Abstract
The main objective of this study is to assess the effects of CYP2C9 and VKORC1 polymorphisms on warfarin sensitivity and responsiveness in a Jordanian population during the stabilization phase of treatment. This study was conducted at the Queen Alia Heart Institute (QAHI) anticoagulation clinic in Amman, Jordan. We assessed three CYP2C9 (rs1799853, rs1057910, rs4086116) and four VKORC1 (rs10871454, rs8050894, rs9934438, rs17708472) polymorphisms in 139 Jordanian cardiovascular patients. Demographic and clinical data were also collected. Of the 139 patients in the cohort, 80% had the VKORC1 polymorphisms rs10871454 and rs9934438, while 22.3% and 24.5% of patients had the rs1799853 and rs1057910 CYP2C9 alleles, respectively. Carriers of the CYP2C9 polymorphisms rs1057910 and rs4086116 had an increased risk of warfarin sensitivity compared to subjects with no or only one polymorphism. Similarly, carriers of all four VKORC1 variants had an increased risk of warfarin sensitivity (over anticoagulation) compared to those with no or only one polymorphism. Patients with a CYP2C9 or VKORC1 polymorphism required significantly lower doses than patients with no polymorphisms. The presence of any of CYP2C9 or VKORC1 polymorphisms is associated with sensitivity to warfarin during the stabilization period. Being a CYP2C9 or VKORC1 polymorphism carrier is associated with a variation in doses required to achieve the therapeutic INR compared to non-carrier patients.
Keywords: CYP2C9, VKORC1, Warfarin, Warfarin maintenance phase of therapy, INR
1. Introduction
Anticoagulants are effective medications for the treatment and prevention of thromboembolic disorders. These anticoagulants work by maintaining the international normalized ratio (INR), a measure of thrombotic status, within the therapeutic range. Indeed, several studies have shown that the higher the INR, the greater the risk of hemorrhage, while a lower INR is associated with increased risk of thromboembolism or stroke (Hirsh et al., 2001). Although warfarin is the most commonly used anticoagulant in patients with stroke, the use of this drug is complicated by the wide interindividual variability in response and the dose required to achieve the target INR. This variability may be explained in part by age, use of concomitant medications, dietary vitamin K intake, impaired liver function, and heart failure (Hirsh et al., 2001, Glasheen et al., 2005, Visser et al., 2004; and Ansell et al., 2004) as well as individual genetic variability (Higashi et al., 2002, Rost et al., 2004, Veenstra et al., 2005).
Polymorphisms of the CYP2C9 gene, which encodes warfarin’s major metabolizing enzyme, have been extensively studied. Genetic polymorphisms in CYP2C9 alter its catalytic activity; rs1799853 and rs1057910 variants decrease enzyme activity (Linder et al., 2009). Several studies have demonstrated that, an association between patients carrying of at least one of the rs1057910 or rs1799853 variant alleles and reduced warfarin doses, delayed stabilization of warfarin and a risk of major bleeding (Higashi et al., 2002, Freeman et al., 2000; and Scordo et al., 2002).
The vitamin K epoxide reductase (VKOR) enzyme plays a significant role in the clotting pathway via the reduction of vitamin K-2,3 epoxide to vitamin K-hydroquinone, the latter of which is biologically active and enhances the production of clotting factors II, VII, IX and X. Warfarin inhibits VKOR activity by inhibiting the activity of subunit 1 of the vitamin K epoxide reductase complex (VKORC1) encoded by the VKORC1 gene (Li et al., 2004, Rost et al., 2004). Several studies have found an association between the presence of VKORC1 gene polymorphisms and a reduced dose of warfarin. Most of these studies demonstrated that VKORC1 polymorphisms accounted for most of the variation in warfarin dose requirements compared to its CYP2C9 counterpart (Veenstra et al., 2005, Wadelius et al., 2005; and Sconce et al., 2005).
In order to keep warfarin therapy efficient and safe, the INR must remain within the target range (i.e., the therapeutic range of about 2–3). Therefore, the dosage must be adapted to the INR. At the initial phase of therapy, the warfarin dose is prescribed by the physician using the trial and error method and based on patients’ clinical factors to obtain the therapeutic INR. While during the stabilization phase of therapy, the patients reached the target INR of at least two consecutive visits (Gage and Lesko, 2007, Kuruvilla and Gurk-Turner, 2001). Therefore, the objective of this study was to evaluate the effects of CYP2C9 and VKORC1 polymorphisms on warfarin dose requirements during the stabilization phase of therapy in the Jordanian population.
2. Material and methods
2.1. Study design and patients
The study population consisted of 139 unrelated warfarin patients recruited from the Queen Alia Heart Institute (QAHI) anticoagulation clinic in Amman, Jordan. We included patients who started their therapy between January 2014 to November 2015. The inclusion criteria included 18 years or older, warfarin intake patients for at least 3 months and visit the anticoagulation clinic regularly. We excluded patients who have lost clinical data or patients with pharmacokinetics interacting drugs from the analysis during the follow-up period. These drugs have been identified according to Dutch standards for the treatment of coumarin interactions (Standaard Af handeling Cumarine Interacties, 2003). This study was approved by the human ethics committees with ethical code number 13/78/2014 at Jordan University of Science and Technology. All patients who met the inclusion criteria were informed about the objectives of the study and asked for their written consent.
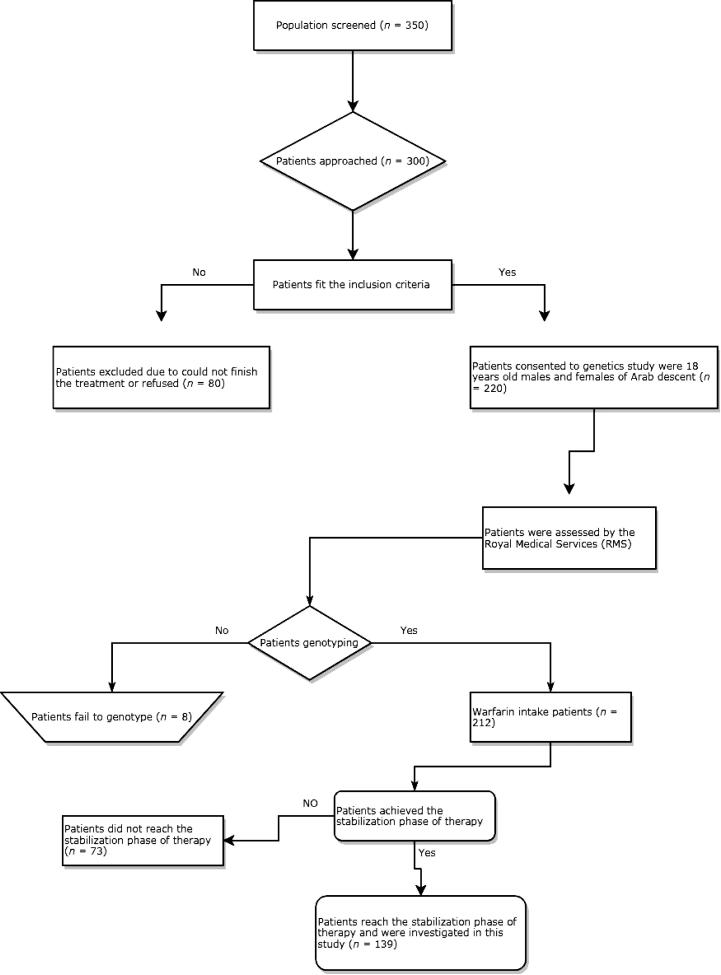
Initially, 350 patients were examined, 300 of which were referred for participation in this study. 80 patients were subsequently excluded based on inclusion and exclusion criteria. Of the remaining patients, 220 agreed to participate in this study. Thereafter, eight more patients were excluded from the final analysis due to failure of genotyping (our method failed to determine the genotype of these patients due to low DNA content or degradation of DNA samples). Of these 212 patients, only 139 patients achieved the stabilization phase of therapy. Therefore, all data were obtained from 139 patients with cardiovascular disease who were treated with warfarin. The study design summarized in Fig. 1.
Fig. 1.
Flow chart depicting study design. INR: international normalized ratio.
2.2. Data collection and follow-up time
Data was collected on a number of demographic characteristics such as gender, age, body mass index, smoking, and eating habits. Information was also collected on patient characteristics, including anticoagulant indication, appropriate therapeutic range of INR, INR measurements, warfarin dose, co-morbidity, co-medications, hemorrhage and lipid profile (e.g. cholesterol, low density lipoprotein (LDL), and high density lipoprotein (HDL)).
2.3. SNP selection and genotyping
Candidate SNPs associated with warfarin metabolism or warfarin targets were obtained from a public database such as the SNP database of the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/SNP/) and the Applied Biosystems SNP database (http://www.appliedbiosystems). We selected 17 SNPs in the CYP2C9 and VKORC1 genes. Table S1 lists the genes, their SNP IDs, and their loci. After DNA extraction using the Wizard Genomic DNA Purification Kit (Promega), samples that met the quantitative requirements for this study were shipped to the Australian Genome Research Facility (AGRF) to determine genotype using the MassARRAY® System (iPLEX GOLD) (Sequenom, San Diego, CA, USA). The MassARRAY™ system protocol and the primers information used for the CYP2C9 and VKORC1 genes are available upon request.
2.4. Outcome measure
The primary point of our study was based on evaluation of the sensitivity to warfarin during the stabilization phase of therapy. Therefore, patients were divided into the following three groups based on Gordon (2009) study:
-
1.
Extensive metabolizer or warfarin resistance group (high dose required > 49 mg/week)
-
2.
Moderate metabolizer or warfarin response group (average dose required between 21 and 49 mg/week)
-
3.
Poor metabolizer group or warfarin-sensitive group (minimum required dose < 21 mg/week).
The secondary point of the study was focused on warfarin responsiveness during the stabilization phase of therapy. Therefore, patients were classified according to Higashi et al. (2002) study into:
-
1.
Good responders who have an INR value in the target range (therapeutic range)
-
2.
Poor responders (INR value below target)
-
3.
Ultra responders (INR over target)
Finally, the maintenance dose was defined as the average of all doses administered to a patient during stable anticoagulation. The stable maintenance dose was calculated from all weekly doses that remained unchanged for at least two consecutive visits under therapeutic INR.
2.5. Statistical analysis
The minor allele frequency (MAF) and the Hardy-Weinberg equilibrium (HWE) p-values were calculated. The Pearson χ2 test was used to evaluate the deviation of the HWE. Various genetic association analyses (Chi-square test, non-parametric correlation Kruskal Wallis test, unidirectional analysis of variance, and Tukey HSD post hoc multiple comparison test) were also performed to test which of the selected SNPs are associated with warfarin response. The Statistical Package for the Social Sciences (SPSS) version 21.0 was used for all statistical analyses.
3. Results
A total of 139 patients met the criteria of our study. The demographic and clinical characteristics, mean dose and INR values from the first visits to the clinic and after achieving stabilization were collected and summarized in Table 1. Ten of the 17 selected SNPs were not polymorphic, and only seven SNPs were included in this study, the latter of which passed quality control tests with high accuracy and low discrepancy rate. Genotypic and allelic frequencies for the 139 patients are summarized in Table S2.
Table 1.
Analytical analysis of demographics and clinical characteristics of 139 cardiovascular patient treated with warfarin at the Queen Alia Heart Institute.
| Category | Subcategory | Extensive Metabolizera | Good Metabolizersb | Poor Metabolizersc | P value* |
|---|---|---|---|---|---|
| Demographics | Patients (N, %) | (25/139) 18.0% | (88/139) 63.3% | (26/139) 18.7% | |
| Aged [years] | 47.6 [17.7] | 53.2 [15.3] | 54.2 [18.4] | 0.457 | |
| BMId | 26.7 [4.0] | 27.6 [4.8] | 28.0 [6.6] | 0.641 | |
| Smoking (%) | (12/25) 48.0% | (18/88) 31.8% | (12/26) 46.2% | 0.043 | |
| Male | (17/25) 68.0% | (49/88) 55.7% | (17/26) 65.4% | 0.304 | |
| Female | (8/25) 32.0% | (39/88) 44.3% | (9/26) 34.6% | ||
| Concomitant Disease | Co morbidity | (15/25) 60.0% | (54/88) 61.4% | (13/26) 50.0% | 0.578 |
| Hypertension | (6/25) 24.0% | (35/88) 39.8% | (10/26) 38.5% | 0.425 | |
| Diabetes mellitus | (9/25) 36.0% | (16/88) 18.2% | (6/26) 23.1% | 0.124 | |
| CHDe | (8/25) 32.0% | (17/88) 19.3% | (6/26) 23.1% | 0.329 | |
| Thyroid | (1/25) 4.0% | (2/88) 2.3% | (0/26) 0.0% | 0.596 | |
| Lipid | (1/25) 4.0% | (7/88) 8.0% | (1/26) 3.8% | 0.673 | |
| Medication | Aspirin | (18/25) 72.0% | (57/88) 64.8% | (17/26) 65.4% | 0.600 |
| Indication of Treatment | MVRf | (5/25) 20.0% | (9/88) 10.3% | (6/26) 23.1% | 0.127 |
| AVRg | (7/25) 28.0% | (31/88) 35.2% | (2/26) 7.7% | ||
| AFh | (6/25) 24.0% | (14/88) 15.9% | (5/26) 19.2% | ||
| DVRi | (6/25) 24.0% | (15/88) 17.0% | (9/26) 34.6% | ||
| Others | (1/25) 4.0% | (19/88) 21.6% | (4/26) 15.4% | ||
| Target INR | 2–3 | (10/25) 40.0% | (37/88) 42.0% | (11/26) 42.3% | 0.998 |
| 2.5–3.5 | (15/25) 60.0% | (51/88) 58.0% | (15/26) 57.7% | ||
| Mean INRd | 2.4 [0.53] | 2.6 [0.64] | 2.9 [0.56] | 0.021 | |
P value < 0.05 is considered significant.
Extensive metabloizer or warfarin resistance group (required high warfarin dose > 49 mg/week).
Moderate metabolizer or warfarin response group (required average warfarin dose between 21 and 49 mg/week).
Poor metabolizer group or warfarin-sensitive group (required minimum warfarin dose < 21 mg/week).
Mean Standard deviation in square brackets.
CHD: Chronic heart disease.
MVR:Mitral valve replacement.
AVR: Aortic valve replacement.
AF: Atrial Fibrillation.
DVR: Double valve replacement.
3.1. Association of SNPs with warfarin sensitivity
Significant association was found between all four VKORC1 SNPs and sensitivity to warfarin (p < 0.001). Two of the CYP2C9 SNPs, rs4086116 (P = 0.007) and rs1057910 (P < 0.001), were also found to be significantly associated with warfarin sensitivity.
In fact, carriers of the VKORC1 polymorphism rs10871454 (TT) had a significantly higher risk (33.3%) of being sensitive to warfarin compared to wild-type (CC) subjects (3.6%) or carriers of only one polymorphism (CT) (7.2%) (Table 2). Moreover, carriers of the CYP2C9 rs1057910 (AC) polymorphism had a significantly higher risk (32.3%) of being sensitive to warfarin (over-anticoagulation) compared with carriers of (AA) polymorphism (10.2%) (Table 2). However, only the CYP2C9 rs1799853 SNP showed no significant differences among the three warfarin-sensitive groups (Sensitive, moderate, resistant) (P = 0.654) (Table 2).
Table 2.
Association of VKORC1 and CYP2C9 SNPs with Warfarin Sensitivity during the Stabilization Phase of Therapy of 139 Cardiovascular Patients Treated with Warfarin.
| Gene | SNP ID | Genotype | Sensitivea | Moderateb | Resistancec | P-value* |
|---|---|---|---|---|---|---|
| VKORC1 | rs10871454 | CC | (1/28) 3.6% | (10/28) 35.7% | (17/28) 60.7% | <0.001 |
| CT | (5/69) 7.2% | (51/69) 73.9% | (13/69) 18.8% | |||
| TT | (14/42) 33.3% | (25/42) 59.5% | (3/42) 7.1% | |||
| rs8050894 | CC | (1/26) 3.8% | (9/26) 34.6% | (16/26) 61.5% | <0.001 | |
| CG | (5/69) 7.2% | (50/69) 72.5% | (14/69) 20.3% | |||
| GG | (14/44) 31.8% | (27/44) 61.4% | (3/44) 6.8% | |||
| rs9934438 | CC | (2/28) 7.2% | (10/28) 35.7% | (16/28) 57.1% | <0.001 | |
| CT | (5/71) 7.0% | (53/71) 74.6% | (13/71) 18.3% | |||
| TT | (14/40) 35.0% | (23/40) 57.5% | (3/40) 7.5% | |||
| rs17708472 | CC | (20/106) 18.8% | (68/106) 64.2% | (18/106) 17.0% | 0.003 | |
| CT | (1/30) 3.3% | (17/30) 56.7% | (12/30) 40.0% | |||
| TT | (1/3) 33.3% | (0/3) 0.0% | (2/3) 66.7% | |||
| CYP2C9 | rs1799853 | CC | (14/105) 13.3% | (65/105) 61.9% | (26/105) 24.8% | 0.654 |
| CT | (7/32) 21.9% | (18/32) 56.2% | (7/32) 21.9% | |||
| TT | (0/2) 0.0% | (2/2) 100.0% | (0/2) 0.0% | |||
| rs4086116 | CC | (6/77) 7.8% | (46/77) 59.7% | (25/77) 32.5% | 0.007 | |
| CT | (13/53) 24.5% | (32/53) 60.4% | (8/53) 15.1% | |||
| TT | (2/9) 22.2% | (7/9) 77.8% | (0/9) 0.0% | |||
| rs1057910 | AA | (11/108) 10.2% | (66/108) 61.1% | (31/108) 28.7% | 0.001 | |
| AC | (10/31) 32.3% | (20/31) 64.5% | (1/31) 3.2% | |||
Chi-square test with p value < 0.05 is considered significant.
Warfarin Sensitive group (required minimum warfarin dose < 21 mg/week).
Warfarin Moderate response group (required average warfarin dose between 21 and 49 mg/week).
Warfarin Resistance group (required high warfarin dose > 49 mg/week).
3.2. Impact of VKORC1 and CYP2C9 SNPs on dose requirements
Significant effect was observed when compared the doses collected at the beginning of the treatment with the doses after reaching the stabilization phase of the therapy, P < 0.0001, except rs1799853 CYP2C9 polymorphism (Table 3).
Table 3.
Association of VKORC1 and CYP2C9 SNPs with variability on warfarin required doses.
| Gene | SNP ID | Genotype | Initiation dose | P-value * | Maintenance dose | P-value* |
|---|---|---|---|---|---|---|
| VKORC1 | rs10871454 | CC | 51.35 [15.55] | <0.0001 | 53.02 [18.02] | <0.0001 |
| CT | 35.33 [14.42] | 39.48 [17.06] | ||||
| TT | 26.56 [10.89] | 27.41 [10.33] | ||||
| rs8050894 | CC | 51.84 [15.32] | <0.0001 | 53.06 [18.39] | <0.0001 | |
| CG | 35.76 [14.81 | 39.96 [17.33] | ||||
| GG | 26.73 [10.58] | 27.80 [10.22] | ||||
| rs9934438 | CC | 51.35 [15.55] | <0.0001 | 53.02 [18.02] | <0.0001 | |
| CT | 35.56 [14.28] | 39.47 [16.83] | ||||
| TT | 25.72 [10.46] | 26.82 [10.19] | ||||
| rs17708472 | CC | 33.67 [15.68] | 0.012 | 36.37 [18.13] | 0.003 | |
| CT | 41.64 [15.53] | 44.20 [15.04] | ||||
| TT | 57.80 [8.08] | 59.63 [9.79] | ||||
| CYP2C9 | rs1799853 | CC | 37.22 [16.37] | 0.154 | 40.00 [18.30] | 0.189 |
| CT | 32.54 [15.11] | 34.68 [16.23] | ||||
| TT | 21.25 [0.354] | 25.00 [0.00] | ||||
| rs4086116 | CC | 40.61 [16.81] | 0.0002 | 43.86 [19.10] | 0.0002 | |
| CT | 30.90 [13.38] | 32.55 [14.39] | ||||
| TT | 25.23 [11.56] | 28.57 [8.76] | ||||
| rs1057910 | AA | 39.06 [16.32] | <0.0001 | 41.75 [18.48] | <0.0001 | |
| AC | 24.95 [9.34] | 27.43 [9.29] | ||||
One-way ANOVA test with P-value < 0.05 is considered significant, Mean Standard deviation in square brackets.
3.3. Association of VKORC1 and CYP2C9 SNPs with warfarin responsiveness
Patients were divided into three groups (Poor-, good-, and ultra-responders). No significant differences were found between the investigated SNPs of VKORC1 and CYP2C9 (Table 4).
Table 4.
Association of VKORC1 and CYP2C9 SNPs with warfarin response during the stabilization phase of therapy of 139 cardiovascular patients treated with warfarin.
| Gene | SNP ID | Genotype | Poor respondera | Good responderb | Ultra responderc | P-value* |
|---|---|---|---|---|---|---|
| VKORC1 | rs10871454 | CC | (0/28) 0.0% | (27/28) 96.4% | (1/28) 3.6% | 0.621 |
| CT | (6/69) 8.7% | (60/69) 87.0% | (3/69) 4.3% | |||
| TT | (3/42) 7.1% | (37/42) 88.1% | (2/42) 4.8% | |||
| rs8050894 | CC | (0/26) 0.0% | (25/26) 96.2% | (1/26) 3.8% | 0.659 | |
| CG | (6/69) 8.7% | (60/69) 87% | (3/69) 4.3% | |||
| GG | (3/44) 6.8% | (39/44) 88.6% | (2/44) 4.5% | |||
| rs9934438 | CC | (0/28) 0.0% | (27/28) 96.4% | (1/28) 3.6% | 0.527 | |
| CT | (6/71) 8.6% | (61/71) 85.9% | (4/71) 5.5% | |||
| TT | (3/40) 7.5% | (36/40) 90.0% | (1/40) 2.5% | |||
| rs17708472 | CC | (6/106) 5.7% | (94/106) 88.7% | (6/106) 5.6% | 0.593 | |
| CT | (3/30) 10.0% | (27/30) 90.0% | (0/0) 0.0% | |||
| TT | (0/3) 0.0% | (3/3) 100.0% | 0.0% | |||
| CYP2C9 | rs1799853 | CC | (8/105) 7.6% | (93/105) 88.6% | (4/105) 3.8% | 0.851 |
| CT | (1/32) 3.1% | (29/32) 90.6% | (2/32) 6.3% | |||
| TT | (0.0) 0.0% | (2/2) 100.0% | 0.0% | |||
| rs4086116 | CC | (6/77) 7.8% | (68/77) 88.3% | (3/77) 3.9% | 0.806 | |
| CT | (3/53) 5.7% | (47/53) 88.6% | (3/53) 5.7% | |||
| TT | (0/9) 0.0% | (9/9) 100.0% | 0.0% | |||
| rs1057910 | AA | (8/108) 7.4% | (94/108) 87.0% | (6/108) 5.6% | 0.269 | |
| AC | (1/31) 3.2% | (30/31) 96.8% | 0.0% | |||
Chi-Square Test with p value < 0.05 is considered significant.
Poor responders (INR value below target).
Good responders who have an INR value in the target range (therapeutic range).
Ultra responders (INR over target).
3.4. Impact of VKORC1 and CYP2C9 SNPs on INR values
No significant associations were observed when compared the INR values were measure at the beginning of treatments and the INR values measured after reaching the stabilization phase of the therapy (Table 5).
Table 5.
Association of VKORC1 and CYP2C9 SNPs with INR Treatment Outcome.
| Gene | SNP ID | Genotype | Initiation INR | P-value* | Maintenance INR | P-value* |
|---|---|---|---|---|---|---|
| VKORC1 | rs10871454 | CC | 2.73 [0.58] | 0.346 | 2.63 [0.40] | 0.297 |
| CT | 2.60 [0.40] | 2.68 [0.63] | ||||
| TT | 2.54 [0.65] | 2.74 [0.36] | ||||
| rs8050894 | CC | 2.71 [0.58] | 0.362 | 2.63 [0.39] | 0.246 | |
| CG | 2.63 [0.67] | 2.67 [0.39] | ||||
| GG | 2.52 [0.58] | 2.77 [0.38] | ||||
| rs9934438 | CC | 2.73 [0.58] | 0.253 | 2.63 [0.40] | 0.312 | |
| CT | 2.61 [0.67] | 2.68 [0.40] | ||||
| TT | 2.51 [0.57] | 2.76 [0.36] | ||||
| rs17708472 | CC | 2.59 [0.67] | 0.901 | 2.71 [0.41] | 0.517 | |
| CT | 2.67 [0.45] | 2.64 [0.33] | ||||
| TT | 2.53 [0.50] | 2.50 [0.17] | ||||
| CYP2C9 | rs1799853 | CC | 2.51 [0.61] | 0.159 | 2.69 [0.38] | 0.381 |
| CT | 2.63 [0.69] | 2.69 [0.42] | ||||
| TT | 3.20 [0.14] | 3.00 [0.0] | ||||
| rs4086116 | CC | 2.67 [0.61] | 0.621 | 2.67 [0.40] | 0.389 | |
| CT | 2.54 [0.63] | 2.70 [0.37] | ||||
| TT | 2.49 [0.73] | 2.83 [0.43] | ||||
| rs1057910 | AA | 2.66 [0.66] | 0.046 | 2.67 [0.40] | 0.190 | |
| AC | 2.41 [0.46] | 2.77 [0.35] | ||||
One-way ANOVA test with p-value < 0.05 is considered significant, Mean Standard deviation in square brackets.
4. Discussion
The results of our study, which examined the role of the CYP2C9 and VKORC1 polymorphisms in warfarin response and sensitivity in cardiovascular patients during the stabilization phase of therapy, strongly suggest an association between certain CYP2C9 and VKORC1 variant alleles and sensitivity to warfarin. However, no such correlation was found between the VKORC1 and CYP2C9 polymorphisms and warfarin response and INR measurements.
The findings of the current research are consistent with previous studies. For example, the allelic frequency of the CYP2C9 (rs1799853) T allele (12.9%) was found at similar levels for Jordanians (Yousef et al., 2012), Americans and Europeans (Abecasis et al., 2012). While, the CYP2C9 (rs1057910) C allele (11.2%) was more common in our study than Yousef et al. (2012) study with 6.8% frequency, and more than Americans (6%), Europeans (6%), Asians (4%) and Africans (1%) frequencies (Abecasis et al., 2012). Moreover, the allelic frequency of the rs10871454 T allele of VKORC1 is 55% and is drastically different from those found in the African (6%) and East-Asian (89%) populations. Similarly, the allelic frequency of the rs8050894C allele of VKORC1 in our population (43.5%) is also different from the African (74%) and East-Asian (12%) populations (Abecasis et al., 2012).
Takahashi and Echizen (2001) reported that CYP2C9 polymorphisms rs1799853 and rs1057910 decrease the rates of warfarin clearance, reducing its activity will lead to a lower required dose, therefore, increase warfarin sensitivity risk (Takahashi and Echizen, 2001). In accordance with that, we found a significant association between rs1057910 (A > C) and sensitivity to warfarin during the stabilization phase of therapy (P = 0.001). In fact, 32% of CA carriers and only 10% of the wild-type carriers were sensitive to warfarin, meaning that patients with this variant require lower doses than the wild-type (Table 2). In addition, the CYP2C9 variant (rs4086116 C > T) also showed a significant association with warfarin sensitivity during the stabilization phase (P = 0.007) (Table 2). Therefore, patients carrying this variant require lower warfarin doses than wild-type carriers. Although numerous studies have reported that patients who carry one rs1799853 allele results in a dose reduction of 6–45% compared with the wild type dose (Higashi et al., 2002, Limdi et al., 2007), our results showed no significant association between CYP2C9 rs1799853 and warfarin sensitivity (P = 0.654). This could be explained by the impact of genotypic frequency on the genetic associations, since in this study of the 139 patients there were only two patients who are homozygous for the variant allele and 105 patients who are homozygous for wild-type allele (Table 2). Additionally, Limdi et al., 2008, Shrif et al., 2011 found an association between rs8050894 (1542G > C) and rs1057910 CYP2C9 SNPs and with a low warfarin dose in European American and Sudanese patients, respectively (Limdi et al., 2008, Shrif et al., 2011). Our findings exhibit a significant association between rs1057910 and rs8050894 SNPs and variation in warfarin required dose with a 34.9% reduction in maintenance dose for carriers of homozygous variant allele TT of rs4086116 with P = 0.0002 (Table 3).
Moreover, the VKORC1 rs9934438 SNP was particularly associated with the low-dose warfarin phenotype, causing patients with this SNP to be classified as warfarin-sensitive patients (D'Andrea et al., 2005). Accordingly, our results show that 33% of patients carrying homozygous variants are more sensitive to warfarin than wild-type carriers (9.7%) (Table 2). Furthermore, Schelleman et al., 2007, Wadelius et al., 2005 reported that patients with the VKORC1 rs9934438 (1173T) allele had a lower dose of warfarin (average dose of 25 mg/week) compared with carriers wild type (35 mg/week) (Schelleman et al., 2007, Wadelius et al., 2005). Accordingly, our study showed that patients with the homozygous to T allele had an average dose of 26.82 mg/week and carriers of wild type CC an average dose of 53.02 mg/week (P < 0.0001).
During the stabilization phase of therapy, patients carrying different CYP2C9 genotypes were not significantly distributed through the different responder groups. For example, 87% of patients carrying the CYP2C9 rs1057910 wild-type allele CC were good responders, 7.4% were poor responders, and 5.6% were ultra-responders, while 96.8% of CT heterozygote carrier were good responders and 3.2% were poor responders (p = 0.269) (Table 4). Schelleman et al. (2007) reported that, although, genotypic and allelic variants affected warfarin sensitivity and causing lower warfarin required dose, once a stable anticoagulation is established in patients, their response is constant and they are no longer having a risk of over anticoagulation (Schelleman et al., 2007). In accordance with previous research, our results show that patients carrying different VKORC1 genotypes were not significantly associated with warfarin responsiveness during the stabilization phase of therapy. For example, 88.7% of patients having the VKORC1 rs17708472 wild type allele (CC) were good responders and 5.7% of them were poor responders. On a similar note, 90% of CT heterozygote carriers were good responders and 10.0% were poor responders (P = 0.593) (Table 4).
When we compared the measurement of INR values in the first weeks of treatment and when patients reached the stabilization phase and then investigated the influence of different alleles of CYP2C9 and VKORC1 on the average INR, no significant differences were observed in all SNPs studied. Therefore, the CYP2C9 and VKORC1 polymorphisms are not associated with the development of a high or low INR in Jordanian cardiovascular patient (Table 5).
5. Conclusion
In the present study, we examined the incidence of CYP2C9 and VKORC1 polymorphisms in our population and then investigated whether they have an effect on warfarin sensitivity and responsiveness during the stabilization phase of therapy. Conclusively, our study indicates that the presence of VKORC1 or CYP2C9 polymorphisms is associated with sensitivity to warfarin during the stabilization phase. Moreover, being a VKORC1 or CYP2C9 polymorphism carrier is associated with a variation in the warfarin dose required to achieve a therapeutic INR compared to that required in wild-type patients. While, these variants lack associated with the responsiveness to warfarin and the outcome measure of INR in our population. In order to confirm our findings, further studies with larger sample sizes are required in addition to investigating the combined effect of these SNPs on the sensitivity and responsiveness of warfarin. Finally, expanded pharmacogenetic study is required to evaluate the effects of other clinical and genetic factors (such as the coagulation factors and metabolizing warfarin genes) on warfarin sensitivity and responsiveness during the stabilization phase of therapy and their effects on adverse drug events.
Acknowledgments
Acknowledgments
This work was supported by the Deanship of Research at Jordan University of Science and Technology under grant number (203/2014).
Conflict of interest
The Authors declare no conflict of interest, financial or otherwise.
Footnotes
Peer review under responsibility of King Saud University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jsps.2019.01.011.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- Ansell J., Hirsh J., Poller L., Bussey H., Jacobson A., Hylek E. The pharmacology and management of the vitamin K antagonists: the Seventh ACCP Conference on Antithrombotic and Thrombolytic Therapy. Chest. 2004;126:204S–233S. doi: 10.1378/chest.126.3_suppl.204S. [DOI] [PubMed] [Google Scholar]
- D'Andrea G., D'Ambrosio R.L., Di Perna P., Chetta M., Santacroce R., Brancaccio V., Grandone E., Margaglione M. A polymorphism in the VKORC1 gene is associated with an interindividual variability in the dose-anticoagulant effect of warfarin. Blood. 2005;105:645–649. doi: 10.1182/blood-2004-06-2111. [DOI] [PubMed] [Google Scholar]
- Freeman B.D., Zehnbauer B.A., McGrath S., Borecki I., Buchman T.G. Cytochrome P450 polymorphisms are associated with reduced warfarin dose. Surgery. 2000;128:281–285. doi: 10.1067/msy.2000.107283. [DOI] [PubMed] [Google Scholar]
- Gage B., Lesko L. Pharmacogenetics of warfarin: regulatory, scientific, and clinical issues. J. Thromb. Thrombolysis. 2007;25:45–51. doi: 10.1007/s11239-007-0104-y. [DOI] [PubMed] [Google Scholar]
- Genomes Project, Consortium, Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A. An integrated map of genetic variation from 1092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasheen J.J., Fugit R.V., Prochazka A.V. The risk of overanticoagulation with antibiotic use in outpatients on stable warfarin regimens. J. Gen. Int. Med. 2005;20:653–656. doi: 10.1111/j.1525-1497.2005.0136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon M. Estimation of the warfarin dose with clinical and pharmacogenetic data. N. Engl. J. Med. 2009;360:753–764. doi: 10.1056/NEJMoa0809329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi M.K., Veenstra D.L., Kondo L.M., Wittkowsky A.K., Srinouanprachanh S.L., Farin F.M., Rettie A.E. Association between CYP2C9 genetic variants and anticoagulation related outcomes during warfarin therapy. JAMA. 2002;287:1690–1698. doi: 10.1001/jama.287.13.1690. [DOI] [PubMed] [Google Scholar]
- Hirsh J., Dalen J., Anderson D.R., Poller L., Bussey H., Ansell J., Deykin D. Oral anticoagulants: mechanism of action, clinical effectiveness and optimal therapeutic range. Chest. 2001;119:8S–21S. doi: 10.1378/chest.119.1_suppl.8s. [DOI] [PubMed] [Google Scholar]
- Kuruvilla M., Gurk-Turner C.A. Review of warfarin dosing and monitoring. Proc. (BaylUniv. Med. Cent.) 2001;14:305–306. doi: 10.1080/08998280.2001.11927781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Chang C.Y., Jin D.Y., Lin P.J., Khvorova A., Stafford D.W. Identification of the gene for vitamin K epoxide reductase. Nature. 2004;427:541–544. doi: 10.1038/nature02254. [DOI] [PubMed] [Google Scholar]
- Limdi N., McGwin G., Goldstein J., Beasley T., Arnett D., Adler B., Baird M., Acton R. Influence of CYP2C9 and VKORC1 1173C/T genotype on the risk of hemorrhagic complications in African-American and European-American Patients on Warfarin. Clin. Pharmacol. Ther. 2007;83:312–321. doi: 10.1038/sj.clpt.6100290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limdi N., Arnett D., Goldstein J., Beasley T., McGwin G., Adler B., Acton R. Influence of CYP2C9 and VKORC1 on warfarin dose, anticoagulation attainment and maintenance among European Americans and African Americans. Pharmacogenomics. 2008;9:511–526. doi: 10.2217/14622416.9.5.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder M.W. Interactive modeling for ongoing utility of pharmacogenetic diagnostic testing: application for warfarin therapy. Clin. Chem. 2009;55:1861–1868. doi: 10.1373/clinchem.2009.125898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ncbi.nlm.nih.gov. dbSNP Home Page, 2018 [Online]. Available at: http://www.ncbi.nlm.nih.gov/SNP (accessed 15 Sep. 2018).
- Rost S., Fregin A., Ivaskevicius V., Conzelmann E., Hortnagel K., Pelz H.J., Lappegard K., Seifried E., Scharrer I., Tuddenham E.G., Muller C.R., Strom T.M., Oldenburg J. Mutations in VKORC1 cause warfarin resistance and multiple coagulation factor deficiency type 2. Nature. 2004;427:537–541. doi: 10.1038/nature02214. [DOI] [PubMed] [Google Scholar]
- Schelleman H., Chen Z., Kealey C., Whitehead A., Christie J., Price M., Brensinger C., Newcomb C., Thorn C., Samaha F., Kimmel S. Warfarin response and vitamin k epoxide reductase complex 1 in African Americans and Caucasians. Clin. Pharmacol. Ther. 2007;81:742. doi: 10.1038/sj.clpt.6100144. [DOI] [PubMed] [Google Scholar]
- Sconce E.A., Khan T.I., Wynne H.A., Avery P., Monkhouse L., King B.P., Wood P., Kesteven P., Daly A.K., Kamali F. The impact of CYP2C9 and VKORC1 genetic polymorphism and patient characteristics upon warfarin dose requirements: proposal for a new dosing regimen. Blood. 2005;106:2329–2333. doi: 10.1182/blood-2005-03-1108. [DOI] [PubMed] [Google Scholar]
- Scordo M.G., Pengo V., Spina E., Dahl M.L., Gusella M., Padrini R. Influence of CYP2C9 and CYP2C19 genetic polymorphisms on warfarin maintenance dose and metabolic clearance. Clin. Pharmacol. Ther. 2002;72:702–1013. doi: 10.1067/mcp.2002.129321. [DOI] [PubMed] [Google Scholar]
- Shrif N., Won H., Lee S., Park J., Kim K., Kim M., Kim S., Lee S., Ki C., Osman I., Rhman E., Ali I., Idris M., Kim J. Evaluation of the effects of VKORC1 polymorphisms and haplotypes, CYP2C9 genotypes, and clinical factors on warfarin response in Sudanese patients. Eur. J. Clin. Pharmacol. 2011;67:1119–1130. doi: 10.1007/s00228-011-1060-1. [DOI] [PubMed] [Google Scholar]
- Standaard Af handeling Cumarine Interacties, 2003. Available from: URL: www.fnt.nl (accessed Dec 23, 2005).
- Takahashi H., Echizen H. Pharmacogenetics of warfarin elimination and its clinical implications. Clin. Pharmacokinet. 2001;40:587–603. doi: 10.2165/00003088-200140080-00003. [DOI] [PubMed] [Google Scholar]
- Thermofisher.com. Applied Biosystems | Thermo Fisher Scientific – US, 2018 [Online]. Available at: https://www.thermofisher.com/us/en/home/brands/applied-biosystems.html (Accessed 15 Sep. 2018).
- Veenstra D.L., You J.H., Rieder M.J., Farin F.M., Wilkerson H.W., Blough D.K., Cheng G., Rettie A.E. Association of vitamin K epoxide reductase complex 1 (VKORC1) variants with warfarin dose in a Hong Kong Chinese patient population. Pharmacogenet. Genom. 2005;15:687–691. doi: 10.1097/01.fpc.0000174789.77614.68. [DOI] [PubMed] [Google Scholar]
- Visser L.E., Bleumink G.S., Trienekens P.H., Vulto A.G., Hofman A., Stricker B.H. The risk of overanticoagulation in patients with heart failure on coumarin anticoagulants. Br. J. Haematol. 2004;127:85–89. doi: 10.1111/j.1365-2141.2004.05162.x. [DOI] [PubMed] [Google Scholar]
- Wadelius M., Chen L.Y., Downes K., Ghori J., Hunt S., Eriksson N., Wallerman O., Melhus H., Wadelius C., Bentley D., Deloukas P. Common VKORC1 and GGCX polymorphisms associated with warfarin dose. Pharmacogenom. J. 2005;5:262–270. doi: 10.1038/sj.tpj.6500313. [DOI] [PubMed] [Google Scholar]
- Yousef A.M., Bulatova N.R., Newman W., Hakooz N., Ismail S., Qusa H., Al-Diab O. Allele and genotype frequencies of the polymorphic cytochrome P450 genes (CYP1A1, CYP3A4, CYP3A5, CYP2C9 and CYP2C19) in the Jordanian population. Mol. Biol. Rep. 2012;39:9423–9433. doi: 10.1007/s11033-012-1807-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.