Abstract
Aims
The pyrosequencing (PSQ) has been regarded as the gold standard for MGMT promoter methylation testing in gliomas. However, various CpG combinations are currently used in clinical practice. We aimed to clarify how and how many CpGs combined is robust enough to predict MGMT mRNA expression and therapeutic prognosis of patients.
Methods
Total 223 patients with WHO III/IV gliomas were enrolled from Chinese Glioma Genome Atlas, including two independent cohorts, the eight‐site cohort (with CpGs 75‐82 tested) and the seven‐site cohort (with CpGs 72‐78 tested). Spearman’s correlation and ROC curves were employed to investigate the value of different CpG combinations on predicting MGMT mRNA expression. The ROC curves and Kaplan‐Meier steps were performed to compare the TMZ therapeutic prognostic values of different CpG combinations.
Results
The methylation level of all individual CpG and CpG combinations for the eleven CpGs (CpGs 72‐82), significantly correlated to MGMT mRNA expression (Spearman, all P < 0.0001), could effectively predict the mRNA expression (AUC, 0.86‐0.91 in the eight‐site cohort, 0.83‐0.90 in the seven‐site cohort). Moreover, the correlation coefficients and the predictive values presented equivalent when four or more CpGs combinedly used (AUC, 0.88‐0.90 in the eight‐site cohort, 0.87‐0.88 in the seven‐site cohort). Finally, similar results were also observed when using selected CpG combinations to predict therapeutic prognosis of patients.
Conclusions
Four‐CpG combinations of pyrosequencing are sufficient for evaluating the methylation status of MGMT and predicting therapeutic prognosis in gliomas.
Keywords: CpGs, glioma, MGMT, pyrosequencing, temozolomide
1. INTRODUCTION
O6‐methylguanine‐DNA methyltransferase (MGMT), a ubiquitous DNA repair enzyme, can rapidly reverse alkylation at the O6 position of guanine by transferring the alkyl group to its cysteinyl residue.1, 2 Temozolomide (TMZ), a typical alkylating agent, is the first‐line chemotherapy drug for glioma. And the expression deficiency of MGMT in glioma has been acknowledged as a predictive marker for TMZ sensitivity.2 The cysteine‐phosphate‐guanine (CpG) island (CGI) in the promoter region of MGMT is susceptible to DNA methylation, which is highly related to the MGMT transcription suppression.2, 3 Thus, MGMT promoter methylation implies a TMZ sensitivity status of glioma patients, confirmed in several subsequent studies and clinical trials.4, 5, 6, 7, 8, 9, 10
Given the difficulty of detecting MGMT mRNA or protein expression directly in glioma,11 MGMT promoter methylation testing is now wildly employed in clinical practice. Total 98 CpGs situated in the MGMT CGI,12 named CpGs 1‐98 in this study according to whose location in the 762 bps (chr10: 131264949‐131265710) from the 5′‐end to the 3′‐end. In early clinical trials, the methylation‐specific PCR(MSP) was mainly used to determine the methylation status, and the primers of which were designed specifically to CpGs 76‐80 and CpGs 84‐87 fully methylated sequences, respectively.4, 5 However, along with the high heterogeneity of the CpGs methylation gradually identified,13, 14 MSP‐based methods were unable to reflect such heterogeneity.12, 15, 16 Currently, pyrosequencing (PSQ) has been developed to be a stable technique, offering a valid, reliable and quick evaluation of both fresh frozen and formalin fixed paraffin embedded (FFPE) specimens.11, 13, 14, 17, 18, 19, 20, 21, 22
Similar to the MSP method, PSQ determines the MGMT promoter methylation by the mean methylation level of several selected consecutive CpGs.11, 15, 23 The methylation statuses of CpGs 25‐50 and 72‐90 are proved highly correlated to MGMT transcription, and CpGs 72‐90 seems to play a more critical role.12, 24 However, the high heterogeneity of different CpGs’ methylation level is also recognized,12, 16, 24 how many CpGs in the MGMT CGI is robust enough to reflect the transcription status is still a controversial issue for MGMT methylation PSQ testing.14, 15
Various number of CpGs from four to eighteen has been used as CpG combinations in reported studies (Figure 1).11, 13, 14, 15, 17, 20, 25, 26 Among which, the commonly adopted combinations are CpGs 76‐79 and 74‐78, which are currently used in Qiagen commercial kits. Thus, it is critical to clarify the following issues for MGMT methylation PSQ testing. (a) whether four CpGs is robust enough in the MGMT promoter methylation PSQ testing; (b) whether different CpG combinations can provide equivalent predictive value on MGMT transcription; (c) for the commonly utilized combinations of CpGs 76‐79 and 74‐78 should be analyzed separately or in combination?
Figure 1.
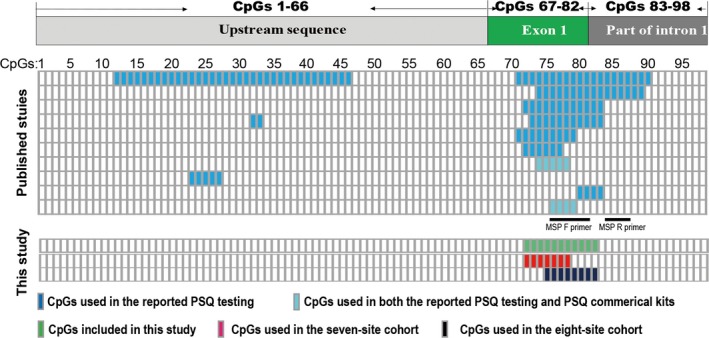
Schematic diagram of CpG studied in previous and current studies. The distribution of CpG in the MGMT 5′ CpG island is shown in the upper panel. CpGs that had been used in pyrosequencing (PSQ) testing of the published literature are shown in the middle panel. The CpGs tested in the current study are shown in the lower panel
The aim of this study was to clarify the issues mentioned above with patients from two independent cohorts, the eight‐site cohort with CpGs 75‐82 tested and the seven‐site cohort with CpGs 72‐78 tested. We used Spearman’s correlation analysis and ROC curve to compare the predictive values of individual CpG and different CpG combinations on the mRNA expression of MGMT. Moreover, we also compared the therapeutic prognosis value of several selected CpG combinations, including CpGs 76‐79 and CpGs 74‐78, in patients received TMZ treatment. Finally, our study indicated that combinations including four CpGs were robust enough in the MGMT methylation PSQ testing.
2. MATERIALS AND METHODS
2.1. Patients and samples
To study the relationship between different MGMT promoter CpGs methylation status and MGMT mRNA expression level, 159 cases were totally enrolled according to the following criteria: (a) diagnosed with WHO grade III or IV glioma; (b) containing MGMT promoter methylation PSQ testing data in detail; (c) including exact MGMT mRNA sequencing data. Within the 159 cases, of 82 contain the CpGs 75‐82 methylation information, and the other 77 contain the CpGs 72‐78 methylation information (Supporting Information Table S1).
To compare the therapeutic prognostic value of several selected CpG combinations, 86 cases with CpGs 75‐82 methylation information and 48 cases with CpGs 72‐78 methylation information were included. More than detailed CpGs methylation information, the inclusion criteria include the following: (a) diagnosed with WHO grade III or IV glioma; (b) having received radiotherapy (RT) + temozolomide (TMZ) treatment; (c) containing overall survival (OS) information (Table 1).
Table 1.
Characteristics of patients received Radiotherapy (RT) and Temozolomide (TMZ) treatment
| WHO grade | Eight‐site cohort | Seven‐site cohort | |||||
|---|---|---|---|---|---|---|---|
| Total | IV | III | Total | IV | III | ||
| Total | 86 | 51 | 35 | 48 | 24 | 24 | |
| Gender | Male | 52 (60.5%) | 33 (64.7%) | 19 (54.3%) | 32 (66.7%) | 14 (58.3%) | 18 (75.0%) |
| Female | 34 (39.5%) | 18 (35.3%) | 16 (45.7%) | 16 (33.3%) | 10 (41.7%) | 6 (25.0%) | |
| Age | Median | 43 (19‐72) | 44 (19‐72) | 42 (25‐62) | 48 (24‐79) | 55 (29‐79) | 43 (24‐65) |
| KPS | <80 | 28 (32.6%) | 16 (31.4%) | 12 (34.3%) | 16 (33.3%) | 11 (45.8%) | 5 (20.8%) |
| ≥80 | 35 (40.7%) | 21 (41.2%) | 14 (40.0%) | 20 (41.7%) | 9 (37.5%) | 11 (45.8%) | |
| NA | 26 (30.2%) | 14 (27.4%) | 12 (34.3%) | 12 (25.0%) | 4 (16.7) | 8 (33.3%) | |
| Resection | Total | 58 (67.4%) | 36 (70.6%) | 22 (62.9%) | 23 (47.9%) | 12 (50.0%) | 11 (45.8%) |
| Sub | 27 (31.4%) | 15 (29.4%) | 12 (34.3%) | 24 (50.0%) | 12 (50.0%) | 12 (50.0%) | |
| NA | 1 (1.2%) | 0 (0.0%) | 1 (2.9%) | 1 (2.1%) | 0 (0.0%) | 1 (4.2%) | |
| IDH status | Mutant | 36 (41.9%) | 12 (23.5%) | 24 (68.6%) | 15 (31.3%) | 5 (20.8%) | 10 (41.7%) |
| Wild type | 46 (53.5%) | 36 (70.6%) | 10 (28.6%) | 32 (66.7%) | 18 (75.0%) | 14 (58.3%) | |
| NA | 4 (4.6%) | 3 (5.9%) | 1 (2.9%) | 1 (2.0%) | 1 (4.2%) | 0 (0.0%) | |
The pathological grade was determined by two independent pathologists, and the freshly frozen samples with >80% tumor cells were used to determine the MGMT promotor methylation status. All the cases included in this study were assigned to the eight‐site (CpGs 75‐82) or the seven‐site (CpGs 72‐78) cohort according to the CpG sites tested. All the case information included was collected from the Chinese Glioma Genome Atlas (CGGA) database. This study was approved by the institutional review board of Beijing Tiantan Hospital.
2.2. DNA isolation and bisulfite modification
Genomic DNA was extracted from freshly frozen tumor tissue with the QIAamp DNA Mini Kit (Qiagen, Stockach, Germany). The DNA concentration and quality were determined by a Nano‐Drop ND‐1000 spectrophotometer (NanoDrop Technologies, Houston, TX, USA). And then the bisulfite conversion used 100 ng DNA and an Epitect Bisulfite kit (Qiagen) according to the manufacturer’s protocol.
2.3. PSQ testing
The template used in the MGMT PSQ testing was prepared as previously described.17, 27 Briefly, bisulfite‐treated DNA was preamplified with the primers (a) F‐primer 5′‐GTT TYG GAT ATG TTG GGA TAG TT‐3′; (b) biotinylated R‐primer 5′‐biotin‐ACR ACC CAA ACA CTC ACC AA‐3′. Different samples were analyzed with two independent assays, and the PSQ primers were (a) 5′‐GAT ATG TTG GGA TAG T‐3′ (for CpGs 72‐78); (b) 5′‐GTT TTT AGA AYG TTT TG‐3′ (for CpGs 75‐82). The sequence of CpGs 72‐78 and CpGs 75‐82 for analysis is TYG YGT TTT TAG AAY GTT TTG YGT TTY GAY GTT YGT AGG T and YGT TTT GYG TTT YGA YGT TYG TAG GTT TTY GYG GTG YGT A, respectively. PSQ testing was performed on a PyroMarker Q96 instrument, and the results were analyzed with PyroMarker Q96 software (Qiagen).
Mutations of the isocitrate dehydrogenase (IDH) 1/2 genes were also determined by PSQ, and the templates were prepared with following primers (a) F‐primer for IDH1, 5′‐GCT TGT GAG TGG ATG GGT AAA AC‐3′; (b) biotinylated R‐primer for IDH1, 5′‐biotin‐TTG CCA ACA TGA CTT ACT TGA TC‐3′; (c) F‐primer for IDH2, 5′‐ATC CTG GGG ACT GTC TT‐3′; and (d) biotinylated R‐primer for IDH2, 5′‐biotin‐CTC TCC ACC CTG GCC TAC CT‐3′. PSQ testing was performed with the primers 5′‐TGG ATG GGT AAA ACC T‐3′ for IDH1 and 5′‐AGC CCA TCA CCA TTG‐3′ for IDH2.
2.4. RNA sequencing (RNA‐seq), quality control, and mRNA expression calculation
RNA sequencing library was constructed as previously published study.28 Briefly, RNA was extracted from the frozen tissue sample, and the RNA‐seq library was constructed and subsequently sequenced on the Illumina HiSeq 2000 platform (Illumina, San Diego, CA, USA) using 101‐bp pair‐end sequencing strategy. Image data were converted into sequence data using base calling software (Illumina pipeline CASAVA v1.8.2, CA, USA) and then further estimated by standard quality control criteria.
Sequencing reads were excluded when any one of the following parameters fitted: (a) the read is aligned to adaptor or primer with no more than two mismatches; (b) the read contains over 10% unknown bases; and (c) the read includes more than 50% of low‐quality bases (quality value ≤5). The detailed information of the quality control for the whole RNA sequencing database had been described in our previous study.29
The MGMT mRNA expression level was calculated following the listed procedures: (a) mapping the reads to the Ref Seq‐RNA reference sequence set Hg 19 (RNA sequences, GRCh37, available online: https://genome.ucsc.edu); (b) evaluating mRNA expression level according to the number of reads per kilobase transcriptome per million; and (c) normalizing of the estimated gene expression between different samples.
2.5. Statistical analysis
Statistical analysis was performed with R (https://www.r-project.org/, v3.4.1), and GraphPad Prism 7 (GraphPad Software, the state of California, USA). 0.05 was chosen to be the significance cutoff for P‐value. The relationship between methylation level of CpGs and MGMT mRNA expression was analyzed by Spearman’s correlation analysis. The patients were stratified into two groups according to their MGMT mRNA expression (using the median expression as the cutoff), and the receiver operating characteristic (ROC) analysis was employed to evaluate the predictive value of MGMT promoter methylation status on the mRNA expression level by calculating the area under the curve (AUC).
The ROC analysis was also used to evaluate the predictive value of MGMT promotor methylation status for patients’ survival. The median OS was used to stratified the patients. Kaplan‐Meier step was used to estimating the survival of subgrouped patients, and the value between groups was compared by the log‐rank test.
3. RESULTS
3.1. Strategy for CpG combinations selection, and correlation of MGMT mRNA expression with single CpG or selected CpG combinations
In the eight‐site cohort, CpGs 75‐82 were tested, and the CpGs 76‐79 combination is currently used in the commercial kit (Qiagen, 970061). We aimed to (a) compare the predictive values of each individual CpG and CpG combinations, which are composed by a different number of consecutive CpGs, on MGMT mRNA expression; (b) study whether total or part of CpGs within CpGs 76‐79 should be analyzed. Thus, all the individual CpG, all the two or three CpGs composed CpG combinations within CpGs 76‐79, and all consecutive four, five, seven, and eight CpG combinations were selected for the analysis in this study (Figure 2A and Supporting Information Table S2).
Figure 2.
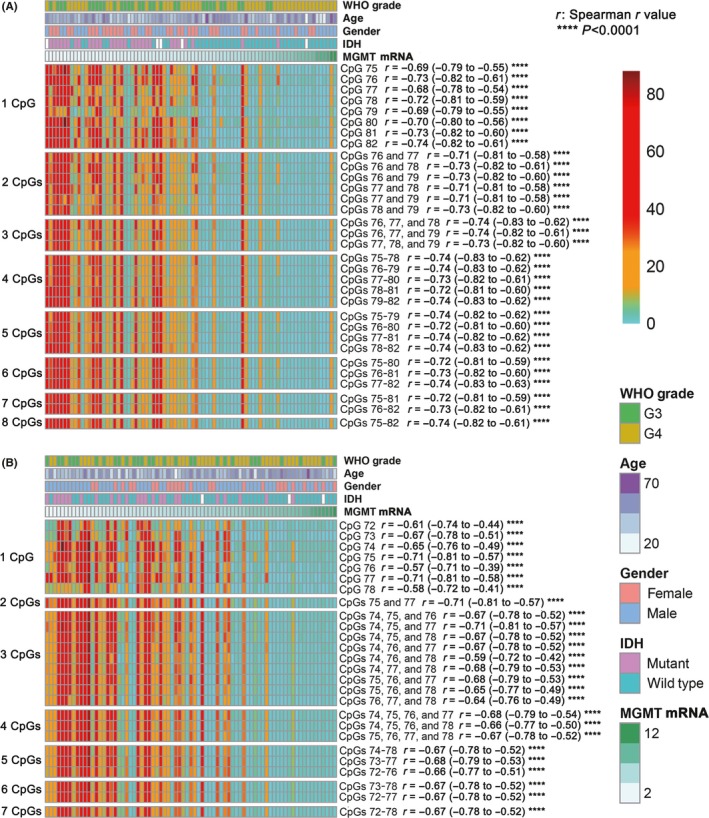
Correlation of MGMT mRNA expression to the methylation level of MGMT promoter individual CpG or CpG combinations. A and B, The methylation level of individual CpG or CpG combinations is applied to heatmap in the eight‐site cohort (A) and the seven‐site cohort (B). The methylation level of a CpG combination is the average methylation level of all CpGs included in this combination. “r” is the Spearman r value, and the 95% confidence interval of the “r” is shown in the following bracket, respectively. ****P < 0.0001
In the seven‐site cohort, CpGs 72‐78 were tested, and the CpGs 74‐78 combination is currently used in the commercial kit (Qiagen, 972032, and 970032). Based on the similar strategy used in the eight‐site cohort, all individual CpG, representative two‐CpG combination (CpGs 75 and 77), all three or four CpG combinations within CpGs 74‐78 combination, and all consecutive five, six, and seven CpG combinations were finally selected for investigation (Figure 2B and Supporting Information Table S2).
We presented the methylation level of each individual CpG and selected CpG combinations of patients included as heatmaps, in ascending order of MGMT mRNA expression (Figure 2). In the eight‐site cohort, although the methylation levels of all individual CpG and selected CpG combinations were significantly associated with the MGMT mRNA expression (P < 0.0001), the heterogeneity among individual CpG and selected CpG combinations methylation level could also be observed, and the Spearman r value deviated from ‐0.68 to ‐0.74. Meanwhile, we noticed that there was a tendency that the heterogeneity decreases among CpG combinations which included four or more CpGs, and the Spearman r values also converged to be around ‐0.73 (from ‐0.72 to ‐0.74) when the including four or more CpGs (Figure 2A). Similarly, consistent phenomena were observed in the seven‐site cohort (Figure 2B). Moreover, the combinations with only part of CpGs within CpGs 76‐79 or CpGs 74‐78 showed worse correlation with MGMT mRNA expression than that of total combination, respectively. These findings indicated that the CpG combinations with four or more than four CpGs are enough to eliminate the influence of heterogeneity among individual CpG within CpGs 72‐82. Thus, we inferred that CpG combinations with more four or more CpGs have similar predictive value on MGMT mRNA expression.
3.2. Predictive value of each individual CpG and selected CpG combinations methylation level for MGMT mRNA expression
To verify our inference, we further compared the predictive value of each individual CpG and selected CpG combinations methylation level for the MGMT mRNA expression with ROC curve (Supporting Information Table S2). The calculated AUC of each individual CpG and selected CpG combinations to range from 0.86 to 0.91 but converge to around 0.89 (from 0.88 to 0.90) when three or more CpGs combined in the eight‐site cohort (Figure 3A). Similar results could also be observed in the seven‐site cohort, and the AUCs ranging from 0.83 to 0.90 converged to 0.87 or 0.88 when four or more CpGs combined (Figure 3B). This finding confirmed our inference that CpG combinations with four or more CpGs have similar predictive value on the MGMT mRNA expression.
Figure 3.
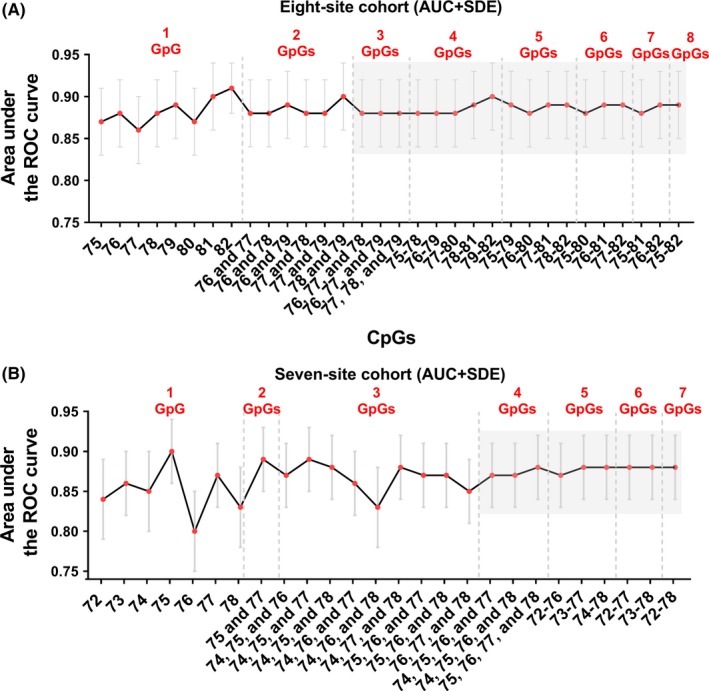
Predictive value of different individual CpG or selected CpG combinations methylation levels for MGMT mRNA expression. A and B, The area under the ROC (AUC) and corresponding Standard error (SDE) of different individual CpG or selected CpG combinations in the eight‐site cohort (A), and the seven‐site cohort (B) is arranged along with the CpG numbers contained in each combination. The CpG combinations with convergent AUCs are highlighted with the gray background
3.3. Therapeutic prognostic effects of the selected CpG combinations in glioblastoma
Based on the above findings, we further compared the therapeutic prognostic effect of the MGMT promoter methylation status determined by the several‐CpG combinations mean methylation level, and the selected CpG combinations were CpGs 75‐82, 76‐79, and 75‐78 from the eight‐site cohort and CpGs 72‐78, 74‐78, and 75‐78 from the seven‐site cohort. In the eight‐site cohort, the therapeutic prognostic effects of CpGs 75‐82 (Figure 4A), 76‐79 (Figure 4B), and 75‐78 (Figure 4C) were evaluated by ROC curve within 51 glioblastoma patients. And the respective AUCs were 0.7624, 0.7609, and 0.7741, which indicated that the three CpG combinations had similar therapeutic prognostic effects for glioblastoma patients (Figure 4A‐C). The similar result was observed in the 24 glioblastoma patients from the seven‐site cohort, and the AUCs of CpGs 72‐78, 74‐78, and 75‐78 were 0.7851, 0.7714, and 0.7893, respectively (Figure 4D‐F).
Figure 4.
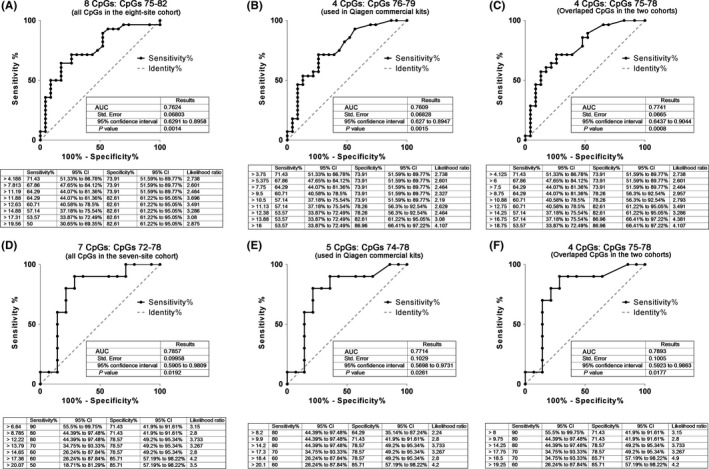
Comparison of the therapeutic prognostic value of different CpG combinations for glioblastoma patient survival. A‐C, Therapeutic prognostic effects of the MGMT methylation statuses determined by CpGs 75‐82 (A), CpGs 76‐79 (B), and CpGs 75‐78(C) for glioblastoma patient OS were evaluated by ROC in the eight‐site cohort. D‐F,Therapeutic prognostic effects of the MGMT methylation statuses determined by CpGs 72‐78 (D), CpGs 74‐79 (E), and CpGs 75‐78(F) for glioblastoma patient OS were evaluated by ROC in the seven‐site cohort
The Kaplan‐Meier steps showed that the OS of patients is significantly different, separated by the calculated cutoff of mean methylation level of CpGs 75‐82 (Figure 5A), 76‐79 (Figure 5B), and 75‐78 (Figure 5C). The results also showed that the methylation statuses of different CpG combinations had similar stratification ability for patient survival. In the seven‐site cohort, the methylation statuses determined by the mean methylation level of CpGs 72‐78 (Figure 5D), 74‐78 (Figure 5E), and 75‐78 (Figure 5F) also showed approximative predictive values. Then we compared the stratification ability of these CpG combinations in WHO grade III glioma patients by Kaplan‐Meier steps (Supporting Information Figure S1). For the predictive and prognostic roles of MGMT methylation in WHO grade III glioma patients have been reported,9 the results indicated that all the selected CpG combinations in the eight‐site and the seven‐site cohort had similar predictive value.
Figure 5.
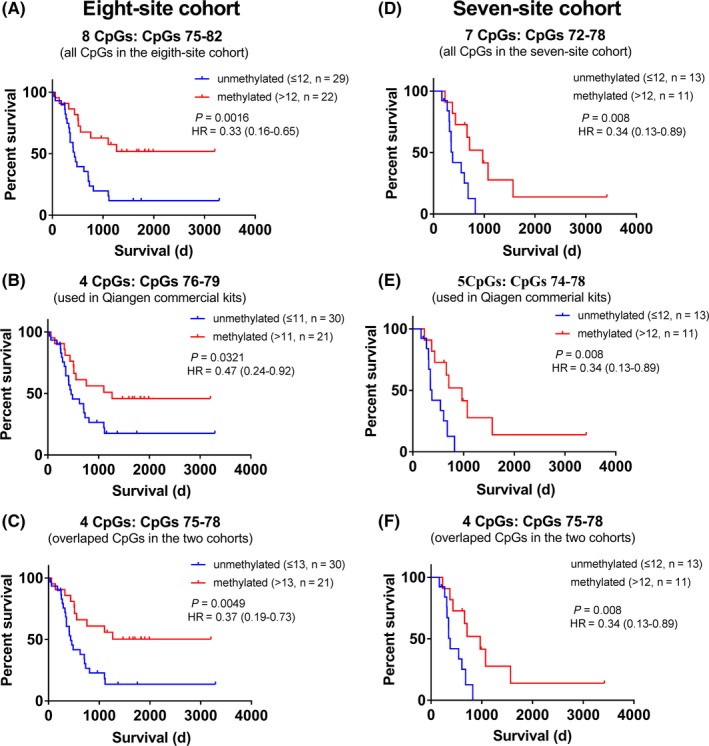
Kaplan‐Meier steps for the OS of glioblastoma patients stratified by the methylation level of different CpG combinations. A‐C, Kaplan‐Meier step for the OS of glioblastoma patients in the eight‐site cohort. The MGMT methylation statuses determined by CpGs 75‐82 (A), CpGs 76‐79 (B), and CpGs 75‐78(C) were used as the stratification reference, respectively. D‐F, Kaplan‐Meier curves for the OS of glioblastoma patients in the eight‐site cohort. The MGMT methylation statuses determined by CpGs 72‐78 (D), CpGs 74‐79 (E), and CpGs 75‐78(F) were used as the stratification reference, respectively
4. DISCUSSION
The PSQ testing has been regarded as the gold standard forMGMT promoter methylation testing and wildly used in reported studies and clinical practice.12, 16, 17, 21, 30, 31 However, which CpGs should be included in the MGMT methylation PSQ testing and analysis is still a controversial issue. In this study, we demonstrated that CpG combinations with four CpGs were robust enough to be adopted in the MGMT methylation PSQ testing, for the CpG combinations with four or more consecutive CpGs had the similar correlation with the MGMT mRNA expression, and they also showed alike predictive value on the MGMT mRNA expression. We also found that the predictive value of CpGs 75‐78 was close to that of CpGs 76‐79 and 74‐78 in the eight‐site cohort and seven‐site cohort, respectively, indicating that the commercial kits utilized CpGs combinations, CpGs 76‐79 and 74‐78, had almost equal predictive value for the MGMT mRNA expression and survival of patients received TMZ.
Several studies have tried to compare the predictive values of individual CpG and CpG combinations on MGMT mRNA expression and survival of TMZ treated patients, but the result is still controversial. Watts et al identified that three distinct regions of MGMT CGI were highly related to the MGMT expression level.3 Among the three regions, two overlap the CpGs 25‐50 and CpGs 73‐90, which were proved to be crucial for MGMT promoter activity.12 In glioblastoma tissues, individual CpG 27, 32, 73, 75, 79 and 80 were reported to be significantly correlated to the MGMT mRNA expression, and combinations of CpGs 32‐33 and CpGs 72‐83 were concordant with MGMT mRNA expression.24 But, there was no consensus on which individual CpG or CpG combinations should be used in MGMT methylation PSQ testing, and the combinations of CpGs 72‐83, 72‐80, 72‐77, 74‐78, 74‐89, 76‐79, and 80‐83 were used in distinct studies.12, 14, 15, 16, 17, 21, 23, 26, 32, 33, 34 Here, we systematically compared the predictive value of all combinations within the CpGs 72‐82 on the MGMT mRNA expression through analyzing paired samples with both the MGMT methylation PSQ testing and mRNA expression data. We indicated that the predictive value differences among combinations with four or more CpGs within CpGs 72‐82 were marginal, which may explain why controversial results got from different studies.
A study had compared the prognostic value of different CpGs on patients clinical outcome and showed that CpG 89, 84 and the combination of CpGs 84‐88 was the optimal predictive models within CpGs 74‐89 for the survival of glioblastoma patients, but the predictive values of which were merely improved, compared with the CpGs 74‐78 combination.13 In this study, we directly evaluated the predictive value of different CpG combinations and individual CpG for MGMT mRNA expression and verified the findings with the survival of TMZ treated glioblastoma patients as well. Besides, the strategy for CpG combination selection in our study is more comprehensive and targeted to the frequently used combinations in commercial kits, such as CpGs 76‐79 and 74‐78. Our results suggested that combinations with part of CpGs within CpGs 76‐79 or CpGs 74‐78 presented poorer predictive value than totally included, respectively. Given the convenience of standardization between different laboratories, it is a proper choice to analyze results with included CpGs in commercial kits totally.
The cutoff value is another important issue for theMGMT PSQ testing. Here, we found that the optimal cutoff for the mean methylation level of different CpG combinations varied obviously. This is consistent with previous studies, where the cutoff varied from 2.68% to 30% for different CpG combinations.14, 17, 23, 24, 25, 26 Some studies even suggested that there may be a “gray zone” between the true methylated status and true un‐methylated status, and the cutoff value around 10% used in most studies was overestimated.25, 26 We also noticed that the cutoff of the mean methylation level cannot be easily determined, and the different cutoff for the same CpG combination was used in different studies.13, 14, 17, 33 Recently, a study indicated that the optimal cutoff should not only be determined by the ROC likelihood value but also by the sensitivity and specificity.17 Thus, we determined the cutoff in this study by similar strategy, comprehensively considering the ROC likelihood value, sensitivity, specificity, and cutoffs used in reported studies. However, a more comprehensive and targeted study is still required for the optimal cutoff determination for MGMT methylation PSQ testing.
5. CONCLUSIONS
In summary, our study demonstrates that (a) combinations of four CpGs within CpGs 72‐82 are robust enough in MGMT promoter methylation PSQ testing; (b) CpG combinations with four or more consecutive CpGs within CpGs 72‐82, including the combinations of CpGs 76‐79 and CpGs 74‐78 used in commercial kits, are equally effective to predict the MGMT mRNA expression and the survival of TMZ treated glioma patients; (c) for the commonly used combinations of CpGs 76‐79 and CpGs 74‐78, it is proper to analyze the final methylation status of MGMT promoter with included CpGs entirely. All above‐mentioned indicated that four‐CpG combinations are sufficient for MGMT methylation testing by the PSQ approach.
CONFLICT OF INTEREST
There is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Supporting information
Chai R‐C, Zhang K‐N, Liu Y‐Q, et al. Combinations of four or more CpGs methylation present equivalent predictive value for MGMT expression and temozolomide therapeutic prognosis in gliomas. CNS Neurosci Ther. 2019;25:314–322. 10.1111/cns.13040
The first two authors contributed equally to this work
Funding information
This work was supported by the National Natural Science Foundation of China (81773208); the Beijing Nova Program (Z16110004916082).
REFERENCES
- 1. Gerson SL. MGMT: its role in cancer aetiology and cancer therapeutics. Nat Rev Cancer. 2004;4(4):296‐307. [DOI] [PubMed] [Google Scholar]
- 2. Esteller M, Garcia‐Foncillas J, Andion E, et al. Inactivation of the DNA‐repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343(19):1350‐1354. [DOI] [PubMed] [Google Scholar]
- 3. Watts GS, Pieper RO, Costello JF, Peng YM, Dalton WS, Futscher BW. Methylation of discrete regions of the O6‐methylguanine DNA methyltransferase (MGMT) CpG island is associated with heterochromatinization of the MGMT transcription start site and silencing of the gene. Mol Cell Biol. 1997;17(9):5612‐5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hegi ME, Diserens AC, Godard S, et al. Clinical trial substantiates the predictive value of O6‐methylguanine‐DNA methyltransferase promoter methylation in glioblastoma patients treated with temozolomide. Clin Cancer Res. 2004;10(6):1871‐1874. [DOI] [PubMed] [Google Scholar]
- 5. Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997‐1003. [DOI] [PubMed] [Google Scholar]
- 6. Barault L, Amatu A, Bleeker FE, et al. Digital PCR quantification of MGMT methylation refines prediction of clinical benefit from alkylating agents in glioblastoma and metastatic colorectal cancer. Ann Oncol. 2015;26(9):1994‐1999. [DOI] [PubMed] [Google Scholar]
- 7. van den Bent MJ, Dubbink HJ, Sanson M, et al. MGMT promoter methylation is prognostic but not predictive for outcome to adjuvant PCV chemotherapy in anaplastic oligodendroglial tumors: a report from EORTC Brain Tumor Group Study 26951. J Clin Oncol. 2009;27(35):5881‐5886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wick W, Hartmann C, Engel C, et al. NOA‐04 randomized phase III trial of sequential radiochemotherapy of anaplastic glioma with procarbazine, lomustine, and vincristine or temozolomide. J Clin Oncol. 2009;27(35):5874‐5880. [DOI] [PubMed] [Google Scholar]
- 9. Wick W, Meisner C, Hentschel B, et al. Prognostic or predictive value of MGMT promoter methylation in gliomas depends on IDH1 mutation. Neurology. 2013;81(17):1515‐1522. [DOI] [PubMed] [Google Scholar]
- 10. Kim J, Lee SH, Jang JH, Kim MS, Lee EH, Kim YZ. Increased expression of the histone H3 lysine 4 methyltransferase MLL4 and the histone H3 lysine 27 demethylase UTX prolonging the overall survival of patients with glioblastoma and a methylated MGMT promoter. J Neurosurg. 2017;126(5):1461‐1471. [DOI] [PubMed] [Google Scholar]
- 11. Wick W, Weller M, van den Bent M, et al. MGMT testing–the challenges for biomarker‐based glioma treatment. Nat Rev Neurol. 2014;10(7):372‐385. [DOI] [PubMed] [Google Scholar]
- 12. Malley DS, Hamoudi RA, Kocialkowski S, Pearson DM, Collins VP, Ichimura K. A distinct region of the MGMT CpG island critical for transcriptional regulation is preferentially methylated in glioblastoma cells and xenografts. Acta Neuropathol. 2011;121(5):651‐661. [DOI] [PubMed] [Google Scholar]
- 13. Quillien V, Lavenu A, Sanson M, et al. Outcome‐based determination of optimal pyrosequencing assay for MGMT methylation detection in glioblastoma patients. J Neurooncol. 2014;116(3):487‐496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bienkowski M, Berghoff AS, Marosi C, et al. Clinical Neuropathology practice guide 5–2015: MGMT methylation pyrosequencing in glioblastoma: unresolved issues and open questions. Clin Neuropathol. 2015;34(5):250‐257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Weller M, Stupp R, Reifenberger G, et al. MGMT promoter methylation in malignant gliomas: ready for personalized medicine? Nat Rev Neurol. 2010;6(1):39‐51. [DOI] [PubMed] [Google Scholar]
- 16. Mulholland S, Pearson DM, Hamoudi RA, et al. MGMT CpG island is invariably methylated in adult astrocytic and oligodendroglial tumors with IDH1 or IDH2 mutations. Int J Cancer. 2012;131(5):1104‐1113. [DOI] [PubMed] [Google Scholar]
- 17. Yuan G, Niu L, Zhang Y, et al. Defining optimal cutoff value of MGMT promoter methylation by ROC analysis for clinical setting in glioblastoma patients. J Neurooncol. 2017;133(1):193‐201. [DOI] [PubMed] [Google Scholar]
- 18. Cheng W, Ren X, Cai J, et al. A five‐miRNA signature with prognostic and predictive value for MGMT promoter‐methylated glioblastoma patients. Oncotarget. 2015;6(30):29285‐29295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rapkins RW, Wang F, Nguyen HN, et al. The MGMT promoter SNP rs16906252 is a risk factor for MGMT methylation in glioblastoma and is predictive of response to temozolomide. Neuro Oncol. 2015;17(12):1589‐1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhao H, Wang S, Song C, Zha Y, Li L. The prognostic value of MGMT promoter status by pyrosequencing assay for glioblastoma patients' survival: a meta‐analysis. World J Surg Oncol. 2016;14(1):261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Quillien V, Lavenu A, Karayan‐Tapon L, et al. Comparative assessment of 5 methods (methylation‐specific polymerase chain reaction, MethyLight, pyrosequencing, methylation‐sensitive high‐resolution melting, and immunohistochemistry) to analyze O6‐methylguanine‐DNA‐methyltranferase in a series of 100 glioblastoma patients. Cancer. 2012;118(17):4201‐4211. [DOI] [PubMed] [Google Scholar]
- 22. Cheng W, Li M, Jiang Y, et al. Association between small heat shock protein B11 and the prognostic value of MGMT promoter methylation in patients with high‐grade glioma. J Neurosurg. 2016;125(1):7‐16. [DOI] [PubMed] [Google Scholar]
- 23. Havik AB, Brandal P, Honne H, et al. MGMT promoter methylation in gliomas‐assessment by pyrosequencing and quantitative methylation‐specific PCR. J Transl Med. 2012;10:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Everhard S, Tost J, El Abdalaoui H, et al. Identification of regions correlating MGMT promoter methylation and gene expression in glioblastomas. Neuro Oncol. 2009;11(4):348‐356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dunn J, Baborie A, Alam F, et al. Extent of MGMT promoter methylation correlates with outcome in glioblastomas given temozolomide and radiotherapy. Br J Cancer. 2009;101(1):124‐131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Brigliadori G, Foca F, Dall'Agata M, et al. Defining the cutoff value of MGMT gene promoter methylation and its predictive capacity in glioblastoma. J Neurooncol. 2016;128(2):333‐339. [DOI] [PubMed] [Google Scholar]
- 27. Zhang W, Zhang J, Hoadley K, et al. miR‐181d: a predictive glioblastoma biomarker that downregulates MGMT expression. Neuro Oncol. 2012;14(6):712‐719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang C, Cheng W, Ren X, et al. Tumor purity as an underlying key factor in glioma. Clin Cancer Res. 2017;23(20):6279‐6291. [DOI] [PubMed] [Google Scholar]
- 29. Zhao Z, Meng F, Wang W, Wang Z, Zhang C, Jiang T. Comprehensive RNA‐seq transcriptomic profiling in the malignant progression of gliomas. Sci Data. 2017;4:170024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jiang T, Mao Y, Ma W, et al. CGCG clinical practice guidelines for the management of adult diffuse gliomas. Cancer Lett. 2016;375(2):263‐273. [DOI] [PubMed] [Google Scholar]
- 31. Chai R, Zhang K, Wang K, et al. A novel gene signature based on five glioblastoma stem‐like cell relevant genes predicts the survival of primary glioblastoma. J Cancer Res Clin Oncol. 2018;144(3):439‐447. [DOI] [PubMed] [Google Scholar]
- 32. Cai J, Zhang W, Yang P, et al. Identification of a 6‐cytokine prognostic signature in patients with primary glioblastoma harboring M2 microglia/macrophage phenotype relevance. PLoS One. 2015;10(5):e0126022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Reifenberger G, Hentschel B, Felsberg J, et al. Predictive impact of MGMT promoter methylation in glioblastoma of the elderly. Int J Cancer. 2012;131(6):1342‐1350. [DOI] [PubMed] [Google Scholar]
- 34. Karayan‐Tapon L, Quillien V, Guilhot J, et al. Prognostic value of O6‐methylguanine‐DNA methyltransferase status in glioblastoma patients, assessed by five different methods. J Neurooncol. 2010;97(3):311‐322. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


