Abstract
SOX2 is a stem cell-associated pluripotency transcription factor whose role in neuronal populations is undefined. Here we present the RNA-sequencing based transcriptome profiles of control (Sox2fl/fl) and SOX2 conditional knock-out (Vgat-cre;Sox2fl/fl) mice at four time points in one 24-h circadian cycle. The raw sequencing data were deposited to ArrayExpress database at EMBL-EBI (https://www.ebi.ac.uk/arrayexpress) under the accession number E-MTAB-7496. Results of rhythmicity analysis, differential expression analysis, network prediction, and potential target identification stemming from the RNA-sequencing dataset are also given in this article. The interpretation and discussion of these data can be found in the related research article entitled “SOX2-dependent transcription in clock neurons promotes the robustness of the central circadian pacemaker.” Cheng et al. 2019.
Keywords: Transcriptomics, Circadian rhythms, SOX2, Suprachiasmatic nucleus, Mice
Specifications table
| Subject area | Biology |
| More specific subject area | Circadian rhythm; chronobiology |
| Type of data | Tables, figures and graphs |
| How data were acquired | RNA sequencing using Illumina HiSeq4000 platform |
| Data format | Raw and analyzed |
| Experimental factors | Organism: Mus musculus Strain background: C57BL6 x FVB x 129/SvEv Developmental stage: Adult Tissue: Suprachiasmatic nucleus Genotype: Sox2fl/fl (control) and Vgat-cre;Sox2fl/fl mice |
| Experimental features | Mice were maintained on a fixed 12-h light: 12-h dark schedule and released into constant darkness for two consecutive cycles prior to tissue harvest. RNA from the suprachiasmatic nuclei was extracted and libraries were prepared and sequenced. |
| Data source location | Department of Biology, University of Toronto Mississauga, Canada |
| Data accessibility | The RNA-seq data are publicly available at the ArrayExpress database at EMBL-EBI (https://www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-7496 |
| Related research article | A.H. Cheng, P. Bouchard-Cannon, S. Hegazi, C. Lowden, S.W. Fung, C.-K. Chiang, R.W. Ness, H.-Y.M. Cheng, SOX2-dependent transcription in clock neurons promotes the robustness of the central circadian pacemaker, Cell Rep, 26, 2019, 3191–3202, https://doi.org/10.1016/j.celrep.2019.02.068. [1] |
Value of the data
|
1. Data
To understand the effects of Sox2 ablation within the central circadian pacemaker in mammals, the suprachiasmatic nucleus (SCN), we sequenced the SCN transcriptomes of Sox2fl/fl (control) and Vgat-cre;Sox2fl/fl (SOX2 conditional knock-out) mice at 4 circadian times (CT0, 6, 12, and 18; n = 5 per CT). Raw RNA sequencing data were deposited in the ArrayExpress database at EMBL-EBI (E-MTAB-7496). Alignment metrics can be found in Data S1.
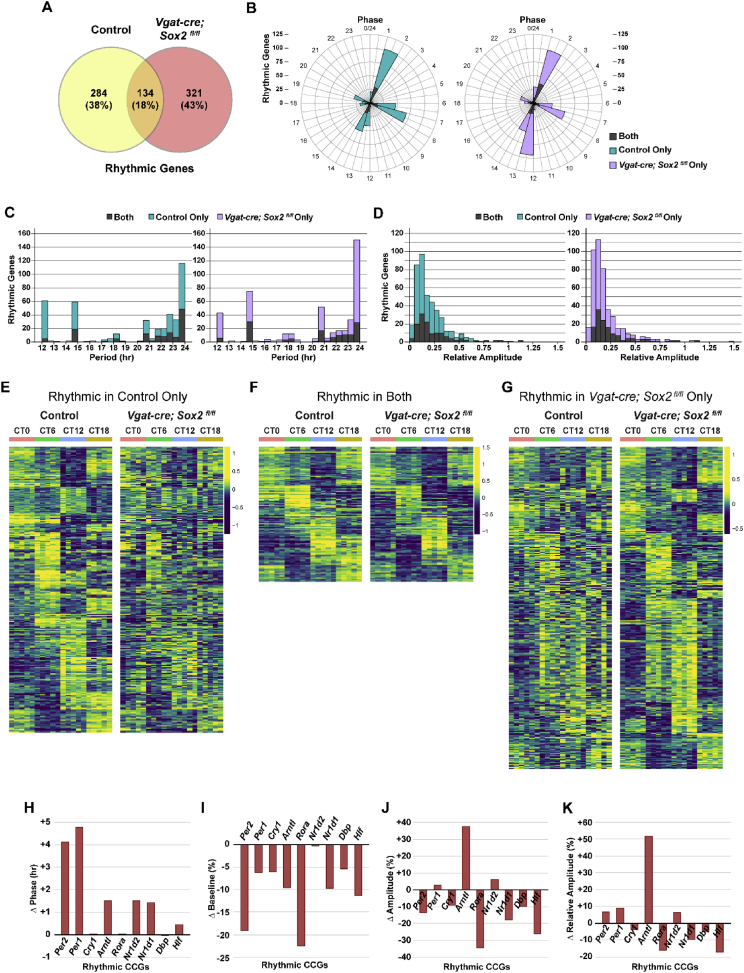
Rhythmicity analysis of the SCN transcriptome of control and Vgat-cre;Sox2fl/fl mice was conducted with MetaCycle and detailed results are summarized in Fig. 1 and Data S2. Phase, period, and relative amplitude of the rhythmic transcriptomes were compiled and shown in Fig. 1B–D. Expression profile of rhythmic genes are visualized by heatmaps in Fig. 1E–G. The changes in rhythmic component of rhythmic core clock genes (CCGs) were calculated and depicted in Fig. 1H–K.
Fig. 1.
MetaCycle analysis of control and Vgat-cre;Sox2fl/flSCN transcriptomes. (A) Venn diagram illustrating the overlap between the sets of rhythmic transcripts in the control (yellow) and Vgat-cre;Sox2fl/fl (pink) SCN. 418 and 455 rhythmic genes were identified in the SCN of control and Vgat-cre;Sox2fl/fl mice, respectively. (B) Rose plots illustrating the phase distribution of rhythmic genes in the control (left) and Vgat-cre;Sox2fl/fl (right) SCN. The phase distribution of the common, rhythmic genes was similar in both genotypes (Fig. 1B, gray bars). The phase distribution of genotype-specific rhythmic genes was significantly different (Kolmogorov-Smirnov [K–S] test, p = 0.009). (C, D) Histograms showing the distribution of (C) period and (D) relative amplitude of rhythmic genes in the control (left) and Vgat-cre;Sox2fl/fl (right) SCN. There was no significant difference between control and Vgat-cre;Sox2fl/fl SCN in terms of the period for the entire set of rhythmic genes (Fig. 1C, gray + teal vs. gray + lilac bars). Among the set of common, rhythmic genes, there was a significant reduction in median period in Vgat-cre;Sox2fl/fl animals (Mann-Whitney [M - W] U test, p = 0.011) (Fig. 1C, gray bars). For the genotype-specific rhythmic genes, the median period was significantly longer (M - W U test, p = 0.003), and the distribution of period was altered (K-S test, p = 0.004), in Vgat-cre;Sox2fl/fl animals (Fig. 1C, teal vs. lilac bars). The distribution of relative amplitude for the genotype-specific rhythmic genes was different between control and Sox2-deficient mice (K-S test, p = 0.012) (Fig. 5D, teal vs. lilac bars). (E–G) Heat map representation of genes that are rhythmic in the SCN of (E) control mice only, (F) both genotypes, or (G)Vgat-cre;Sox2fl/fl mice only, arranged according to phase. Each column represents one biological replicate. (H–K) Difference in (H) phase, (I) baseline, (J) amplitude, or (K) relative amplitude of expression of rhythmic clock genes between Vgat-cre;Sox2fl/fl mice and controls (knockout value – control value). For (H), positive values indicate a delayed phase, whereas negative values denote an advanced phase in the Vgat-cre;Sox2fl/fl animals.
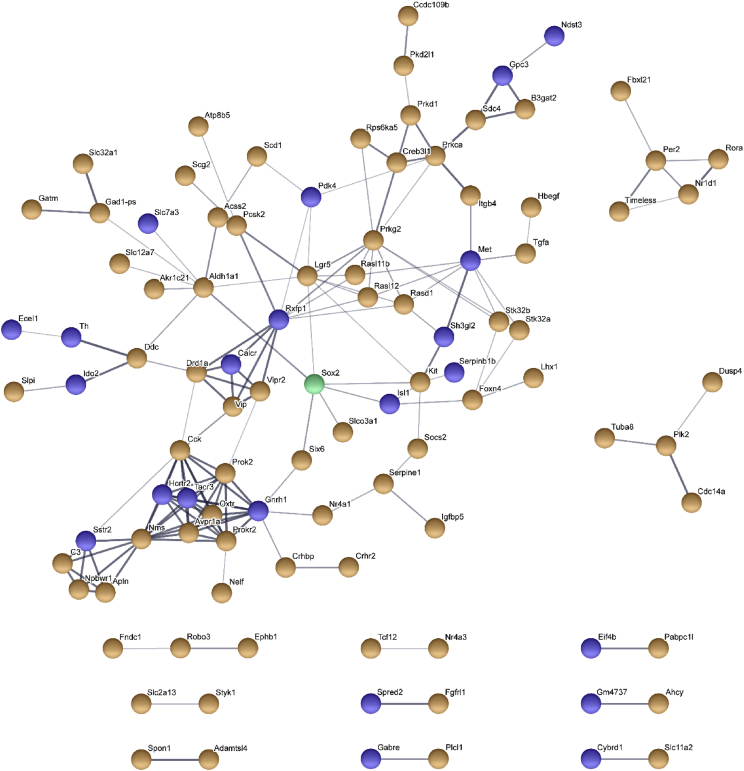
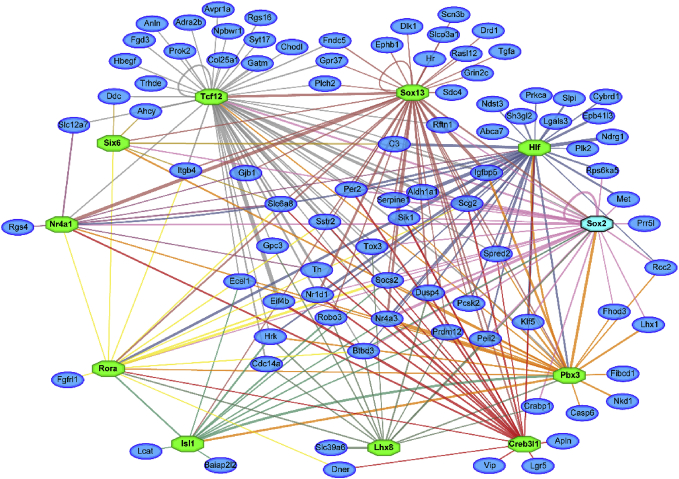
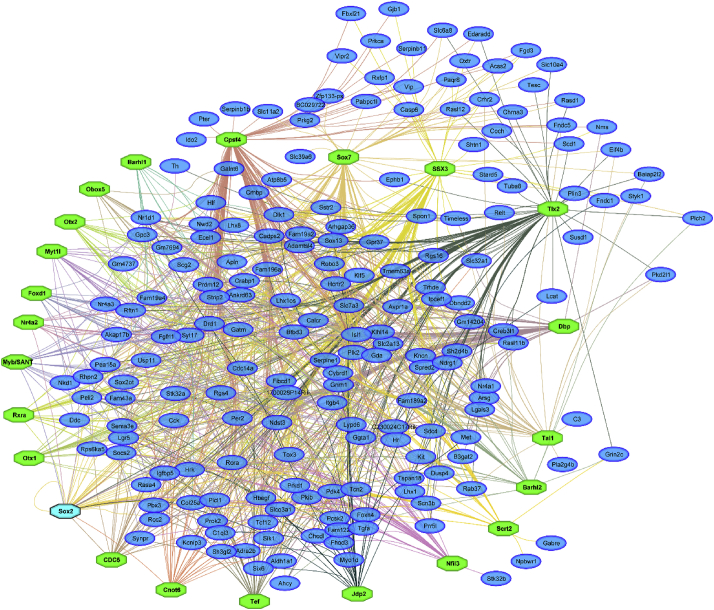
Differential expression analysis for all transcripts was performed with DESeq2, yielding a list of 233 differentially expressed genes (DEGs) with FDR ≤0.05 (Data S3). Protein-protein interaction networks among the set of 233 DEGs was constructed with STRING and presented in Fig. 2 and Data S4. Potential targets of 11 differentially expressed transcription factors were identified with iRegulon and the resulting amalgamated metatargetome is shown in Fig. 3 and Data S5. Transcription factor networks with enriched regulatory features among the set of 233 DEGs were combined with the metatargetome of SOX2 and presented in Fig. 4 and Data S6.
Fig. 2.
Protein-protein interaction networks among the set of differentially expressed genes. 101 DEGs that had at least one predicted interacting partner among the set of 211 mapped DEGs are shown. The largest interaction network is comprised of 73 proteins. Orange denotes DEGs that are downregulated in Vgat-cre;Sox2fl/fl SCN, and blue indicates upregulated DEGs. SOX2 is shown in green.
Fig. 3.
Amalgamated metatargetome of 11 differentially expressed transcription factors. Using iRegulon, predicted targets (blue) of transcription factors (green, turquoise) that are differentially expressed in the SCN of Vgat-cre;Sox2fl/fl mice were identified among the set of DEGs. The network consists of 99 DEGs (out of 233, 42.5%).
Fig. 4.
Transcription factor networks with significantly enriched DNA-binding motifs among the set of differentially expressed genes. Transcription factors (green) recognizing DNA-binding motifs that are significantly enriched among the set of 213 mapped DEGs (blue) were identified with i-cisTarget. 32 TF regulatory motifs were significantly enriched (Data S5), distributed across 22 TFs.
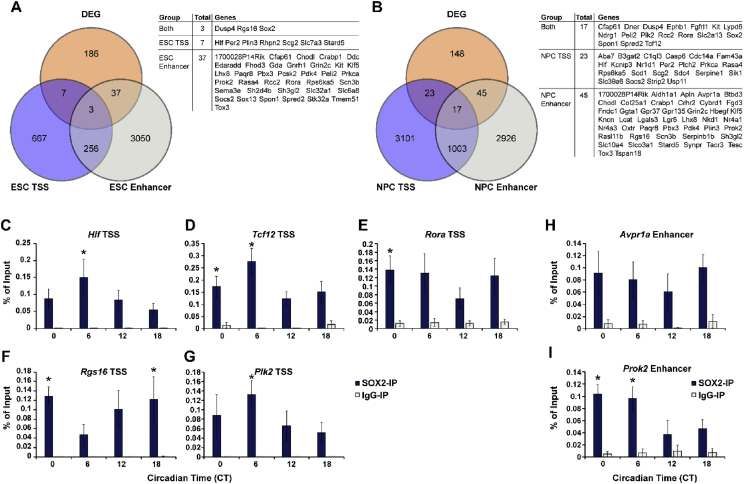
The list of DEGs was then compared with a previously reported SOX2 ChIP-seq dataset from murine embryonic stem cells (ESCs) and ESC-derived neural progenitor cells (NPC). The gene overlap from this cross-referencing is documented in Fig. 5A–B and Data S7. The binding of SOX2 to 7 predicted genomic regions was validated in SCN tissues by ChIP-qPCR (Fig. 5C–I).
Fig. 5.
Identification of potential SOX2 target genes among the set of DEGs and their validation by ChIP-qPCR. (A–B) Venn diagrams illustrating the overlap between our DEG dataset and the SOX2 ChIP-seq datasets obtained from (A) ES cells and (B) NPCs, as reported in Lodato et al. (2013). The Lodato study distinguishes between SOX2 binding to TSS regions vs. binding to distal enhancer elements of the same gene. The tables list the genes that show SOX2 binding to both TSS and enhancer regions, TSS regions only, or enhancer regions only. (C–I) ChIP-qPCR analyses of the relative binding of SOX2 to the TSS regions of (C)Hlf, (D)Tcf12, (E)Rora, (F)Rgs16, and (G)Plk2, or to the distal enhancer regions of (H)Avpr1a and (I)Prok2 in SCN tissues harvested at 4 CTs. Values are represented as mean ± SEM. n = 3–6 per condition. *p<0.05 vs. IgG-IP.
2. Experimental design, materials, and methods
2.1. Sample collection and RNA sequencing
All animal handling and experimental procedures were performed at the University of Toronto Mississauga (UTM) Animal Facility and were approved by the UTM Animal Care Committee, complying with guidelines established by the University of Toronto Animal Care Committee and the Canadian Council on Animal Care. Detailed procedures for tissue harvesting, RNA extraction, library preparation, high throughput sequencing, read trimming, read mapping, and transcript annotation were described in the related research article [1]. Briefly, SCN tissues from control and Vgat-cre;Sox2fl/fl mice were harvested at circadian time (CT) 0, 6, 12, and 18. RNA were extracted and then submitted to the Génome Québec Innovation Centre at McGill University (Montréal, Canada) for RNA-seq library preparation. RNA libraries were sequenced on an Illumina HiSeq4000 platform and the reads were trimmed using Trimmomatic v.0.36. Reads for each replicate were aligned to the Mus musculus reference genome (GRCm38) using Bowtie2. The Mus musculus gene annotation from Ensembl (release 90) was loaded as gene model using makeTxDbFromGFF from the GenomicFeatures package. The function summarizeOverlaps from the GenomicAlignments package was used to generate a SummarizedExperiment object that contained metadata and number of reads per gene. Genes with counts-per-million (cpm) below 0.1 in at least 35 of the 40 libraries were not considered to be expressed in the SCN and were thus discarded from further analysis. Expression data of the remaining 26,798 genes were used for downstream analysis.
2.2. Bioinformatics and in silico analysis
To identify rhythmic genes, meta2d algorithm from the R package MetaCycle was used on the normalized counts of 26,798 expressed genes in the SCN [3]. Minimum and maximum period were set to 12 and 24 hours, respectively. “JTK” and “LS” integrated period, phase, amplitude, relative amplitude, and benjamini-hochberg q (BH.Q) values were used to compare control and Vgat-cre;Sox2fl/fl SCN transcriptomes. Genes showing a significant profile (BH.Q < 0.05) were classified as rhythmic genes.
For constructing heatmaps and hierarchical clustering of rhythmic genes, variance stabilizing transformations (VST) were first applied to the normalized counts to remove the dependence of the variance on the mean [4]. The amount by which each gene deviates in a specific sample from the average VST transformed values across all samples was used to construct clustered heat maps.
Differential expression analysis was performed using the DESeq2 package with the assumption of negative binomial distribution for RNA-seq data [2]. For each time point, genes were considered differentially expressed in Vgat-cre;Sox2fl/fl SCN compared to controls when the reported benjamini-hochberg adjusted p values (PADJ) is less than 0.05.
Protein-protein interaction networks among the set of 233 DEGs was constructed with STRING [5]. Out of 211 mapped DEGs, 101 were predicted to interact with at least one other DEG. There was a total of 163 interactions, with the largest network consisting of 73 DEGs.
Using Cytoscape plugin iRegulon [6], we amalgamated the metatargetome of 11 differentially expressed transcription factors with positive queries. The occurrence count threshold and number of nodes to return were set to 5 and 1000, respectively. GeneSigDB, Ganesh clusters, and MSigDB were used as the databases of signatures/gene sets.
The 233 DEGs were submitted to i-cisTarget regulatory features and cis-regulatory modules prediction [7]. Minimum fraction of overlap, normalized enrichment score (NES) threshold, and AUC threshold were set to 0.4, 3.0, and 0.005, respectively. Enriched predicted regulatory motifs and the associated transcription factor were documented in Data S6. Number of potential regulatory features were calculated for each regulator-target pair and represented by the stroke weight in the regulatory network. DEGs with potential SOX2 binding sites were identified with iRegulon and mapped onto the same network.
ChIP-seq data of bound promoters, active enhancers, and poised enhancers for SOX2 in ESCs and NPCs were extracted from Table S2 of Lodato et al. (2013) [8]. Active enhancers and poised enhancers were grouped together for comparison. The list of SOX2 binding sites were cross-checked with our list of DEGs. Seven genes from the set of DEGs that overlapped with the Lodato dataset were selected and binding of SOX2 to their reported regulatory region was confirmed with ChIP-qPCR using SCN tissues extracted at four CTs. Comprehensive procedure on tissue harvesting, ChIP, and qPCR were described in the related research article [1].
2.3. Statistical analysis
Data were analyzed using two-way ANOVA, Mann-Whitney U test, and Kolmogorov-Smirnov test with R version 3.5.0. Post hoc significance of pairwise comparisons was assessed using Tukey's Honest Significant Difference test with α set at 0.05.
Acknowledgments
The authors wish to thank Chris Lowden, Samuel W. Fung, and Sara Hegazi for advice on the manuscript. This work was supported by operating grants to H.-Y.M.C. from the Canadian Institutes of Health Research (CIHR) and the Natural Sciences and Engineering Research Council (NSERC) of Canada. H.-Y.M.C. was supported by an NSERC Tier II Canada Research Chair in Molecular Genetics of Biological Clocks. R.W.N. was supported by an operating grant from NSERC and the Canadian Foundation for Innovation John R. Evans Leaders fund. A.H.C. and P.B.-C. were supported by NSERC post-graduate scholarships.
Footnotes
Transparency document associated with this article can be found in the online version at https://doi.org/10.1016/j.dib.2019.103909.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.dib.2019.103909.
Transparency document
The following is the transparency document related to this article:
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Multimedia component 2
Multimedia component 3
Multimedia component 4
Multimedia component 5
Multimedia component 6
Multimedia component 7
Multimedia component 8
References
- 1.Cheng A.H., Bouchard-Cannon P., Hegazi S., Lowden V., Fung S.W., Chiang C.-K., Ness R.W., Cheng H.-Y.M. SOX2-dependent transcription in clock neurons promotes the robustness of the central circadian pacemaker. Cell Rep. 2019;26:3191–3202. doi: 10.1016/j.celrep.2019.02.068. [DOI] [PubMed] [Google Scholar]
- 2.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:1–21. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu G., Anafi R.C., Hughes M.E., Kornacker K., Hogenesch J.B. MetaCycle: an integrated R package to evaluate periodicity in large scale data. Bioinformatics. 2016;32:3351–3353. doi: 10.1093/bioinformatics/btw405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Szklarczyk D., Morris J.H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N.T., Roth A., Bork P., Jensen L.J., Von Mering C. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Janky R., Verfaillie A., Imrichová H., van de Sande B., Standaert L., Christiaens V., Hulselmans G., Herten K., Naval Sanchez M., Potier D., Svetlichnyy D., Kalender Atak Z., Fiers M., Marine J.C., Aerts S. iRegulon: from a gene list to a gene regulatory network using large motif and track collections. PLoS Comput. Biol. 2014;10 doi: 10.1371/journal.pcbi.1003731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Imrichová H., Hulselmans G., Kalender Atak Z., Potier D., Aerts S. i-cisTarget 2015 update: generalized cis-regulatory enrichment analysis in human, mouse and fly. Nucleic Acids Res. 2015;43:W57–W64. doi: 10.1093/nar/gkv395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lodato M.A., Ng C.W., Wamstad J.A., Cheng A.W., Thai K.K., Fraenkel E., Jaenisch R., Boyer L.A. SOX2 Co-occupies distal enhancer elements with distinct POU factors in ESCs and NPCs to specify cell state. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003288. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multimedia component 2
Multimedia component 3
Multimedia component 4
Multimedia component 5
Multimedia component 6
Multimedia component 7
Multimedia component 8