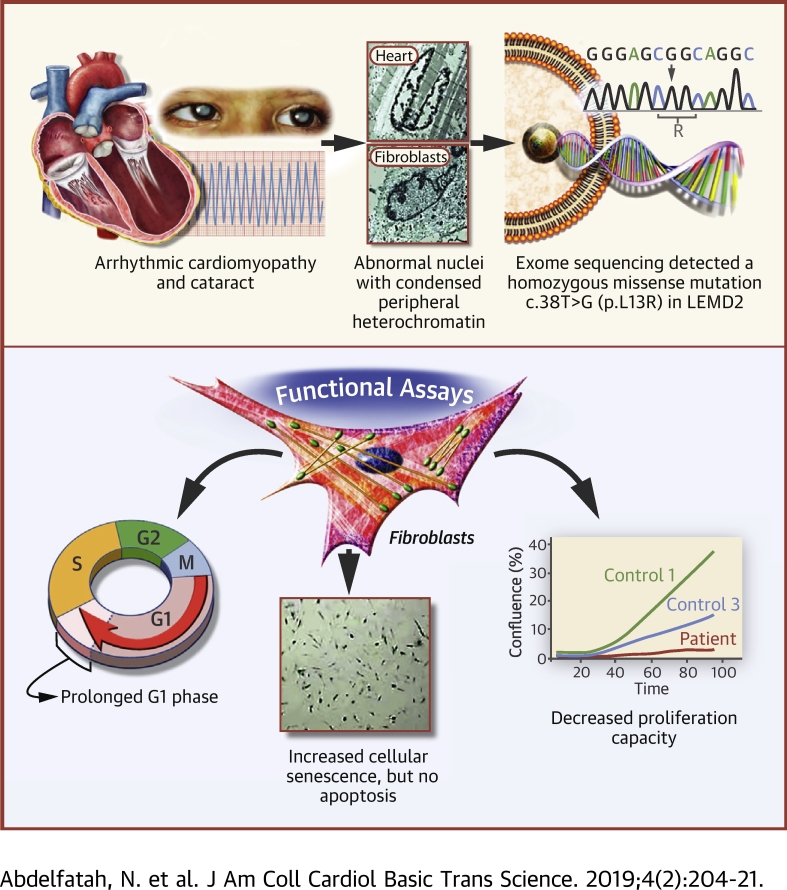
Visual Abstract
Key Words: chromatin remodeling, dilated cardiomyopathy, inner nuclear membrane, LEMD2, sudden death
Abbreviations and Acronyms: ACM, arrhythmogenic cardiomyopathy; BANF, barrier to autointegration factor; CMR, cardiac magnetic resonance; DAPI, 4′,6′-diamidino-2-phenylindole; DCM, dilated cardiomyopathy; DNA, deoxyribonucleic acid; eGFP, enhanced green fluorescent protein; EMD, emerin; ICD, implantable cardioverter-defibrillator; LEMD2, LEM domain containing protein 2; LGE, late gadolinium enhancement; LMNA, lamin A/C; LV, left ventricular; PBS, phosphate-buffered saline; NE, nuclear envelope; P, passage; SAHF, senescence-associated heterochromatin foci; SNV, single nucleotide variant
Highlights
-
•
The homozygous c.38T>G mutation in the LEMD2 gene causes arrhythmic cardiomyopathy with bilateral juvenile cataract in the Hutterite population.
-
•
The cardiac phenotype is characterized by localized inferior and inferolateral fibrosis of the left ventricle and mild impairment of left ventricular systolic function but severe ventricular arrhythmias leading to sudden cardiac death.
-
•
Affected heart tissue and fibroblasts exhibit abnormally shaped nuclei with condensed peripheral heterochromatin.
-
•
Functional assays on affected fibroblasts show decreased proliferation capacity, cellular senescence, and a prolonged G1 phase, suggesting premature aging and cellular senescence in proliferating cells.
Summary
Nuclear envelope proteins have been shown to play an important role in the pathogenesis of inherited dilated cardiomyopathy. Here, we present a remarkable cardiac phenotype caused by a homozygous LEMD2 mutation in patients of the Hutterite population with juvenile cataract. Mutation carriers develop arrhythmic cardiomyopathy with mild impairment of left ventricular systolic function but severe ventricular arrhythmias leading to sudden cardiac death. Affected cardiac tissue from a deceased patient and fibroblasts exhibit elongated nuclei with abnormal condensed heterochromatin at the periphery. The patient fibroblasts demonstrate cellular senescence and reduced proliferation capacity, which may suggest an involvement of LEM domain containing protein 2 in chromatin remodeling processes and premature aging.
Dilated cardiomyopathy (DCM) is characterized by left ventricular (LV) or biventricular dilatation and systolic dysfunction that often leads to heart failure and sudden cardiac death (1). Genetic forms of DCM are heterogeneous, which became more evident with the widespread use of next-generation sequencing panels or exome sequencing (2). To date, >50 disease-related genes have been reported, although relatively few are supported by robust segregation analyses or experimental data. Mutations in genes encoding inner nuclear membrane (INM) proteins lamin A/C (LMNA) and emerin (EMD) have been shown to be involved in the pathogenesis of DCM. LMNA encodes A-type lamins, which are intermediate filaments that, together with B-type lamins, form a filamentous structure that underlies the nuclear envelope (NE) (3). Dominant LMNA mutations cause a variety of phenotypes involving skeletal muscle, cardiac muscle, adipose tissue, and peripheral nerves, including a form of Emery-Dreifuss muscular dystrophy and isolated DCM. LMNA mutations account for 6% to 10% of genetically determined DCM and are frequently associated with arrhythmias and conduction system disturbances (4). EMD encodes an LEM domain protein located in the nuclear lamina and has a role in assembly of the nuclear lamina and structural organization of the NE. EMD mutations are associated with an X-linked form of Emery-Dreifuss muscular dystrophy, which usually includes a severe form of cardiomyopathy (5). Moreover, variants in genes encoding other nuclear membrane components have also been implicated in cardiomyopathy, including SYNE1, SYNE2, and LAP2α (1).
LEM domain containing protein 2 (LEMD2) is a member of the group II LEM domain proteins, which contain ∼40 conserved amino acids representing the LEM domain, a domain discovered previously in other INM proteins 6, 7, 8, 9. It is ubiquitously expressed in the INM of the NE with an increase during telophase and lower expression in the endoplasmic reticulum 10, 11, 12, 13. LEMD2 binds the deoxyribonucleic acid (DNA)-binding protein barrier to autointegration factor (BANF) mediated by the LEM domain and interacts with nuclear lamins by its N-terminal and transmembrane domains (14). Complete disruption in mice causes embryonic lethality by embryonic day 11.5, leading to reduced sizes of most tissues. Neural and heart structures appeared to be less developed and/or abnormal. Studies of knock-out embryos exhibited thin myocardium with underdeveloped trabeculae, consistent with a role for LEMD2 in cardiac development. Moreover, knockdown of it in an immortalized mouse myoblast cell line (C2C12) causes a myogenesis defect 13, 15.
Recently, a mutation in LEMD2 has been associated with juvenile cataract and a risk for sudden cardiac death in the Hutterite population (16). The Hutterite population is a genetic isolate who originated in Europe in the 16th century and emigrated to the United States and Canada in the 1870s. They can be traced back to <100 founders and are divided into 3 branches: Dariusleut, Lehrerleut (L-leut), and Schmiedeleut (S-leut) 17, 18.
In the present study, we clinically and genetically characterized 2 large Hutterite families with LEMD2-associated disease from the L-leut and S-leut branches. Individuals carrying the homozygous mutation (c.38T>G; p.L13R) in the LEMD2 gene exhibited a new form of arrhythmic cardiomyopathy with localized inferior and inferolateral myocardial scarring and severe arrhythmias but only mild impairment of systolic LV function. Cardiac tissue and fibroblasts from affected patients exhibited abnormally shaped and elongated nuclei as well as heterochromatin disorganization. Mutant fibroblasts showed a proliferation defect and cell senescence but no increased apoptosis, suggesting an involvement of mutant LEMD2 in chromatin remodeling processes and premature aging.
Methods
Patient characterization
The study conformed to the principles outlined in the Declaration of Helsinki and was approved by institutional review boards of the University of Calgary and University of Manitoba (ID-E23515, ID-E20729, HS16978). All participating individuals provided written informed consent. Clinical evaluation included 12-lead electrocardiography, signal-averaged electrocardiography, and exercise testing according to the Bruce protocol, 24-h Holter monitoring, and 2-dimensional transthoracic echocardiography and/or cardiac magnetic resonance (CMR) imaging. In some cases, additional investigations and/or records such as reports from implantable cardioverter-defibrillators (ICDs) were obtained. Medical records of deceased individuals have been collected when possible to reconstruct their phenotypes. Tissue blocks of heart and liver of the deceased individual 600, II-16 have been further investigated. A skin biopsy was taken from individual 600, II-18, indicated as “Pat” and 2 volunteers at an age of 40 years (age-matched control subjects: Ctrl1 and Ctrl2) and 61 years (“old” senescent control: Ctrl3) for isolating dermal fibroblasts.
Genetic studies
DNA from 4 affected individuals of family 600 (II-2, II-3, II-18, and III-4) were extracted and underwent exome sequencing. Sequence enrichment was performed by using the Agilent SureSelect Whole Human Exon 50 Mb XT (version 3) kit (Agilent Technologies, Santa Clara, California) and sequenced on a 5500xl SOLiD system (Thermo Fisher Scientific, Waltham, Massachusetts). Raw color space sequences were error-corrected and aligned to the human reference genome version hg19 with version 2.5 of the 5500xl manufacturer's LifeScope software version 2.5, using the “targeted.resequencing.pe” workflow (19). The aligned sequences were deduplicated, realigned, and genotypes called by using multiple third-party programs, using the validated workflow described previously (20). Nonsynonymous single nucleotide variants (SNVs) and insertions and deletions in coding regions were retained for the downstream analysis, which were further annotated with functional impact. Minor allele frequencies of the control subjects were evaluated by using the Genome Aggregation Database (21). Rare homozygous variants (population allele frequency <0.05) that were shared among all 4 individuals were identified as potential causal variants. Fine mapping was undertaken in 4 affected family members of family 600 and 2 affected family members of family 290 (III-12 and III-13) using 8 SNVs that flanked the 18.3 Mb homozygous region at chr.6p21.3 containing LEMD2.
Molecular modeling
The PyMOL Molecular Graphics System, version∼1.3r1 (August 2010) by Schrödinger LLC (Cambridge, Massachusetts), was used to introduce the p.L13R mutation into the LEMD2 domain with the atomic structure PDB ID 2ODC (22). The Swiss Model Homology Server was used to generate a model of this domain, including the mutant p.L13R using LEM domain template (PDB ID 2ODC).
Histology and transmission electron microscopy
Heart tissue was embedded in paraffin, and 5-μm thick slices were deparaffinized and stained with hematoxylin and eosin, Masson’s trichrome, and picro sirius red (1% sirius red in saturated aqueous picric acid) as previously described (23). An Olympus Bx54 microscope equipped with an UPlanSApo 100x/1.4NA objective (Olympus, Tokyo, Japan) was used for imaging. Samples for the transmission electron microscopy were fixed, dehydrated, and embedded in resin blocks. Blocks were split to 50- to 70-nm thin sections and imaged by using a Hitachi H-7650 transmission electron microscope (Hitachi High Technologies America, Inc., Schaumburg, Illinois).
Plasmid generation and cell culture
A plasmid containing the full-length human LEMD2 complementary DNA fused to an enhanced green fluorescent protein (eGFP)-tag on the C terminus was purchased (GeneCopoeia, Rockville, Maryland). The mutation c.38T>G was introduced by using the QuikChange Lightning Kit (Agilent Technologies). The mCherry-tagged Lamin A plasmid (Addgene, Cambridge, Massachusetts) was purchased and used for co-transfection. C2C12 and HEK293 cells were cultured in Dulbecco’s modified Eagle’s medium (L-glutamine, 4.5 g/l glucose; Lonza Group, Basel, Switzerland) supplemented with10% fetal calf serum and 500 U/ml penicillin and 500 μg/ml streptomycin. Fibroblast cell lines were cultured in gelatin 0.01% (MilliporeSigma, Burlington, Massachusetts) coated plates and incubated in fibroblast media consisting of Dulbecco’s modified Eagle’s medium (L-glutamine, 4.5 g/l glucose; Lonza Group) supplemented with 10% fetal calf serum, 500 U/ml penicillin, 500 μg/ml streptomycin, and MEM Non-Essential Amino Acids Solution 100× (Thermo Fisher Scientific).
Immunocytochemistry, immunohistochemistry, and confocal microscopy
C2C12 cells were transfected with either eGFP-LEMD2 or mCherry-Lamin A/C or co-transfected with both by using Lipofectamine 2000 (Thermo Fisher Scientific). Forty-eight hours after transfection, cells were fixed for 20 min in 4% paraformaldehyde at 4°C, washed 3 times with phosphate-buffered saline (PBS) for 10 min each) and blocked for 1 h with goat serum. Afterwards, the slides were washed, and Prolong Gold Antifade Mountant with 4′,6′-diamidino-2-phenylindole (DAPI) (Thermo Fisher Scientific) was added. Fibroblast cells were spread on 0.01% gelatin-coated cover slides and incubated at 37°C for 48 h and fixed with 4% paraformaldehyde for 20 min at 4°C. Paraffin-embedded tissue slides (5 μm) were deparaffinized by using xylene and ethanol. Tissue and fixed fibroblast cells were washed with PBS, blocked with goat serum, and stained overnight with primary antibodies followed by secondary antibodies conjugated with Alexa-488 and Alexa-555 dyes for 2 h at room temperature (Supplemental Table 1 and Supplemental Figure 1 for isotype control staining) and embedded in ProLong Gold Antifade Mountant with DAPI. The LSM5 Exciter (Carl Zeiss AG, Oberkochen, Germany) was used for confocal imaging. All images were processed with Zen software v6,0,0,303 (MDaemon Technologies, Ltd., Grapevine, Texas).
Western blot analysis
Transfected HEK293 cells were collected 48 h after transfection. Fibroblasts were harvested after reaching 80% confluence. Both were collected in radio-immunoprecipitation assay buffer supplemented with proteinase inhibitor cocktail (Roche, Mannheim, Germany) and homogenized. Protein samples were separated by using sodium dodecyl sulfate polyacrylamide gel electrophoresis, transferred to the membrane and incubated with primary antibodies overnight at 4°C followed by the horseradish peroxidase–conjugated secondary antibody at room temperature for 2 h (Supplemental Table 1). Bands were detected on the ChemiDoc equipment (Bio-Rad, Hercules, California) after adding Amersham ECL Western Blotting Detection Reagent (GE Healthcare, Life Sciences, Chicago, Illinois). Bands were quantified by using Image Lab Touch Software version 6.0 (Bio-Rad, Hercules, California).
Fibroblast cell proliferation assay
Fibroblast cells from a 38-year-old patient (family 600, II-18), a 40-year-old age-matched control subject (Ctrl1), and a 61-year-old male control subject (Ctrl3) at different passage numbers were used for the proliferation assay. Cells were diluted to 20,000 cells/ml with fibroblast medium and seeded into a 96-well flat bottom plate coated with gelatin and incubated at 37°C for 2 h before scanning. The plate was placed into the IncuCyte ZOOM system (Essen BioScience Inc., Ann Arbor, Michigan) and scanned under the phase 4× object with a 2-h interval. Collected data were analyzed by using the IncuCyte ZOOM’s confluence processing analysis tool (basic analysis).
Apoptosis assay from tissue
Paraffinized sectioned slices from heart and liver of the deceased patient (family 600, II-16) and a control were stained with 5-bromo-2′-deoxyuridine 5′-triphosphate according to the APO-BRDU-IHC Colorimetric kit (MilliporeSigma) instructions (24). Images were conducted by using the Olympus Bx54 microscope equipped with an UPlanSApo 100x/1.4NA objective (Olympus, Tokyo, Japan).
Terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling and cell senescence assay of fibroblasts
Fibroblasts were harvested and fixed with freshly prepared 2% paraformaldehyde for 1 h at room temperature. Cells were suspended in permeabilization solution and incubated with terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling reaction mixture for 1 h at 37°C in the dark. For the staining, the in situ cell death detection kit (Roche) was used. Cells were sorted with the fluorescence-activated cell sorting (FACS) Calibur (Becton Dickinson) flow cytometer, and the results were analyzed with FlowJo software version 7.6.1 (FlowJo, Ashland, Oregon). Cultured fibroblasts with identical passage number were subjected to β-galactosidase staining by using a senescence β-galactosidase staining kit (CS0030, MilliporeSigma) following the manufacturer’s instructions.
Cell cycle analysis
Harvested fibroblasts were fixed by –20°C pre-cooled 75% ethanol and stored at –20°C overnight. Fixed cells were rinsed twice with PBS, re-suspended in PBS containing 100 μg/ml RNase A (Takara Bio, Kusatsu, Japan), and incubated at 37°C for 30 min. Propidium iodide 40 μg/ml plus 0.25% Triton X-100 was added to the cell suspension, and cells were incubated in the dark at 37°C for 30 min. Stained DNA was analyzed by using a FACS Calibur flow cytometer and FlowJo software version 7.6.1.
Statistical analysis
All datasets are expressed as mean ± SEM. The replicates and numbers (N) of experiments are indicated with each experiment. Two-way analysis of variance was used for all the assays with multiple samples. Statistical analyses were performed by using Prism 7 software (version 7.0a; GraphPad Software, La Jolla, California), if not otherwise indicated.
Results
Arrhythmic cardiomyopathy with regional inferior and inferolateral myocardial scarring is associated with juvenile cataract
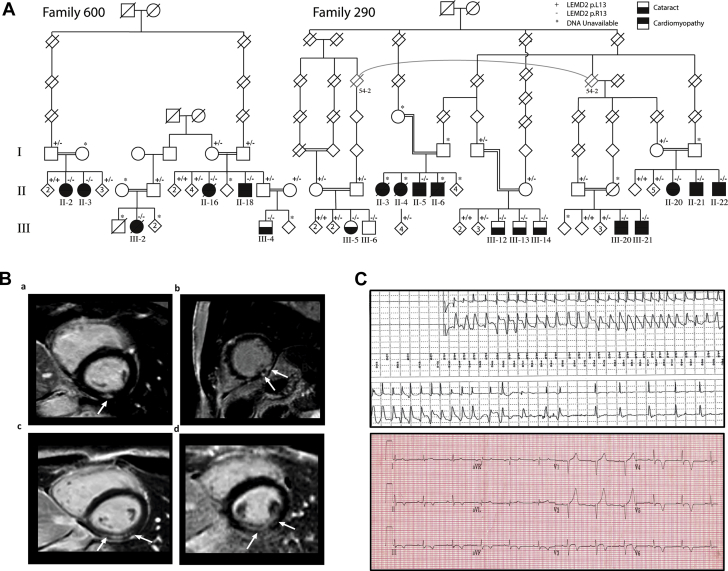
We clinically investigated 20 members of 2 extended families (family 600 and family 290) of the L-leut and S-leut branches of the Hutterite population who have a history of bilateral juvenile cataract (Figure 1A, Table 1). The phenotype of bilateral juvenile cataract was reported previously in family 600 16, 25, 26; however, it became obvious that cardiac disease may also be associated with it after 2 individuals from the original publications died suddenly at the ages of 28 and 43 years (family 600, III-2 and II-16).
Figure 1.
Clinical Features of 2 Extended Hutterite Families With Arrhythmic Cardiomyopathy and Juvenile Cataract
(A) Pedigrees of 2 multigenerational Hutterite families of L-leut (family 600) and S-leut (family 290) descendants. Filled black squares (male subjects) and circles (female subjects) refer to affected individuals with cataract and arrhythmic cardiomyopathy. Filled upper half symbols indicate individuals diagnosed with arrhythmic cardiomyopathy. Filled lower half symbols refer to individuals with juvenile cataract. Diagonal lines indicate deceased individuals. Double lines refer to known consanguinity. The genotype indicated by a “+” is the p.L13 (wild-type) allele and that indicated by a “-” is the mutant p.R13 allele. (B) Short-axis views of late gadolinium enhancement cardiac magnetic resonance imaging of family 290, II-20 (a), II-22 (b), III-20 (c), and III-21 (d) confirming nearly transmural delayed enhancement of the inferior/inferolateral walls as indicated by the arrows. (C) Rhythm strip of individual 600, II-18 recorded by the implantable cardioverter-defibrillator before delivering an appropriate shock (upper panel). Representative 12-lead electrocardiogram of the same individual showing deep T-wave inversions inferior and lateral corresponding to areas of fibrosis in the cardiac magnetic resonance image (lower panel). DNA = deoxyribonucleic acid; LEMD2 = LEM domain containing protein 2.
Table 1.
Clinical Characteristics of Individuals With Bilateral Juvenile Cataract
| Age of Investigation (yrs) | CMR |
Echocardiography | ECG | Arrhythmias (24 Holter) | SA-ECG | ICD or ILR | Cataract | Clinical History | Other | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tissue Characterization (Scar) and Wall Motion Abnormalities | RVEDVi | RVEF | LVEDVi | LVEF | ||||||||||
| Family 600 | ||||||||||||||
| II-2 | 31 | Inferior and inferolateral scar, thinned and a kinetic, fibrosis subendocardial and subepicardial | 74 ml/m2 | 52% | 84 ml/m2 | 47% | LA dilated, mildly reduced LV function, hypokinetic apex, LVEDD 5.0 cm (normal), normal RV function | SR, T-wave inversions III, aVF, V4-V6 | 2,757 isolated PVCs, 51 couplets, slow runs of nsVT, no pauses | 3/3 positive for late potential | ILR: no significant arrhythmias | Yes | No symptoms, no fainting | NA |
| II-3 | 29 | Fibrosis inferior and inferolateral with corresponding hypokinesis, fibrosis subendocardial and subepicardial | 72 ml/m2 | 57% | 84 ml/m2 | 48% | Normal LA, LVEF 45%, inferolateral wall akinetic, LVEDD 4.7 cm (normal), normal RV function | SR, incomplete RBBB, T-wave inversions II, III, aVF, V5, V6, delayed R V2-V4 | 1,984 isolated PVCs, 38 couplets, no nsVT, no pauses | 3/3 positive | ILR: no significant arrhythmias | Yes | No symptoms, no fainting | NA |
| III-2 | 28 deaths | NA | NA | NA | NA | NA | Moderate decreased LV and RV function, LVEF 36% | SR, RBBB, abnormal R-wave progression V3-V5, T-wave inversion across all leads | NA | NA | NA | Yes | Died suddenly during work; felt unwell few hours before, 3 yrs ago diagnosed with peripartum cardiomyopathy | NA |
| II-16 AR-900A 62† |
42 deaths | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | Yes | Died suddenly during kitchen work; no previous symptoms | Autopsy∗ |
| II-18 AR-900A 31† |
38 | Basal and mid inferolateral and apical lateral hypokinetic, apex dyskinetic, same segments transmural scarring | 101 ml/m2 | 54% | 94 ml/m2 | 43% | Normal LA, LVEF 45%, mild global hypokinesis, apex akinetic, LVEDD 4.2 cm (normal), normal RV function | SR, T-wave inversions III, aVF, V3-V6, delayed R-wave progression V2-V6 | nsVT runs of 10 and 13 beats, monomorphic, frequent PVCs (2286), no pauses | NA | ICD: 1× shock fast polymorphic VT | Yes | Syncope associated with ICD shock | CT angiogram: no coronary artery disease |
| III-4 AR-900A 37† |
9 | NA | NA | NA | NA | NA | Normal | Normal for age | None | 3/3 negative | NA | Yes | No symptoms, no fainting | Normal exercise stress test |
| Family 290 | ||||||||||||||
| III-12 | 21 | None | 84 ml/m2 | 63% | 97 ml/m2 | 56% | Normal | Early repolarization | 1 isolated PAC, no nsVT | 1/3 positive | NA | Yes | No symptoms, no fainting | NA |
| III-13 | 19 | None | 108 ml/m2 | 51% | 100 ml/m2 | 58% | Normal | Normal for age | Normal | 1/3 positive | NA | Yes | No symptoms, no fainting | NA |
| III-14 | 10 | None | 84 ml/m2 | 55% | 78 ml/m2 | 56% | Normal | Normal for age | Normal | 2/3 positive | NA | Yes | No symptoms, no fainting | NA |
| III-20 | 20 | Delayed enhancement inferolateral wall | 62 ml/m2 | 51% | 67 ml/m2 | 64% | Normal | Normal for age | 650 PVCs, 1 couplet, no nsVT | 1/3 positive | NA | Yes | No symptoms, no fainting | Normal Exercise stress test |
| III-21 | 15 | Delayed enhancement inferolateral wall | 57.5 ml/m2 | 58% | 64 ml/m2 | 57% | Normal | Normal for age | Normal | Normal | NA | Yes | No symptoms, no fainting | NA |
| II-20 | 46 | Delayed enhancement, epi-mid myocardial, inferolateral wall mid-level | 66 ml/m2 | 58% | 57 ml/m2 | 68% | Normal | Normal | Normal | Normal | NA | Yes | No symptoms, no fainting | Normal exercise stress test |
| II-21 | 43 | Delayed enhancement, epi-mid myocardial, inferolateral wall at base | 64 ml/m2 | 54% | 68 ml/m2 | 57% | Normal | Normal | 4,800 PVCs | Normal | NA | Yes | No symptoms, no fainting | Normal exercise stress test |
| II-22 | 36 | LV basal inferolateral wall akinesis, LBBB (abnormal septal motion), delayed enhancement basal inferior lateral walls | 38 ml/m2 | 70% | 93 ml/m2 | 45% | Regional wall motion abnormalities, moderately dilated LA | LBBB, T inversions V4-V6 | NA | NA | ICD, no shocks | Yes | Collapsed with exertion, required resuscitation, cardioversion for sustained VT | Normal coronary angiogram |
| III-5 | 8 | NA | NA | NA | NA | NA | Normal | Normal for age | Normal | Normal | NA | Yes | No symptoms, no fainting | NA |
| III-6 | 13 | None | 109.3 ml/m2 | 56% | 79.6 ml/m2 | 56% | Normal | SR RBBB | Rare PVCs of 2 dominant morphologies | Abnormal repolarization | NA | Not at age 13 yrs | No symptoms, no fainting | Homozygous for LEMD2 p.L13R |
| II-5 | 25 | NA | NA | NA | NA | NA | Dilatation of both ventricles, severe MV regurgitation, severe systolic dysfunction | LBBB with left-axis deviation | NA | NA | NA | Yes | Followed up from age 25 yrs, heart transplant at age 27 yrs, today age 44 yrs | No cardiac pathology available |
| II-4 | 44 deaths | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | Yes | Died suddenly at kitchen table at age 44 yrs, history of “enlarged heart” | No autopsy done |
| II-6 | 30 deaths | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | Yes | Died suddenly at age 30 yrs while skating | No autopsy done |
| II-3 | 51 deaths | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | Yes | Died suddenly at age 51 yrs, was followed up for “cardiac condition” | No autopsy done |
All living individuals are homozygous mutation carriers of the LEMD2 mutation.
CMR = cardiac magnetic resonance imaging; ICD = implantable cardioverter-defibrillator; ILR = internal loop recorder; LA = left atrium; LBBB = left bundle branch block; LVEDi = left ventricular end-diastolic volume index; LVEF = left ventricular ejection fraction; LVEDD = left ventricular end-diastolic diameter; MV = mitral valve; nsVT = no sustained ventricular tachycardia; NA = not available; RV = right ventricular; RBBB = right bundle branch block; RVEDVi = right ventricular end-diastolic volume index; RVEF = right ventricular ejection fraction; SA-ECG = signal averaged electrocardiogram; SR = sinus rhythm; PAC = premature atrial beat; PVCs = premature ventricular complexes.
Autopsy report summary: Heart was not enlarged but showed area of scarring at left ventricular (LV) free wall. Microscopic findings: Extensive transmural scar with neovascularization. Myocytes at the edge of scar are hypertrophied. The fibrosis extends around the myocytes and appears to expand interstitial spaces from the endocardium to the epicardial surface. There was no gross or microscopic vascular disease as well as no acute inflammation (no myocarditis).
Corresponding individual described by Boone et al. (16).
Eighteen additional individuals from both families with a history of bilateral juvenile cataract were identified. Those who were available for study underwent detailed cardiac investigations (Table 1). Six individuals aged 8 to 22 years had normal clinical findings. The remainder exhibited some form of cardiac disease that appeared to be age dependent. A group of 4 people (family 290, II-20, II-21, III-20, and III-21) aged 15 to 46 years demonstrated a subclinical phenotype with normal cardiac function and no arrhythmias but a pattern of fibrosis detected by late gadolinium enhancement (LGE) in the cardiac magnetic resonance imaging (MRI) (Figure 1B, a,c,d). Interestingly, the LGE pattern and location appeared to be very specific and was also seen in more severely affected individuals (Figure 1B, b).
In the entire cohort, 5 individuals died of sudden cardiac death ranging in ages from 28 to 51 years (family 600, III-2 and II-16; family 290, II-3, II-4, and II-6). One additional individual had a sudden cardiac arrest on exertion at the age of 36 years and was resuscitated (family 290, II-22). This patient along with 3 other individuals between 29 and 38 years of age (family 600, II-2, II-3, and II-18) also exhibited a significant pattern of scarring in the CMR imaging in the inferior and lateral wall segments with transmural LGE. All 4 individuals had wall motion abnormalities and mildly reduced systolic LV function ranging between 43% and 48% on CMR imaging. A similar pattern of LGE can also be observed in inferior/inferolateral myocardial infarction. Therefore, 2 of the patients underwent a computed tomography angiogram (family 600, II-18) or a coronary angiogram (family 290, II-22), and normal coronary arteries were found. Patient 600, II-18 had an ICD placed for primary prevention and had ventricular arrhythmias terminated by an ICD shock ∼1 year later (Figure 1C).
Two patients had phenotypes more consistent with progressive dilation and heart failure rather than having a sudden arrhythmogenic event (family 290, II-3 and II-5). Patient 290, II-5 presented at 25 years of age due to shortness of breath and was found to have a phenotype of DCM with severe mitral regurgitation and significant systolic dysfunction. His disease progressed over the next 2 years despite medical therapy, and he eventually received a heart transplant. He is currently doing well in his 40s but unfortunately many of his medical records reporting the clinical phenotype were not available for this study, including cardiac pathology findings. His sisters (family 290, II-3 [mentioned earlier] and II-4) reportedly had similar clinical phenotypes and were being medically managed when they died suddenly of presumed arrhythmias.
Although not assessed by a neuromuscular specialist, none of the homozygous carriers demonstrated clinically evident skeletal muscle disease. Of note, several heterozygous carriers of family 600 underwent clinical investigations with electrocardiography and echocardiography (e.g., all siblings and the parents of patient 600, II-16); no cardiac abnormalities have been observed.
Overall, the cardiac phenotype can be summarized as a LV cardiomyopathy with or without dilatation with a localized inferior and inferolateral pattern of fibrosis and subsequent wall motion abnormalities in this region. The main clinical phenotype is characterized by ventricular arrhythmias and sudden death, suggesting the term arrhythmic cardiomyopathy although a subset presented with a phenotype more consistent with classic DCM. The earliest signs of the disease were detected in individual 290, III-21 with mild changes in the CMR imaging at 15 years of age (Figure 1B, d), whereas 6 individuals, all ≤22 years of age, have not yet shown any signs of the disease (Table 1).
Genetic analyses confirmed a homozygous mutation in LEMD2 (c.38T>G; p.L13R) in all clinically affected individuals
Recently, a homozygous mutation in the inner nuclear membrane protein LEMD2 (c.38T>G; p.L13R) was reported by Boone et al. (16) in individuals of the Hutterite population with juvenile cataract and sudden death. The cohort we report here consists of 3 affected individuals reported from the original publication by Boone et al. (II-16, II-18, and III-4 in family 600) (Table 1) and an additional 17 unique affected individuals. We independently confirmed the region of homozygosity on chromosome 6p21.31 between genomic positions 24,784,436 and 43,044,640, covering 18.3 Mb. Eight SNVs with a minor allele frequency <0.01 located in the shared region were used for fine mapping of 4 affected family members of family 600 and 2 affected family members (III-12 and III-13) of family 290, which narrowed down the region of homozygosity on chr.6p.21 to 6.8 Mb containing ∼60 genes (Supplemental Table 2). LEMD2 was the only gene containing a novel homozygous missense mutation c.38T>G; p.L13R (chr.6: 33,772.202), which was not found in the Genome Aggregation Database (21). All individuals from both families with juvenile cataracts were found to be homozygous for the c.38T>G mutation, except 1 carrier, who we believe is pre-symptomatic at 13 years of age (family 290, III-6).
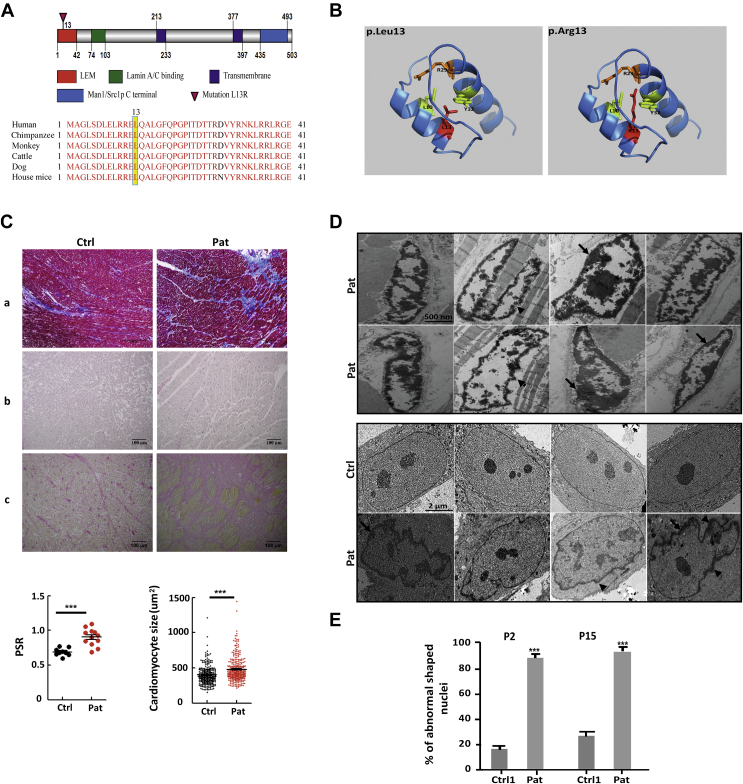
The p.L13R mutation is located in the LEM domain at a highly evolutionary conserved position (Figure 2A). Three-dimensional protein remodeling of the mutated protein showed that the amino acid residue substitution R13 leads to steric hindrance to form a proper three-dimensional structure (Figure 2B).
Figure 2.
The Mutation p.L13R in LEMD2 Causes Interstitial Fibrosis and Abnormal Nuclei in Affected Myocardial Tissue and Fibroblasts
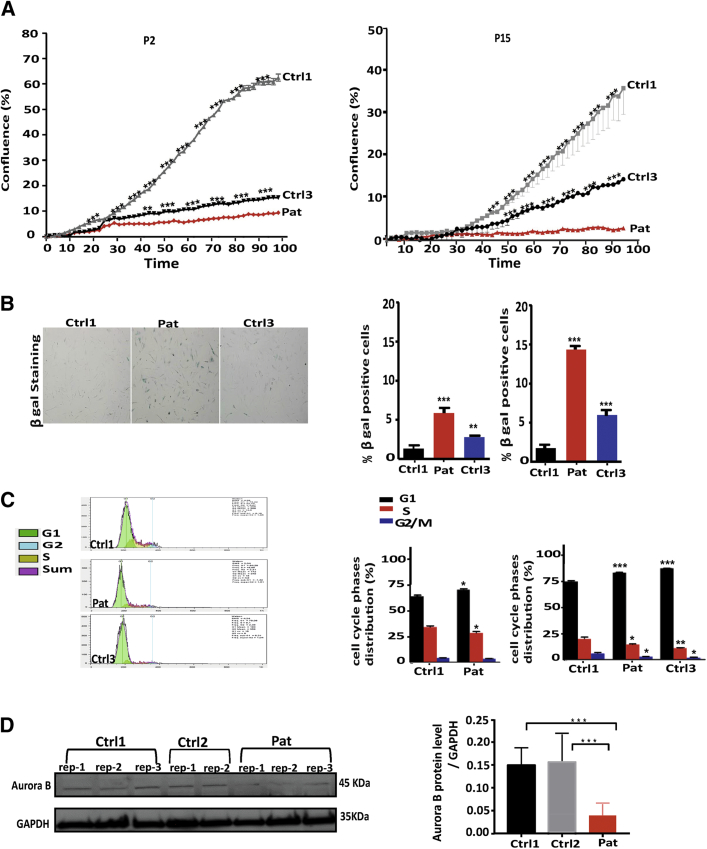
(A) Protein domain structure of human LEM domain containing protein 2 (LEMD2) and conserved motifs. (Top) the LEMD2 protein consists of an LEM domain, lamin A/C–binding domain, 2 transmembrane domains, and a Man1/Src1p C-terminal domain. Domain information was obtained from UniProt (63) and Brachner et al. (10). The red arrow indicates the location of the human mutation. (Bottom) The leucine residue at position 13 (yellow shadow) of LEMD2 is conserved across species. (B) Predicted 3-dimensional structure of the N-terminal domain of LEMD2 with wild-type leucine (Leu) in the left panel and the replaced arginine (Arg) at position 13 in the right. (C) Histology of cardiac tissue from patient 600, II-16 (Pat) showing extensive interstitial fibrosis and myocyte hypertrophy compared with the control (Ctrl). a, Masson’s trichrome (scale bar: 500 μm) staining shows fibrotic tissue in blue; b, hematoxylin and eosin staining (scale bar: 100 μm) and (c) picro sirius red (PSR) demonstrate collagen deposits (scale bar: 100 μm). Myocyte size in hematoxylin and eosin and collagen deposits in PSR were significantly increased in Pat vs. Ctrl; ***p < 0.001. (D) Representative images of affected myocardial tissue (upper panel) recorded by transmission electron microscopy revealed elongation and bizarre shapes/invagination of the membrane (arrowheads) of nuclei with clumping of peripheral heterochromatin (arrows). Transmission electron microscope images of fibroblasts (lower panel) from patient 600, II-18 (Pat) and age-matched control (Ctrl1). Note the abnormal morphology (arrowhead) of the nuclei and the condensed heterochromatin (arrows). (E) Quantification of abnormal nuclei in patient and age-matched control fibroblasts at passage 2 (P2) and passage 15 (P15); n = 300 nuclei, respectively. There is a significant increase of abnormal nuclei in the patient cells (Pat) compared with the control (Ctrl1); n = 300 nuclei; ***p < 0.001.
To determine the carrier frequency of the p.L13R mutation in the Hutterite population, we used a previously published resource available through the University of Chicago 27, 28. Briefly, this resource consists of a database of 98 healthy Hutterite individuals from the S-leut branch who have had full genome sequencing (i.e., complete allele ascertainment) and another 1,317 healthy Hutterites from the S-leut branch who had a more limited analysis but were imputed for every possible genotype in the genome. The imputed genotypes were all calculated with high confidence by using linkage disequilibrium and pedigree analysis as previously described (28). The database was interrogated for the disease allele, and 8 individuals from the fully sequenced cohort were found to carry 1 copy, yielding an estimated carrier frequency of 1 of 13 (0.0769) assuming Hardy-Weinberg equilibrium. However, this frequency was even higher at 1 of 8 (0.125) when the larger cohort with imputed genotypes was used, suggesting this disease may have a very large impact in this population.
Affected heart tissue of a deceased patient and fibroblasts show elongated and invaginated nuclei with condensed peripheral heterochromatin
We next analyzed heart tissue of the left ventricle from the deceased patient (family 600, II-16). Histology from specimens from patient 600, II-16 and a control LV heart sample demonstrated significantly more myocyte hypertrophy and interstitial fibrosis, as well as an increase in collagen deposits in the deceased patient tissue compared with control myocardium (Figure 2C). It has been previously reported that muscle cell nuclei from patient tissue with DCM due to mutations in the NE protein LMNA exhibit an alteration of nuclear morphology (29). To examine the structure of the nuclei, we performed transmission electron microscopy from cardiac tissue as well as from patient fibroblasts (family 600, II-18) compared with an aged-matched control (Ctrl1) at 2 different passage numbers (P2 and P15). The nuclei of the affected heart tissue appeared to be very abnormal in shape and heterochromatin structure. They were elongated and showed an invagination of the nuclear membrane as well as condensed clumping of peripheral heterochromatin (Figure 2D). All observed nuclei in cardiac tissue demonstrated those abnormalities (n = 100 nuclei examined). In patient fibroblast cells, the invagination of the nuclear membrane was also frequently noticed, but the peripheral chromatin was less condensed compared with the nuclei of the heart tissue. Abnormal fibroblast nuclei were detected in 80% of the nuclei (n = 300) at P2 and were increased to 90% at P15. Only a few abnormal nuclei (∼20%) at P2 and 30% at P15 were detected in 300 nuclei of the age-matched control (Figure 2E).
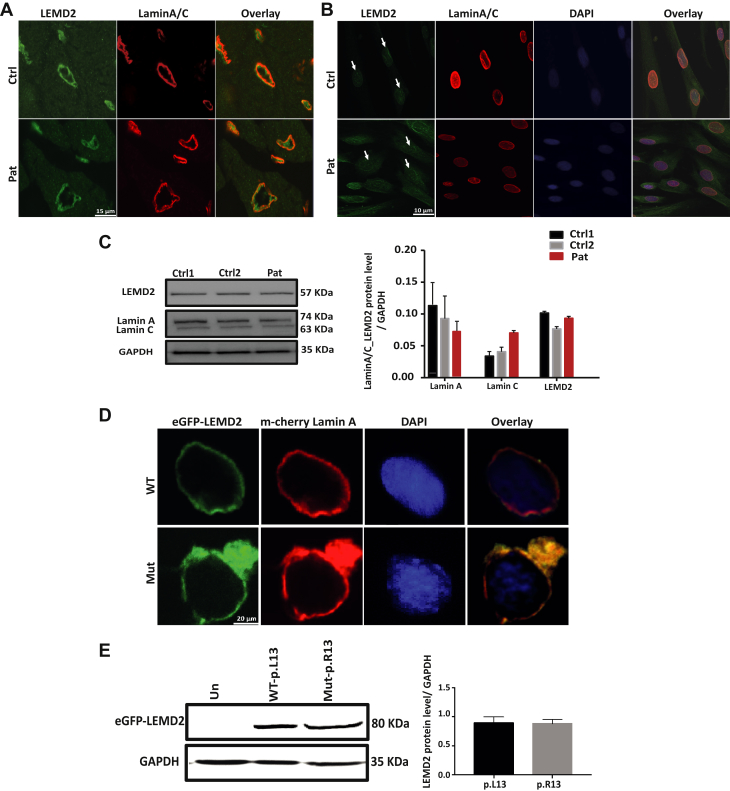
To investigate a potential mislocalization of mutant LEMD2, immunohistochemistry experiments were conducted of affected cardiac tissue, affected fibroblasts, and in vitro cellular expression studies of recombinant wild-type and mutant LEMD2 proteins. We detected LEMD2 at the expected localization at the nuclear membrane in cardiac tissue from patient 600, II-16 and fibroblasts from patient 600, II-18 by using a C-terminal LEMD2-specific antibody. There was no difference compared with the control subjects in either experiment. Furthermore, staining of cardiac tissue and fibroblast cells showed co-localization of both LMNA and LEMD2 in patients and control subjects (Figures 3A and 3B, Supplemental Figure 1). We measured the expression of LEMD2 and LMNA by Western blot analysis using whole protein lysates from fibroblasts; no significant difference was noted between the patient and control subjects (Figure 3C).
Figure 3.
Unchanged LEMD2 Localization and Expression in Patient Cells, Cardiac Tissue, and Transfected C2C12 Cells
(A and B) Representative confocal images of patient and control myocardial tissue as well as fibroblasts showing normal localization of LEMD2 (green, arrow) co-localizing with lamin A/C (red) at the nuclear membrane; 4′,6′-diamidino-2-phenylindole (DAPI) (blue) indicates nuclei. Scale bars represent 20 μm. (C) Western blot showing LEMD2 and lamin A/C protein expression in fibroblasts in the left panel. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as loading control. Quantification by densitometry (n = 3 experiments with 2 to 3 replicates) of LEMD2; quantification of lamin A and C and LEMD2 protein expression in the right panel shows no difference. (D) Representative confocal images of transfected C2C12 cells with recombinant enhanced green fluorescent protein (eGFP)-tagged wild-type (WT) (p.L13) or mutant (p.R13) LEMD2 (green) co-transfected with mCherry–lamin A/C (LMNA) (red) demonstrated co-localization. Scale bar represents 20 μm. (E) Western blot analysis of eGFP-LEMD2 proteins after transfection into HEK293 cells. GAPDH was used for loading control. Quantification by densitometry (n = 3) reveals no difference in the protein level between mutant (p.R13) and WT (p.L13) LEMD2. Un = un-transfected; other abbreviations as in Figure 2.
We subsequently introduced the c.38T>G mutation into a full-length human LEMD2 complementary DNA clone and expressed either mutant or wild-type eGFP-tagged LEMD2 in C2C12 cells. The wild-type (p.L13) as well as the mutant (p.R13) protein showed normal localization at the nuclear membrane (Figure 3D), consistent with the finding in cardiac tissue and fibroblast cells. Furthermore, co-transfection with LMNA complementary DNA clone showed co-localization of both proteins in vitro. In addition, HEK293 cells expressing wild-type or mutant eGFP-LEMD2 demonstrated similar expression levels, indicating a stable mutant protein (Figure 3E).
The LEMD2 mutation induces a proliferation defect and cell senescence in patient fibroblasts
To test the effect of mutant LEMD2 on cell proliferation, fibroblast cells from patient and 2 control subjects (age matched, younger control) underwent live cell counting using the IncuCyte microscope at different passage numbers (P2 and P15). We found that patient fibroblasts demonstrated a significantly diminished proliferation rate at P2 compared with both control subjects; however, the age-matched fibroblasts also showed slower rates of proliferation compared with the younger-aged control. At P15, patient cells did not proliferate but remained viable in culture, whereas both controls still proliferated (Figure 4A). Because this finding is a feature of premature senescence, we stained for β-galactosidase activity as a marker for cell senescence and found a significant increase in the number of cells positive for β-galactosidase at P6 and P15 in patient cells compared with both control subjects (Figure 4B). Remarkably, even fibroblasts from an individual 20 years older (Ctrl3) exhibited fewer β-galactosidase–positive cells compared with patient fibroblasts. We concluded that cells expressing mutant LEMD2 undergo premature senescence.
Figure 4.
The LEMD2 p.L13R Mutation Inhibits Proliferation, Induces Cell Senescence, and Cell Cycle Arrest in Fibroblasts
(A) Cell proliferation assay based on confluence from passage 2 (P2) in the left panel and P15 in the right panel in patient (Pat) fibroblasts, Ctrl1, and an older age (“high-senescence”) control (Ctrl3). There was a significant difference in the rate of proliferation detected between both control subjects and patient at P2 and P15. The cells were imaged every 2 h for 90 h. n = 8 wells, mean ± SD, ***p < 0.001. (B) Images of β-galactosidase (β gal)-stained fibroblasts from patient and Ctrl1 as well as Ctrl3 at passage 6 (P6) and P15 in the left panel. Note that the amount of blue-stained cells is visibly higher in patient cells. (Right) Quantification of β gal–stained cells revealed increased cell senescence in patient cells compared with both control subjects at passages as indicated (n = 3; **p < 0.01; ***p < 0.001). (C) Fibroblasts stained with propidium iodide and measurements of the deoxyribonucleic acid content for each phase of the cell cycle by flow cytometry. Cells were taken from Ctrl1 at P6 and 9, from the patient (Pat) at passage 8 and passage 11, and from Ctrl3 at passage 12. Representative diagrams of each phase of the cell cycle are shown in the left panel. (Right) Quantification of the deoxyribonucleic acid content showed a potential arrest in the G1 phase in patient fibroblasts compared with Ctrl1 and similar to Ctrl3 at later passage. n = 3; ∗∗p < 0.01; ∗p < 0.05; ***p < 0.001. (D) Western blot of Aurora B protein expression in patient and control cells on the left panel. GAPDH was used as loading control. (Right) Quantification by densitometry of Aurora B protein expression revealed a significant difference between both control subjects and patient fibroblasts. n = 3 experiments with each 2 to 3 replicates; ***p < 0.001. Abbreviations as in Figures 2 and 3.
LEMD2 has been shown to be involved in DNA replication and mitosis. To understand the effects of mutant LEMD2 on cell cycle progression, we stained fibroblasts from the patient and 2 control subjects by using propidium iodide and examined cell cycle phase distribution. A larger portion of patient cells and the older age control cells were unable to progress from the G1 phase to the S phase and resulted in a prolonged G1 phase, which was even more significant at higher passage numbers. These data indicate that mutant LEMD2 may induce a G1 arrest (Figure 4C). Furthermore, we examined levels of cell cycle regulatory protein (Aurora B), a well-established G2/M phase marker. Whole protein lysates from patient cells exhibited decreased levels of Aurora B expression compared with both control subjects, consistent with the result from the cell cycle experiment (Figure 4D).
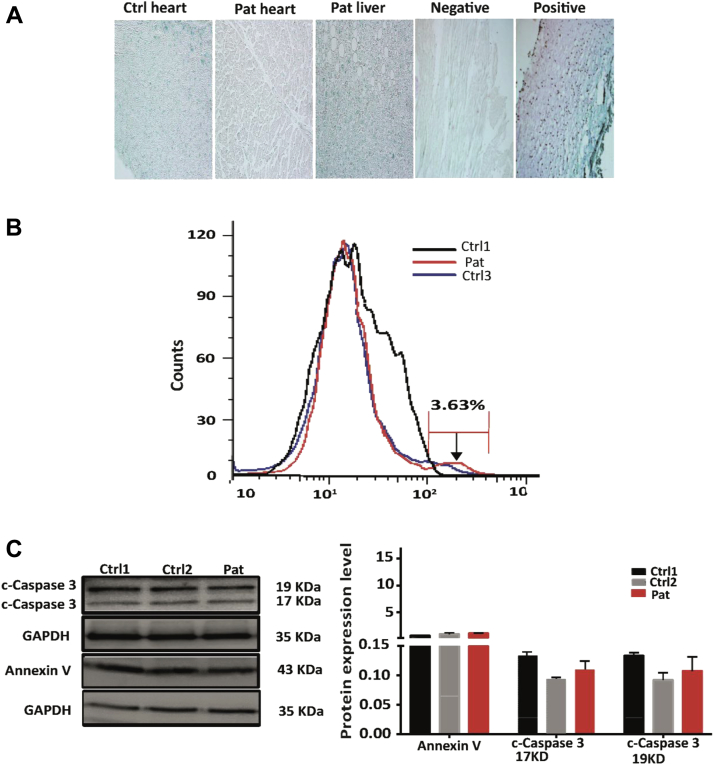
The abnormal proliferation profile seen in mutant fibroblasts could be a consequence of the imbalance between proliferation and apoptosis. We therefore assessed increased apoptosis in patient tissue and fibroblasts. First, affected heart and liver tissue were stained with 5-bromo-2′-deoxyuridine 5′-triphosphate, and the number of brown apoptotic bodies was counted. No obvious apoptotic bodies were detected in the patient and 2 control tissues (Figure 5A). We used the same assay in the patient and control fibroblasts and measured broken DNA ends by using flow cytometry. No significant difference in the apoptotic signals between the patient and the 2 control subjects were detected (Figure 5B). Finally, we tested the expression of caspase 3 and annexin V in whole protein lysates from fibroblasts and found no difference between the patient and control cells (Figure 5C). These data suggest that the mutation in LEMD2 does not affect the apoptotic pathway.
Figure 5.
The Mutation in LEMD2 Does Not Induce Significant Apoptosis
(A) Apoptosis assays were conducted by labeling the deoxyribonucleic acid with 5-bromo-2′-deoxyuridine 5′-triphosphate) in heart and liver tissue from the affected patient (family 600, II-16) and control heart samples. A positive control was created by adding 1 μg/μl deoxyribonucleic acids. Apoptotic cells were detected as brown dots. No signs of apoptosis were detected in all tested tissue. (B) Terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling staining of fibroblasts and flow cytometry from patient and Ctrl1 and Ctrl3. The histograms of the fluorescein signal collected by using the terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling method are shown. There is no difference in the apoptotic signals between the 2 control subjects and patient cells. (C) Western blot analysis of apoptotic markers (annexin V, caspase 3 [2 subunits: 17 and 19 KD]) in patient and control fibroblasts on the left panel. GAPDH was used as loading control. Quantification of apoptotic markers by densitometry does not show a significant difference in expression levels (n = 3 experiments with each 2 to 3 replicates). Abbreviations as in Figures 2, 3, and 4.
Discussion
Genes-encoding INM proteins, most importantly LMNA and EMD, have been shown to be involved in the pathogenesis of human inherited DCMs. Here, we report a new homozygous missense mutation in the INM protein LEMD2 that leads to juvenile cataract and a severe form of arrhythmic cardiomyopathy with variable onset in people of the Hutterite founder population. We clinically and genetically characterized a cohort of 18 individuals from 2 different branches of the Hutterite population and found a primarily arrhythmic phenotype. Importantly, frequent ventricular arrhythmias and focal LV fibrosis preceded LV dilation and depressed myocardial performance, which are the classic hallmarks of DCM (30). However, the progression of LEMD2-associated disease may also lead to the common features of DCM: a hypokinetic left ventricle with decreased LV function as seen in one of the patients who required heart transplantation (family 290, II-5). However, our data support the requirement to recognize a broader phenotypic spectrum of DCM as recently outlined by Pinto et al. (31), in particular for early stages of genetically determined cardiomyopathies. Because ventricular arrhythmias are often life-threatening before other clinical symptoms occur and precede LV dysfunction in most of our cases, we decided to call it “arrhythmic cardiomyopathy,” which should also be distinguished from arrhythmogenic cardiomyopathy (ACM). In ACM, myocardium of the right, and often of both, ventricles are replaced by fibrotic and fatty tissue. Our patients did not exhibit any right ventricular involvement by cardiac imaging, we observed no fat tissue replacement in the histology of autopsy tissue, and none of the patients fulfilled the diagnostic Task Force Criteria for ACM (32).
Interestingly, the delayed enhancement by CMR imaging detected subepicardial, subendocardial, and at later stages transmural patchy fibrosis affecting mainly the inferior and inferolateral wall segments. Although the LGE pattern was suggestive of an ischemic etiology, there was no evidence of obstructive coronary artery disease on cardiac computed tomography imaging or coronary angiography. Localized fibrotic patterns have also been shown in other forms of cardiomyopathy; for example, storage diseases such as Fabry and Danon disease exhibiting predominantly inferolateral scarring 33, 34. Scar formation in DCM is often a sign of poor prognosis due to malignant arrhythmias and sudden death, which is common in our LEMD2 population. More than one-third (7 of 18) of patients died suddenly or required resuscitation and/or an ICD shock for malignant arrhythmias, indicating that clinical risk assessment should be attentive to arrhythmias and late enhancement in the CMR images rather than focus on the LV function, which was either normal or mildly reduced in most cases.
Even if not directly comparable, the outcome of LMNA-related DCM is known as less favorable compared with other forms of DCM 35, 36, 37 . Myocardial fibrosis with minor systolic dysfunction and dilatation, but significant atrioventricular conduction disease, is a common feature of LMNA mutation carriers, particularly in early disease (38). In addition, EMD-related cardiomyopathy often starts with sinus bradycardia and conduction disease (39). Similar to LEMD2 disease, in LMNA and EMD cardiomyopathy, fibrosis and arrhythmias usually preceded the DCM phenotype, but in contrast, LEMD2 cardiomyopathy did not present with major conduction defects, at least not in our patients. Further clinical characterization of a bigger patient cohort and longer follow-ups will help to define the phenotype and outcome of LEMD2 cardiomyopathy more precisely.
LEMD2 is one of the Group II LEM domain proteins that contain an amino-terminal LEM domain in the nucleoplasm and 2 interior transmembrane domains. It interacts with LMNA by its N-terminal and transmembrane domains. The localization of LEMD2 in a lamin-dependent manner is mediated by the retention domain inside the N-terminal of LEMD2 (Figure 1D). Deficiency or loss of the A-type lamins (LMNA) causes mislocalization of the LEMD2 protein to the endoplasmic reticulum (10). If and how our missense mutation affects LMNA association was investigated by co-localization studies in cardiac tissue and fibroblast cells, as well as transfected C2C12 cells. Our results showed that the localization of both LEMD2 and LMNA at the NE was not altered, indicating that the mutation may not affect the retention signal and thus LMNA binding.
A remarkable finding was the bizarre nuclear morphology and the abnormally condensed peripheral chromatin in affected heart tissue and fibroblast cells. It is known that deficiency of LEMD2 disrupts the NE structure and causes a misshaped nucleus with invaginations and lobulations 40, 41, resulting in lower nuclear circularity in the Caenorhabditis elegans mutant (42). LEMD2 has an important indirect role in chromatin organization because it is a binding component of the peripheral heterochromatin A-tether through formation of a complex with the N-terminus of LMNA (43). It also binds the DNA-binding protein BANF, which can influence histone posttranslational modifications (44). BANF plays important structural roles in nuclear assembly and chromatin architecture during interphase, and undefined roles in gene regulation. It regulates specific gene expression through its interaction with LEM proteins. It is a common feature of many LEM proteins to interact directly with transcriptional regulators (45). We hypothesize that the L13R mutation may disrupt the interaction of BANF and LEMD2, which in turn leads to abnormal downstream transcriptional regulation. Interestingly, in Schizosaccharomyces pombe, the chromatin association and tethering of centromeres to the periphery are mediated by the LEM domain of yeast LEMD2, whereas the C-terminal MSC domain is required and sufficient to mediate the proper localization of telomeres (46) and contributes to heterochromatin silencing (47). We speculate that the abnormal chromatin condensation was either caused by a binding defect to BANF or an increased affinity for the LEM domain to bind with centromeres because of the mutated positive charged “arginine,” which may lead to heterochromatin augmentation.
We investigated cell proliferation, cell cycle arrest, and β-galactosidase activity in patient fibroblasts. Mutated fibroblasts showed a trend toward disturbed cell proliferation, cell cycle arrest in G1 phase, and increased β-galactosidase activity, which is a consequence of an increased number of lysosomes (48). In combination with the abnormally condensed peripheral chromatin, these affected cells enter into senescence. The condensed punctate foci of peripheral chromatin are termed senescence-associated heterochromatin foci (SAHF) 49, 50. SAHF formation occurs relatively late during the onset of senescence (51) and is prominent in acute senescence models, such as oncogene-induced senescent cells, and infrequently detected in replicative senescent cells 52, 53. SAHFs are generated by a spatial rearrangement of pre-existing heterochromatin involving a clustering of regions that share specific histone modifications 54, 55, 56. They appear to maintain senescence through repression of growth-promoting genes (57), a process that might be important in the pathogenesis of LEMD2-associated cardiac disease.
We observed that a high percentage of patient fibroblasts exhibited premature cell senescence compared with control subjects, whereas apoptosis did not play a major role. Recently, several studies have shown that senescent cells actively suppress apoptosis 58, 59; Chang et al. (60) and Zhu et al. (61) used the antiapoptotic protein inhibitors to trigger the senescent cells into apoptotic death. Apoptotic cells are eliminated by phagocytes in a manner that does not stimulate inflammation (62); senescent cells are viable, however, and have the potential to influence neighboring cells through pro-inflammatory secretion of growth factors and cytokines, which are well known as the senescence-associated secretory phenotype. These phenotypes may influence the development of cardiomyopathy due to inflammation followed by fibrosis.
There are number of unanswered questions in this disease, including: Why is there a phenotype restricted to the heart and lens despite ubiquitous LEMD2 expression? How does the mutation disturb the interaction to potential binding partners? How does the abnormal nuclear morphology affect gene transcription and signal transduction? More mechanistic research needs to be conducted to understand the complex mechanisms involved in the pathogenesis of the p.L13R LEMD2 mutation in the development of cardiomyopathy.
Conclusions
Mutation in LEMD2 leads to a syndromic form of early-onset cataract and an arrhythmic DCM in which ventricular arrhythmias often precede dilatation and LV dysfunction. Mutant LEMD2 leads to remarkable changes in the shape of nuclei with condensed heterochromatin formation, reduced proliferation capacity, and cell senescence in fibroblasts suggesting the involvement of LEMD2 in chromatin remodeling and premature aging.
Perspectives.
COMPETENCY IN MEDICAL KNOWLEDGE: Familial DCM can be caused by mutated proteins involved in many subcellular systems, but the NE and its interacting proteins play an emerging role in the pathogenesis of DCM. LEMD2 is a novel disease gene for DCM with an arrhythmic phenotype preceding LV dysfunction similar to LMNA- and EMD-related cardiomyopathies. Our analysis presents for the first time a detailed description of the cardiac phenotype associated with LEMD2 disease, as well as pathogenetic and molecular findings observed in patient cells and cardiac tissue indicating chromatin remodeling, reduced proliferation capacity, and cell senescence. LEMD2 disease expands the spectrum of clinical laminopathies and may account for a proportion of cardiomyopathy of undetermined cause in populations outside the Hutterites.
TRANSLATIONAL OUTLOOK: Despite decades of discovery, the complex mechanisms of laminopathies leading to cardiac disease and premature aging (progeria) are still not fully understood. LEMD2 as a new player, and its role in cardiomyopathy, may help to further unravel the interactions of INM proteins of the INM, nucleoplasm, and cytoplasm. It may also aid in determining the roles of INM proteins in the maintenance of cellular integrity, in regulation of signaling pathways, and in chromatin organization and regulation of gene expression. Future research should focus on the role of LEMD2 in health and disease and may add a piece to the puzzle of understanding the complex network involved in laminopathies.
Acknowledgments
The authors thank the family members for their help and participation in this study. They also thank Dr. Yong Xiang Chen (University of Calgary) for his help with the histology and Kristina Martens (University of Calgary) for her technical help in the laboratory. They are also thankful to Dr. Wei-Xiang Dong (University of Calgary) for his help performing the transmission electron microscopy, as well as Dr. Rima-Marie Wazen and Dr. Pina Colarusso (University of Calgary) for helping with the IncuCyte ZOOM system. The IncuCyte ZOOM system has been purchased by the International Microbiome Centre, which is supported by the Cumming School of Medicine, University of Calgary, Western Economic Diversification, and Alberta Economic Development and Trade, Canada.
The authors thank the Genome Aggregation Database and the groups that provided exome and genome variant data to this resource. A full list of contributing groups can be found at http://gnomad.broadinstitute.org/about.
Footnotes
This research was supported by the Care4Rare Canada Consortium funded by Genome Canada, the Canadian Institutes of Health Research, and the Ontario Genomics Institute with additional funding provided to the predecessor of Care4Rare, FORGE Canada, by Genome Quebec, Genome British Columbia, and the McLaughlin Centre to Dr. Frosk. The study was further supported by Alberta Innovates Health Solutions (grant no. 201200822), the Canadian Institutes of Health Research (FRN: 123351), and the Libin Cardiovascular Institute of Alberta to Drs. Gerull and Abdelfatah. Support was also provided by the German Ministry of Education and Research, Berlin, Germany (grant no. 01EO1504 to Dr. Gerull). All other authors have reported that they have no relationships relevant to the contents of this paper to disclose.
All other authors attest they are in compliance with human studies committees and animal welfare regulations of the authors’ institutions and Food and Drug Administration guidelines, including patient consent where appropriate. For more information, visit the JACC: Basic to Translational Scienceauthor instructions page.
Appendix
References
- 1.Dellefave L., McNally E.M. The genetics of dilated cardiomyopathy. Curr Opin Cardiol. 2010;25:198–204. doi: 10.1097/HCO.0b013e328337ba52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haas J., Frese K.S., Peil B. Atlas of the clinical genetics of human dilated cardiomyopathy. Eur Heart J. 2015;36:1123–1135. doi: 10.1093/eurheartj/ehu301. [DOI] [PubMed] [Google Scholar]
- 3.Schreiber K.H., Kennedy B.K. When lamins go bad: nuclear structure and disease. Cell. 2013;152:1365–1375. doi: 10.1016/j.cell.2013.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fatkin D., MacRae C., Sasaki T. Missense mutations in the rod domain of the lamin A/C gene as causes of dilated cardiomyopathy and conduction-system disease. N Engl J Med. 1999;341:1715–1724. doi: 10.1056/NEJM199912023412302. [DOI] [PubMed] [Google Scholar]
- 5.Bione S., Maestrini E., Rivella S. Identification of a novel X-linked gene responsible for Emery-Dreifuss muscular dystrophy. Nat Genet. 1994;8:323–327. doi: 10.1038/ng1294-323. [DOI] [PubMed] [Google Scholar]
- 6.Cai M., Huang Y., Ghirlando R. Solution structure of the constant region of nuclear envelope protein LAP2 reveals two LEM–domain structures: one binds BAF and the other binds DNA. EMBO J. 2001;20:4399–4407. doi: 10.1093/emboj/20.16.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee K.K., Haraguchi T., Lee R.S. Distinct functional domains in emerin bind lamin A and DNA-bridging protein BAF. J Cell Sci. 2001;114:4567–4573. doi: 10.1242/jcs.114.24.4567. [DOI] [PubMed] [Google Scholar]
- 8.Shumaker D.K., Lee K.K., Tanhehco Y.C. LAP2 binds to BAF.DNA complexes: requirement for the LEM domain and modulation by variable regions. EMBO J. 2001;20:1754–1764. doi: 10.1093/emboj/20.7.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mansharamani M., Wilson K.L. Direct binding of nuclear membrane protein MAN1 to emerin in vitro and two modes of binding to barrier-to-autointegration factor. J Biol Chem. 2005;280:13863–13870. doi: 10.1074/jbc.M413020200. [DOI] [PubMed] [Google Scholar]
- 10.Brachner A., Reipert S., Foisner R. LEM2 is a novel MAN1-related inner nuclear membrane protein associated with A-type lamins. J Cell Sci. 2005;118:5797–5810. doi: 10.1242/jcs.02701. [DOI] [PubMed] [Google Scholar]
- 11.Ikegami K., Egelhofer T.A., Strome S. Caenorhabditis elegans chromosome arms are anchored to the nuclear membrane via discontinuous association with LEM-2. Genome Biol. 2010;11:R120. doi: 10.1186/gb-2010-11-12-r120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gruenbaum Y., Lee K.K., Liu J. The expression, lamin-dependent localization and RNAi depletion phenotype for emerin in C. elegans. J Cell Sci. 2002;115:923–929. doi: 10.1242/jcs.115.5.923. [DOI] [PubMed] [Google Scholar]
- 13.Tapia O., Fong L.G., Huber M.D. Nuclear envelope protein Lem2 is required for mouse development and regulates MAP and AKT kinases. PloS One. 2015;10:e0116196. doi: 10.1371/journal.pone.0116196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Segura-Totten M., Wilson K.L. BAF: roles in chromatin, nuclear structure and retrovirus integration. Trends Cell Biol. 2004;14:261–266. doi: 10.1016/j.tcb.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 15.Huber M.D., Guan T., Gerace L. Overlapping functions of nuclear envelope proteins NET25 (Lem2) and emerin in regulation of extracellular signal-regulated kinase signaling in myoblast differentiation. Mol Cell Biol. 2009;29:5718–5728. doi: 10.1128/MCB.00270-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boone P.M., Yuan B., Gu S. Hutterite-type cataract maps to chromosome 6p21.32-p21.31, cosegregates with a homozygous mutation in LEMD2, and is associated with sudden cardiac death. Mol Genet Genomic Med. 2016;4:77–94. doi: 10.1002/mgg3.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hostetler J.A. History and relevance of the Hutterite population for genetic studies. Am J Med Genet. 1985;22:453–462. doi: 10.1002/ajmg.1320220303. [DOI] [PubMed] [Google Scholar]
- 18.Martin A.O. The founder effect in a human isolate: evolutionary implications. Am J Phys Anthropol. 1970;32:351–367. doi: 10.1002/ajpa.1330320305. [DOI] [PubMed] [Google Scholar]
- 19.UCSC Gene Bioinformatics. Available at: http://genome.ucs.edu/FAQ.FAQreleases.html. Accessed February 1, 2019.
- 20.Genome Aggregate Database. Available at: http://gnomad.broadinstitute.org/. Accessed February 1, 2019.
- 21.Lek M., Karczewski K.J., Minikel E.V. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berman H.M., Westbrook J., Feng Z. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.IHC WORLD. Available at: http://www.ihcworld.com. Accessed February 1, 2019.
- 24.APO-BRDU-IHC Colorimetric kit. Phoenix Flow Systems. Available at: http://www.phnxflow.com/APO-BrdU-IHC.protocol.pdf. Accessed February 1, 2019.
- 25.Shokeir M.H., Lowry R.B. Juvenile cataract in Hutterites. Am J Med Genet. 1985;22:495–500. doi: 10.1002/ajmg.1320220307. [DOI] [PubMed] [Google Scholar]
- 26.Pearce W.G., Mackay J.A., Holmes T.M. Autosomal recessive juvenile cataract in Hutterites. Ophthalmic Paediatr Genet. 1987;8:119–124. doi: 10.3109/13816818709028527. [DOI] [PubMed] [Google Scholar]
- 27.Uricchio L.H., Chong J.X., Ross K.D. Accurate imputation of rare and common variants in a founder population from a small number of sequenced individuals. Genetic Epidemiol. 2012;36:312–319. doi: 10.1002/gepi.21623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Livne O.E., Han L., Alkorta-Aranburu G. PRIMAL: fast and accurate pedigree-based imputation from sequence data in a founder population. PLoS Comput Biol. 2015;11:e1004139. doi: 10.1371/journal.pcbi.1004139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cattin M.E., Bertrand A.T., Schlossarek S. Heterozygous LmnadelK32 mice develop dilated cardiomyopathy through a combined pathomechanism of haploinsufficiency and peptide toxicity. Hum Mol Genet. 2013;22:3152–3164. doi: 10.1093/hmg/ddt172. [DOI] [PubMed] [Google Scholar]
- 30.Bozkurt B., Colvin M., Cook J. Current diagnostic and treatment strategies for specific dilated cardiomyopathies: a scientific statement from the American Heart Association. Circulation. 2016;134:e579–e646. doi: 10.1161/CIR.0000000000000455. [DOI] [PubMed] [Google Scholar]
- 31.Pinto Y.M., Elliott P.M., Arbustini E. Proposal for a revised definition of dilated cardiomyopathy, hypokinetic non-dilated cardiomyopathy, and its implications for clinical practice: a position statement of the ESC working group on myocardial and pericardial diseases. Eur Heart J. 2016;37:1850–1858. doi: 10.1093/eurheartj/ehv727. [DOI] [PubMed] [Google Scholar]
- 32.Marcus F.I., McKenna W.J., Sherrill D. Diagnosis of arrhythmogenic right ventricular cardiomyopathy/dysplasia: proposed modification of the Task Force criteria. Circulation. 2010;121:1533–1541. doi: 10.1161/CIRCULATIONAHA.108.840827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deva D.P., Hanneman K., Li Q. Cardiovascular magnetic resonance demonstration of the spectrum of morphological phenotypes and patterns of myocardial scarring in Anderson-Fabry disease. J Cardiovasc Magn Reson. 2016;18:14. doi: 10.1186/s12968-016-0233-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Etesami M., Gilkeson R.C., Rajiah P. Utility of late gadolinium enhancement in pediatric cardiac MRI. Pediatr Radiol. 2016;46:1096–1113. doi: 10.1007/s00247-015-3526-2. [DOI] [PubMed] [Google Scholar]
- 35.van Spaendonck-Zwarts K.Y., van Rijsingen I.A., van den Berg M.P. Genetic analysis in 418 index patients with idiopathic dilated cardiomyopathy: overview of 10 years' experience. Eur J Heart Fail. 2013;15:628–636. doi: 10.1093/eurjhf/hft013. [DOI] [PubMed] [Google Scholar]
- 36.Jansweijer J.A., Nieuwhof K., Russo F. Truncating titin mutations are associated with a mild and treatable form of dilated cardiomyopathy. Eur J Heart Fail. 2017;19:512–521. doi: 10.1002/ejhf.673. [DOI] [PubMed] [Google Scholar]
- 37.van Tintelen J.P., Tio R.A., Kerstjens-Frederikse W.S. Severe myocardial fibrosis caused by a deletion of the 5' end of the lamin A/C gene. J Am Coll Cardiol. 2007;49:2430–2439. doi: 10.1016/j.jacc.2007.02.063. [DOI] [PubMed] [Google Scholar]
- 38.Holmstrom M., Kivisto S., Helio T. Late gadolinium enhanced cardiovascular magnetic resonance of lamin A/C gene mutation related dilated cardiomyopathy. J Cardiovasc Magn Reson. 2011;13:30. doi: 10.1186/1532-429X-13-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanna T., Dello Russo A., Toniolo D. Cardiac features of Emery-Dreifuss muscular dystrophy caused by lamin A/C gene mutations. Eur Heart J. 2003;24:2227–2236. doi: 10.1016/j.ehj.2003.09.020. [DOI] [PubMed] [Google Scholar]
- 40.Liu J., Lee K.K., Segura-Totten M. MAN1 and emerin have overlapping function(s) essential for chromosome segregation and cell division in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2003;100:4598–4603. doi: 10.1073/pnas.0730821100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gonzalez Y., Saito A., Sazer S. Fission yeast Lem2 and Man1 perform fundamental functions of the animal cell nuclear lamina. Nucleus. 2012;3:60–76. doi: 10.4161/nucl.18824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morales-Martinez A., Dobrzynska A., Askjaer P. Inner nuclear membrane protein LEM-2 is required for correct nuclear separation and morphology in C. elegans. J Cell Sci. 2015;128:1090–1096. doi: 10.1242/jcs.164202. [DOI] [PubMed] [Google Scholar]
- 43.Thanisch K., Song C., Engelkamp D. Nuclear envelope localization of LEMD2 is developmentally dynamic and lamin A/C dependent yet insufficient for heterochromatin tethering. Differentiation. 2017;94:58–70. doi: 10.1016/j.diff.2016.12.002. [DOI] [PubMed] [Google Scholar]
- 44.de Montes Oca R., Andreassen P.R., Wilson K.L. Barrier-to-autointegration factor influences specific histone modifications. Nucleus. 2011;2:580–590. doi: 10.4161/nucl.2.6.17960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berk J.M., Tifft K.E., Wilson K.L. The nuclear envelope LEM-domain protein emerin. Nucleus. 2013;4:298–314. doi: 10.4161/nucl.25751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barrales R.R., Forn M., Georgescu P.R. Control of heterochromatin localization and silencing by the nuclear membrane protein Lem2. Genes Develop. 2016;30:133–148. doi: 10.1101/gad.271288.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Braun S., Barrales R.R. Beyond tethering and the LEM domain: MSCellaneous functions of the inner nuclear membrane Lem2. Nucleus. 2016;7:523–531. doi: 10.1080/19491034.2016.1252892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dimri G.P., Lee X., Basile G. A biomarker that identifies senescent human-cells in culture and in aging skin in-vivo. Proc Natl Acad Sci U S A. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Narita M., Nunez S., Heard E. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003;113:703–716. doi: 10.1016/s0092-8674(03)00401-x. [DOI] [PubMed] [Google Scholar]
- 50.Zhang R., Zhang R.G., Poustovoitov M.V. Formation of macroH2A-containing senescence associated heterochromatin foci (SAHF) and senescence driven by ASF1A and HIRA. Gerontologist. 2005;45:46. doi: 10.1016/j.devcel.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 51.Swanson E.C., Rapkin L.M., Bazett-Jones D.P. Unfolding the story of chromatin organization in senescent cells. Nucleus. 2015;6:254–260. doi: 10.1080/19491034.2015.1057670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun L., Yu R., Dang W. Chromatin architectural changes during cellular senescence and aging. Genes. 2018;9 doi: 10.3390/genes9040211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kosar M., Bartkova J., Hubackova S. Senescence-associated heterochromatin foci are dispensable for cellular senescence, occur in a cell type- and insult-dependent manner and follow expression of p16(ink4a) Cell Cycle. 2011;10:457–468. doi: 10.4161/cc.10.3.14707. [DOI] [PubMed] [Google Scholar]
- 54.Parry A.J., Narita M. Old cells, new tricks: chromatin structure in senescence. Mamm Genome. 2016;27:320–331. doi: 10.1007/s00335-016-9628-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chandra T., Kirschner K., Thuret J.Y. Independence of repressive histone marks and chromatin compaction during senescent heterochromatic layer formation. Molecular Cell. 2012;47:203–214. doi: 10.1016/j.molcel.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chandra T., Ewels P.A., Schoenfelder S. Global reorganization of the nuclear landscape in senescent cells. Cell Reports. 2015;10:471–483. doi: 10.1016/j.celrep.2014.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Corpet A., Stucki M. Chromatin maintenance and dynamics in senescence:a spotlight on SAHF formation and the epigenome of senescent cells. Chromosoma. 2014;123:423–436. doi: 10.1007/s00412-014-0469-6. [DOI] [PubMed] [Google Scholar]
- 58.Childs B.G., Baker D.J., Kirkland J.L. Senescence and apoptosis:dueling or complementary cell fates? EMBO Reports. 2014;15:1139–1153. doi: 10.15252/embr.201439245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.He S., Sharpless N.E. Senescence in health and disease. Cell. 2017;169:1000–1011. doi: 10.1016/j.cell.2017.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chang J., Wang Y., Shao L. Clearance of senescent cells by ABT263 rejuvenates aged hematopoietic stem cells in mice. Nature Med. 2016;22:78–83. doi: 10.1038/nm.4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhu Y., Tchkonia T., Fuhrmann-Stroissnigg H. Identification of a novel senolytic agent, navitoclax, targeting the Bcl-2 family of anti-apoptotic factors. Aging Cell. 2016;15:428–435. doi: 10.1111/acel.12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Erwig L.P., Henson P.M. Clearance of apoptotic cells by phagocytes. Cell Death Differentiation. 2008;15:243–250. doi: 10.1038/sj.cdd.4402184. [DOI] [PubMed] [Google Scholar]
- 63.UniProt. Available at: http:www.uniprot.org/uniprot/Q8NC56. Accessed February 1, 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.