Abstract
Aims
Immune checkpoint blockade has made breakthroughs in immunotherapy for glioma. However, current immunotherapy has therapeutic benefits only in a subset of patients and accompanied by immune‐related side effects. SLAMF8 is a costimulatory molecule that affects the activation of macrophages in inflammation. The study of SLAMF8 may provide new information for immunological research and treatment of glioma.
Methods
CGGA and TCGA cohorts of 946 patients with RNA sequencing data and full clinical information were analyzed using R language and GraphPad Prism 7.
Results
SLAMF8 was overexpressed along with malignancy progression and was a biomarker of mesenchymal subtype. As an independent prognostic factor, high SLAMF8 conferred reduced overall survival and chemotherapy resistance. SLAMF8 implied lower proportion of cancer cells along with increasing enrichment of monocytic lineage, myeloid dendritic cells. Functional analysis showed higher SLAMF8 indicated activation of antigen processing and presenting and the IFN‐γ/TNF/TLR‐mediated signaling. Meanwhile, coexpressing with classical checkpoint SLAMF8 aggravated immunosuppression and enhanced inflammation response.
Conclusion
Our study highlighted the important role of SLAMF8 in malignancy progression, shortened survival, and immune disorders. Further research on SLAMF8 in immunosuppression and inflammation response to glioma cells could aid immunotherapy for glioma.
Keywords: checkpoint, glioma, microenvironment, prognosis, SLAMF8
1. INTRODUCTION
Glioma is the most common and aggressive type of primary tumor in the central nervous system (CNS).1 The current standard of care treatment is debulking surgery followed by radiotherapy with concomitant and adjuvant temozolomide.2 Despite this multimodal approach, patients with glioblastoma (GBM), the most aggressive type of glioma, still have a limited median survival of 15 months.3 Recently, immunotherapy, including chimeric antigen receptor (CAR) T‐cell therapy, dendritic cell (DC) therapy, and checkpoint blockade, has brought new light to patients with glioma.4 Further exploration of tumor immunity is ongoing for developing effective immunotherapy approaches for glioma.
Immune checkpoints are costimulatory or coinhibitory molecules required for a productive immune response.5 Blockade of coinhibitory checkpoint molecules such as CTLA‐4, PD‐1, and PD‐L1 has resulted in breakthroughs for multiple malignancies,6 including glioma.7 However, checkpoint blockade has therapeutic benefit only for subsets of glioma.8 Treatment by checkpoint blockade is often accompanied by inflammation that is responsible for immune‐related side effects such as dermatitis, colitis, inflammatory endocrinopathies, and even fatal cerebral edema.8, 9, 10 Given the limited regenerative capacity of neuronal tissue, adverse events from CNS inflammation of brain parenchyma are particularly deleterious.8 Therefore, balancing immunization activation and inflammation inhibition is a top priority during glioma immunotherapy. From this point of view, learning about immune checkpoints involved in inflammation may have implications for current checkpoint treatment.
Signaling lymphocytic activation molecule family 8 (SLAMF8, CD353) is the eighth member of SLAMF costimulatory receptors, which regulate development and function of many immune cells including T lymphocytes, B cells, neutrophils, dendritic cells, macrophages, and eosinophils.11, 12, 13 SLAMF8 is reported to activate macrophages during inflammation, which are a major cellular component of glioma tissue.14 Tumor‐associated macrophages (TAMs) contribute to a supportive microenvironment for glioma expansion and influence the local immune and inflammation response.15 In addition, SLAMF8 is overexpressed in autoimmune inflammation such as inflammatory bowel disease16 and human kidney transplants,17 suggesting SLAMF8 plays significant role in immune‐related inflammation response. However, few studies have investigated the role of SLAMF8 in cancer. Accordingly, we hypothesized that SLAMF8 might be important in the immune and inflammation response of glioma. Here, we analyzed RNA sequencing (RNA‐seq) data on 946 patients in Chinese Glioma Genomic Atlas (CGGA) and The Cancer Genome Atlas (TCGA) to explore the clinical and functional effects of SLAMF8, which may provide novel insights into immune checkpoints in glioma treatment.
2. METHODS
2.1. Patient sample
A total of 946 glioma samples from CGGA and TCGA were included in our study. Samples from 310 patients with detailed clinical information were obtained from the CGGA database. The TCGA cohort consisted of 636 patients with detailed clinical and molecular information that was downloaded from public databases (https://tcga-data.nci.nih.gov/tcga/tcgaDownload.jsp). Overall survival (OS) was estimated from date of diagnosis to death or final follow‐up. Methods for sequencing, detecting IDH mutation, and MGMT promoter methylation state were described previously.18, 19
2.2. Bioinformatics analysis
Stromal score, immune score, and glioma purity were calculated as described previously to evaluate nontumor cells with the microenvironment.20 Microenvironment cell populations were used to quantify the absolute abundance of eight immune and two stromal cell populations from transcriptomic data as reference.21 Pearson correlation analysis was used to obtain genes for functional annotation. GO analysis was performed via DAVID (https://david.abcc.ncifcrt.gov/home.jsp). GSEA (https://www.broadinstitute.org/gsea/index.jsp) was performed to find differential phenotypes between patients with low and high SLAMF8. Function gene sets were obtained from the Amigo2 Web portal (https://amigo.geneontology.org/amigo/landing) and reference articles.22 Principal components analysis (PCA) and gene set variation analysis (GSVA) were used to profile patterns of the transcriptome, immune function, and inflammation attributed to low or high SLAMF8 expression.
2.3. Statistical analysis
SPSS, GraphPad Prism 7, and R 3.3.3 (https://www.r-project.org/) software were used for statistical analysis. Student's t test was used to assess differences in expression, and Pearson correlation was used to calculate correlations. Low‐ and high‐expression groups were classified according to median expression. Survival distribution was estimated using Kaplan‐Meier survival analysis, and the log‐rank test was applied to evaluate differences between stratified groups. To identify independent prognostic factors, we used univariate and multivariate Cox regression analyses. Receiver operating characteristic (ROC) curve was made using MedCalc software. Other statistical computations and figures were built with R (ggplot2, corrplot, pheatmap). Statistical significance was defined as a 2‐tailed p value <0.05.
3. RESULTS
3.1. SLAMF8 was highly expressed in glioblastoma and mesenchymal subtypes
Based on expression profiles, we found that SLAMF8 increased with malignancy progression and that higher grade gliomas expressed higher levels of SLAMF8 in CGGA. According to histopathologic classifications, GBM had the highest SLAMF8. In five molecular entities, as previously reported,20 SLAMF8 was enriched in tumors of patients with wild‐type IDH lower grade glioma (LGG) and IDH wild‐type GBM (Figure 1A). These expression trends of SLAMF8 were validated in the TCGA cohort (Figure S1A).
Figure 1.
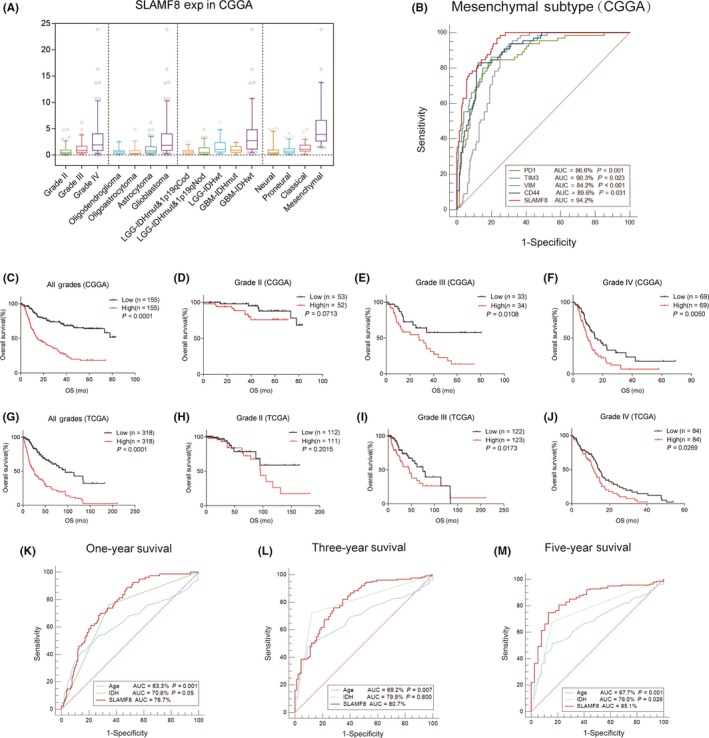
Expression profile and prognostic value of SLAMF8 in glioma. Expression of SLAMF8 differed by grade, histopathologic classification, and molecular and TCGA subtype (A). SLAMF8 was specifically enriched in mesenchymal subtype (B) in CGGA. SLAMF8 influenced glioma prognosis in CGGA (C‐F) and TCGA (G‐J) cohorts. SLAMF8 predicted 1‐year, 3‐year, and 5‐year of survival in CGGA (K‐M)
The TCGA subtyping scheme classifies glioma into four subtypes.23 We found that SLAMF8 had the highest expression in the mesenchymal subtype compared with the other subtypes in the CGGA and TCGA cohorts (Figures 1A and S1A). To validate this finding, we used ROC curves to test the predictive ability of SLAMF8 to determine mesenchymal subtype. The area under curve (AUC) was up to 94.2% in CGGA data (Figure 1B) and 93.1% in TCGA data (Figure S1B), which indicated SLAMF8 was more accurate than the mesenchymal subtype determination of PD‐1, TIM‐3, CD44, and VIM. These results indicated that SLAMF8 was highly expressed specifically in gliomas with aggressive phenotypes.
3.2. High SLAMF8 expression indicates unfavorable prognosis in glioma
To examine the prognostic value of SLAMF8, patients were separated based on median expression and survival curves were generated. Patients with higher SLAMF8 generally had shorter survival than those with lower SLAMF8 levels (Figure 1C). Stratified survival analysis was conducted based on glioma grade. Although tests for grade II failed to achieve statistical significance, high SLAMF8 still suggested inferior outcome in most analyses (Figures 1D‐F and S1C). Similar analysis using the TCGA cohort verified that high SLAMF8 conferred poor prognosis (Figures 1G‐J and S1D). Considering the prognostic significance of SLAMF8, we generated ROC curves to assess the predictive value of SLAMF8 on 1‐year, 3‐year, and 5‐year survival of the CGGA cohort. AUC was 76.7% for 1 year, 80.7% for 3 years, and 85.1% for 5 years, which was more accurate than prediction using age or IDH status (Figure 1K‐M).
Cox regression analysis was used to test the prognostic independence of SLAMF8. Univariate and multivariate analysis indicated that SLAMF8 expression level was an independent prognostic factor in glioma (HR = 1.074, P = 0.008; Table 1). SLAMF8 also independently indicated poor prognosis in the TCGA cohort (HR = 1.195, P = 0.001; Table S1). These findings suggested SLAMF8 had an important function in determining glioma prognosis.
Table 1.
Univariate and multivariate analysis of clinical prognostic parameters in CGGA
| Variable | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
| HR | P value | HR | P value | |
| SLAMF8 expression | 1.165 | <0.0001 | 1.074 | 0.008 |
| Age at diagnosis | 1.038 | <0.0001 | 1.012 | 0.119 |
| WHO grade | 3.480 | <0.0001 | 2.407 | <0.0001 |
| KPS score | 0.972 | <0.0001 | 1.001 | 0.530 |
| IDH status | 0.228 | <0.0001 | 0.953 | 0.848 |
| 1p19q codeletion | 0.135 | <0.0001 | 0.324 | 0.002 |
| MGMT promoter status | 0.526 | 0.0003 | 1.172 | 0.618 |
| Radiotherapy | 0.429 | <0.0001 | 0.606 | 0.119 |
3.3. SLAMF8 has different values for predicting glioma prognosis for different molecular subtypes
The status of IDH mutations, MGMT promoter methylation, and 1p19q codeletion has significant biological and clinical value in glioma.24, 25, 26 We investigated the prognostic role of SLAMF8 in LGG and GBM subtypes stratified by these characteristics. In LGG, no matter patients were IDH‐mutant or wild‐type, and increased SLAMF8 indicated shorter survival time (CGGA Figure 2A,B, TCGA Figure S2A,B). For LGG with intact 1p19q, higher SLAMF8 expression level meant shorter survival time. In patients with 1p19q codeletion, the prognostic value of SLAMF8 was not significant (Figures 2C,D, S2C,D).
Figure 2.
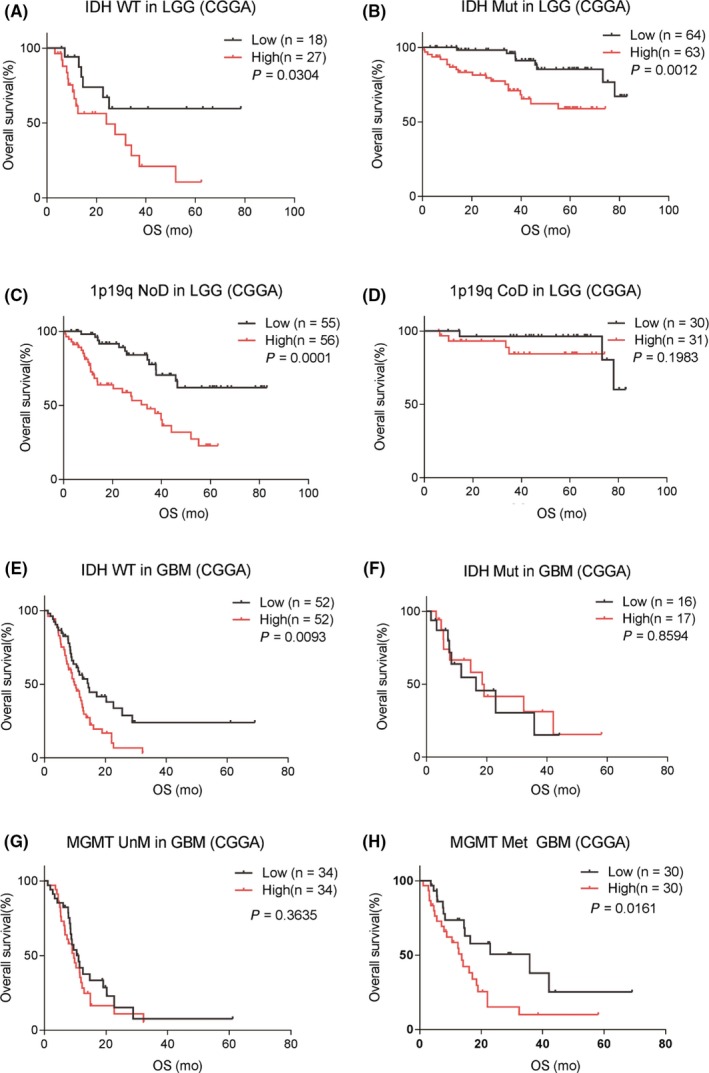
Prognostic value of SLAMF8 in molecule subtypes in CGGA. In LGG, SLAMF8 was prognostically significant for IDH wild‐type and mutant groups (A and B). SLAMF8 correlated with prognosis in patients with intact but not codeleted 1p19q (C and D). In GBM, high SLAMF8 indicated poor prognosis with IDH wild‐type and MGMT methylation status, but not IDH mutation and MGMT unmethylation status (E‐H)
In GBMs, there was an obvious difference in survival time between SLAMF8 low and high groups in IDH wild‐type patients, but not in IDH‐mutant patients (CGGA Figure 2E,F, TCGA Figure S2E,F). In GBM with unmethylated MGMT promoter, no survival difference was observed between low and high SLAMF8 expression. But for MGMT‐methylated GBM, the prognosis was different attributing to various SLAMF8 expression (CGGA Figure 2G,H, TCGA Figure S2G,H).
3.4. SLAMF8 is associated with chemotherapy in GBM
Radiotherapy and chemotherapy are standard adjuvant treatments for GBM. Using GBM in the CGGA cohort, we investigated the relationship between SLAMF8 and prognosis under various treatments. For patients without radiotherapy or chemotherapy, the prognosis was similar between low‐ and high‐SLAMF8 groups. However, SLAMF8 showed prognostic significance for patients who had radiotherapy or chemotherapy (Figure S3I‐M). We divided patients with GBM based on SLAMF8 expression. We found that in low‐SLAMF8 group patients survived significantly longer with radiochemotherapy compared to patients who received radiation alone. However, this therapeutic benefit was not significant in patients with higher SLAMF8 (Figure 3A,B).
Figure 3.
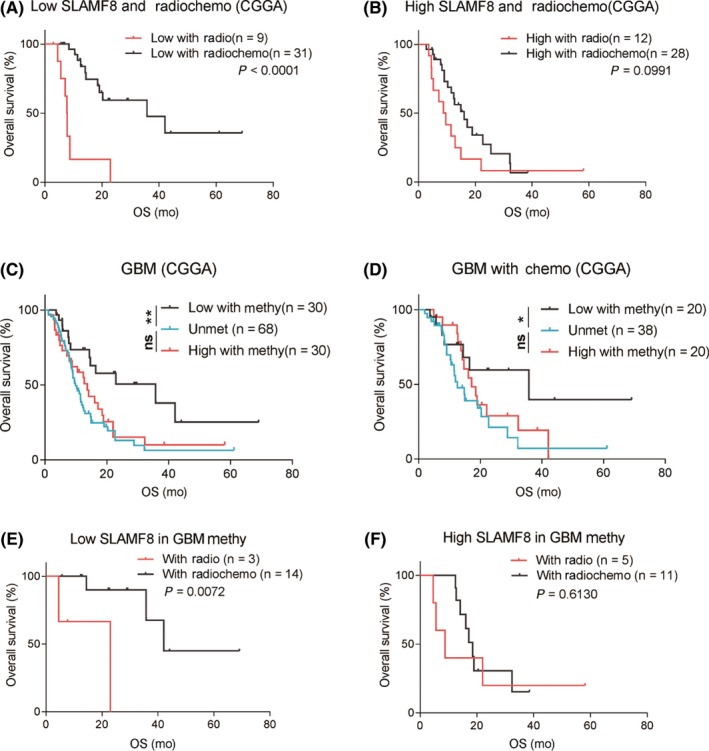
Influence of SLAMF8 on response to radiotherapy or chemotherapy. Patients receiving radiochemotherapy had better prognosis than those with radiotherapy in the low‐SLAMF8 but not high‐SLAMF8 group (A and B). High‐SLAMF8 patients with tumors with MGMT methylation had similar survival to patients with tumors with MGMT unmethylation. Patients with tumors with low SLAMF8 survived longer than patients with GBM with MGMT unmethylation (C). Similar results for GBM patients with chemotherapy (D). For GBM with MGMT methylation and low SLAMF8, patients had better prognosis with radiochemotherapy than radiotherapy (E). Similar overall survival was seen in high‐SLAMF8 group (F)
GBMs with methylated MGMT promoter are more likely to have better prognosis and benefit from chemotherapy.27, 28 We generated survival curves based on MGMT promoter status, treatment, and SLAMF8. For all GBMs or GBMs who received chemotherapy, only the MGMT promoter‐methylated patients with lower expression of SLAMF8 had a survival advantage over the unmethylated ones. Nevertheless, survival time of MGMT promoter‐methylated patients with higher SLAMF8 was just similar to that of unmethylated patients (Figure 3C,D). Further analysis showed that even GBMs with methylated MGMT promoters, only patients with lower SLAMF8 benefited from chemotherapy (Figure 3E,F). These results indicated that high SLAMF8 may confer resistance to chemotherapy.
3.5. SLAMF8 influenced glioma purity and local immune cell populations
To determine the influence of SLAMF8 on the tumor microenvironment, we calculated glioma immune score, stromal score, and purity in CGGA and TCGA cohorts. SLAMF8 positively correlated with immune score and stromal score, but negatively correlated with glioma purity (Figures 4A and S3G‐L). Even stratified by grade, their relationship remained significant (Figure S3A‐F).
Figure 4.
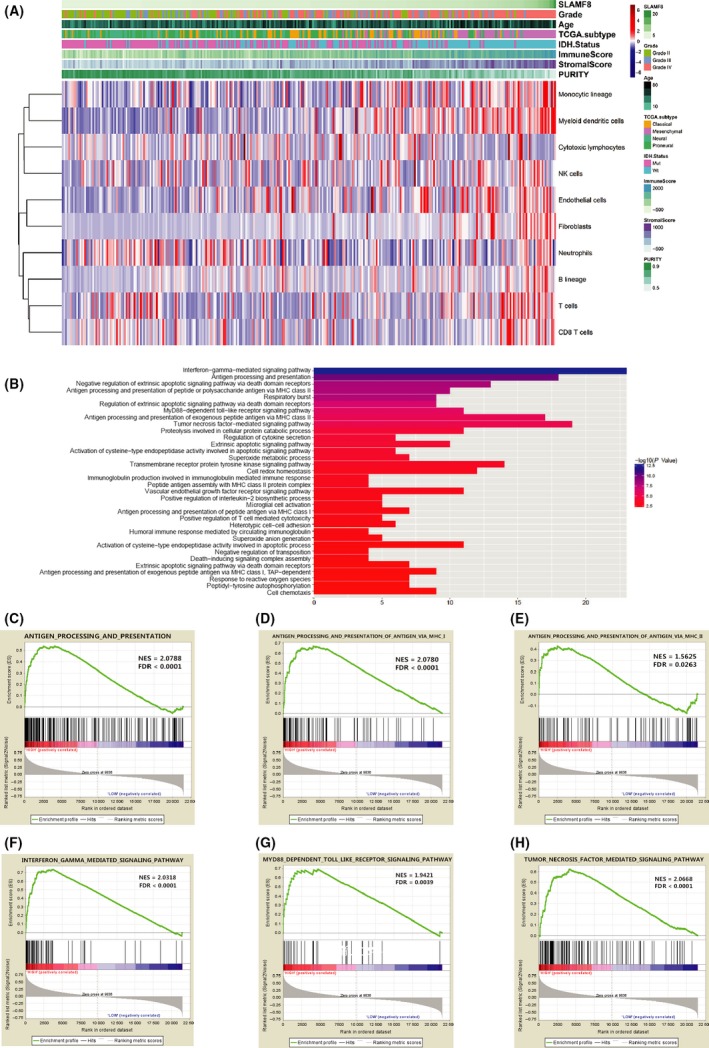
Microenvironment and biological function of SLAMF8. High SLAMF8 correlated with low purity, monocytic lineage, and myeloid dendritic cells and fibroblasts (A). DAVID analysis showed biological functions of genes correlated with SLAMF8 (B). GSEA validated biological processes related to SLAMF8 in CGGA (C‐H)
To explore differences in recruited nontumor cells leading to various microenvironment compositions, we used the microenvironment cell population counter method described by Becht.21 SLAMF8 was strongly correlated with enrichment of monocytic lineage (r = 0.462), myeloid dendritic cells (r = 0.490), and fibroblasts (r = 0.444) (Figure 4A). This indicated that high‐SLAMF8 gliomas developed more stroma composition and recruited more mononuclear macrophages and dendritic cells leading to decreasing purity of glioma cells.
3.6. SLAMF8 is involved in immunity and inflammation‐related functions
To explore the functional implications of SLAMF8, we carried out PCA to study the transcriptomic features associated with SLAMF8. Whole transcriptome expression profiles were different between patients with tumors with high and low SLAMF8 (Figure S4A‐B), implying distinct general biological phenotypes attributed to SLAMF8 expression.
To clarify the most important biological implications attributing to various SLAMF8 expression, we investigated 1365 genes that were strongly correlated with SLAMF8 (Pearson |r| ≥ 0.4) in both CGGA and TCGA databases. These genes were enriched in antigen processing and presenting and IFN‐γ/TNF/TLR‐mediated signaling pathways that were involved in generating immune and inflammation responses (Figure 4B). GSEA was used to verify the biological function of SLAMF8, showing that high SLAMF8 group had an activated phenotype for antigen processing and presenting and IFN‐γ/TNF/TLR‐mediated signaling pathways (Figure 4C‐H and S4C‐J). These results suggested that SLAMF8 was important in immune and inflammation responses in glioma.
3.7. SLAMF8 reinforced immunosuppression of glioma
To distinguish immune responses attributed to SLAMF8 expression, we analyzed gene sets on T cell‐mediated immune response and immune response to tumor cell (Tables S2 and S3). PCA analysis showed that these sets of immune response were generally different based on SLAMF8 expression status (Figure 5A‐D). GSEA confirmed that high SLAMF8 correlated with an enhanced T cell‐mediated immune phenotype (Figure S5A‐F).
Figure 5.
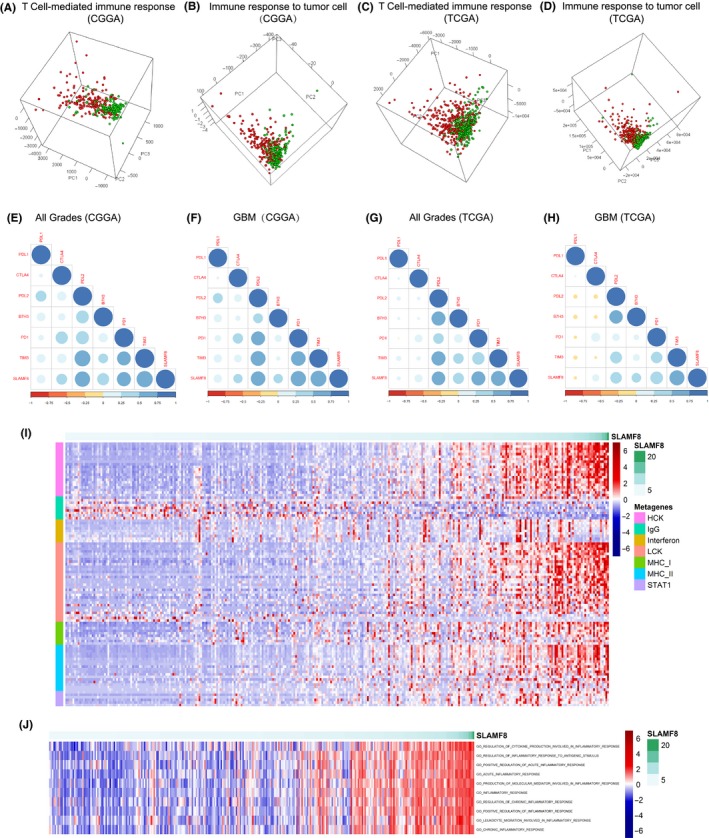
Immune and inflammation response related to SLAMF8. T cell‐mediated immune response and immune response to tumor cell were dissimilar in SLAMF8 low and high groups in CGGA and TCGA (A‐D). Classical checkpoints correlated with SLAMF8 (E‐H).Genes involved in inflammation response correlated with SLAMF8 in CGGA cohort (I). Inflammation process, including acute and chronic inflammation, was affected by SLAMF8 in CGGA (J)
To further clarify the relationship between SLAMF8 and antitumor immune reactions, we analyzed the correlations between SLAMF8 and several coinhibitory checkpoints, including PD‐L1, CTLA‐4, PD‐1, PDL‐2, B7‐H3, and TIM‐3. SLAMF8 had a strong positive correlation with CTLA‐4, PD‐1, PD‐L2, B7‐H3, and TIM‐3 in CGGA and TCGA cohorts. However, SLAMF8 had little correlation with PD‐L1. The same analysis was carried out for GBM, with similar results (Figure 5E‐H). These results suggested SLAMF8 involved in suppressing effective antitumor immune reactions, which differed from the function of PD‐L1.
3.8. High expression of SLAMF8 aggravates inflammation activity in glioma
As inflammation responses might reinforce immunosuppression and affect tumor progress,29 we examined inflammation status for different expression levels of SLAMF8. We used metagenes to represent different types of inflammation and immune responses.22 Genes of most clusters, HCK, LCK, MHC‐I, MHC‐II, STAT1, and interferon were overexpressed in high‐SLAMF8 gliomas. Only IgG, which associated with activities of B lymphocytes, was enriched in low‐SLAMF8 glioma (CGGA Figure 5I, TCGA Figure S5G). Based on GO terms involved in the inflammation response, GSVA was performed to determine the relationship between SLAMF8 and inflammation more clearly. SLAMF8 had a close relationship to acute inflammation and chronic inflammation (CGGA Figure 5J, TCGA Figure S5H). The results indicated that SLAMF8 expression was decisive for promoting the inflammation response involved in glioma.
4. DISCUSSION
Immunotherapy is a promising treatment with challenges.10, 30 We found a costimulatory checkpoint molecule, SLAMF8, was important in clinical, molecular, and biological situations of glioma. SLAMF8 was highly expressed in gliomas with malignant entities and specifically enriched in IDH wild‐type and mesenchymal subtypes, which are recognized as more aggressive subtypes of glioma with enhanced immune responses.31, 32 These findings indicated that SLAMF8 was commonly highly expressed in glioma with active immune and inflammation responses. Compared with some markers of mesenchymal subtype, SLAMF8 was more accurate in discriminating patients of mesenchymal tumor. Therefore, SLAMF8 could be an accurate molecular predictor of mesenchymal subtype and play important role in facilitating the malignant phenotype of glioma.
Previously, our team found an immune‐related risk signature has important prognostic value in glioma.33 Similarly, as an independent prognostic risk factor, the immune checkpoint SLAMF8 influenced the overall survival of glioma that higher SLAMF8 correlated with shorter survival. IDH mutation is an important molecular event in glioma management that is not frequent in primary GBM.34, 35 We found that SLAMF8 remained prognostic significant for GBMs with wild‐type IDH but not mutant IDH. IDH wild‐type GBM is known to have more immune cell infiltration and enhanced immune responses than IDH‐mutant ones.32, 35 Therefore, SLAMF8 may have less influence on immune and inflammation responses for IDH mutants than IDH wild‐type GBMs. These findings suggested that the influence of SLAMF8 depended on an active immune phenotype.
Combining immunotherapy with current treatment is a promising approach for gliomas. More information is needed to facilitate individualized treatments. Our data showed that SLAMF8 determined survival only for patients with radiotherapy or chemotherapy. This result may be because radiotherapy or chemotherapy killed tumor cells to release tumor antigens that were essential for immune response and inflammation.36, 37 SLAMF8 would then promote immune and inflammation responses, resulting in poor prognosis. We also found that high‐SLAMF8 gliomas did not benefit from adjuvant chemotherapy. This finding may be due to the enhanced inflammatory phenotype resulting from elevated SLAMF8. Therefore, patients with high‐SLAMF8 tumors need aggressive treatment and might benefit from chemotherapy combined with blockade against SLAMF8.
Glioma purity and infiltrated immune cell components are reported to have important clinical and biological implications. Lower purity means malignancy progression, poor prognosis, and enhanced immune phenotype.20 SLAMF8 was closely related to immune score, stromal score, and glioma purity. This result suggested that high‐SLAMF8 gliomas exist in tissues with more complex microenvironments. Consistently, we found that SLAMF8 positively correlated with monocytic lineage (including monocytes and macrophages) and myeloid dendritic cells in the microenvironment. TAMs are the most abundant immune cells within glioma tissue. They create a supportive stroma for neoplastic cell expansion and invasion.17, 38 High TAM enrichment in gliomas often indicates poor prognosis.36 This result might explain why high‐SLAMF8 gliomas had malignant progress and adverse outcomes.
By analyzing the biological functions of SLAMF8 in gliomas, we found that SLAMF8 was important in antigen processing and presenting and IFN‐γ/TNF/TLR‐mediated signaling pathways, which are involved in tumor immunity and inflammation. Antigen processing and presenting are the main immunologic function of dendritic cells and macrophages and are necessary processes for promoting the immune response.39 Higher SLAMF8 was associated with more active T cell‐mediated immune response, which was in line with our previous findings that high‐risk GBM correlated with enhanced local immune phenotype.33 However, SLAMF8 was also positively associated with several suppressive checkpoints such as TIM‐3, CTLA‐4, PD‐1, B7‐H3, and PD‐L2, which led to an impaired antitumor immune phenotype. Meanwhile, patients with higher SLAMF8 recruited more TAMs to disturb effective antitumor immunity and to generate inflammation. Therefore, even though SLAMF8 facilitated antigen presenting and local immune response, effective antitumor immunity was still suppressed. An overloaded and ineffective immune response may lead to inflammation and immune‐related side effects.
Inflammation accompanying an immune response is a major challenge in applying immune checkpoint inhibitors in glioma. Enhanced inflammation aggravates malignant progression, reduces the effectiveness of adjuvant treatment, and can lead to fatal cerebral damage.40, 41, 42 Therefore, we assumed that synergistically targeting coinhibitory and costimulatory checkpoints could be a reasonable approach to promoting antitumor immunity without stimulating excessive inflammation. SLAMF8 could be a suitable adjuvant target, especially with inhibitors targeting PD‐L1, which showed an immunosuppressive function distinct from SLAMF8.
5. CONCLUSION
We examined large glioma cohorts and conducted systematic analyses in multidimensional conditions. We found that SLAMF8 was associated with malignancy progression, unfavorable prognosis, and chemotherapy resistance. As a costimulatory checkpoint, SLAMF8 has an immune function distinct from coinhibitory checkpoints, implying a promising strategy might target both coinhibitory and costimulatory checkpoints in glioma.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
DISCLOSURE
No benefits in any form have been or will be received from a commercial party related directly or indirectly to the subject of this manuscript.
Supporting information
ACKNOWLEDGEMENTS
We thank all the members in Dr. Wu AH's laboratory for help with our study.
Zou C‐Y, Guan G‐F, Zhu C, et al. Costimulatory checkpoint SLAMF8 is an independent prognosis factor in glioma. CNS Neurosci Ther. 2019;25:333–342. 10.1111/cns.13041
Funding information
This work was supported by grants from the National Natural Science Foundation of China (Grant Numbers: 81172409, 81472360, and 81402045) and the Science and Technology Department of Liaoning Province (Grant Numbers: 2011225034).
Contributor Information
Wen Cheng, Email: cmu071207@163.com.
An‐Hua Wu, Email: wuanhua@yahoo.com.
REFERENCES
- 1. Ostrom QT, Bauchet L, Davis FG, et al. The epidemiology of glioma in adults: a “state of the science” review. Neuro Oncol. 2014;16:896‐913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987‐996. [DOI] [PubMed] [Google Scholar]
- 3. Thomas AA, Brennan CW, DeAngelis LM, Omuro AM. Emerging therapies for glioblastoma. JAMA Neurol. 2014;71:1437‐1444. [DOI] [PubMed] [Google Scholar]
- 4. Boussiotis VA, Charest A. Immunotherapies for malignant glioma. Oncogene. 2017;37:1121‐1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Korman AJ, Peggs KS, Allison JP. Checkpoint blockade in cancer immunotherapy. Adv Immunol. 2006;90:297‐339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Riess JW, Lara PN Jr, Gandara DR. Theory Meets Practice for Immune Checkpoint Blockade in Small‐Cell Lung Cancer. J Clin Oncol. 2016;34:3717‐3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Preusser M, Lim M, Hafler DA, Reardon DA, Sampson JH. Prospects of immune checkpoint modulators in the treatment of glioblastoma. Nat Rev Neurol. 2015;11:504‐514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lucca LE, Hafler DA. Co‐inhibitory blockade while preserving tolerance: checkpoint inhibitors for glioblastoma. Immunol Rev. 2017;276:9‐25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhu X, McDowell MM, Newman WC, Mason GE, Greene S, Tamber MS. Severe cerebral edema following nivolumab treatment for pediatric glioblastoma: case report. J Neurosurg Pediatr. 2017;19:249‐253. [DOI] [PubMed] [Google Scholar]
- 10. Curry WT, Lim M. Immunomodulation: checkpoint blockade etc. Neuro Oncol 2015;17(Suppl. 7):vii26–vii31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Detre C, Keszei M, Romero X, Tsokos GC, Terhorst C. SLAM family receptors and the SLAM‐associated protein (SAP) modulate T cell functions. Semin Immunopathol. 2010;32:157‐171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Romero X, Sintes J, Engel P. Role of SLAM family receptors and specific adapter SAP in innate‐like lymphocytes. Crit Rev Immunol. 2014;34:263‐299. [DOI] [PubMed] [Google Scholar]
- 13. De Calisto J, Wang N, Wang G, Yigit B, Engel P, Terhorst C. SAP‐Dependent and ‐Independent Regulation of Innate T Cell Development Involving SLAMF Receptors. Front Immunol. 2014;5:186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang G, Abadia‐Molina AC, Berger SB, et al. Cutting edge: Slamf8 is a negative regulator of Nox2 activity in macrophages. J Immunol. 2012;188:5829‐5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hambardzumyan D, Gutmann DH, Kettenmann H. The role of microglia and macrophages in glioma maintenance and progression. Nat Neurosci. 2016;19:20‐27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. de Lange KM, Moutsianas L, Lee JC, et al. Genome‐wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat Genet. 2017;49:256‐261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Venner JM, Famulski KS, Badr D, Hidalgo LG, Chang J, Halloran PF. Molecular landscape of T cell‐mediated rejection in human kidney transplants: prominence of CTLA4 and PD ligands. Am J Transplant. 2014;14:2565‐2576. [DOI] [PubMed] [Google Scholar]
- 18. Cheng W, Li M, Jiang Y, et al. Association between small heat shock protein B11 and the prognostic value of MGMT promoter methylation in patients with high‐grade glioma. J Neurosurg. 2016;125:7‐16. [DOI] [PubMed] [Google Scholar]
- 19. Cheng W, Li M, Cai J, et al. HDAC4, a prognostic and chromosomal instability marker, refines the predictive value of MGMT promoter methylation. J Neurooncol. 2015;122:303‐312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang C, Cheng W, Ren X, et al. Tumor Purity as an Underlying Key Factor in Glioma. Clin Cancer Res. 2017;23:6279‐6291. [DOI] [PubMed] [Google Scholar]
- 21. Becht E, Giraldo NA, Lacroix L, et al. Estimating the population abundance of tissue‐infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016;17:218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Motaln H, Koren A, Gruden K, Ramsak Z, Schichor C, Lah TT. Heterogeneous glioblastoma cell cross‐talk promotes phenotype alterations and enhanced drug resistance. Oncotarget. 2015;6:40998‐41017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li G, Wang Z, Zhang C, et al. Molecular and clinical characterization of TIM‐3 in glioma through 1,024 samples. Oncoimmunology. 2017;6:e1328339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lu G, Chang JT, Liu Z, Chen Y, Li M, Zhu JJ. Phospholipase C Beta 1: a Candidate Signature Gene for Proneural Subtype High‐Grade Glioma. Mol Neurobiol. 2016;53:6511‐6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Turkalp Z, Karamchandani J, Das S. IDH mutation in glioma: new insights and promises for the future. JAMA Neurol. 2014;71:1319‐1325. [DOI] [PubMed] [Google Scholar]
- 26. Lin T, Wang M, Liang HS, Liu EZ. The expression of p53, mgmt and egfr in brain glioma and clinical significance. J Biol Regul Homeost Agents. 2015;29:143‐149. [PubMed] [Google Scholar]
- 27. Hu X, Martinez‐Ledesma E, Zheng S, et al. Multigene signature for predicting prognosis of patients with 1p19q co‐deletion diffuse glioma. Neuro Oncol. 2017;19:786‐795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Melguizo C, Prados J, Gonzalez B, et al. MGMT promoter methylation status and MGMT and CD133 immunohistochemical expression as prognostic markers in glioblastoma patients treated with temozolomide plus radiotherapy. J Transl Med. 2012;10:250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cheng W, Ren X, Cai J, et al. A five‐miRNA signature with prognostic and predictive value for MGMT promoter‐methylated glioblastoma patients. Oncotarget. 2015;6:29285‐29295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shalapour S, Karin M. Immunity, inflammation, and cancer: an eternal fight between good and evil. J Clin Invest. 2015;125:3347‐3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Platten M, Bunse L, Wick W, Bunse T. Concepts in glioma immunotherapy. Cancer Immunol Immunother. 2016;65:1269‐1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Carro MS, Lim WK, Alvarez MJ, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463:318‐325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Amankulor NM, Kim Y, Arora S, et al. Mutant IDH1 regulates the tumor‐associated immune system in gliomas. Genes Dev. 2017;31:774‐786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cheng W, Ren X, Zhang C, et al. Bioinformatic profiling identifies an immune‐related risk signature for glioblastoma. Neurology. 2016;86:2226‐2234. [DOI] [PubMed] [Google Scholar]
- 35. Cohen AL, Holmen SL, Colman H. IDH1 and IDH2 mutations in gliomas. Curr Neurol Neurosci Rep. 2013;13:345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cheng W, Ren X, Zhang C, Cai J, Han S, Wu A. Gene Expression Profiling Stratifies IDH1‐Mutant Glioma with Distinct Prognoses. Mol Neurobiol. 2017;54:5996‐6005. [DOI] [PubMed] [Google Scholar]
- 37. Tanaka F, Yamaguchi H, Ohta M, et al. Intratumoral injection of dendritic cells after treatment of anticancer drugs induces tumor‐specific antitumor effect in vivo. Int J Cancer. 2002;101:265‐269. [DOI] [PubMed] [Google Scholar]
- 38. Shin JY, Lee SK, Kang CD, et al. Antitumor effect of intratumoral administration of dendritic cell combination with vincristine chemotherapy in a murine fibrosarcoma model. Histol Histopathol. 2003;18:435‐447. [DOI] [PubMed] [Google Scholar]
- 39. Squadrito ML, De Palma M. A niche role for periostin and macrophages in glioblastoma. Nat Cell Biol. 2015;17:107‐109. [DOI] [PubMed] [Google Scholar]
- 40. Vyas JM, Van der Veen AG, Ploegh HL. The known unknowns of antigen processing and presentation. Nat Rev Immunol. 2008;8:607‐618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Boye K, Pujol N, D Alves, I . et al. The role of CXCR40/LRP1 cross‐talk in the invasion of primary brain tumors. Nat Commun. 2017;8;1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stanzani E, Martinez‐Soler F, Mateos TM, et al. Radioresistance of mesenchymal glioblastoma initiating cells correlates with patient outcome and is associated with activation of inflammatory program. Oncotarget. 2017;8:73640‐73653. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


