated lncRNAs were thought to be differentially expressed lncRNAs. Specifically, we found that 242 differentially expressed grapevine lncRNAs were predicted to target 326 protein-coding genes in cis-regulatory relationships, and many differentially expressed grapevine lncRNAs targeted stress response-related genes such as CBF4 transcription factor genes, late embryogenesis abundant protein genes, peroxisome biogenesis protein genes, and WRKY transcription factor genes (Table S6). The expression levels of some target genes in cis-regulatory relationships were positively related to lncRNAs. For example, VIT_216s0100n00030, LXLOC_027751, LXLOC_010422, and VIT_202s0025n00100 were up-regulated under cold stress compared to the CK. Based on our RNA-seq data, their target genes VIT_216s0100g00380 (CBF4 transcription factor), VIT_208s0058g00960 (transcription factor bHLH61), VIT_215s0046g02110 (late embryogenesis abundant protein Lea14-A), and VIT_202s0025g01280 (WRKY transcription factor 41) respectively, were also up-regulated under cold stress compared to the CK. The RNA-seq data was validated by the qRT-PCR results (Fig. 4). The expression levels of some target genes in cis-regulatory relationships were negatively related to lncRNAs. For example, compared to the CK, LXLOC_013001 was down-regulated under cold stress, but its target gene in the cis-regulatory relationship, VIT_217s0000g06350 (chlorophyll a-b binding protein 4), was up-regulated under cold stress when compared to the control. LXLOC_019156 was up-regulated under cold stress, but its target gene in the cis-regulatory relationship, VIT_202s0154g00610 (peroxisome biogenesis protein), was down-regulated under cold stress (Table S6, Fig. 5A). We calculated the correlation coefficient between the expression changes of lncRNAs and their target genes in cis-regulatory relationships under cold stress. As shown in Fig. 5B, the values of the x-axis are the log2fold change of lncRNAs (fold change = FPKM value of genes in the cold treatment/FPKM value of lncRNA genes in the control). The values along the y-axis are the log2fold change of their target genes in cis-regulatory relationships (fold change = FPKM value of genes in the cold treatment/FPKM value of genes in the control). The correlation coefficient was 0.53 (t-test, P < 0.05), indicating that the expression of overall target genes with a cis-regulatory relationship was positively related to the expression of related lncRNAs in grapevine under cold stress (Fig. 5B). The heatmap of expression of lncRNAs and their target genes in cis-regulatory relationships under cold stress based on the log2fold change value also showed that the expression of the overall target genes with cis-regulatory relationships were positively related to the expression of related lncRNAs in grapevine under cold stress (Fig. 5A).
Figure 4.
Expression level of cold inducible grapevine lncRNAs and their target genes validated by qRT-PCR. T-test p-values < 0.05 are considered to be significantly different, and “*” represents a p-value < 0.05.
Figure 5.
Comparison of the expression changes of differentially expressed lncRNAs, related target genes, and the correlation between them. The heatmap was generated from the fold change values in the RNA-seq data and was used to visualize the lncRNAs and cis-regulated relation target expression changes (A) and the lncRNAs and trans-regulated relation target expression changes (C). (B) The correlation between the expression changes of lncRNAs and cis-regulated relation target. The values along the x-axis are the log2fold change of lncRNAs (fold change = FPKM value of genes in the cold treatment/FPKM value of lncRNA genes in the control). The values along the y-axis are the log2fold change of their target genes in cis-regulatory relationships (fold change = FPKM value of genes in the cold treatment/FPKM value of genes in the control). (D) The correlation between the expression changes of lncRNAs and trans-regulated relation target. The values along the x-axis are the log2fold change of lncRNAs (fold change = FPKM value of genes in the cold treatment/FPKM value of lncRNA genes in the control). The values along the y-axis are the log2fold change of their target genes in trans-regulatory relationships (fold change = FPKM value of genes in the cold treatment/FPKM value of genes in the control).
Analysis of target genes of cold-related lncRNAs in trans-regulatory relationships
To investigate the possible functions of grapevine lncRNAs, we predicted the potential targets of lncRNAs in trans-regulatory relationships. RNAplex software42 was used to identify the lncRNA (parameters: >−30 binding energy) as described in a previous study41. The Pearson and Spearman correlation coefficients between the expression of these genes identified using RNAplex and the expression of related lncRNAs must be ≥0.6 or ≤0.6 and P < 0.05, or will be filtered out41.
In grapevine, we predicted a total of 574 target genes in trans-regulatory relationships with 422 lncRNAs (Table S6). The results showed that 62 differentially expressed grapevine lncRNAs were predicted to target 100 protein-coding genes in trans-regulatory relationships such as NADH dehydrogenase subunit genes, UDP-glycosyltransferase genes, calcium-transporting ATPase genes, disease resistance protein genes, and glutamate receptor genes. However, most target genes in trans-regulatory relationships were unknown protein coding genes (Table S6). The expression levels of some target genes in trans-regulatory relationships were positively related with lncRNAs. For example, VIT_200s0225n00020 was down-regulated under cold stress, and its target gene, VIT_200s0246g00150 (NADH dehydrogenase subunit 5), in the trans-regulatory relationship was up-regulated under cold stress. Some target genes with trans-regulatory relationships were negatively related to lncRNAs (Fig. 5C).
We calculated the correlation coefficient between the expression changes of lncRNAs and their target genes in trans-regulatory relationships in the cold stress treatment. As shown in Fig. 5D, the values along the x-axis are the log2fold change of lncRNAs (fold change = FPKM value of genes in the cold treatment/FPKM value of lncRNA genes in the CK). The values along the y-axis are the log2fold change of their target genes in trans-regulatory relationships (fold change = FPKM value of genes in the cold treatment/FPKM value of genes in the control). The correlation coefficient was 0.71 (t-test, P < 0.05), indicating that the expression levels of overall target genes with trans-regulatory relationships were positively related to lncRNAs in grapevine under cold stress (Fig. 5D). The heatmap of expression of lncRNAs and their target genes in trans-regulatory relationships under cold stress based on the log2fold change value also showed that the expression of overall target genes with a trans-regulatory relationship were positively related to the expression of lncRNAs in grapevine under cold stress (Fig. 5C).
GO enrichment and KEGG pathway analyses for differentially expressed lncRNA targets
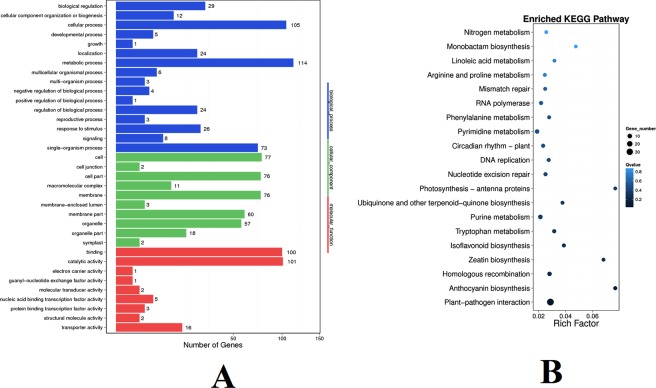
The potential function of grapevine lncRNAs in response to cold stress was studied using gene ontology (GO) annotation and enrichment analysis. Targets of differentially expressed cultivated grapevine lncRNAs were classified into three categories, 438 in biological processes, 231 in molecular functions, and 455 in cellular components. Biological processes contained 16 sub-categories with 299 terms, including the regulation of jasmonic acid mediated signaling pathway (GO: 2000022), regulation of defense response (GO: 0031347), regulation of signal transduction (GO: 0009966), hormone metabolic process (GO: 0042445), regulation of hormone levels (GO: 0010817), transmembrane transport (GO: 0055085), lipid metabolic process (GO: 0006629), chloroplast organization (GO: 0009658), flavonoid biosynthetic process (GO: 0009813), and flavonoid metabolic process (GO: 0009812). Molecular functions contained nine sub-categories with 156 terms, including chlorophyll binding (GO: 0016168), transcription factor activity, sequence-specific DNA binding (GO: 0003700), signal transducer activity (GO: 0004871), transcription factor activity, and transcription factor binding (GO: 0000989). Cellular components contained 10 sub-categories with 85 terms, including chloroplast (GO: 0009507), photosystem (GO: 0009521), chloroplast stroma (GO: 0009570), chloroplast envelope (GO: 0009941), and photosystem I (GO: 0009535) (Fig. 6A, Table S7). In molecular functions, the significantly enriched (P < 0.05) GO term was calcium ion transmembrane transporter activity (GO: 0015085). In target genes of differentially expressed grapevine lncRNAs, 87 KEGG (The Kyoto Encyclopedia of Gene and Genome) pathways were obtained and significantly enriched (P < 0.05) KEGG pathways included plant-pathogen interaction (ko04626), anthocyanin biosynthesis (ko00942), homologous recombination (ko03440), and zeatin biosynthesis (ko00908) (Table 1, Fig. 6B).
Figure 6.
GO annotation and KEGG enrichment analysis of the differentially expressed target genes of lncRNAs. (A) The GO terms of the target genes of differentially expressed grapevine lncRNAs. (B) The top 20 enriched target genes of differentially expressed grapevine lncRNAs.
Table 1.
Significantly enriched KEGG pathway of differential expressed grape lncRNAs.
| KEGG Pathway | Pvalue | Pathway ID |
|---|---|---|
| Plant-pathogen interaction | 1.9E-07 | ko04626 |
| Anthocyanin biosynthesis | 0.00072 | ko00942 |
| Homologous recombination | 0.00595 | ko03440 |
| Zeatin biosynthesis | 0.01217 | ko00908 |
| Isoflavonoid biosynthesis | 0.01363 | ko00943 |
| Tryptophan metabolism | 0.01814 | ko00380 |
| Purine metabolism | 0.02709 | ko00230 |
| Ubiquinone and other terpenoid-quinone biosynthesis | 0.02882 | ko00130 |
| Photosynthesis - antenna proteins | 0.03233 | ko00196 |
| Nucleotide excision repair | 0.04746 | ko03420 |
| DNA replication | 0.04943 | ko03030 |
Validation of lncRNA expression using qRT-PCR
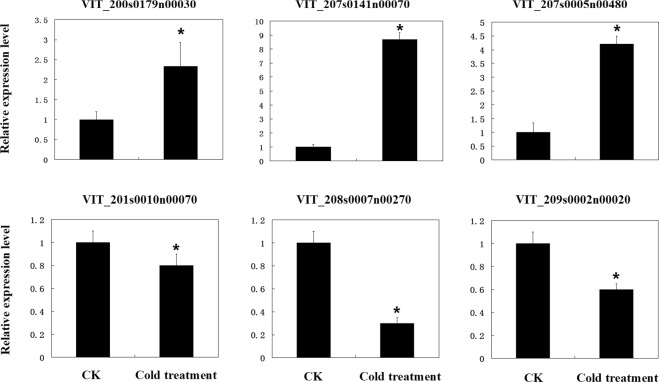
We performed qRT-PCR analyses to validate the RNA-seq results from six randomly selected grapevine lncRNAs, VIT_201s0010n00070, VIT_209s0002n00020, VIT_200s0179n00030, VIT_207s0141n00070, VIT_208s0007n00270, and VIT_207s0005n00480. The primers for qRT-PCR are listed in Table S8. The expression results were similar to the deep sequencing data (Fig. 7). VIT_200s0179n00030, VIT_207s0141n00070, and VIT_207s0005n00480 were shown to be up-regulated by the qRT-PCR data, showing a positive correlation with the deep sequencing results. VIT_201s0010n00070, VIT_208s0007n00270 and VIT_209s0002n00020 were down-regulated in both the qRT-PCR and RNA-seq results (Fig. 7, Table S5).
Figure 7.
Expression level of select grapevine lncRNAs validated by qRT-PCR. T-test p-values < 0.05 are considered to be significantly different, and “*” represents a p-value < 0.05.
LncRNAs as potential miRNA precursors
By aligning miRNA precursors to grapevine lncRNAs, we identified 31 known lncRNAs as 34 grapevine miRNA precursors, including vvi-MIR169h, vvi-MIR399a, vvi-MIR394b, vvi-MIR166a, and vvi-MIR156c (Table 2). We identified 25 novel lncRNA transcripts (19 lncRNA genes) as 22 grapevine miRNA precursors, including vvi-MIR162, vvi-MIR168, vvi-MIR535, vvi-MIR403a, vvi-MIR3623, and vvi-MIR3630. Some miRNAs may be derived from multiple lncRNAs. For example, vvi-MIR396b may be derived from VIT_211s0016n00330 and LXLOC_003224 (Table 2).
Table 2.
grape miRNAs and the lncRNAs as their precursors.
| MiRNA and lncRNA as precursor | MiRNA and lncRNA as precursor | ||
|---|---|---|---|
| vvi-MIR156c | VIT_204s0008n00030 | vvi-MIR162 | LXLOC_012888 |
| vvi-MIR159a | VIT_215s0046n00070 | vvi-MIR162 | LXLOC_012888 |
| vvi-MIR159b | VIT_215s0046n00080 | vvi-MIR164c | LXLOC_029337 |
| vvi-MIR160c | VIT_210s0092n00020 | vvi-MIR167a | LXLOC_000093 |
| vvi-MIR164b | VIT_209s0002n00040 | vvi-MIR167b | LXLOC_009023 |
| vvi-MIR166a | VIT_208s0032n00030 | vvi-MIR167b | LXLOC_007999 |
| vvi-MIR166b | VIT_212s0034n00230 | vvi-MIR167b | LXLOC_007999 |
| vvi-MIR166c | VIT_215s0048n00320 | vvi-MIR168 | LXLOC_019119 |
| vvi-MIR166d | VIT_216s0098n00100 | vvi-MIR168 | LXLOC_019119 |
| vvi-MIR166e | VIT_202s0025n00230 | vvi-MIR169g | LXLOC_028343 |
| vvi-MIR166f | VIT_207s0031n00260 | vvi-MIR169r | LXLOC_003511 |
| vvi-MIR167d | VIT_200s0179n00030 | vvi-MIR169t | LXLOC_003511 |
| vvi-MIR167e | VIT_205s0020n00290 | vvi-MIR169u | LXLOC_003511 |
| vvi-MIR169y | VIT_201s0146n00060 | vvi-MIR396b | LXLOC_003224 |
| vvi-MIR169m | VIT_211s0103n00100 | vvi-MIR396d | LXLOC_003867 |
| vvi-MIR169r | VIT_211s0103n00110 | vvi-MIR398a | LXLOC_000033 |
| vvi-MIR169t | VIT_211s0103n00110 | vvi-MIR535a | LXLOC_033356 |
| vvi-MIR169u | VIT_211s0103n00110 | vvi-MIR535a | LXLOC_033356 |
| vvi-MIR171a | VIT_214s0068n00210 | vvi-MIR535a | LXLOC_033356 |
| vvi-MIR171b | VIT_212s0059n00020 | vvi-MIR535b | LXLOC_033356 |
| vvi-MIR394b | VIT_218s0001n00020 | vvi-MIR535b | LXLOC_033356 |
| vvi-MIR394b | VIT_218s0001n00020 | vvi-MIR535b | LXLOC_033356 |
| vvi-MIR396b | VIT_211s0016n00330 | vvi-MIR535c | LXLOC_033356 |
| vvi-MIR396d | VIT_211s0016n00340 | vvi-MIR535c | LXLOC_033356 |
| vvi-MIR399a | VIT_210s0003n00240 | vvi-MIR535c | LXLOC_033356 |
| vvi-MIR399b | VIT_216s0100n00020 | vvi-MIR403a | LXLOC_022332 |
| vvi-MIR169h | VIT_211s0103n00060 | vvi-MIR403c | LXLOC_022332 |
| vvi-MIR169i | VIT_211s0103n00070 | vvi-MIR477a | LXLOC_033061 |
| vvi-MIR169l | VIT_211s0103n00080 | vvi-MIR477a | LXLOC_016789 |
| vvi-MIR169n | VIT_211s0103n00100 | vvi-MIR3623 | LXLOC_014879 |
| vvi-MIR169o | VIT_211s0103n00090 | vvi-MIR3630 | LXLOC_013003 |
| vvi-MIR319e | VIT_211s0016n00290 | vvi-MIR3630 | LXLOC_013632 |
| vvi-MIR394c | VIT_218s0001n00230 | vvi-MIR3630 | LXLOC_013003 |
| vvi-MIR828a | VIT_216s0098n00140 | vvi-MIR3633a | LXLOC_012920 |
| vvi-MIR3636 | VIT_216s0013n00110 | vvi-MIR3633b | LXLOC_012920 |
The relationships between grape mRNAs, lncRNAs, and miRNAs
We predicted the lncRNAs as targets or target mimics of miRNAs. In some previous studies, lncRNAs were found as both targets and target mimics of miRNAs43,44. As target mimics, lncRNAs could bind to miRNAs with a three-nucleotide bulge43. In our study, we only found lncRNAs that paired with miRNAs without any bulges. These lncRNA may be targets of miRNAs but not target mimics of miRNAs. Here, 212 lncRNAs as targets of miRNAs in grapevine were involved with 150 miRNAs (Table S9). Additionally, 120 predicted grapevine genes were both the target of grapevine miRNAs and lncRNAs (Table S10).
Gene clusters show the same trends
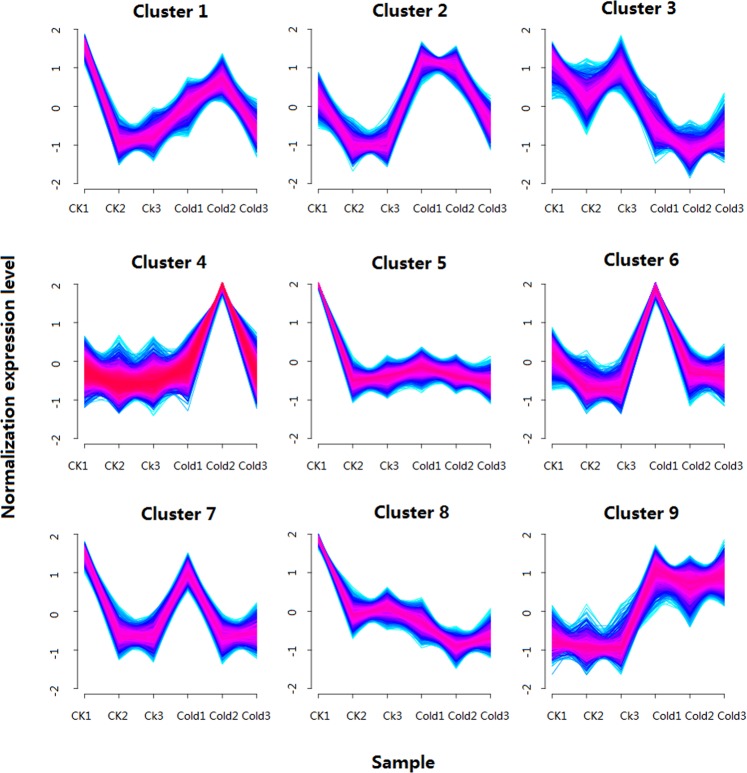
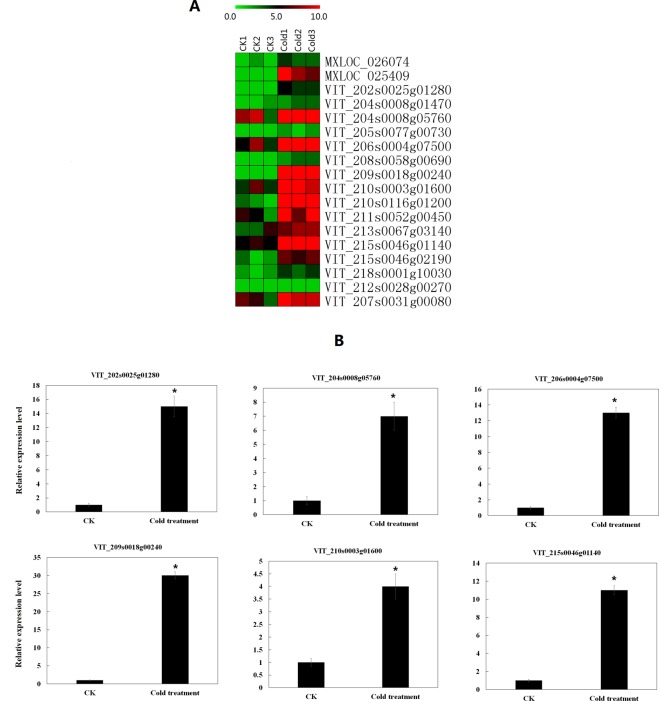
The Mufzz software45 was used to cluster grapevine genes into gene clusters showing similar expression trends based on the expression changes of genes in cold treated grapevines. Nine clusters were identified and the genes with the same expression trend were clustered together (Fig. 8). Cluster 5 only contained 19 lncRNAs, Cluster 1 contained one lncRNAs, and Cluster 7 did not contain any lncRNAs. Clusters 2, 3, 4, 6, 8, and 9 contained more lncRNAs and their target genes than other clusters (Table S11). For example, Cluster 9 contained 137 lncRNAs and their 89 target genes. In cold treated grapevines, Cluster 9 showed an up-regulated expression pattern (Fig. 8), and in this cluster, 45 lncRNAs were significantly up-regulated under cold stress. In addition, 12 of their target genes were in Cluster 9, and the 12 target genes, which were significantly up-regulated under cold stress, contained LRR receptor-like serine/threonine-protein kinase, hydroxyacyl glutathione hydrolase, prolyl 4-hydroxylase subunit alpha-1, calcium-transporting ATPase 2, and some unnamed proteins (Table 3). Cluster 9 contained many ethylene-responsive transcription factor genes, such as two ERF5s and four ABSCISIC ACID-INSENSITIVE 5-like protein genes. Cluster 9 also contained NAM/ATAF/CUC (NAC) trans-transcription factor genes, such as NAC 68 and 94, as well as Hsf transcription factor genes, such as HsfA3, MYBA1, flavanone 7-O-glucoside 2′-O-beta-L-rhamnosyltransferase, isoflavone-7-O-methyltransferase 9, and WD repeat-containing protein (Table S11). Cluster 9 also contained 19 WRKY transcription factor genes including WRKY 3, 7, 11, 22, 28, 33, 40, 41, 46, 47, 48, and 50 (Table 4). RNA-seq data showed that 17 of these WRKY genes were significantly up-regulated (Fig. 9A), which was confirmed by the qRT-PCR (Fig. 9B). In cold-treated grapevines, Cluster 3 showed a down-regulated expression pattern (Fig. 8). Cluster 3 contained some ABSCISIC ACID-INSENSITIVE protein genes, auxin response factor genes, proline synthase co-transcribed bacterial homolog protein genes, NAC domain-containing protein genes, basic helix-loop-helix DNA-binding super family protein genes, cold-inducible RNA-binding protein genes, and WRKY transcription factor genes (Table S11).
Figure 8.
The expression pattern of genes from nine clusters in the control and cold treated samples.
Table 3.
Up-regulated target genes belonged to cluster 9.
| LnRNAs ID | Target genes ID | Gene annotation |
|---|---|---|
| LXLOC_000552 | MXLOC_000552 | solute carrier family 35 member B1 |
| LXLOC_001158 | MXLOC_000261 | DUF246 domain-containing protein |
| LXLOC_001364 | MXLOC_001364 | unnamed protein product |
| LXLOC_001364 | MXLOC_001363 | unnamed protein product |
| LXLOC_001969 | MXLOC_001969 | LRR receptor-like serine/threonine-protein kinase |
| LXLOC_007796 | MXLOC_008787 | exopolyphosphatase |
| LXLOC_008601 | MXLOC_008600 | GPI ethanolamine phosphate transferase |
| LXLOC_011465 | MXLOC_011465 | unnamed protein product |
| LXLOC_019669 | MXLOC_019669 | unnamed protein product |
| LXLOC_024063 | MXLOC_024063 | hydroxyacylglutathione hydrolase 2 |
| LXLOC_033142 | MXLOC_033143 | prolyl 4-hydroxylase subunit alpha-1 |
| VIT_207s0129n00010 | MXLOC_026393 | calcium-transporting ATPase 2 |
Table 4.
WRKY genes in cluster 9.
| Genes ID | Gene annotation | Expressed change |
|---|---|---|
| MXLOC_026074 | WRKY47 | Up-regulated significantly |
| MXLOC_025409 | WRKY33 | Up-regulated significantly |
| VIT_202s0025g01280 | WRKY41 | Up-regulated significantly |
| VIT_204s0008g01470 | WRKY50 | Up-regulated significantly |
| VIT_204s0008g05760 | WRKY3 | Up-regulated significantly |
| VIT_205s0077g00730 | WRKY48 | Up-regulated significantly |
| VIT_206s0004g07500 | WRKY33 | Up-regulated significantly |
| VIT_207s0031g00080 | WRKY7 | No change |
| VIT_208s0058g00690 | WRKY33 | Up-regulated significantly |
| VIT_209s0018g00240 | WRKY40 | Up-regulated significantly |
| VIT_210s0003g01600 | WRKY65 | Up-regulated significantly |
| VIT_210s0116g01200 | WRKY6 | Up-regulated significantly |
| VIT_211s0052g00450 | WRKY11 | Up-regulated significantly |
| VIT_212s0028g00270 | WRKY28 | No change |
| VIT_213s0067g03140 | WRKY70 | Up-regulated significantly |
| VIT_215s0046g01140 | WRKY46 | Up-regulated significantly |
| VIT_215s0046g02190 | WRKY22 | Up-regulated significantly |
| VIT_218s0001g10030 | WRKY7 | Up-regulated significantly |
Figure 9.
Expression level of WRKY genes in Cluster 9. (A) The heatmap was generated from the FPKM value of WRKY genes in Cluster 9 in each set of replicates. (B) Expression levels of some WRKY genes in Cluster 9 validated by qRT-PCR. T-test p-values < 0.05 are considered to be significantly changed, and “*” represents a p-value < 0.05.
Discussion
A previous study reported the existence of lncRNAs in plants37. As next generation sequencing technology developed, it became possible to identify lncRNAs including those identified in Arabidopsis, rice, maize, cassava4,8,28,46, and grapevine (http://genomes.cribi.unipd.it/DATA/V2/V2.1/lncRNA/). However, few studies have been conducted on the roles of lncRNAs involved in abiotic and biotic stress responses. In addition, there has been limited research conducted on the roles of lncRNA involved in abiotic stress response, such as response to cold stress, in grapevine. In this study, we detected the expression changes of lncRNAs in grapevine exposed to cold treatment and found 2 088 novel grapevine lncRNAs. Previous studies have also identified novel lncRNAs in other plant taxa including 6 500 novel lncRNAs in Arabidopsis thaliana8, 1 704 novel lncRNAs in maize45, and 682 novel lncRNAs in cassava28.
Here, we found that the average expression level of the total lncRNAs was lower than the average expression level of mRNAs in grapevine in both the control and cold treatment conditions (Fig. 2C,D). This indicates that the expression levels of total lncRNAs should be lower than mRNAs in grapevine. In A. thaliana, approximately 300 lncRNAs were evidenced to be differentially expressed under abiotic stressors27,31, and 318 cassava lncRNAs were differentially expressed under cold and drought conditions28. Here, we found 813 differentially expressed grapevine lncRNAs in the cold stress treatment, showing that more grapevine lncRNAs were differentially expressed under cold stress. We hypothesize that many grapevine lncRNAs may be related to cold stress and may play important roles in cold stress response. Though the expression levels in many lncRNAs changed in the cold treatment, the average expression levels of the total lncRNAs in the cold treatment were similar to the average expression levels of the total lncRNAs under control conditions (Fig. 2B).
We predicted the target genes of cold inducible grape lncRNAs, finding more target genes of cold inducible grapevine lncRNAs in cis-regulatory relationships than in trans-regulatory relationships. This indicated that the target genes in cis-regulatory relationships may be more related to cold stress response. We also analyzed the expression correlation between the total cold inducible grapevine lncRNAs and their target genes, and our results showed that the expression patterns were positively related.
The expression correlation between cold inducible grapevine lncRNAs and their target genes in trans-regulatory relationships were higher than in cis-regulatory relationships. However, some of the expression patterns of lncRNAs were negatively related to their target genes (Fig. 3). A previous study showed that lncRNAs could act as enhancers of gene expression47. In kiwifruit, the expression of both protein-coding genes and lncRNA genes tended to be more positively correlated than negatively correlated in trans-regulatory relationships48. Here, we found that the expression of the overall target genes with a cis-regulatory relationship was also positively related to the expression of related lncRNAs in grapevine under cold stress.
Some target genes of cold inducible grapevine lncRNAs in cis-regulatory relationships may be involved in abiotic stress response such as VIT_216s0100g00380 (CBF4 transcription factor), VIT_215s0046g02110 (late embryogenesis abundant protein Lea14-A), and VIT_202s0025g01280 (WRKY transcription factor 41). These genes were also up-regulated in the cold stress treatment. Previous research has shown that CBF family genes play critical roles related to control of an important pathway in the cold acclimation process49,50. CBF4 is one of the most important members for the over-wintering of grapevines50. Some LEA proteins have been shown to be involved in the freezing tolerance of plants51. Additionally, some WRKY transcription factors have been shown to be involved in modulating gene expression in plants during cold stress52. These cold stress-related genes were also up-regulated under cold stress. Therefore, we hypothesize that these cold stress-related genes could be regulated by related lncRNAs under cold stress. These lncRNAs may play important roles in cold stress tolerance and may be related to the regulation of these cold stress-related genes.
The GO analysis showed that the biological process terms that are related to cold stress lncRNAs contained the regulation of jasmonic acid (JA) mediated signaling pathway (GO: 2000022), regulation of defense response (GO: 0031347), regulation of signal transduction (GO: 0009966), hormone metabolic process (GO: 0042445), and regulation of hormone levels (Fig. 6A, Table S7). Jasmonic acid is related to cold stress response in plants53. Other hormones, such as abscisic acid (ABA), are related to abiotic responses in plants54. Molecular function terms of genes that are related to cold stress lncRNAs contained transcription factor activity, sequence-specific DNA binding (GO: 0003700), and transcription factor binding (GO: 0000989) (Fig. 6A, Table S7), indicating that many target genes were transcription factors or were related to transcription factors. These transcription factors may be involved in cold response and the regulation of other downstream genes involved in cold response. Cellular component terms of genes that were related to the cold-related lncRNAs contained photosystem (GO: 0009521) and photosystem I (GO: 0009535) (Fig. 6A, Table S7), showing that many target genes may be related to photosystems. Under cold stress, the photosystems have been shown to be related to cold tolerance55.
We identified 31 known lncRNAs as 34 grapevine miRNA precursors, including vvi-MIR169h, vvi-MIR399a, vvi-MIR394b, vvi-MIR166a, and vvi-MIR156c (Table 2). In cassava, 12 lncRNAs were identified as 11 known cassava miRNA precursors, including miR156g, miR160d, miR166h, miR167g, and miR169d28. The lncRNAs that are precursors of miR156 and miR169 family members were identified in both grape and cassava28, indicating that some lncRNAs from different species might have been derived from same ancestral genes.
A previous study has shown that the lncRNAs that acted as target mimics could bind to miRNAs with three-nucleotide bulges43. However, our data did not predict similar target mimics that have been found in previous studies43, but the data did predict some targets that could bind to miRNAs without three-nucleotide bulges. These lncRNA may be targets of miRNAs. Similarly, a previous report has shown that lncRNAs acting as target genes could bind to miRNAs without bulges44. We found that lncRNAs and protein coding genes shared common miRNAs, which could target both lncRNAs and protein coding genes, and miRNAs and lncRNAs shared common target genes in grapevine. We hypothesize that the lncRNAs may regulate protein coding genes via complex pathways in grapevines.
The genes with the same expression trends were clustered together, and the genes in the same cluster may be involved in the same biological process45. We identified one cluster (Cluster 9) that showed an up-regulated expression pattern under cold treatment (Fig. 8). In this cluster, many genes may be involved in abiotic stress response such as WRKY transcription factor genes51, Hsf transcription factor genes16, and NAC transcription factor genes56. In Cluster 9, we also found 19 WRKY transcription factor genes, most of which were significantly up-regulated. Cluster 9 contained many lncRNAs and many protein coding genes that are the target genes of the lncRNAs in this cluster. Therefore, we suggest that the cluster may contain one or more pathways related to cold stress response and that lncRNAs may play important roles in cold stress response in this pathway. Because many WRKYs were found in Cluster 9, WRKY family members may play important roles in the key cold stress response pathway. Although none of the WRKY genes in Cluster 9 was a target gene of the lncRNAs, they may still be indirectly regulated by lncRNAs or regulated by the expression of lncRNAs; however, this requires further study. Additionally, in this cluster, there are some genes related to anthocyanin or flavonoid biosynthesis such as VvMYBA1, flavanone 7-O-glucoside 2′-O-beta-L-rhamnosyltransferase, isoflavone-7-O-methyltransferase 9, and WD repeat-containing protein (Table 11)57–59. Previous studies have shown that abiotic stressors (such as cold or heat stress) may regulate anthocyanin or flavonoid biosynthesis-related genes57,58. Anthocyanins have been shown to be synthesized as protective compounds in response to cold stress60. Our cluster analysis showed that some key anthocyanin biosynthesis related genes may be located in pathways involved in cold stress response; therefore, lncRNAs in pathways involved in cold stress response are related to these anthocyanin biosynthesis related genes. Supporting our findings, previous studies have shown that biotic or abiotic stressors are related to the biosynthesis of anthocyanins or flavonols in grapevine58,59,61. Further studies should be conducted on the relationship between anthocyanin/flavonoid biosynthesis pathways and cold stress as additional results will positively impact viticulture and breeding.
Materials and Methods
Plant materials
One-year-old self-rooted seedlings of the grapevine cv. Cabernet Sauvignon were grown and maintained in the greenhouse under a 16 h light/8 h dark photoperiod at 26 °C. For the cold stress treatment, plant materials under a 16-h light/8-h dark photoperiod were transferred to 4 °C for 4 hours. For the control (CK), plants were kept under a 16-h light/8-h dark photoperiod at 26 °C for 4 hours. The shoot apices with well-developed leaves from these plant materials were collected. Each treatment consisted of three independent replicates. RNA was isolated for the construction of RNA-seq libraries and real-time PCR analysis.
Transcriptome library construction and high-throughput sequencing
Extracted RNA was sent to BGI (Shenzhen, China) for transcriptome library construction. In this process, RNA was treated with a Ribo-Zero™ Magnetic Kit to degrade rRNA. First-strand cDNA is generated by First Strand Master Mix and Super Script II reverse transcription (Invitrogen). High-throughput sequencing was performed using a HiSeq 2500 instrument. The clean reads generated by high-throughput sequencing were mapped on the grape genome (http://genomes.cribi.unipd.it/grape/) using the HISAT software (V2.0.4)62, and the reads mapped on the genome were assembled into transcripts using the stringTie software (V1.0.4)63.
Identification of lncRNA
To identify novel grapevine lncRNA transcripts, we first filtered out all mRNA transcripts, transcripts with a length < 200 nt, and known lncRNA transcripts predicted in data from the grape genome database (http://genomes.cribi.unipd.it/grape/)28. Then, we predicted the protein coding ability of the remaining transcripts using the CPC64, txCdsPredict, and CNCI software65. The transcripts without protein coding ability were subsequently employed in the remainder of the study. If transcripts without protein coding ability were not found in any known domain using the pfam database66, we considered them lncRNA transcripts. The known lncRNAs were annotated in the grape genome database (http://genomes.cribi.unipd.it/grape/). Finally, the transcripts with FPKM < 0.5 were removed28.
LncRNAs that were not found near any protein-coding locus (within < 10 kb) are considered lincRNAs5,6. LncRNAs transcribed from introgenic regions are long intronic RNAs, which can be transcribed in any orientation relative to coding genes. LncNAT are those that overlap with protein-coding regions or ncRNAs on the opposite strand and antisense RNA5–7.
Analysis of differentially expressed lncRNAs and mRNAs
DEGseq67 was used to identify the differentially expressed lncRNAs and mRNAs based on an MA-plot68. The lncRNAs significantly up-regulated (fold change > 2, P < 0.05) and down-regulated (fold change < −2, P < 0.05) under cold stress were considered the differentially expressed known lncRNAs.
Quantitative real time PCR validation of lncRNA and protein-coding genes
Quantitative RT-PCR (qRT-PCR) was performed to analyze the expression of lncRNAs following the methods outlined by a previous study69. Primers used in all qRT-PCR experiments are listed in Table S8.
Prediction of lncRNAs as miRNA targets
Target genes (lncRNAs and protein-coding genes) of grape miRNAs were identified using psRobot software set to moderate parameters (penalty score threshold = 2.5, five-prime boundary of essential sequence = 2, three-prime boundary of essential sequence = 17, maximal number of permitted gaps = 1, and position after gaps permitted = 17)70. Grape miRNAs were downloaded from the miRBase database (http://www.mirbase.org/).
GO and KEGG pathway analysis
Target genes were annotated based on the GO database (http://www.geneontology.org/). Pathway analyses of target genes were performed using the KEGG database (http://www.genome.jp/kegg/kegg1.html)71.
Analysis of miRNAs derived from lncRNAs
The grape precursors of the miRNAs dataset from miRbase were downloaded. The miRNAs were mapped to lncRNAs using the STAR program. The miRNAs were thought to be derived from lncRNAs if the identification between precursors of the miRNAs could be mapped to lncNRAs28.
Supplementary information
Acknowledgements
This work was supported by the Agricultural scientific and technological innovation project of Shandong Academy of Agricultural Sciences (CXGC2016D01, CXGC2018E17, CXGC2018F04), Agricultural scientific and technological innovation project of Shandong Academy of Agricultural Sciences-cultivating project for National Natural Science Foundation of China in 2018 “identification and function research of Vitis vinifera and Vitis amurensis cold stress response-related microRNAs”, Major Agricultural Application Technology Innovation Project of Shandong Province “Research and Application of Precision Control of Maturation and Product Innovation of Featured Brewing Grape”, Major Agricultural Application Technology Innovation Project of Shandong Province “Development of Landmark Wines and Integrated Application of Key Technologies in Shandong Province” and Fruit innovation team of modern agricultural industry technology system in Shandong Province-Jinan comprehensive test station (SDAIT-06-21).
Author Contributions
P.W., Y.W. and F.R. designed the study, P.W. wrote the manuscript. P.W., J.A. and L.D. carried out most of the experiment, data analysis, and wrote the method section of the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Pengfei Wang, Email: fengqiaoyouzi@126.com.
Yongmei Wang, Email: wangym228@126.com.
Fengshan Ren, Email: rensd65@163.com.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-43269-5.
References
- 1.Chekanova JA, et al. Genome-wide high-resolution mapping of exosome substrates reveals hidden features in the Arabidopsis transcriptome. Cell. 2007;131:1340–1353. doi: 10.1016/j.cell.2007.10.056. [DOI] [PubMed] [Google Scholar]
- 2.Kapranov P, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 3.Jin J, et al. PLncDB: plant long non-coding RNA database. Bioinformatics. 2013;29:1068–1071. doi: 10.1093/bioinformatics/btt107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang YC, et al. Genome-wide screening and functional analysis identify a large number of long noncoding RNAs involved in the sexual reproduction of rice. Genome biology. 2014;15:512. doi: 10.1186/s13059-014-0512-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 7.Knauss JL, Sun T. Regulatory mechanisms of long noncoding RNAs in vertebrate central nervous system development and function. Neuroscience. 2013;235:200–214. doi: 10.1016/j.neuroscience.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu J, et al. Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell. 2012;24:4333–4345. doi: 10.1105/tpc.112.102855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim DH, Sung S. Vernalization-triggered intragenic chromatin loop formation by long noncoding RNAs. Dev Cell. 2017;40:302–312.e4. doi: 10.1016/j.devcel.2016.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuan J, et al. Systematic characterization of novel lncRNAs responding to phosphate starvation in Arabidopsis thaliana. BMC Genomics. 2016;17:655. doi: 10.1186/s12864-016-2929-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li S, et al. High-resolution expression map of the Arabidopsis root reveals alternative splicing and lincRNA regulation. Dev Cell. 2016;39:508–522. doi: 10.1016/j.devcel.2016.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berry S, Dean C. Environmental perception and epigenetic memory: mechanistic insight through FLC. Plant J. 2015;83:133–148. doi: 10.1111/tpj.12869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matzke MA, Mosher RA. RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat Rev Genet. 2014;15:394–408. doi: 10.1038/nrg3683. [DOI] [PubMed] [Google Scholar]
- 14.Bardou F, et al. Long noncoding RNA modulates alternative splicing regulators in Arabidopsis. Dev Cell. 2014;30:166–176. doi: 10.1016/j.devcel.2014.06.017. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y, et al. Arabidopsis noncoding RNA mediates control of photomorphogenesis by red light. PNAS. 2014;111:10359–10364. doi: 10.1073/pnas.1409457111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang D, et al. Transposable elements (TEs) contribute to stress-related long intergenic noncoding RNAs in plants. Plant J. 2017;90:133–146. doi: 10.1111/tpj.13481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wierzbicki AT, Haag JR, Pikaard CS. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell. 2008;135:635–648. doi: 10.1016/j.cell.2008.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zilberman D, Cao X, Jacobsen SE. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science. 2003;299:716–719. doi: 10.1126/science.1079695. [DOI] [PubMed] [Google Scholar]
- 19.Xie Z, et al. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng X, Zhu J, Kapoor A, Zhu JK. Role of Arabidopsis AGO6 in siRNA accumulation, DNA methylation and transcriptional gene silencing. EMBO J. 2007;26:1691–1701. doi: 10.1038/sj.emboj.7601603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao Z, et al. An RNA polymerase II- and AGO4-associated protein acts in RNA-directed DNA methylation. Nature. 2010;465:106–109. doi: 10.1038/nature09025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matzke M, et al. RNA-mediated chromatin-based silencing in plants. Curr Opin Cell Biol. 2009;21:367–376. doi: 10.1016/j.ceb.2009.01.025. [DOI] [PubMed] [Google Scholar]
- 23.Franco-Zorrilla JM, et al. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet. 2007;39:1033–1037. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- 24.Zhang YC, Chen YQ. Long noncoding RNAs: new regulators in plant development. Biochemical and biophysical research communications. 2013;436:111–114. doi: 10.1016/j.bbrc.2013.05.086. [DOI] [PubMed] [Google Scholar]
- 25.Charon C, Moreno AB, Bardou F, Crespi M. Non-Protein-Coding RNAs and their Interacting RNA-Binding Proteins in the Plant Cell Nucleus. Molecular Plant. 2010;3:729–739. doi: 10.1093/mp/ssq037. [DOI] [PubMed] [Google Scholar]
- 26.Werner A, Berdal A. Natural antisense transcripts: sound or silence? Physiological Genomics. 2005;23:125–131. doi: 10.1152/physiolgenomics.00124.2005. [DOI] [PubMed] [Google Scholar]
- 27.Di C, et al. Characterization of stress-responsive lncRNAs in Arabidopsis thaliana by integrating expression, epigenetic and structural features. Plant J. 2014;80:848–861. doi: 10.1111/tpj.12679. [DOI] [PubMed] [Google Scholar]
- 28.Li S, et al. Genome-wide identification and functional prediction of cold and/or drought-responsive lncRNAs in cassava. Sci Rep. 2017;7:45981. doi: 10.1038/srep45981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu X, et al. Genome-Wide Analysis of Long Noncoding RNAs and Their Responses to Stress in Cotton (Gossypium hirsutum L.) PLoS One. 2016;11:e0156723. doi: 10.1371/journal.pone.0156723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heo JB, Sung S. Vernalization-Mediated Epigenetic Silencing by a Long Intronic Noncoding RNA. Science. 2011;331:76–79. doi: 10.1126/science.1197349. [DOI] [PubMed] [Google Scholar]
- 31.Ben Amor B, et al. Novel long non-protein coding RNAs involved in Arabidopsis differentiation and stress responses. Genome Res. 2009;19:57–69. doi: 10.1101/gr.080275.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ding JH, et al. A long noncoding RNA regulates photoperiod-sensitive male sterility, an essential component of hybrid rice. PNAS. 2012;109:2654–2659. doi: 10.1073/pnas.1121374109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sunkar R, Zhu JK. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell. 2004;16:2001–2019. doi: 10.1105/tpc.104.022830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Babar IA, Slack FJ, Weidhaas JB. miRNA modulation of the cellular stress response. Future Oncol. 2008;4:289–298. doi: 10.2217/14796694.4.2.289. [DOI] [PubMed] [Google Scholar]
- 35.Khraiwesh B, Zhu JK, Zhu J. Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim Biophys Acta. 2012;1819:137–148. doi: 10.1016/j.bbagrm.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones-Rhoades MW, Bartel DP. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell. 2004;14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 37.Chekanova JA, et al. Long non-coding RNAs and their functions in plants. Current opinion in plant biology. 2015;27:207–216. doi: 10.1016/j.pbi.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 38.Mattick JS, Rinn JL. Discovery and annotation of long noncoding RNAs. Nat Struct Mol Biol. 2015;22:5–7. doi: 10.1038/nsmb.2942. [DOI] [PubMed] [Google Scholar]
- 39.van Werven FJ, et al. Transcription of two long noncoding RNAs mediates mating-type control of gametogenesis in budding yeast. Cell. 2012;150:1170–1181. doi: 10.1016/j.cell.2012.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Flynn RA, Chang HY. Long noncoding RNAs in cell-fate programming and reprogramming. Cell stem cell. 2014;14:752–761. doi: 10.1016/j.stem.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang SY. The lncRNAs involved in mouse airway allergic inflammation following induced pluripotent stem cell-mesenchymal stem cell treatment. Stem Cell Research & Therapy. 2017;8:2. doi: 10.1186/s13287-016-0456-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tafer H, Hofacker IL. RNAplex: a fast tool for RNA–RNA interaction search. Bioinformatics. 2008;24:2657–2663. doi: 10.1093/bioinformatics/btn193. [DOI] [PubMed] [Google Scholar]
- 43.Wu HJ, et al. Widespread long noncoding RNAs as endogenous target mimics for microRNAs in plants. Plant Physiol. 2013;161:1875–1884. doi: 10.1104/pp.113.215962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shuai P, et al. Genome-wide identification and functional prediction of novel and drought-responsive lincRNAs in Populus trichocarpa. J Exp Bot. 2014;65:4975–4983. doi: 10.1093/jxb/eru256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kumar L, Futschik ME. Mfuzz: a software package for soft clustering of microarray data. Bioinformation. 2007;2:5–7. doi: 10.6026/97320630002005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li L, et al. Genome-wide discovery and characterization of maize long non-coding RNAs. Genome Biol. 2014;15:R40. doi: 10.1186/gb-2014-15-2-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ørom UA, Derrien T, Guigo R, Shiekhattar R. Long noncoding RNAs as enhancers of gene expression. Cold Spring Harb Symp Quant Biol. 2010;75:325–31. doi: 10.1101/sqb.2010.75.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Z, et al. Whole transcriptome sequencing of Pseudomonas syringae pv. actinidiae-infected kiwifruit plants reveals species-specific interaction between long non-coding RNA and coding genes. Sci Rep. 2017;7:4910. doi: 10.1038/s41598-017-05377-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thomashow M. F. Molecular basis of plant cold acclimation: insights gained from studying the CBF cold response pathway. Plant Physiol. 2010;154:571–577. doi: 10.1104/pp.110.161794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiao H, et al. CBF4 is a unique member of the CBF transcription factor family of Vitis vinifera and Vitis riparia. Plant Cell Environ. 2008;31:1–10. doi: 10.1111/j.1365-3040.2007.01741.x. [DOI] [PubMed] [Google Scholar]
- 51.Sasaki K, Christov NK, Tsuda S, Imai R. Identification of a novel LEA protein involved in freezing tolerance in wheat. Plant Cell Physiol. 2014;55:136–147. doi: 10.1093/pcp/pct164. [DOI] [PubMed] [Google Scholar]
- 52.Zou CS, Jiang WB, Yu DQ. Male gametophyte-specific WRKY34 transcription factor mediates cold sensitivity of mature pollen in Arabidopsis. J Exp Bot. 2010;61:3901–3914. doi: 10.1093/jxb/erq204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yuan HM, et al. Overexpression of Hevea brasiliensis HbICE1 Enhances Cold Tolerance in Arabidopsis. Front. Plant Sci. 2017;8:146. doi: 10.3389/fpls.2017.01462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang GT, et al. Signal transduction during cold, salt, and drought stresses in plants. Mol Biol Rep. 2012;39:969–987. doi: 10.1007/s11033-011-0823-1. [DOI] [PubMed] [Google Scholar]
- 55.Su LG, Dai ZW, Li SH, Xin HP. A novel system for evaluating drought-cold tolerance of grapevines using chlorophyll fluorescence. BMC Plant Biol. 2015;15:82. doi: 10.1186/s12870-015-0459-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.An JP, et al. An apple NAC transcription factor negatively regulates cold tolerance via CBF-dependent pathway. J Plant Physiol. 2017;221:74–80. doi: 10.1016/j.jplph.2017.12.009. [DOI] [PubMed] [Google Scholar]
- 57.Malacarne G, et al. Regulation of flavonol content and composition in (Syrah × Pinot Noir) mature grapes: integration of transcriptional profiling and metabolic quantitative trait locus analyses. Journal of experimental botany. 2015;66:4441–4453. doi: 10.1093/jxb/erv243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Azuma A, Yakushiji H, Koshita Y, Kobayashi S. Flavonoid biosynthesis-related genes in grape skin are differentially regulated by temperature and light conditions. Planta. 2012;236:1067–1080. doi: 10.1007/s00425-012-1650-x. [DOI] [PubMed] [Google Scholar]
- 59.Rienth M, et al. Day and night heat stress trigger different transcriptomic responses in green and ripening grapevine (vitis vinifera) fruit. BMC Plant Biol. 2014;14:108. doi: 10.1186/1471-2229-14-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Costantini L. New candidate genes for the fine regulation of the colour of grapes. J. Exp. Bot. 2015;66:4427–4440. doi: 10.1093/jxb/erv159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang P, et al. Genome-Wide Characterization of bHLH Genes in Grape and Analysis of their Potential Relevance to Abiotic Stress Tolerance and Secondary Metabolite Biosynthesis. Front. Plant Sci. 2018;9:64. doi: 10.3389/fpls.2018.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim D, Langmead B, Salzberg S. L.. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pertea M, et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kong L, et al. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic acids research. 2007;35:W345–349. doi: 10.1093/nar/gkm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sun L, et al. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic acids research. 2013;41:e166. doi: 10.1093/nar/gkt646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Finn RD, et al. Pfam: the protein families database. Nucleic acids research. 2014;42:D222–230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang LK, et al. DEGseq: an R package for identifying differentially expressed genes from RNAseq data. Bioinformatics. 2010;26:136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- 68.Yang YH, et al. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang P, et al. Genome-Wide Dissection of the Heat Shock Transcription Factor Family Genes in Arachis. Front Plant Sci. 2017;8:106. doi: 10.3389/fpls.2017.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang J, et al. Genome-wide analysis of tomato long non-coding RNAs and identification as endogenous target mimic for microRNA in response to TYLCV infection. Sci Rep. 2015;5:16946. doi: 10.1038/srep16946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kanehisa M, et al. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014;42:D199–D205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.