Abstract
Natural antimicrobial peptides have been shown as one of the important tools to combat certain pathogens and play important role as a part of innate immune system in plants and, also adaptive immunity in animals. Defensin is one of the antimicrobial peptides with a diverse nature of mechanism against different pathogens like viruses, bacteria and fungi. They have a broad function in humans, vertebrates, invertebrates, insects, and plants. Plant defensins primarily interact with membrane lipids for their biological activity. Several antimicrobial peptides (AMPs) have been overexpressed in plants for enhanced disease protection. The plants defensin peptides have been efficiently employed as an effective strategy for control of diseases in plants. They can be successfully integrated in plants genome along with some other peptide genes in order to produce transgenic crops for enhanced disease resistance. This review summarizes plant defensins, their expression in plants and enhanced disease resistance potential against phytopathogens.
Keywords: Plant defensins, Genetic engineering, Disease resistance, Pathogens
Introduction
The production of natural antimicrobial peptides by the plants and other organisms has been an important mechanism to counteract certain pathogens and play key role as a part of innate immunity in the plants and also adaptive immunity in the animals. Defensin is one of the antimicrobial peptides with a diverse nature of mechanism against a vast number of pathogens including viruses, bacteria and fungi. They have a broad function in humans (Ganz et al. 1985), insects (Hoffmann et al. 1992), vertebrates (Ericksen et al. 2005), invertebrates (Rodriguez et al. 2005), and plants (Lay et al. 2005). Firstly, defensins were isolated from human and rabbit neutrophils, where these polypeptides contributed to host’s immunity against various microbial pathogens and viral infections (Ganz et al. 1985). These antimicrobial peptides are also a part of adaptive immunity and regulate different processes such as expression of the cytokines and chemokines, the enhancement of antibody responses against invading pathogens and the production of some neurotransmitters such as histamine (Ganz et al. 2003, 2004, 2005). In fusion with non-immunogenic tumor antigens, they also help to induce and boost antitumor immunity (Yang et al. 2002). Defensins also regulate the signal transduction pathways and inflammatory effects, wound healing, control proliferation and chemotaxis (Kim and Kaufmann 2006; Shi 2007).
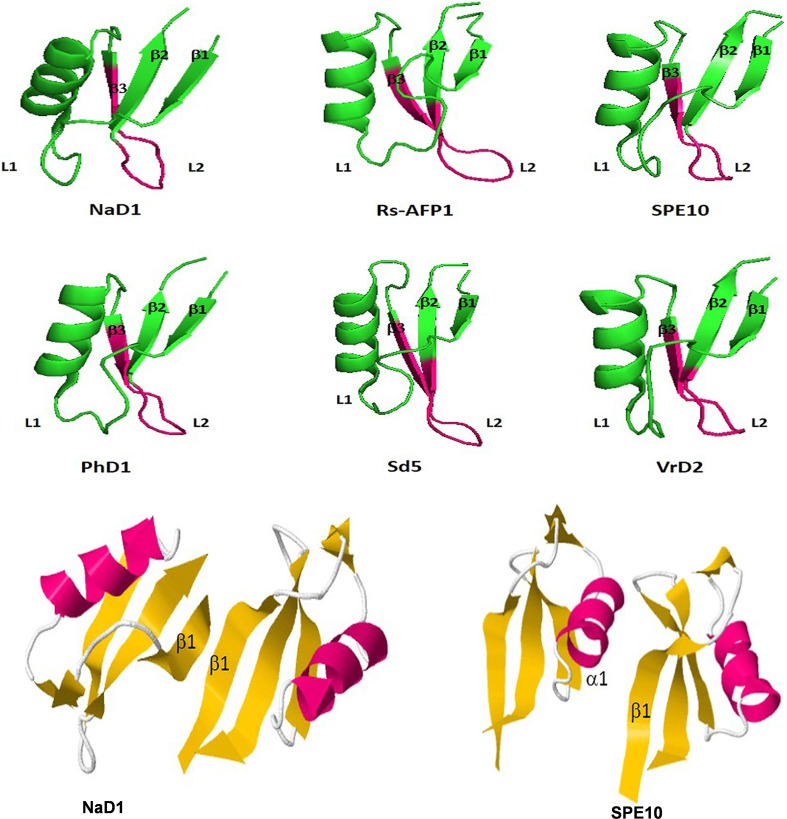
Defensins make an important class of antimicrobial peptides consisting of highly conserved structural scaffold, with cysteine stabilized αβ conformation that binds and interacts with negatively charged microbial cell membranes. So, it could be assumed that all genes for defensins have evolved from single ancestral gene. Two independent types of defensins, cis-defensins (like from plants and insects) and trans-defensins (mammalian defensins), with similar structures as indicated by their disulphide topology have originated through convergent evolution (Pairisi et al. 2018). In general, defensins consist of three to five disulfide bonds (Fig. 1a), stabilizing the antiparallel β-sheet conformation flanked by α-helical segment forming a compact structure that confers resistance to protease-mediated degradation, extreme temperature and pH (Bullet et al. 2005; Tavares et al. 2008; Batta et al. 2009). Plant defensins commonly have a three-dimensional structure consisting of triple-stranded antiparallel β-sheets and one α-helix lying parallel to the β-sheet. They consist of a cystine-stabilized (Cs) motif (C1XnC2X3C3XnC4XnC5X1C6) (Lay et al. 2005). The Cs-motif can also be found in toxins and defensins from other organisms such as scorpions and insects, respectively (Garcia-Olmedo et al. 1998; Lay et al. 2003). Interestingly, two defensins, PhD1 and PhD2, from Petunia hybrida, were found with a different structural pattern; presenting an extra disulfide bond, replacing a conserved H-bond present in the 8C-defensin peptides (C1m,XnC2XnC3XnC4X3C5XnC6XnC7X1C8) (Pelegrini et al. 2008).
Fig. 1.
Three-dimensional structure of six antifungal defensins from plants. Pink (darker) region indicates the γ-core motif of each peptide. β1: β-sheet 1, β2: β-sheet 2 and β3: β-sheet3. L1: Loop1 and L2; Loop2; NaD1 from Nicotiana alata defensin1, Rs-AFP1 from Raphanus sativus antifungal peptide 1, SPE10 from Pachyrrihizus erosu peptide, PhD1 from Petunia hybrida defensin1, Sd5 from Saccharum officinarum defensin5 and VrD2 from Vigna radiata defensin2 (Lacerda et al. 2014)
Defensins have a range of activities against bacteria, both gram-positive and gram-negative, and kill them in a number of ways. Some defensins create voltage-dependent channels in bacterial membranes that allow the influx of water. Increased osmotic pressure ruptures the bacterial membranes. Other defensins move through bacterial cell walls, bind to target cells, and disrupt normal metabolism. Defensins have exhibited enhanced resistance against a number of fungi (Thevissen et al. 2000a, b; Thomma et al. 2002; Lay and Anderson 2005; Khan et al. 2006; Wong et al. 2007, 2011a, b, 2014).
Types of defensins
Among the all host defense peptides (HDP), defensins are the first peptides to be identified. First cationic and cysteine-rich α-defensin was isolated from the mammalian neutrophils phagocytes in the early 1980. Lehrer et al. (2004) coined the term “defensin” for the first time. Similarly, some analogous molecules termed “cryptdins” were identified from host defense cells of the intestinal crypts. Another term corticostatin was coined due to the ability of inhibiting adrenocortical steroidogenesis (Zue et al. 1989). Similarly, β-defensins were discovered in the epithelial and white blood cells of the mammals in the early 1990s and then in the avian leukocytes and more recently in the reptiles and fish (Dalla et al. 2012; Zou et al. 2007), having a slight difference in the cysteine bridges connectivity. Most of the animal defensins are antibacterial, which show their effect by disrupting the integrity of host’s cell membrane leading to leakage of intracellular contents and cell lysis (Park et al. 2018).
The term “insect defensin” was coined in the late 1980s, when an inducible peptide from insect hemolymph was identified having significant similarity to the mammalian defensin (Dimarcq et al. 1998). Mammalian defensins consist of three structural subfamilies, alpha, beta and theta defensins. Theta defensins, derived from Old World monkeys, are produced by binary ligation of two truncated alpha defensins (Selsted 2004). In plants, same antimicrobial peptides were identified in the early 1990s and named as γ-thionins. Subsequently, the proteins from plants homologous to the γ-thionins were identified as plant defensins in 1995 because of their structural resemblance with the animal and insect defensins (Lay et al. 2005). Finally, their discovery in fungi shows the antiquity of defensin and defensin-like peptides in innate immune responses (Zhu et al. 2008). So, the term “defensin” is not a single word but a group of several peptides with the same structure of cysteine stabilized β-sheet and host defense function. Evolutionary evidences show that all defensins from invertebrates, fungi, insects, plants and animals share an evolutionarily related group (Zhu et al. 2008; Rehaume et al. 2008).
Plant defensins
The origin of plant defensins traces back to the prokaryotic genera (Myxobacterium, Anaeromyxobacter dehalogenans and Stigmatella aurantiaca) because of their structural resemblance with the insect defensin as both have similar conserved γ-core motif. It has been supposed that these peptides were present as endosymbionts of eukaryotes which then transferred the gene from prokaryotes to the eukaryotic lineage and over a course of evolution this peptide underwent some alteration and modification which gave rise to modern day defensin (Phoenix et al. 2013). Only in Arabidopsis genome, about 317 genes are found to be coding for defensin and defensin-like peptides (Silverstein et al. 2005; Stotz 2009). Similarly, 778 potential sequences in Medicago truncatula genome (Graham et al. 2004), and 79 such sequences in Vitis vinifera genome (Nanni et al. 2014) have been found. Such a large number of defensin and defensin-like peptides expressed in various parts of plants shows their importance in innate host resistance triggered by these peptides.
The first plant defensin was isolated from seeds of monocot and dicot plant species as reported by Terras et al. (1995). Plant defensins, small cationic peptides of 45–54 amino acids, consist of βαββ pattern, whereas mammalian defensins consist of an N-terminal α-helix with αβββ-folding. Seventy eight percent of defensins and defensin-like peptides isolated from Arabidopsis consist of cysteine-stabilized αβ-motif (Silverstein et al. 2005).
Although plant defensins are antibacterial like human defensin and show their effect against gram positive bacteria thus playing a role in innate immunity (Bulet et al. 2004; Lacerda et al. 2014), but plants are often attacked by fungal pathogen, so plant’s defensins are antifungal in function (Mith et al. 2015) unlike human defensins which are all antibacterial. This discrepancy may attribute to a difference in the structural scaffold of a plant (βαββ) and human defensin (αβββ) which make it specific for the respective pathogen type. For example, a human β-defensin gene (HBD-2) of about 2.0 kb consisting of two small exons and one intron, was induced by inflammation (Liu et al. 1998). These changes in specificity of pathogen type are due to positive changes at the gene level coding for this peptide. So, the plant defensin was evolved from the prokaryotic peptides with some positive modification in the gene, thus showing a strong defense mechanism against fungal pathogens (Bulet et al. 2005; Zhu et al. 2008).
Plant defensin genes make precursor proteins that have a mature defensin domain with endoplasmic reticulum targeting a signal at the amino terminal and pro-peptide at c-terminal (CTPP). This CTPP is optional, it may be present in some defensins while not in others. Those peptides having a c-terminal pro-peptide signal (CTPP) of 27–33 amino acids residues are classified as class II plant defensins. These amino acid residues are rich in glutamic acid and aspartic acid that give minus charge and neutralizes the positive charge of the defensin domain as reported by Lay et al. (2014). The other class lacking such signals is classified as class I. Plant defensins of class II are found to be abundantly expressed in both reproductive and vegetative parts of the plant (Lay et al. 2003) and class I defensins are found to be present in seeds only. The expression of class II defensins with no CTPP in plants caused retarded growth which indicated the phytotoxicity of these defensin peptides (Lay et al. 2014; Francisco and Georgina 2017).
Plant defensins, abundantly present in the stomatal and peripheral cells, the entry point of plant pathogens, show their primary effect on the invading pathogens (Broekaert et al. 1995). Some of the plant defensins are found to be involved in signal transduction pathways and show induced expression during certain abiotic stress conditions (Lay and Anderson 2005). In growing seeds, they are expressed to prevent the newly formed radical tissues from fungal invasion (Stotz et al. 2009). Some defensins, as reported, also inhibited protein formation (Mendez et al. 1990). In addition, overexpression of a tomato defensin, DEF2 in transgenic tomato caused decreased pollen viability and low seed yield (Stotz et al. 2009).
Structural conformation of plant defensins
Plant defensins have a well conserved CSαβ-motif arranged in a three-dimensional pattern of one alpha helix and three antiparallel beta sheets. Similarly, amino acid residues of plant defensins are also conserved, showing four to five disulfide bridges. According to Lay and Anderson (2005), the sequence of these disulfide bonds is as Cys1–Cys8, Cys2–Cys5, Cys3–Cys6, and Cys4–Cys7. The peptides having four disulfide bridges are termed as 8C- plants e.g. NaD1 (defensin from Nicotiana alata), VrD1 (defensin from Vigna radiate), AlfAFP (antifungal protein from alfalfa), Ms-Def1 (defensin from Medicago sativa), ω-hordothionin, Psd1 (defensin from Pisum sativum) and Rs-AFPs (antifungal proteins from Raphanus sativus), while those having five sulfide bridges are termed as 10C- plants eg. PhDs (defensins from Petunia hybrida). The NaD1 (47 aa) was first isolated from flowering parts of Nicotiana alata as it provides protection to the reproductive organs. NaD1 inhibited the growth of Fusarium oxysporum and Botrytis cinerea (Lay et al. 2003). The mung bean (Vigna radiata) contains an antimicrobial peptide (VrD1) of 46 amino acid residues which is a protein synthesis inhibitor and insect (bruchids)-resistant (Chen et al. 2002). Rs-AFPs from Raphanus sativus, released in response to fungal invasions where they created fungal suppressing microenvironment in nearly matured and matured seeds (Terras et al. 1995). SPE10, isolated from seeds of Pachyrrhizus erosus, had a conserved hydrophobic patch on the molecular head which was found responsible for their antifungal activity (Song et al. 2004). PhD1 and PhD2, the antifungal peptides from Petunia hybrida flowers, are 10-C peptides consisting of a disulfide bond between alpha-helix and the loop after β1 (Lay et al. 2003). It does not change the side-chain orientation of substituted residues and the typical CSαβ topology (Janssen et al. 2003) but only change the corresponding hydrophobic interaction due to change of hydrogen bond to a covalent disulfide bond. Structural studies of many plant defensins using nuclear magnetic resonance (NMR) and crystallography have been carried out extensively in the last decades. The antifungal peptides with known structures are given in Fig. 1.
Mode of action
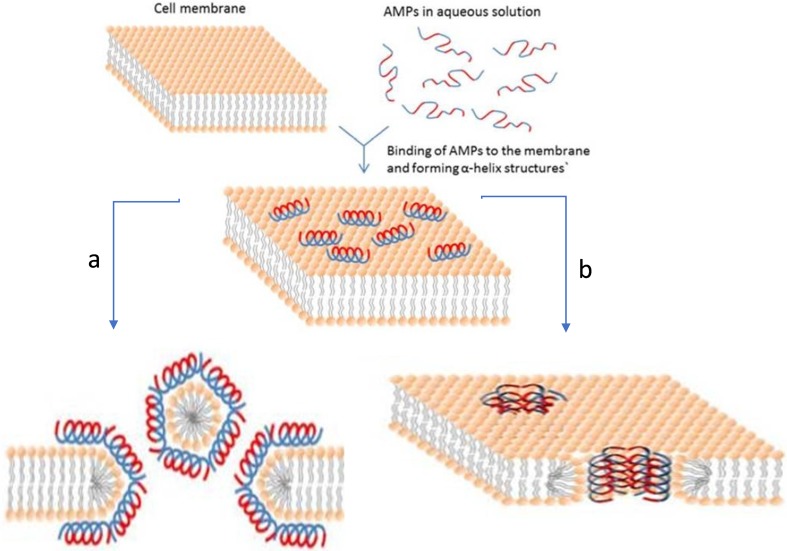
Two models were presented for explaining the mechanism of action of defensin. One is the carpet model and the other is the Toroidal pore model. In both these models, the interaction of defensin with the negatively charged membrane is common which causes increase in permeability of the cell membrane and its leakage thus causing cell death. According to the carpet model small pores are formed due to inward movement of hydrophobic sides of AMPs molecules at the surface of cell-membrane, while the latter shows that peptides first form oligomers which then, create multiple pores in the cell membrane (Fig. 2). Järvå et al. (2018) determined the crystal structure of the plant defensin NaD1 bound to PA (phosphatidic acid). The X-ray structure indicated a 20-mer that adopts carpet-like topology in which the NaD1 dimers make one face and the PA acyl chains form the other face of the sheet.
Fig. 2.
Schematic representation of some action mechanisms of membrane-active AMPs. Red color shows hydrophilic part while blue indicates the hydrophobic portions. a Carpet model. The hydrophobic sides of AMP molecules facing inward make pores in the membrane. b Toroidal pore model. AMPs are always in contact with phospholipid head groups of the membrane (Bahar and Ren 2013)
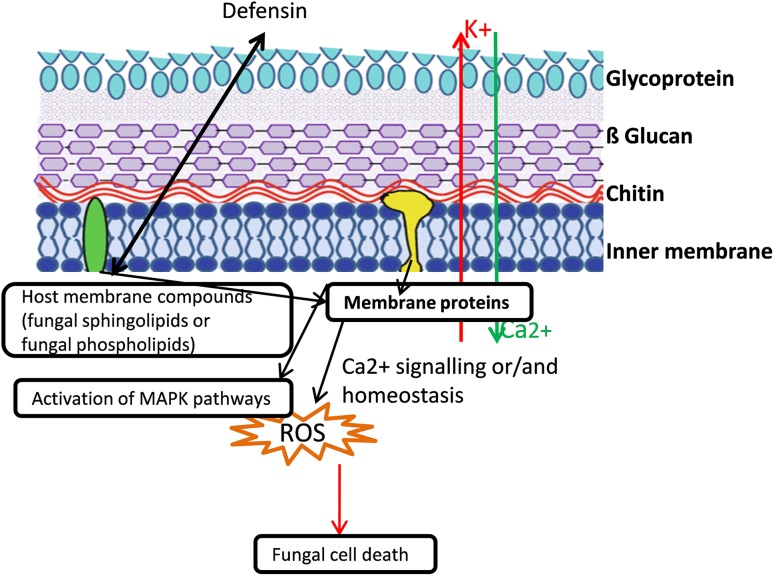
According to another hypothesis, defensins only interact with cell membrane and cause increased ion permeability and transportation access to intracellular environment (Fig. 3). Defensins can also accelerate production of reactive oxygen species (ROS) and hence, activate programmed cell death (PCD) (Lacerda et al. 2014). Plant defensins may first bind to specific binding sites called receptors of the microbial membranes as found in Rs-AFP2, Hs-AFP1 (AFP from Heuchera sanguine), and Dm-AMP1 (AMP from Dahlia merckii) (Thevissen et al. 2000a, b, 2004) which results in the ion leakage and inflow, outflow of the positive ions like Ca2+ and K+ (Thevissen et al. 1996) (Fig. 3).
Fig. 3.
A generalized schematic diagram showing mode of action of plant defensins. Interaction of defensins with fungal membrane may results in ions leakage, Ca ions signaling, MAPK activation, production of reactive oxygen species and ultimately death of fungal cells
The mechanism of action of plant defensins, PhD1, Rs-AFP1, and VrD2 involves the electrostatic interaction as they contain an γ-core region in the second loop which is highly positively charged and an important site for antifungal activity. Antifungal activity of NaD1 involves the membrane permeabilization of hyphae of the pathogen, Fusarium oxysporum and upon getting into cytoplasm of the cell, it induces production of reactive oxygen species. NaD1 requires a cell specific receptor rather than causing membrane permeabilization of the membrane (Van der Weerden et al. 2008). Whereas, Hayes and his colleagues reported that if the pathway of high-osmolarity glycerol (HOG) is inhibited, it will increase the NaD1 antimicrobial activity against fungal pathogen, Candida albicans (Hayes et al. 2013; Tam et al. 2015).
Structural analysis of sugarcane defensin (Sd5) showed that hydrophobic core of defensin is an important component for membrane interaction and its permeabilization (de Paula et al. 2011). In addition, further evaluations on conformational dynamics of the Sd5 suggested that these structural changes in the backbone of the peptide may change the membrane interaction, such as the hydrogen bond distance between β-sheet and α-helix of the peptide increase the binding ability with the membrane. Then membrane permeabilization and ion leakage from the membrane occurs due to the interaction of serine residue with glycosyl part of the fungal cell membrane (de Paula et al. 2011). Some defensins show their activity through dimerization in which dimers of peptides are formed by arranging side by side in a manner, in which α-helix of one monomer interact with the β-sheet of the second monomer (Fig. 2). The arrangement like Arg36-Trp42-Arg40 has been found in SPE10 defensin from Pachyrrihizus erosus, which is essential for dimer formation. Studies have revealed that Trp42 is key component of antifungal activity as it was found absent in non-antifungal peptides (Song et al. 2011). Similarly, dimer formation was observed in NaD1 defensin, but the dimer was formed between β-sheets unlike SPE10, but the antifungal activity remained same (Lay et al. 2012). So, plant defensin positively charged coupled dimers interact with negatively charged glycoproteins of the fungal membrane. Several other reviews have been discussed in detail about the plant defensins and their mechanism of action (Wimley 2010; Tam et al. 2015; Jung and Kang 2014; Vriens et al. 2014).
Plant defensins as antimicrobial peptides
Plant defensins possess diverse biological functions including antifungal (Gao et al. 2000; Khan et al. 2006, 2014), antibacterial (Fant et al. 1998; Fujimura et al. 2003; Sitaram 2006; Kaewklom et al. 2018; Velivelli et al. 2018) and α-amylase and trypsin inhibitory activity (Wijaya et al. 2000). BcDef1, isolated from Brugmansia × candida (Bc) exhibited antibacterial activity against both Gram-positive and Gram-negative pathogens with the lowest MIC (15.70 μM) against the Staphylococcus epidermidis. The BcDef1 also showed antioxidant activity and low cytotoxicity against the mouse fibroblast L929 cells (Kaewklom et al. 2018). MtDef5 (defensin from Medicago truncatula) were found effective against plant bacterial pathogen, Xanthomonas campestris (Velivelli et al. 2018). Mendez and his co-workers isolated defensins from the wheat and barley and called them as γ-thionins due to 25% structure similarity i.e. a small size and presence of 8 cysteine residues like other thionins but later on it was discovered that they are different from thionin in the structure aspect and were called as defensins (Mendez et al. 1990). Terras and his colleagues in 1995 coined the term family of plant peptide “plant defensins” when they discovered Rs-AFP1 and Rs-AFP2, antifungal peptides from Rhapanus sativus. According to them, the structure was similar to animal and insect defensins but later on (Thevissen et al. 2000a, b, 2002) noticed that unlike mammalian and insect defensins which are antibacterial, plant defensins are antifungal in function. In addition to being antimicrobial in function, plant defensins also give responses to biotic stress and are involved in plant growth regulation and developmental processes. Defensins have exhibited resistance to broad range of fungal pathogens (Thevissen et al. 2000a,b Thomma et al. 2002; Lay and Anderson 2005; Wong et al. 2007; Khan et al. 2006, 2008, 2010, 2011, 2013, 2014). Plant defensins also inhibited root hair’s growth in A. thaliana (Allen et al. 2008). Defensins have played a great function of defense against pathogens and other developmental processes by altering the growth of several organs in tomato (Stotz 2009). Defensins are the most studied family of plant AMP’s found in all parts of plant. More than 80 genes of defensins have been sequenced from a number of plant species as reported by Thomma et al. (2002).
Although defensins are expressed in each part of the plant, they are found in abundance in seeds which enhance their survival rate and resistance against pathogens like fungi. Defensins have been identified in almost every organ of the plant. One gene is expressed in each organ, but some organs express more than two genes as in Arabidopsis. The ectopic expression of defensins results in growth retardation of some plants as these are organ specific (Hanks et al. 2005). Defensin genes, PDF1.1 and PDF1.2 (plant defensins from Arabidopsis) were expressed in the seeds and in the leaves, respectively during a pathogen infection (Thomma et al. 2002). PDF2.3 was expressed constitutively in all tissues of Arabidopsis except roots. Hanks et al. (2005) reported that the defensins, MsDef1 and MtDef2 isolated from Medicago, could not be expressed in the roots of this plant.
The peripheral cells and stomatal cells of the plant have maximum defensin gene expression for their protection against pathogens. Expression of the defensin gene has been induced upon pathogen infection in tobacco, Arabidopsis and spruce plants (Thomma et al. 2002; Lay and Anderson 2005). The environmental stresses (drought stress, salt stress and cold stress) and certain molecules like ethylene, salicyclic acid and methylene jasmonate also affect the expression of defensin (Lay and Anderson 2005; Hanks et al. 2005). These stresses provide a cross-talk between signal transduction pathway and gene expression.
Most of the defensins are antifungal in function. Different defensins show different mode of responses to different fungi as their activity depends on fungal species (Thevissen et al. 2000a, b). On the basis of their effect on fungi, they have been divided into two groups, one is morphogenetic plant defensins that inhibit the growth of hyphae and decrease their branching, while non-morphogenetic ones inhibit only hyphal growth and do not lead to significant morphological damage (Yamano et al. 1994). For the first time Terras et al. (1992a) isolated two isoforms of antifungal plant defensin, Rs-AFP1 and Rs-AFP2 from radish seed, and their activity was checked against different fungi like Pyricularia Oryza, Phoma betae and Cercospora beticola. Both the isoforms inhibited the growth of these fungi (Terras et al. 1992b). Another defensin, Ms-Def1 showed resistance against fungal pathogens like Verticillium dahlia in transgenic potato (Gao et al. 2000; Lay et al. 2003). Other homologous antifungal proteins which were isolated and reported include proteins from Aesculus hippocastanum, Dahlia merckii (Dm-AMP1), Clitoria ternatea (Ct-AMP1), Heuchera sanguine (Hs-AFP1) and Lens culinaris (Lc-def) (Osborn et al. 1995; Fant et al. 1999). Bloch and Richardson (1991) isolated three defensins, S1α1, S1α2 and S1α3 from seeds of Sorghum bicolor, and checked their α-amylase inhibitory effect. All the three proteins showed inhibitory action against the insect and human saliva α-amylase. Several plant defensins such as γ1-zeathionin and γ2-zeathionin from the maize kernels have been identified which have shown inhibitory action on several ion channels. These defensins inhibited Ca2 + channels while no such inhibitory function was observed on K + channels (de Oliveira Carvalho and Gomes 2007). Selitrennikoff (2001) isolated two homologous peptides, 5459 and 5144 (according to their molecular weight) from Cassia fistula showing sequence similarity with plant defensins. The 5459 showed inhibitory effect against trypsin, whereas the 5144 did not show any inhibitory effect. Huang et al. (2008) reported another defensin peptide, Cp-thionin from seeds of V. uguiculata, which is active against bovine trypsin of the pancreas.
Transgenic plants overexpressing defensins
A variety of plant defensins have been identified, isolated and overexpressed in different plant species to enhance the level of defense against the pathogens. The most thoroughly studied defensins are Rs-AFP1, Rs-AFP2 and Rs-AFP3/4 which were isolated from seeds of Raphanus sativus (Carvalho and Gomes 2009). Rs-AFP2 has been expressed in wheat and showed an enhanced resistance to F. graminearum and R. cerealis (Li et al. 2011). Dm-AMP1, isolated from seeds of Dhalia merckii (Osborn et al. 1995), was expressed in rice and papaya and restricted growth of several fungal pathogens (Zhu et al. 2007; Jha et al. 2010) (Table 1). Br-AMP1, isolated from seeds of Brassica rapa (Terras et al. 1993), has been overexpressed in transgenic rice to increase insect and fungal resistance. Similarly, the antimicrobial peptides, Psd1 from Pisum sativum (Almeida et al. 2002), VrD1 from Vigna radiata (Chen et al. 2004), and MtDef2 from Medicago trunculata (Spelbrink et al. 2004) are seed specific antimicrobial peptides which can inhibit many fungal species. CADEF1 from Capsicum annuum has been expressed in transgenic tomato and showed resistance to Fusarium spp and Phytophthora infestans (Zainal et al. 2009). Another defensin, Ms-Def1 isolated from seeds of Medicago sativa showed resistance against fungal pathogens like Verticillium dahlia in transgenic potato plants (Gao et al. 2000). Defensins isolated from Vigna unguiculata and ω-hordothionin from H. vulgare, also inhibited insect α-amylase (Pelegrini et al. 2008). Wasabi defensin (isolated from Wasabia japonica) was overexpressed in potato, tomato, petunia and exhibited enhanced resistance against phytopathogenic fungi (Khan et al. 2006; 2006b,2011a) (Table 1).
Table 1.
Summary of plant defensins isolated from different plant species and overexpressed in transgenic plants for enhanced resistance against phytopathogens
| AMP/signal | Source of defensin | Transgenic plant | Pathogens tested | References |
|---|---|---|---|---|
| BrD1 | Brassica rapa | Rice | Nilaparvatalugens (brown planthopper insect) | Choi et al. (2009) |
| Fusarium graminearum | ||||
| Rhizoctonia cerealis | Li et al. (2011) | |||
| RsAFP2 | Raphanus sativus | Wheat/rice | Magnaporthe oryza | Jha and Chattoo (2010) |
| Rhizoctonia solani | Terras et al. (1995) | |||
| Alternaria longipes | ||||
| Magnaporthe oryzae | ||||
| Dm‐AMP1 | Dahlia merckii | Rice/papaya | Rhizoctonia solani | Zhu et al. (2007) |
| Phytophthora palmivora | Jha et al. (2010) | |||
| Magnaporthe oryzae | ||||
| MsDef1 | Medicago sativa | Tomato | Fusarium oxysporum | Abdallah et al. (2010) |
| Phytophthora parasitica | ||||
| Peronospora hyoscyami | ||||
| NmDef02 | N. megalosiphon | Tobacco/potato | Phytophthora infestans | Portieles et al. (2010) |
| Alternaria solani | ||||
| WjAMP‐1 | Wasabia japonica | Melon | Fusarium oxysporum | Ntui et al. (2010) |
| Alternaria solani | ||||
| cadef1 | Chili | Tomato | Fusarium sp. | Zainal et al. (2009) |
| Phytophthora infestans | ||||
| DEF2 | Capsicum annum | Tomato | Botrytis cinerea | Stotz et al. (2009) |
| BjD | Mustard | Peanut | Phaeoisariopsis personata | Swathi Anuradha et al. (2008) |
| Cercospora arachidicola | ||||
| ica L | ||||
| alfAFP | Alfalfa | Potato | Verticillium dahliae | Gao et al. (2000) |
| DRR206 | Pea | Canola | Leptosphaeria maculans | Wang et al. (1999) |
| WjAMP-1 | Wasabia japonica | Rice | Magnaporthe grisea | Kanzaki et al. (2002) |
| Wasabi defensin | Wasabia japonica | Potato | Botrytis cinerea | Khan et al. (2006) |
| Tomato | Botrytis cinerea | Khan et al. (2011b,a) | ||
| Alternaria solani | ||||
| Fusarium oxysporum | ||||
| Erysiphe lycopersici | ||||
| Petunia | Botrytis cinerea | Khan et al. (2011b,a) | ||
| Phalaenopsis | Erwinia carotovora | Sjahril et al. (2006) | ||
| MiAMP1 | Macadamia integrifolia | Canola | Leptosphaeria maculans | Kazan et al. (2002) |
| Human β‐defensin 2 | Human | Arabidopsis | Botrytis cinerea | Aerts et al. (2007) |
| NaD1 | Nicotiana alata | Cotton | Fusarium oxysporum | Gaspar et al. (2014) |
| Verticillium dahliae | ||||
| PaDef | Persea americana | Bovine endothelial | E. coli | Guzmán-Rodríguez et al. (2013) |
| Cell line BVE-E6E7 | Staphylococcus aureus | |||
| Candida albicans |
Co-transformation of defensins and other pathogenesis-related proteins
The advantages of gene ‘stacking’ or ‘pyramiding’ are obvious in genetically modified (GM) crops, and several different multi-transgene-stacking methods are available. Using linker peptides for multiple gene transformation is considered to be a good method to meet a variety of needs. As a modern and effective protection strategy against phytopathogens, transgenic approaches are employed to use the natural defense mechanism of plants (Tiwari et al. 2008). Using these strategies may lead to reduced cost of crop protection as well as the potentially hazardous effect of pesticides on ecosystems (Holland et al. 2012). The defensin family, as reported, has quite diverse biological activity and strong potential to be utilized for production of disease resistant transgenic crops (Lay et al. 2003). Researchers have been elucidating the ways to co-transform more than one gene in transgenic plants for enhanced and effective disease resistance. Overexpression of a fusion gene of fenugreek (Trigonella foenumgraecumdefensin 2; Tfgd2) and radish (R. Sativus antifungal protein 2; RsAFP2) resulted in enhanced resistance against fungal pathogens and insects (Vasavirama and Kirti 2011, 2013). Bala et al. (2016) co-transformed a fusion gene, Tfgd2 and RsAFP2 defensins attached by a linker peptide, in transgenic peanut for enhanced resistance to early leaf spot (ELS) and late leaf spot (LLS) diseases. Similarly, Chen et al. (2009) has produced transgenic tomato carrying CHI and AFP genes and reported that co-transforming plants with defense genes can be an effective approach to restrict infection of B. cinerea than individual transformation (Table 2).
Table 2.
Co-transformation of defensin gene with other genes for enhanced disease resistance against phytopathogens
| Co-transformed peptides | Plant species transformed | Target pathogen | References |
|---|---|---|---|
| Dm-AMP1 and Rs-AFP2 | Rice | Magnaporthe oryzae | Jha and chattoo (2009) |
| Rhizoctonia solani | |||
| Chitinase and defensin | Tomato | Botrytis cinerea | Chen et al. (2009) |
| Tfgd2 and RsAFP2 | Peanut | Cercospora arachidicola | Bala et al. (2016) |
| Phaeoisariopsis personate | |||
| Wasabi-def and ipt gene | Eggplant | Alternaria solani | Darwish et al. (2014) |
| Petunia hybrida | Botrytis cinere | Khan et al. (2011b,a) | |
| Tomato | Botrytis cinereal | Khan et al. (2011b,a) | |
| Alternaria solani | |||
| Fusarium oxysporum | |||
| Erysiphe lycopersici | |||
| Wasabi-def and chitinase | Tobacco | Fusarium oxysporum | Ntui et al. (2011) |
| WD and LjNRT2 and | Tobacco, tomato | Fusarium oxysporum | Kong et al. (2014) |
| AtNRT2.1 promoters | |||
| Chitinase and WD | Tomato | Fusarium oxysporum | Khan et al. (2014) |
| Alternaria solani | |||
| (AFP and hpt) | Tomato | Fusarium oxysporum | El-Siddig et al. (2011) |
| Leveilula Taurica, | |||
| Alternaria solani/alternate | |||
| Phytophthora infestans |
Marker-free transgenic plants have also been produced which had enhanced disease resistance against plant pathogens; transgenic tobacco co-transformed with chitinase and wasabi defensin genes exhibited increased resistance to Fusarium oxysporum (Ntui et al. 2011). They concluded that the transgenic tobacco lines co-expressing both the transgenes, ChiC and WD genes, conferred higher resistance against Fusarium oxysporum and Alternaria solani than the non-transformed control, and the transgenic plants transformed with either of the individual genes. Similarly, Khan et al. (2014) have reported that co-transformation of ChiC and WD genes in transgenic potato showed increased resistance against Fusarium oxysporum and Alternaria solani. These findings suggest that disease resistance of the transgenic plants could be enhanced by multiple transformations or transgene stacking. These findings would further elucidate various possible biological activities of fusion proteins leading to the insights into the exact mode of action of these defensins.
Conclusion and future’s prospects
As plant defensins, in general, are not toxic to human cells, exhibit in vivo efficacy and with low resistance occurrence, their therapeutic potential could be used in the fight against fungal infections in future medicine. Plant defensins could be used as potential alternatives to pesticides to control pathogenic fungi. Further investigation is required to evaluate potential of plant defensins as therapeutic agents and biological control agents (biopesticides). Defensin-based agrobioproducts are also expected to be explored for improved crop production.
The plant defensins also could be used against cancer for their potential anticancer and cytotoxicity effects.
Hexima, a biotechnology company working on development of plant-derived peptides and proteins for their potential applications as human therapeutics, has recently started research on application of plant defensin technology to assess its potential in control of medically important Candidaemias and Candida-based biofilms. Hexima is conducting phase I/IIa clinical trials to evaluate safety and preliminary efficacy of its lead therapeutic agent, HXP124 (a small plant defensin) for treatment for fungal nail infections.
Compliance with ethical standards
Conflict of interest
The authors have no conflict of interest.
References
- Abdallah NA, Shah D, Abbas D, Madkour M. Stable integration and expression of a plant defensin in tomato confers resistance to fusarium wilt. GM Crops. 2010;1:344–350. doi: 10.4161/gmcr.1.5.15091. [DOI] [PubMed] [Google Scholar]
- Aerts AM, Thevissen K, Bresseleers SM, Sels J, WoutersP Cammue BPA, François IEJA. Arabidopsis thaliana plants expressing human-defensin-2 are more resistant to fungal attack: functional homology between plant and human defensins. Plant Cell Rep. 2007;26:1391–1398. doi: 10.1007/s00299-007-0329-4. [DOI] [PubMed] [Google Scholar]
- Allen A, Snyder AK, Preuss M, Nielsen EE, Shah DM, Smith TJ. Plant defensins and virally encoded fungal toxin KP4 inhibit plant root growth. Planta. 2008;227:331–339. doi: 10.1007/s00425-007-0620-1. [DOI] [PubMed] [Google Scholar]
- Bala M, Radhakrishnan A, Kumar A, Mishra GP, Dobraia JR, Kirti PB. Overexpression of a fusion defensin gene from radish and fenugreek improves resistance against leaf spot diseases caused by Cercospora arachidicola and Phaeoisariopsis personata in peanut. Turk J Biol. 2016;40:139–149. doi: 10.3906/biy-1412-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batta G, Barna T, Gaspari Z, Sandor S, Kover KE, Binder U, et al. Functional aspects of the solution structure and dynamics of PAF—a highly-stable anti-fungal protein from Penicillium chrysogenum. FEBS J. 2009;276:2875–2890. doi: 10.1111/j.1742-4658.2009.07011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch CJR, Richardson M. A new family of small (5 kD) protein inhibitors of insect alpha-amylases from seeds or sorghum (Sorghum bicolor Moench) have sequence homologies with wheat -purothionins. FEBS Lett. 1991;279:101–104. doi: 10.1016/0014-5793(91)80261-Z. [DOI] [PubMed] [Google Scholar]
- Broekaert WF, Terras FRG, Cammue BPA, Osborn RW. Plant defensins: novel antimicrobial peptides as components of the host defence system. Plant Physiol. 1995;108:1353–1358. doi: 10.1104/pp.108.4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulet P, Stocklin R. Insect antimicrobial peptides: structures, properties and gene regulation. Protein Pept Lett. 2005;12:3–11. doi: 10.2174/0929866053406011. [DOI] [PubMed] [Google Scholar]
- Bulet P, Stöcklin R, Menin L. Anti-microbial peptides: from invertebrates to vertebrates. Immunol Rev. 2004;198:169–184. doi: 10.1111/j.0105-2896.2004.0124.x. [DOI] [PubMed] [Google Scholar]
- Carvalho AO, Gomes VM. Role of plant lipid transfer proteins in plant cell physiology—a concise review. Peptides. 2007;28:1144–1153. doi: 10.1016/j.peptides.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Chen KC, Lin CY, Kuan CC, Sung HY, Chen CS. A novel defensin encoded by a Mungbean cdna exhibits insecticidal activity against bruchid. J Agric Food Chem. 2002;50:7258–7263. doi: 10.1021/jf020527q. [DOI] [PubMed] [Google Scholar]
- Chen JJ, Chen GH, Hsu HC, Li SS, Chen CS. Cloning and Functional Expression of a Mungbean Defensin VrD1 in Pichia pastoris. J Agric Food Chem. 2004;52:2256–2261. doi: 10.1021/jf030662i. [DOI] [PubMed] [Google Scholar]
- Chen SC, Liu AR, Wang FH, Ahammed GJ. Combined overexpression of chitinase and defensin genes in transgenic tomato enhances resistance to Botrytis cinerea. Afr J Biotechnol. 2009;8(20):5182–5188. [Google Scholar]
- Choi MS, Kim YH, Park HM, Seo BY, Jung JK, Kim ST, Kim MC, Shin DB, Yun HT, Choi IS, Kim CK, Lee JY. Expression of Br D1, a plant defensin from Brassica rapa, confers resistance against brown plant hopper (Nilaparvata lugens) in transgenic rice. Mol Cells. 2009;28:131–137. doi: 10.1007/s10059-009-0117-9. [DOI] [PubMed] [Google Scholar]
- Dalla Valle L, Benato F, Maistro S, Quinzani S, Alibardi L. Bioinformatic and molecular characterization of beta-defensins-like peptides isolated from the green lizard Anolis carolinensis. Dev Comp Immunol. 2012;36:222–229. doi: 10.1016/j.dci.2011.05.004. [DOI] [PubMed] [Google Scholar]
- Darwish NA, Khan RS, Ntui VO, Nakamura I, Mii M. Generation of selectable marker-free transgenic eggplant resistant to Alternaria solani using the R/RS site-specific recombination system. Plant Cell Rep. 2014;33:411–421. doi: 10.1007/s00299-013-1541-z. [DOI] [PubMed] [Google Scholar]
- de Paula VS, Razzera G, Barreto-Bergter E, Almeida FCL, Valente AP. Portrayal of complex dynamic properties of sugarcane defensin 5 by NMR: multiple motions associated with membrane interaction. Structure. 2011;19:26–36. doi: 10.1016/j.str.2010.11.011. [DOI] [PubMed] [Google Scholar]
- Dimarcq JL, Bulet P, Hetru C, Hoffmann J. Cysteine-rich antimicrobial peptides in invertebrates. Biopolymers. 1998;47:465–477. doi: 10.1002/(SICI)1097-0282(1998)47:6<465::AID-BIP5>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- El-Siddig MA, El-Hussein AA, Saker MM. Agrobacterium-mediated transformation of tomato plants expressing defensin gene. Inter J of Agric Res. 2011;6:323–334. doi: 10.3923/ijar.2011.323.334. [DOI] [Google Scholar]
- Ericksen B, Wu Z, Lu W, Lehrer RI. Antibacterial activity and specificity of the six human {alpha}-defensins. Antimicrob Agents Chemother. 2005;49:269–275. doi: 10.1128/AAC.49.1.269-275.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fant F, Vranken W, Broekaert W, Borremans F. Determination of the three-dimensional solution structure of Raphanus sativus antifungal protein 1 by 1h nmr. J Mol Biol. 1998;279:257–270. doi: 10.1006/jmbi.1998.1767. [DOI] [PubMed] [Google Scholar]
- Fant F, Vranken WF, Borremans FAM. The three-dimensional solution structure of Aesculus hippocastanum antimicrobial protein 1 determined by 1 H nuclear magnetic resonance. Proteins. 1999;37:388–403. doi: 10.1002/(SICI)1097-0134(19991115)37:3<388::AID-PROT7>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Francisco GCA, Georgina E. Structural motifs in class I and class II plant defensins for phospholipid interactions:intriguing role of ligand binding and modes of action. Francisco and Georgina, J Plant Physiol Pathol. 2017;5:1–7. [Google Scholar]
- Fujimura M, Minami Y, Watanabe K, Tadera K. Purification, characterization, and sequencing of a novel type of antimicrobial peptides, Fa-AMP1 and Fa-AMP2, from seeds of buckwheat (Fagopyrum esculentum Moench.) Biosc Biotechnol Biochem. 2003;67:1636–1642. doi: 10.1271/bbb.67.1636. [DOI] [PubMed] [Google Scholar]
- Ganz T. Defensins antimicrobial peptides of innate immunity. Nat Rev Immunol. 2003;3:710–720. doi: 10.1038/nri1180. [DOI] [PubMed] [Google Scholar]
- Ganz T. Defensins antimicrobial peptides of vertebrates. C R Biol. 2004;327:539–549. doi: 10.1016/j.crvi.2003.12.007. [DOI] [PubMed] [Google Scholar]
- Ganz T. Defensins and other antimicrobial peptides: a historical perspective and an update. Comb Chem High Throughput Screen. 2005;8:209–217. doi: 10.2174/1386207053764594. [DOI] [PubMed] [Google Scholar]
- Ganz T, Selsted ME, Szklarek D, Harwig SS, Daher K, Bainton DF, et al. Defensins: natural peptide antibiotics of human neutrophils. J Clin Invest. 1985;76:1427–1435. doi: 10.1172/JCI112120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao AG, Hakimi SM, Mittanck CA, Woerner BM, Stark DM, Shah DM, Liang J, Rommens CM. Fungal pathogen protection in potato by expression of a plant defensin peptide. Nature biotechnol. 2000;18:1307–1310. doi: 10.1038/82436. [DOI] [PubMed] [Google Scholar]
- Garcia-Olmedo F, Molina A, Alamillo JM, Rodriguez-Palenzuela P. Plant defense peptides. Biopolymers. 1998;47:479–491. doi: 10.1002/(SICI)1097-0282(1998)47:6<479::AID-BIP6>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Gaspar YM, McKenna JA, McGinness BS, Hinch J, Poon S, Connelly AA, Heath RL. Field resistance to Fusarium oxysporum and Verticillium dahliae in transgenic cotton expressing the plant defensin NaD1. J Experiment Bot. 2014;65(6):1541–1550. doi: 10.1093/jxb/eru021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham MA, Silverstein KAT, Cannon SB, VandenBosch KA. Computational identification and characterization of novel genes from legumes. Plant Physiol. 2004;135:1179–1197. doi: 10.1104/pp.104.037531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzmán-Rodríguez JJ, López-Gómez R, Suárez-Rodríguez LM, Salgado-Garciglia R, Rodríguez-Zapata LC, Ochoa-Zarzosa A, López-Meza JE. Antibacterial Activity of Defensin PaDef from Avocado Fruit (Persea americana var. drymifolia) Expressed in Endothelial Cells against Escherichia coli and Staphylococcusaureus. BioMed Research Inter. 2013;5:1–9. doi: 10.1155/2013/986273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks JN, Snyder AK, Graham MA, Shah RK, Blaylock LA, Harrison MJ, Shah DM. Defensin gene family in Medicago truncatula: structure, expression and induction by signal molecules. Plant Mol Biol. 2005;58:385–399. doi: 10.1007/s11103-005-5567-7. [DOI] [PubMed] [Google Scholar]
- Hayes BM, Bleackley MR, Wiltshire JL, Anderson MA, Traven A, van der Weerden NL. Identification and mechanism of action of the plant defensin nad1 as a new member of the antifungal drug arsenal against Candida albicans. Antimicrob Agents Chemother. 2013;57:3667–3675. doi: 10.1128/AAC.00365-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann JA, Hetru C. Insect defensins: inducible antibacterial peptides. Immunol Today. 1992;13:411–415. doi: 10.1016/0167-5699(92)90092-L. [DOI] [PubMed] [Google Scholar]
- Holland JM, Oaten H, Moreby S, Birkett T, Simper J, Southway S, Smith BM. Agri-environment scheme enhancing ecosystem services: a demonstration of improved biological control in cereal crops. Agric Ecosyst Environ. 2012;155:147–152. doi: 10.1016/j.agee.2012.04.014. [DOI] [Google Scholar]
- Huang GJ, Lai HC, Chang YS, Sheu MJ, Lu TL, Huang SS, Lin YH. Antimicrobial, dehydroascorbate reductase, and monodehydroascorbate reductase activities of defensin from sweet potato [ipomoea batatas (l.) lam. ‘Tainong 57’] storage roots. J Agric Food Chem. 2008;56:2989–2995. doi: 10.1021/jf072994j. [DOI] [PubMed] [Google Scholar]
- Janssen BJ, Schirra HJ, Lay FT, Anderson MA, Craik DJ. Structure of Petunia hybrid defensin 1, a novel plant defensin with five disulfide bonds. Biochem. 2003;42:8214–8222. doi: 10.1021/bi034379o. [DOI] [PubMed] [Google Scholar]
- Järvå M, Lay FT, Phan TK, Humble C, Poon IKH, Bleackley MR, Anderson MA, Hulett MD, Kvansakul M. X-ray structure of a carpet-like antimicrobial defensin–phospholipid membrane disruption complex. Nat Commun. 2018;9:1962. doi: 10.1038/s41467-018-04434-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha S, Chattoo BB. Expression of a plant defensin in rice confers resistance to fungal phytopathogens. Transgenic Res. 2010;19:373–384. doi: 10.1007/s11248-009-9315-7. [DOI] [PubMed] [Google Scholar]
- Jung YJ, Kang KK. Application of antimicrobial peptides for disease control in plants. Plant Breed Biotech. 2014;1:1–13. doi: 10.9787/PBB.2014.2.1.001. [DOI] [Google Scholar]
- Kaewklom S, Wongchai M, Petvises S, Hanpithakphong W, Aunpad R. Structural and biological features of a novel plant defensin from Brugmansia x candida. PLoS One. 2018;13(8):201668. doi: 10.1371/journal.pone.0201668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanzaki H, Nirasawa S, Saitoh H. Overexpression of the wasabi defensin gene confers enhanced resistance to blast fungus (Magnaporthe grisea) in transgenic rice. Theor Appl Genet. 2002;105:809–814. doi: 10.1007/s00122-001-0817-9. [DOI] [PubMed] [Google Scholar]
- Kazan K, Rusu A, Marcus JP, Goulter KC, Manners JM. Enhanced quantitative resistance to Laptosphaeria maculans conferred by expression of a novel antimicrobial peptide in canola (Brassica napus L.) Mol Breed. 2002;10:63–70. doi: 10.1023/A:1020354809737. [DOI] [Google Scholar]
- Khan RS, Nishihara M, Yamamura S, Nakamura I, Mii M. Transgenic Potatoes expressing wasabi defensin peptide confer partial resistance to gray mold (Botrytis cinerea) Plant Biotechnol. 2006;23:179–183. doi: 10.5511/plantbiotechnology.23.179. [DOI] [Google Scholar]
- Khan RS, Sjahril R, Nakamura I, Mii M. Production of transgenic potato exhibiting enhanced resistance to fungal infection and herbicide applications. Plant Biotechnol Rep. 2008;2:13–20. doi: 10.1007/s11816-008-0043-x. [DOI] [Google Scholar]
- Khan RS, Ntui VO, Chin DP, Nakamura I, Mii M. Production of marker-Free disease-resistant potato using isopentenyl transferase gene as a positive selection marker. Plant Cell Rep. 2011;30:587–597. doi: 10.1007/s00299-010-0974-x. [DOI] [PubMed] [Google Scholar]
- Khan RS, Nakamura I, Mii M. Development of disease resistant marker-free tomato by R/RS site-specific recombination. Plant Cell Rep. 2011;30:1041–1053. doi: 10.1007/s00299-011-1011-4. [DOI] [PubMed] [Google Scholar]
- Khan RS, Darwish NA, Khattak B, Ntui V, Kong K, Shimomae K, et al. Retransformation of marker-free potato for enhanced resistance against fungal pathogens by pyramiding chitinase and Wasabi Defensin Genes. Mol Biotechnol. 2014;56:814–823. doi: 10.1007/s12033-014-9760-2. [DOI] [PubMed] [Google Scholar]
- Kim C, Kaufmann SH. Defensin a multifunctional molecule lives up to its versatile name. Trends Microbiol. 2006;14:428–431. doi: 10.1016/j.tim.2006.08.001. [DOI] [PubMed] [Google Scholar]
- Kong K, Ntui VO, Makabe S, Khan RS, Mii M, et al. Transgenic tobacco and tomato plants expressing Wasabi defensin genes driven by root-specific LjNRT2 and AtNRT2. 1 promoters confer resistance against Fusarium oxysporum. Plant Biotechnol. 2014;31:89–96. doi: 10.5511/plantbiotechnology.13.1209a. [DOI] [Google Scholar]
- Lacerda AF, Vasconcelos ÉAR, Pelegrini PB, Grossi de Sa MF. Antifungal defensins and their role in plant defense. Front in Microbiol. 2014;5:116. doi: 10.3389/fmicb.2014.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lay FT, Anderson M. Defensins-components of the innate immune system in plants. Curr Pro Pep Sci. 2005;6:85–101. doi: 10.2174/1389203053027575. [DOI] [PubMed] [Google Scholar]
- Lay FT, Brugliera F, Anderson MA. Isolation and properties of floral defensins from ornamental tobacco and petunia. Plant Physiol. 2003;131:1283–1293. doi: 10.1104/pp.102.016626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lay FT, Mills GD, Poon IKH, Cowieson NP, Kirby N, Baxter AA, et al. Dimerization of plant defensin NaD1 enhances its antifungal activity. J Biol Chem. 2012;287:19961–19972. doi: 10.1074/jbc.M111.331009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lay FT, Poon S, McKenna JA, Connelly AA, Barbeta BL, McGinness BS, Fox JL, Daly NL, Craik DJ, Heath RL, et al. The C-terminal propeptide of a plant defensin confers cytoprotective and subcellular targeting functions. BMC Plant Biol. 2014;14:41. doi: 10.1186/1471-2229-14-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrer RI. Primate defensins. Nat Rev Microbiol. 2004;2:727–738. doi: 10.1038/nrmicro976. [DOI] [PubMed] [Google Scholar]
- Li Z, Zhou M, Zhang Z, Ren L, Du L, Zhang B, Xu H, Xin Z. Expression of a radish defensin in transgenic wheat confers increased resistance to Fusarium graminearum and Rhizoctonia cerealis. Funct Integr Genomics. 2011;11:63–70. doi: 10.1007/s10142-011-0211-x. [DOI] [PubMed] [Google Scholar]
- Liu L, Wang L, Jia HP, Zhao C, Heng HHQ, Schutte BC, McCray PB, Ganz T. Structure and mapping of the human β-defensin 2 gene and its expression at sites of inflammation. Gene. 1998;222:237–244. doi: 10.1016/S0378-1119(98)00480-6. [DOI] [PubMed] [Google Scholar]
- Mendez E, Moreno A, Colilla F, Pelaez F, Limas GG, Mendez R, Soriano F, Salinas M, Haro C. Primary structure and inhibition of protein synthesis in eukaryotic cell-free system of a novel thionin, γ-hordothionin, from barley endosperm. Euro J Biochem. 1990;194:533–539. doi: 10.1111/j.1432-1033.1990.tb15649.x. [DOI] [PubMed] [Google Scholar]
- Mith O, Benhamdi A, Castillo T, Bergé M, MacDiarmid CW, Steffen J, Eide DJ, Perrier V, Subileau M, Gosti F, Berthomieu P. The antifungal plant defensin AhPDF1.1b is a beneficial factor involved in adaptive response to zinc overload when it is expressed in yeast cells. Microbiol Open. 2015;3:409–422. doi: 10.1002/mbo3.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanni V, Schumacher J, Giacomelli L, et al. Vv-AMP2, a grapevine flower specific defensin capable of Botrytis cinerea growth inhibition: insights into its mode of action. Plant Pathol. 2014;63:899–910. doi: 10.1111/ppa.12170. [DOI] [Google Scholar]
- Ntui VO, Thirukkumaran G, Azadi P, Khan RS, NakamuraI Mii M. Stable integration and expression of wasabi defensin gene in “Egusi” melon (Colocynthis citrullus L.) confers resistance to Fusarium wilt and Alternaria leaf spot. Plant Cell Rep. 2010;29:943–954. doi: 10.1007/s00299-010-0880-2. [DOI] [PubMed] [Google Scholar]
- Ntui VO, Azadi P, Thirukkumaran G, Khan RS, Chin DP, Nakamura I, Mii M. Increased resistance to fusarium wilt in transgenic tobacco lines co-expressing chitinase and wasabi defensin genes. Plant Pathol. 2011;60:221–231. doi: 10.1111/j.1365-3059.2010.02352.x. [DOI] [Google Scholar]
- Osborn RW, De Samblanx GW, Thevissen K, Goderis I, Torrekens S, Van Leuven F, Attenborough S, Rees SB, Broekaert WF. Isolation and characterisation of plant defensins from seeds of Asteraceae, Fabaceae, Hippocastanaceae and Saxifragaceae. FEBS Lett. 1995;368:257–262. doi: 10.1016/0014-5793(95)00666-W. [DOI] [PubMed] [Google Scholar]
- Parisi K, Shafee TMA, Quimbar P, Van Der Weerden NL, Bleackley MR, Anderson MA. The evolution, function and mechanisms of action for plant defensins. Semin Cell Dev Biol. 2018;5:6. doi: 10.1016/j.semcdb.2018.02.004. [DOI] [PubMed] [Google Scholar]
- Park MS, Kim JI, Lee I, Park Bae JY, Park MS. Towards the Application of Human Defensins as Antivirals. Invited Review Biomol Ther. 2018;5:1–13. doi: 10.4062/biomolther.2017.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelegrini PB, Lay FT, Murad AM, Anderson MA, Franco OL. Novel insights on the mechanism of action of alpha-amylase inhibitors from the plant defensin family. Proteins. 2008;73:719–729. doi: 10.1002/prot.22086. [DOI] [PubMed] [Google Scholar]
- Phoenix DA, Dennison SR, Harris F. Antimicrobial Peptides. Weinheim: Wiley; 2013. Antimicrobial peptides: their history, evolution, and functional promiscuity; pp. 1–38. [Google Scholar]
- Portieles R, Ayra C, Gonzalez E, Gallo A, Rodriguez R, Chacón O, López Y, Rodriguez M, Castillo J, PujolM Enriquez G, Borroto C, Trujillo L, Thomma BP, Borrás-Hidalgo O. NmDef02, a novel antimicrobial gene isolated from Nicotiana megalosiphon confers high-level pathogen resistance under greenhouse and field conditions. P Biotech J. 2010;8:678–690. doi: 10.1111/j.1467-7652.2010.00501.x. [DOI] [PubMed] [Google Scholar]
- Rehaume L, Hancock RE. Neutrophil-derived defensins as modulators of innate immune function. Crit Rev Immunol. 2008;28:185–200. doi: 10.1615/CritRevImmunol.v28.i3.10. [DOI] [PubMed] [Google Scholar]
- Rodriguez de la Vega RC, Possani LD. On the evolution of invertebrate defensins. Trends Genet. 2005;21:330–332. doi: 10.1016/j.tig.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Selitrennikoff CP. Antifungal proteins. Appl Environ Microbiol. 2001;67:2883–2894. doi: 10.1128/AEM.67.7.2883-2894.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selsted ME. Theta-defensins: cyclic antimicrobial peptides produced by binary ligation of truncated alpha-defensins. Curr Protein Pept Sci. 2004;5(5):365–367. doi: 10.2174/1389203043379459. [DOI] [PubMed] [Google Scholar]
- Shi J. Defensins and Paneth cells in inflammatory bowel disease. Inflamm Bowel Dis. 2007;13:1284–1292. doi: 10.1002/ibd.20197. [DOI] [PubMed] [Google Scholar]
- Silverstein KA, Graham MA, Paape TD, VandenBosch KA. Genome organization of more than 300 defensin-like genes in Arabidopsis. Plant Physiol. 2005;138(2):600–610. doi: 10.1104/pp.105.060079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitaram N. Antimicrobial peptides with unusual amino acid compositions and unusual structures. Curr Med Chem. 2006;13:679–696. doi: 10.2174/092986706776055689. [DOI] [PubMed] [Google Scholar]
- Sjahril R, Chin DP, Khan RS, Yamamura S, Nakamura I, Amemiya Y, Mii M. Transgenic Phalaenopsis plants with resistance to Erwinia carotovora produced by introducing wasabi defensin gene using Agrobacterium method. Plant Biotech. 2006;23:191–194. doi: 10.5511/plantbiotechnology.23.191. [DOI] [Google Scholar]
- Song X, Zhou Z, Wang J, Wu F, Gong W. Purification, characterization and preliminary crystallographic studies of a novel plant defensin from Pachyrrhizu serosus seeds. Acta Crystallogr D Biol Crystallogr. 2004;60:1121–1124. doi: 10.1107/S0907444904007395. [DOI] [PubMed] [Google Scholar]
- Song X, Zhang M, Zhou Z, Gong W. Ultra-high resolution crystal structure of a dimeric defensin SPE10. FEBS Lett. 2011;585:300–306. doi: 10.1016/j.febslet.2010.12.039. [DOI] [PubMed] [Google Scholar]
- Spelbrink RG, Dilmac N, Allen A, Smith TJ, Shah DM, Hockerman GH. Differential Antifungal and Calcium Channel-Blocking Activity among Structurally Related Plant Defensins. Plant Physiol. 2004;135(4):2055–2067. doi: 10.1104/pp.104.040873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stotz HU, Thomson J, Wang Y. Plant defensins: defense, development and application. Plant Signal Behav. 2009;4:1010–1012. doi: 10.4161/psb.4.11.9755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swathi Anuradha T, Divya K, Jami SK, Kirti PB. Transgenic tobacco and peanut plants expressing a mustard defensin show resistance to fungal pathogens. Plant Cell Rep. 2008;27:1777. doi: 10.1007/s00299-008-0596-8. [DOI] [PubMed] [Google Scholar]
- Tam JP, Wang S, Wong KH, Tan WL. Antimicrobial peptides from plants. Pharmaceuticals. 2015;8:711–757. doi: 10.3390/ph8040711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares LS, Santos MO, Viccini LF, Moreira JS, Miller RN, Franco OL. Biotechnological potential of antimicrobial peptides from flowers. Peptide. 2008;29:1842–1851. doi: 10.1016/j.peptides.2008.06.003. [DOI] [PubMed] [Google Scholar]
- Terras FR, Goderis IJ, Van Leuven F, Vanderleyden J, Cammue BP, Broekaert WF. In vitro antifungal activity of a radish (Raphanus sativus L.) seed protein homologous to nonspecific lipid transfer proteins. Plant Physiol. 1992;100:1055–1058. doi: 10.1104/pp.100.2.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terras F, Schoofs H, De Bolle M, Van Leuven F, Rees SB, Vanderleyden J, Cammue B, Broekaert WF. Analysis of two novel classes of plant antifungal proteins from radish (Raphanus sativus L.) seeds. J Biol Chem. 1992;267:15301–15309. [PubMed] [Google Scholar]
- Terras FRG, Schoofs HME, Thevissen K, Osborn R, Vanderleyden J, Cammue BPA, Broekaert WF. Synergistic enhancement of the antifungal activity of wheat and barley thionins by radish and oilseed rape 2S albumins and by barley trypsin inhibitors. Plant Physiol. 1993;103:1311–1319. doi: 10.1104/pp.103.4.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terras FRG, Eggermont K, Kovaleva V, Raikhel NV, Osborn RW, Kester A, Rees SB, den Vanderley J, Cammue BPA, Broekaert WF. Small cysteine-rich antifungal proteins from radish: their role in host defence. Plant Cell. 1995;7:573–588. doi: 10.1105/tpc.7.5.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevissen K, Ghazi A, De Samblanx GW, Brownlee C, Osborn RW, Broekaert WF. Fungal membrane responses induced by plant defensins and thionins. J Biol Chem. 1996;271:15018–15025. doi: 10.1074/jbc.271.25.15018. [DOI] [PubMed] [Google Scholar]
- Thevissen K, Osborn RW, Acland DP, Broekaert WF. Specific binding sites for an antifungal plant defensin from dahlia (Dahlia merckii) on fungal cells are required for antifungal activity. Mol Plant Microbe Interact. 2000;13:54–61. doi: 10.1094/MPMI.2000.13.1.54. [DOI] [PubMed] [Google Scholar]
- Thevissen K, Cammue BP, Lemaire K, Winderickx J, Dickson RC, Lester RL, Ferket KK, Van Even F, Parret AH, Broekaert WF. A gene encoding a sphingolipid biosynthesis enzyme determines the sensitivity of Saccharomyces cerevisiae to an antifungal plant defensin from dahlia (Dahlia merckii) Proceed Nat Acad Sci. 2000;97:9531–9536. doi: 10.1073/pnas.160077797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevissen K, Warnecke DC, Francois IE, Leipelt M, Heinz E, Ott C, Zahringer U, Thomma BP, Ferket KK, Cammue BP. Defensins from insects and plants interact with fungal glucosylceramides. J Biol Chem. 2004;279:3900–3905. doi: 10.1074/jbc.M311165200. [DOI] [PubMed] [Google Scholar]
- Thomma BP, Cammue BP, Thevissen K. Plant defensins. Planta. 2002;216:193–202. doi: 10.1007/s00425-002-0902-6. [DOI] [PubMed] [Google Scholar]
- Tiwari S, Mishra DK, Singh A, Singh PK, Tuli R. Expression of a synthetic Cry1EC gene for resistance against Spodoptera litura in transgenic peanut (Arachis hypogaea L.) Plant Cell Rep. 2008;27:1017–1025. doi: 10.1007/s00299-008-0525-x. [DOI] [PubMed] [Google Scholar]
- Van der Weerden NL, Lay FT, Anderson MA. The plant defensin, nad1, enters the cytoplasm of Fusarium oxysporum hyphae. J Biol Chem. 2008;283:14445–14452. doi: 10.1074/jbc.M709867200. [DOI] [PubMed] [Google Scholar]
- Vasavirama K, Kirti PB (2011) Expression, affinity purification, and functional characterization of recombinant fusion gene. World Congress on Biotechnol (21–23 March 2011). J Microbial Biochem Technol S1(013): 35.
- Vasavirama K, Kirti PB. Constitutive expression of a fusion gene comprising Trigonella foenum-graecum defensin (Tfgd2) and Raphanus sativus antifungal protein (RsAFP2) confers enhanced disease and insect resistance in transgenic tobacco. Plant Cell Tiss Org Cult. 2013;115:309–319. doi: 10.1007/s11240-013-0363-6. [DOI] [Google Scholar]
- Velivelli SLS, Islam KT, Hobson E, Shah DM. Modes of Action of a Bi-domain Plant Defensin MtDef5 Against a Bacterial Pathogen Xanthomonas campestris. Front Microbiol. 2018;9:934. doi: 10.3389/fmicb.2018.00934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vriens K, Bruno PA, Cammue BP, Thevissen K. Antifungal Plant Defensins: Mechanisms of Action and Production. Molecules. 2014;19:12280–12303. doi: 10.3390/molecules190812280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Nowak G, Culley D, Hadwiger LA, Fristensky B. Constitutive expression of pea defense gene DRR206 confers resistance to blackleg (Leptosphaeria maculans) disease in transgenic canola (Brassica napus) Mol Plant-Microbe Interact. 1999;12:410–418. doi: 10.1094/MPMI.1999.12.5.410. [DOI] [Google Scholar]
- Wijaya R, Neumann GM, Condron R, Hughes AB, Polya GM. Defense proteins from seed of Cassia fistula include a lipid transfer protein homologue and a protease inhibitory plant defensin. Plant Sci. 2000;159:243–255. doi: 10.1016/S0168-9452(00)00348-4. [DOI] [PubMed] [Google Scholar]
- Wimley WC. Describing the mechanism of antimicrobial peptide action with the interfacial activity model. ACS ChemBiol. 2010;10:905–917. doi: 10.1021/cb1001558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JH, Xia L, Ng T. A review of defensins of diverse origins. Curr Prot Peptide Sci. 2007;8:446–459. doi: 10.2174/138920307782411446. [DOI] [PubMed] [Google Scholar]
- Yamano A, Heo NH, Teeter MM. Crystal structure of Ser-22/Ile-25 form crambin confirms solvent, side chain substate correlations. J Biol Chem. 1997;272:9597–9600. doi: 10.1074/jbc.272.15.9597. [DOI] [PubMed] [Google Scholar]
- Yang D, Biragyn A, Kwak LW, Oppenheim JJ. Mammalian defensins in immunity: more than just microbicidal. Trends Immunol. 2002;23:291–296. doi: 10.1016/S1471-4906(02)02246-9. [DOI] [PubMed] [Google Scholar]
- Zainal Z, Marouf E, Ismail I, Fei CK. Expression ofthe Capsicuum annum (Chili) defensin gene in transgenic tomatoes confers enhanced resistance to fungal pathogens. Am J Physiol. 2009;4:70–79. [Google Scholar]
- Zhu S. Discovery of six families of fungal defensin-like peptides provides insights into origin and evolution of CSαβ defensins. Mol Immunol. 2008;45:828–838. doi: 10.1016/j.molimm.2007.06.354. [DOI] [PubMed] [Google Scholar]
- Zhu Q, Bateman A, Singh A, Solomon S. Isolation and biological activity of corticostatic peptides (anti-ACTH) Endocr Res. 1989;15:129–149. doi: 10.1080/07435808909039093. [DOI] [PubMed] [Google Scholar]
- Zhu YJ, Agbayani R, Moore PH. Ectopic expressionof Dahlia merckii defensin DmAMP1 improves papaya resistance to Phytophthorapalmivora by reducing pathogen vigor. Planta. 2007;226:87–97. doi: 10.1007/s00425-006-0471-1. [DOI] [PubMed] [Google Scholar]
- Zou J, Mercier C, Koussounadis A, Secombes C. Discovery of multiple beta-defensin like homologues in teleost fish. Mol Immunol. 2007;44:638–647. doi: 10.1016/j.molimm.2006.01.012. [DOI] [PubMed] [Google Scholar]