Abstract
A number of microRNAs (miRNAs) are involved in the development and malignant progression of numerous types of human cancer including breast cancer. The underlying regulatory mechanism of miRNA-153 (miR-153) in breast cancer progression remains largely unknown. The present study demonstrated that miR-153 expression levels were significantly reduced in breast cancer tissue samples and cell lines, compared with adjacent healthy tissue samples and normal human breast cell line MCF-10A. In addition, low miR-153 expression was associated with advanced clinical staging and metastasis in patients with breast cancer. However, no association with age, subtype or differentiation was identified. Furthermore, patients with breast cancer with low miR-153 expression had poor prognosis, compared with patients with breast cancer with high miR-153 expression. Overexpression of miR-153 reduced proliferation, migration, invasion and epithelial-mesenchymal transition (EMT) in breast cancer SK-BR-3 and BT-549 cells. Runt-related transcription factor 2 (RUNX2), which was revealed to be significantly upregulated in breast cancer, was verified as a target gene of miR-153 in SK-BR-3 and BT-549 cells by luciferase reporter gene assay. High RUNX2 expression was associated with advanced clinical staging as well as distant and lymph node metastasis in patients with breast cancer. However, no association with age, subtype or differentiation was identified. Additionally, an inverse correlation between miR-153 and RUNX2 mRNA expression levels was observed in breast cancer tissues. RUNX2 overexpression reduced the suppressive effects of miR-153 on the proliferation, migration, invasion and EMT of SK-BR-3 and BT-549 cells. The present study indicated that miR-153 may serve a role in breast tumor growth and metastasis via direct targeting of RUNX2. The miR-153/RUNX2 axis may be used as a potential therapeutic target in breast cancer treatment.
Keywords: breast cancer, microRNA, runt-related transcription factor 2, metastasis
Introduction
Breast cancer is the most commonly diagnosed cancer and the leading cause of cancer-associated mortality among women worldwide (1,2). The prognosis of patients with advanced breast cancer is poor despite having a number of treatment options available, which include surgical resection, chemotherapy and radiotherapy (1,2). Therefore, an understanding of the regulatory mechanisms underlying breast cancer progression is needed for the development of novel and effective treatment strategies.
MicroRNAs (miRNAs) are small non-coding RNAs 18–25 nucleotides in length (3,4). miRNAs can function as regulators of gene expression by pairing with complementary binding sites in the 3′-untranslated region (UTR) of target mRNAs, resulting in mRNA degradation or inhibition of protein translation (3,4). Recently, several miRNAs were revealed to be involved in the development and progression of breast cancer, these include miRNA-148 (5), miRNA-181 (6) and miRNA-200 (7). The role of miRNA-153 (miR-153) in breast cancer has been investigated in several studies (8–11). Wu et al (8) demonstrated that miR-153 induced apoptosis in breast cancer cells by inhibiting the expression of HECT domain E3 ubiquitin ligase 3. In addition, Li et al (9) revealed that miR-153 demonstrated suppressive effects on epithelial-mesenchymal transition (EMT) in human breast cancer cells by inhibiting the expression of metadherin. Furthermore, miR-153 was demonstrated to suppress the expression of the oncogene BRCA1 in breast cancer MCF7 cells (10). Together, these results suggest that miR-153 may serve a tumor suppressive role in breast cancer. However, Anaya et al (11) demonstrated that miR-153 knockdown induced apoptosis in MDA-MB-231 breast cancer cells. In addition, Wang et al (12) revealed that miR-153 could decrease apoptosis and increase colony formation in breast epithelial cells, and following treatment with E2, miR-153 was upregulated in human breast cell lines. Therefore, the exact role of miR-153 in breast cancer growth and metastasis, as well as the underlying molecular mechanism of miR-153 in breast cancer should be further investigated.
Runt-related transcription factor 2 (RUNX2) is an important member of the RUNX family of transcription factors (13–15). It acts as a scaffold for nucleic acids and regulatory factors involved in osteoblastic differentiation and skeletal morphogenesis (13–15). It was recently revealed that RUNX2 can promote breast cancer cell survival under metabolic stress, as well as bone metastases (16,17). Furthermore, the targeting of RUNX2 by miR-135 and miR-203 impairs breast cancer progression and distant metastasis (18). However, whether other miRNAs regulate RUNX2 expression in breast cancer remains unclear.
The present study aimed to investigate the underlying molecular mechanism of miR-153 and RUNX2 in breast cancer growth and metastasis.
Materials and methods
Sample collection
The present study analyzed tissue samples obtained from 67 patients (age range, 31–69 years; mean age, 52.5 years) diagnosed with breast cancer in the Second Xiangya Hospital of Central South University (Changsha, China) from September 2010 and March 2012. Primary breast cancer tissue and adjacent healthy tissue were collected and stored at −80°C until further use, following histopathological evaluation. The follow-up period was 5 years. The current study was conducted with the approval from the Ethics Committee at Second Xiangya Hospital of Central South University (Changsha, China). Written informed consent was obtained from all patients.
Cell culture and transfection
Human breast cancer cell lines BT-549, MCF-7, MDA-MB-453, MDA-MB-231 and SK-BR-3, and a normal human breast epithelial cell line MCF-10A were purchased from Shanghai Institute of Biochemistry and Cell Biology (SIBCB; Shanghai, China). Cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Invitrogen; Thermo Fisher Scientific, Inc.) and maintained at 37°C in a 5% CO2-humidified incubator. Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) was used for cell transfection according to the manufacturer's protocol. SK-BR-3 and BT-549 cells were transfected with miR-NC (100 nM; 4464058; Thermo Fisher Scientific, Inc.), miR-153 mimic (100 nM; 4464066; Thermo Fisher Scientific, Inc.), NC inhibitor (100 nM; 4464076; Thermo Fisher Scientific, Inc.) or miR-153 inhibitor (100 nM; 4464084; Thermo Fisher Scientific, Inc.), or co-transfected with miR-153 mimic and empty pc-DNA3.1 (blank) vector or miR-153 mimic and pc-DNA3.1-RUNX2 plasmid (100 nM; Yearthbio, Changsha, China), respectively. Cells were used for subsequent experimentation 48 h post-transfection.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from tissues or cell lines using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Total RNA (1 µg) was reverse transcribed into cDNA using the miScript Reverse Transcription kit (Qiagen, Inc., Valencia, CA, USA), according to the manufacturer's protocol. qPCR was subsequently performed on an ABI 7500 PCR machine (Thermo Fisher Scientific, Inc.) using the miScript SYBR Green PCR kit (Qiagen, Inc.), according to the manufacturer's protocol. The following thermocycling conditions were used for the qPCR: Initial denaturation at 95°C for 5 min; 40 cycles of 95°C for 10 sec, 60°C for 30 sec and 72°C for 15 sec. The mRNA levels were quantified using the 2−∆∆Cq method (19). The primers used were as follows: miR-153, forward 5′-TTGCATAGTCACAAAAGTGAT-3′ and reverse 5′-CAGTGCGTGTCGTGGAGT-3′; U6, forward 5′-CTCGCTTCGGCAGCACATATACT-3′ and reverse 5′-ACGCTTCACGAATTTGCGTGTC-3′; RUNX2, forward 5′-TGGTTACTGTCATGGCGGGTA-3′ and reverse 5′-TCTCAGATCGTTGAACCTTGCTA-3′; and GAPDH, forward 5′-GGAGCGAGATCCCTCCAAAAT-3′ and reverse 5′-GGCTGTTGTCATACTTCTCATGG-3′.
Western blot analysis
Total protein was extracted from tissues or cells using cold radioimmunoprecipitation assay buffer (Thermo Fisher Scientific, Inc.). Total protein was quantified using a bicinchoninic acid assay kit (Thermo Fisher Scientific, Inc.) and 50 µg protein/lane was separated via SDS-PAGE on a 12% gel. The separated proteins were transferred onto polyvinylidene difluoride membranes and blocked with 5% non-fat milk in Tris-buffered saline with Tween® 20 (Thermo Fisher Scientific, Inc.) for 3 h at room temperature. Membranes were washed with PBS for 10 min, followed by incubation with rabbit anti-human E-cadherin primary antibody (1:200; ab15148; Abcam, Cambridge, MA, USA), rabbit anti-human N-cadherin primary antibody (1:200; ab18203; Abcam), rabbit anti-human vimentin primary antibody (1:100; ab8978; Abcam) or rabbit anti-human GAPDH primary antibody (1:200; ab9485; Abcam) for 3 h at room temperature. Following primary incubation, membranes were incubated with horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (1:5,000; ab6721; Abcam) for 1 h at room temperature. Protein bands were visualized using an ECL Western Blotting kit (Thermo Fisher Scientific, Inc.), and protein expression was quantified using Image-Pro Plus software (version 6.0; Media Cybernetics, Inc., Rockville, MD, USA).
Cell proliferation assay
Cell Counting kit-8 (Dojindo Molecular Technologies, Inc., Kumamoto, Japan) was used to study cell proliferation. SK-BR-3 and BT-549 cells were seeded into 96-well plates at a density of 5×103 cells/well and cultured for 0, 24, 48 and 72 h. The absorbance was measured at a wavelength of 450 nm using a microplate reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Wound healing assay
Transfected SK-BR-3 and BT-549 cells were cultured until confluence reached >90%. Cell layers were scratched with a plastic scriber (~1 mm width) and cells were washed twice with PBS, and incubated at 37°C for 24 h. The wound was observed and images were captured at 0 and 24 h using an inverted microscope (magnification, ×40; Nikon Corporation, Tokyo, Japan).
Cell invasion assay
The cell suspension of transfected SK-BR-3 or BT-549 cells (50,000 cells) in DMEM was added to the upper chamber of 8-mM Transwell inserts which was pre-coated with Matrigel® (BD Biosciences, Franklin Lakes, NJ, USA), and DMEM supplemented with 10% FBS was added to the lower chamber. After incubation for 24 h at 37°C, SK-BR-3 and BT-549 transfected cells remaining in the upper chamber of the inserts were removed using a cotton-tipped swab. SK-BR-3 and BT-549 transfected cells that had migrated to the lower chamber were stained with gentian violet (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) at room temperature for 5 min, and the number of migrated cells was calculated using a light inverted microscope (magnification, ×200).
Bioinformatics analysis and luciferase reporter gene assay
TargetScan 7.2 software (www.targetscan.org) was used to predict the putative target genes of miR-153. The psi-CHECK2 luciferase reporter plasmids containing the wild-type (WT) 3′UTR or the mutant type (MT) 3′UTR of RUNX2 were obtained from Yearthbio. SK-BR-3 and BT-549 cells were co-transfected with miR-153 mimic (or miR-NC) and WT-RUNX2-3′UTR plasmid (or MT-RUNX2-3′UTR plasmid) using Lipofectamine® 2000, according to the manufacturer's protocol. Following incubation for 48 h, cells were collected and the luciferase activity was detected using a Dual Luciferase Reporter Assay system (Promega Corporation, Madison, WI, USA), according to the manufacturer's protocol. The method of normalization was comparison with Renilla luciferase activity.
Statistical analysis
Data are presented as the mean ± standard error. All statistical analyses were performed using SPSS software (version 20.0; IBM Corp., Armonk, NY, USA). Student's t-test was performed for two-group comparisons. One-way analysis of variance followed by Tukey's post hoc test was performed for multiple-group comparisons. Chi-square test was conducted to analyze the association between gene expression and the clinical characteristics of patients with breast cancer. The Kaplan-Meier method and the log-rank rest were used for survival analysis. The Pearson correlation test was used to measure the association between miR-153 and RUNX2 expression in breast cancer tissue samples. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-153 is downregulated in breast cancer
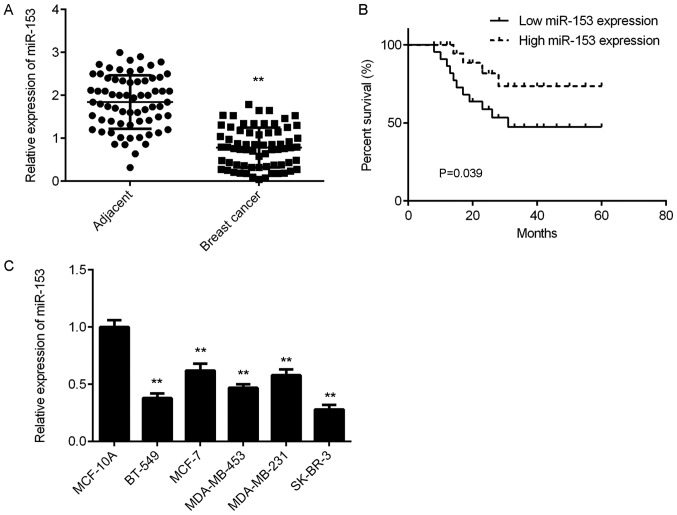
The expression level of miR-153 was significantly decreased in breast cancer tissue samples compared with matched adjacent healthy tissue samples from patients with breast cancer (Fig. 1A). The clinical significance of miR-153 expression in breast cancer was examined. According to the mean miR-153 expression value (0.784), patients with breast cancer were divided into two groups: high miR-153 expression and low miR-153 expression. The low miR-153 expression group was associated with advanced clinical staging as well as metastasis in patients with breast cancer, however no association with age, subtype, or differentiation was identified (Table I). Patients with breast cancer with low miR-153 expression demonstrated a poor prognosis (Fig. 1B). In addition, the mRNA expression levels of miR-153 significantly decreased in several breast cancer cell lines compared with normal human breast cell line MCF-10A (Fig. 1C).
Figure 1.
Downregulation of miR-153 in breast cancer. The mRNA expression level of miR-153 was determined by reverse transcription-quantitative polymerase chain reaction using tissue samples from patients with breast cancer. (A) The miR-153 expression levels were downregulated in breast cancer tissue compared with adjacent healthy tissue. **P<0.01 vs. Adjacent. (B) Patients with breast cancer with low miR-153 expression had a shorter survival time compared with patients with breast cancer with high miR-153 expression. (C) The miR-153 expression levels were downregulated in human breast cancer cell lines compared with normal human breast cell line MCF-10A. **P<0.01 vs. MCF-10A. Adjacent, adjacent healthy tissue; Breast cancer, primary breast cancer tissue; MCF-10A, normal human breast epithelial cell line; BT-549, MCF-7, MDA-MB-453, MDA-MB-231 and SK-BR-3, human breast cancer cell lines; miR, microRNA.
Table I.
Association between miR-153 expression and clinicopathological characteristics of patients with breast cancer.
| Variables | No. of patients (n=67) | Low expression (n=34) | High expression (n=33) | P-value |
|---|---|---|---|---|
| Age, years | 1.00 | |||
| ≤50 | 30 | 18 | 12 | |
| >50 | 37 | 16 | 11 | |
| Subtype | 0.814 | |||
| Lunimal A type | 33 | 15 | 18 | |
| Lunimal B type | 8 | 4 | 4 | |
| HER2 positive | 11 | 6 | 5 | |
| TNBC | 15 | 9 | 6 | |
| Differentiation | 0.136 | |||
| Well and moderately | 40 | 17 | 23 | |
| Poor | 27 | 17 | 10 | |
| Lymph node metastasis | 0.039 | |||
| Present | 45 | 27 | 18 | |
| Absent | 22 | 7 | 15 | |
| Distant metastasis | 0.011 | |||
| Present | 7 | 7 | 0 | |
| Absent | 60 | 27 | 33 | |
| TNM stage | 0.010 | |||
| I–II | 44 | 17 | 27 | |
| III–IV | 23 | 17 | 6 |
HER2, human epidermal growth factor receptor 2; TNBC, triple-negative breast cancer; TNM, tumor, node and metastasis.
Overexpression of miR-153 inhibits the malignant phenotype in breast cancer cell lines
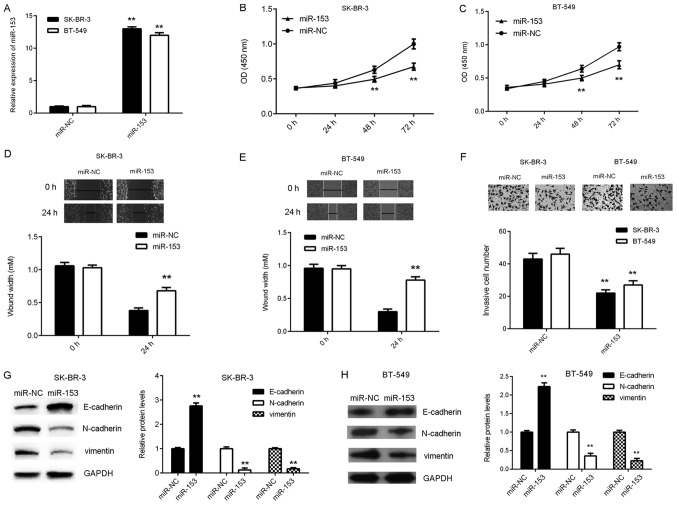
To investigate the function of miR-153 in breast cancer, cellular proliferation, migration and invasion were analyzed in breast cancer cell lines SK-BR-3 and BT-549 following transfection with either miR-153 mimic or miR-NC. The mRNA expression level of miR-153 significantly increased in breast cancer cells transfected with miR-153 mimic compared with miR-NC (Fig. 2A). In addition, SK-BR-3 and BT-549 cell proliferation, migration and invasion significantly decreased following miR-153 overexpression (Fig. 2B-F). The effect of miR-153 expression on EMT in SK-BR-3 and BT-549 cells was subsequently examined. The protein expression level of E-cadherin was significantly increased, whilst the protein expression levels of N-cadherin and vimentin were significantly reduced following overexpression of miR-153 compared with the miR-NC group (Fig. 2G and H). These results indicated that miR-153 may downregulate EMT in breast cancer cell lines.
Figure 2.
Overexpression of miR-153 suppresses the malignant phenotype in SK-BR-3 and BT-549 cells. miR-153 mimic and scramble miR mimic were transiently transfected into SK-BR-3 and BT-549 cells, respectively. (A) The expression level of miR-153 was detected by reverse transcription-quantitative polymerase chain reaction. The effect of miR-153 overexpression on (B) SK-BR-3 and (C) BT-549 cell growth was analyzed by Cell Counting kit-8 assay. The effect of miR-153 overexpression on (D) SK-BR-3 and (E) BT-549 cell migration was analyzed using the wound healing assay (magnification, ×40). (F) The effect of miR-153 overexpression on SK-BR-3 and BT-549 cell invasion was analyzed using the transwell invasion assay (magnification, ×200). The protein expression levels of E-cadherin, N-cadherin and vimentin in (G) SK-BR-3 and (H) BT-549 cells were determined by western blotting. **P<0.01 vs. miR-NC. SK-BR-3 and BT-549, human breast cancer cell lines; miR, microRNA; miR-NC, breast cancer cell lines SK-BR-3 and BT-549 transfected with scramble miR; miR-153, breast cancer cell lines SK-BR-3 and BT-549 transfected with miR-153 mimic.
RUNX2, upregulated in breast cancer, is a novel target of miR-153
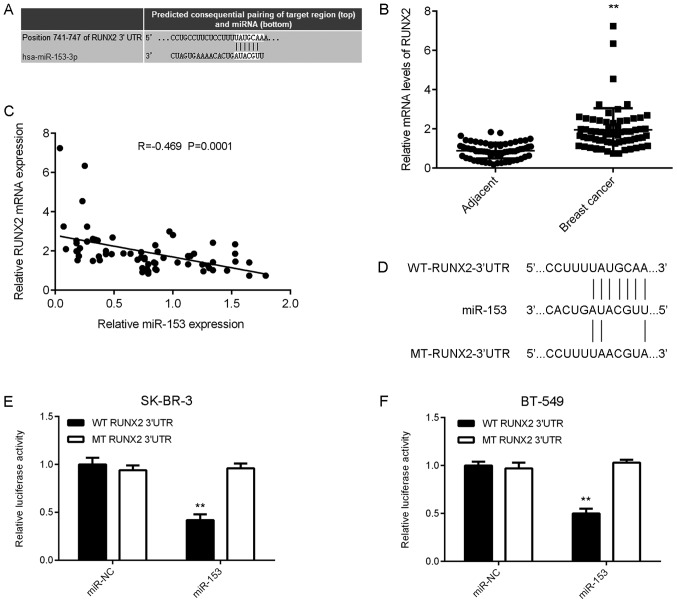
To investigate miR-153 further, the potential targets of miR-153 in breast cancer were examined. TargetScan software was used to identify RUNX2 as a putative target gene of miR-153 in breast cancer (Fig. 3A). To further elucidate the potential association between RUNX2 and miR-153 in breast cancer, RUNX2 expression was examined in tissue samples from patients with breast cancer. The mRNA expression level of RUNX2 was significantly increased in breast cancer tissue samples compared with matched adjacent healthy tissue samples (Fig. 3B). Furthermore, an inverse correlation was observed between miR-153 and RUNX2 expression in breast cancer tissue (Fig. 3C). According to the mean RUNX2 expression value, which was used as the cut-off, patients with breast cancer involved in the current study were divided into two groups: High RUNX2 expression and low RUNX2 expression. The high RUNX2 expression group was associated with advanced clinical staging and metastasis in patients with breast cancer, however no association with age, subtype, or differentiation was identified (Table II). These results suggest that RUNX2 upregulation may contribute to the process of malignant progression in human breast cancer. Luciferase reporter plasmids containing the WT-RUNX2-3′UTR or MT-RUNX2-3′UTR were generated (Fig. 3D) and used in the luciferase reporter gene assays which were conducted in SK-BR-3 and BT-549 cells. Luciferase activity indicated that miR-153 significantly reduced WT RUNX2 expression compared with MT RUNX2, which demonstrated no effect on the luciferase activity, in both breast cancer cell lines (Fig. 3E and F). These results confirm RUNX2 as a novel target gene of miR-153 in SK-BR-3 and BT-549 breast cancer cell lines.
Figure 3.
Upregulated RUNX2 in breast cancer is a target gene of miR-153. (A) Bioinformatics software was used to predict RUNX2 as a putative target gene of miR-153. (B) The mRNA expression level of RUNX2 was detected by reverse transcription-quantitative polymerase chain reaction and found to be upregulated in breast cancer tissue compared with adjacent healthy tissue. **P<0.01 vs. Adjacent. (C) An inverse correlation between miR-153 and RUNX2 mRNA expression in breast cancer tissue samples was identified. (D) WT-RUNX2-3′UTR and MT-RUNX2-3′UTR luciferase reporter plasmids were generated. Following miR-153 overexpression luciferase activity was measured in (E) SK-BR-3 and (F) BT-549 cells. **P<0.01 vs. miR-NC. Adjacent, adjacent healthy tissue; Breast cancer, primary breast cancer tissue; RUNX2, runt-related transcription factor 2; SK-BR-3 and BT-549, human breast cancer cell lines; miR, microRNA; miR-NC, breast cancer cell lines SK-BR-3 and BT-549 transfected with scramble miR; miR-153, breast cancer cell lines SK-BR-3 and BT-549 transfected with miR-153 mimic; WT RUNX2 3′UTR, wild-type 3′UTR of RUNX2 cloned into luciferase reporter plasmid; MT RUNX2 3′UTR, mutant type 3′UTR of RUNX2 cloned into luciferase reporter plasmid.
Table II.
Association between RUNX2 expression and clinicopathological characteristics of patients with breast cancer.
| Variables | No. of patients (n=67) | Low expression (n=43) | High expression (n=24) | P-value |
|---|---|---|---|---|
| Age, years | 0.800 | |||
| ≤50 | 30 | 20 | 10 | |
| >50 | 37 | 23 | 14 | |
| Subtype | 0.175 | |||
| Lunimal A type | 33 | 25 | 8 | |
| Lunimal B type | 8 | 3 | 5 | |
| HER2 positive | 11 | 6 | 5 | |
| TNBC | 15 | 9 | 6 | |
| Differentiation | 0.120 | |||
| Well and moderately | 40 | 29 | 11 | |
| Poor | 27 | 14 | 13 | |
| Lymph node metastasis | 0.014 | |||
| Present | 45 | 24 | 21 | |
| Absent | 22 | 19 | 3 | |
| Distant metastasis | 0.0004 | |||
| Present | 7 | 0 | 7 | |
| Absent | 60 | 43 | 17 | |
| TNM stage | 0.0005 | |||
| I–II | 44 | 35 | 9 | |
| III–IV | 23 | 8 | 15 |
HER2, human epidermal growth factor receptor 2; TNBC, triple-negative breast cancer; TNM, tumor, node and metastasis.
miR-153 negatively regulates RUNX2 expression in breast cancer cells
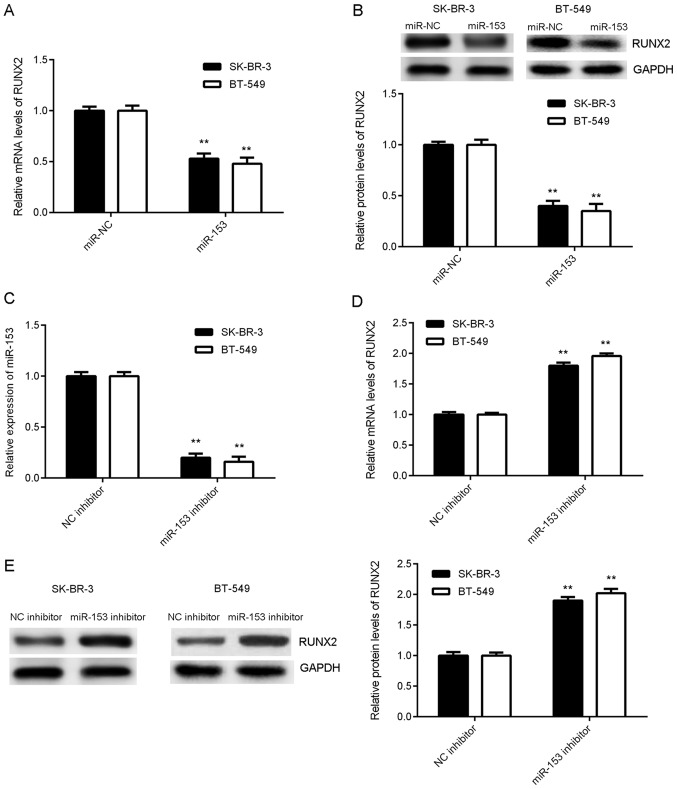
The effect of miR-153 on RUNX2 expression was examined in breast cancer cell lines SK-BR-3 and BT-549 following transfection with either miR-153 mimic or miR-NC. Overexpression of miR-153 significantly decreased the mRNA and protein expression levels of RUNX2 (Fig. 4A and B). To further understand the effect of miR-153, SK-BR-3 and BT-549 cells were transfected with either miR-153 inhibitor or NC inhibitor. The mRNA expression level of miR-153 significantly decreased in breast cancer cells transfected with miR-153 inhibitor compared with NC inhibitor (Fig. 4C). Knockdown of miR-153 significantly increased the mRNA and protein expression levels of RUNX2 (Fig. 4D and E), suggesting that miR-153 negatively regulates RUNX2 expression in breast cancer cells.
Figure 4.
miR-153 negatively regulates RUNX2 expression in SK-BR-3 and BT-549 cells. miR-153 mimic and scramble miR mimic were transiently transfected into SK-BR-3 and BT-549 cells, respectively. (A) The mRNA expression level of RUNX2 in SK-BR-3 and BT-549 cells was detected by RT-qPCR. (B) The protein expression level of RUNX2 in SK-BR-3 and BT-549 cells was determined by western blotting. **P<0.01 vs. miR-NC. miR-153 inhibitor and NC inhibitor were transiently transfected into SK-BR-3 and BT-549 cells, respectively. The mRNA expression level of (C) miR-153 and (D) RUNX2 in SK-BR-3 and BT-549 cells was detected by RT-qPCR. (E) The protein expression level of RUNX2 in SK-BR-3 and BT-549 cells was determined by western blotting. **P<0.01 vs. NC inhibitor. RUNX2, runt-related transcription factor 2; SK-BR-3 and BT-549, human breast cancer cell lines; miR, microRNA; miR-NC, breast cancer cell lines SK-BR-3 and BT-549 transfected with scramble miR; miR-153, breast cancer cell lines SK-BR-3 and BT-549 transfected with miR-153 mimic; NC inhibitor, breast cancer cell lines SK-BR-3 and BT-549 transfected with miRNA NC inhibitor; miR-153 inhibitor, breast cancer cell lines SK-BR-3 and BT-549 transfected with miR-153 inhibitor; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
RUNX2 reduces the suppressive effects of miR-153 in breast cancer cells
RUNX2 was identified in the present study as putative target gene of miR-153 in breast cancer cells. To determine whether RUNX2 was involved in miR-153-mediated breast cancer, SK-BR-3 and BT-549 cells overexpressing miR-153 were transfected with pcDNA3.1-RUNX2 plasmid to increase RUNX2 expression.
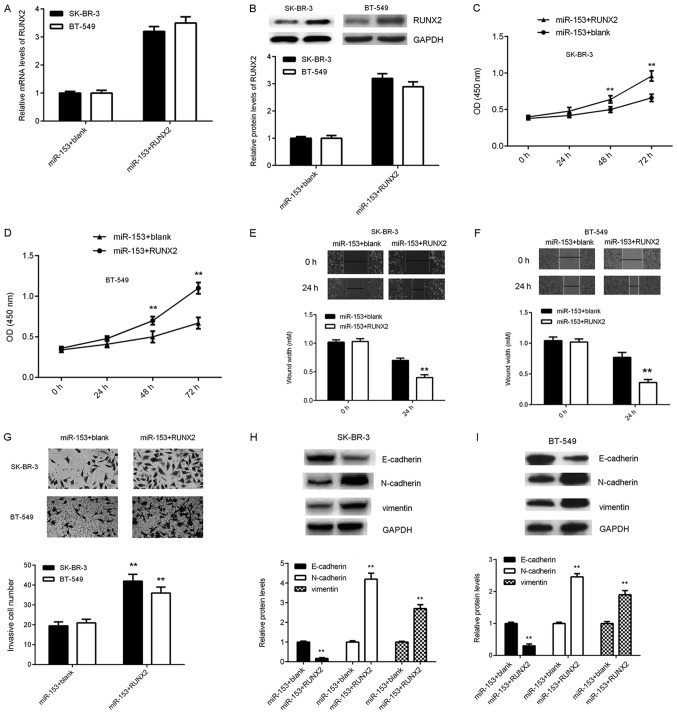
Following transfection, the mRNA and protein expression of levels of RUNX2 were significantly increased in both breast cancer cell lines (Fig. 5A and B). In addition, SK-BR-3 and BT-549 cell proliferation, migration and invasion were significantly increased following transfection with miR-153 + RUNX2 compared with miR-153 + blank (Fig. 5C-G). The effect on EMT in SK-BR-3 and BT-549 cells was subsequently examined. The protein expression level of E-cadherin was significantly reduced, whilst the protein expression levels of N-cadherin and vimentin were significantly increased following transfection with miR-153 + RUNX2 compared with miR-153 + blank (Fig. 5H and I), suggesting that RUNX2 overexpression increased EMT in breast cancer cell lines. Taken together, these results suggested that RUNX2 overexpression impaired the suppressive effects of miR-153 on the malignant phenotype in SK-BR-3 and BT-549 breast cancer cell lines.
Figure 5.
RUNX2 overexpression reduces the suppressive effects of miR-153 on SK-BR-3 and BT-549 cells. SK-BR-3 and BT-549 cells were co-transfected with miR-153 mimic, and pcDNA3.1-RUNX2 plasmid or blank pcDNA3.1 vector, respectively. The (A) mRNA and (B) protein expression levels of RUNX2 were detected by reverse transcription-quantitative polymerase chain reaction and western blotting, respectively. The effect on (C) SK-BR-3 and (D) BT-549 cell growth was analyzed by cell counting kit-8 assay. The effect on (E) SK-BR-3 and (F) BT-549 cell migration was analyzed using the wound healing assay (magnification, ×40). (G) The effect on SK-BR-3 and BT-549 cell invasion was analyzed using the transwell invasion assay (magnification, ×200). The protein expression levels of E-cadherin, N-cadherin and vimentin in (H) SK-BR-3 and (I) BT-549 cells were determined by western blotting. **P<0.01 vs. miR-153 + blank. SK-BR-3 and BT-549, human breast cancer cell lines; miR, microRNA; miR-153 + blank, breast cancer cell lines SK-BR-3 and BT-549 co-transfected with miR-153 mimic and blank pc-DNA3.1 vector; miR-153+RUNX2, breast cancer cell lines SK-BR-3 and BT-549 co-transfected with miR-153 mimic and pc-DNA3.1-RUNX2 plasmid.
Discussion
The underlying regulatory mechanism of miR-153 in breast cancer progression remains unclear. The present study demonstrated that miR-153 expression levels were significantly reduced in breast cancer tissue samples and cell lines, compared with adjacent healthy tissue samples and normal human breast cell line MCF-10A. Furthermore, low miR-153 expression was associated with advanced clinical staging and metastasis in patients with breast cancer, however not association with age, subtype or differentiation was identified. Furthermore, patients with breast cancer with low miR-153 expression had poor prognosis, compared with patients with breast cancer with high miR-153 expression. Overexpression of miR-153 reduced the proliferation, migration, invasion and EMT in breast cancer SK-BR-3 and BT-549 cells. RUNX2, significantly upregulated in breast cancer, was confirmed to be a novel target gene of miR-153 in SK-BR-3 and BT-549 cells by luciferase reporter gene assay. High RUNX2 expression was associated with advanced clinical staging as well as distant and lymph node metastasis in patients with breast cancer; however no association with age, subtype or differentiation was identified. In addition, an inverse correlation between miR-153 and RUNX2 mRNA expression levels was observed in breast cancer tissues. RUNX2 overexpression reduced the suppressive effects of miR-153 upregulation on the proliferation, migration, invasion and EMT of SK-BR-3 and BT-549 cells.
In different types of human cancer, miR-153 appears to be involved in either promoting or suppressing tumor growth and progression (20–22). Wu et al (20) revealed that miR-153 promoted prostate cancer cell proliferation by targeting PTEN. By contrast, miR-153 inhibited the proliferation and invasion of human laryngeal squamous cell carcinoma cells by inhibiting the expression of kruppel-like factor 5 (21). miR-153 may inhibit cell migration and invasion of gastric cancer cells by directly targeting Snail (22). A tumor suppressor function of miR-153 was revealed in glioblastoma (23). The different roles associated with miR-153 in different types of cancer may be as a result of differences in tumor microenvironment and target genes. In the current study, miR-153 expression was significantly reduced in breast cancer tissues and cell lines. Downregulation of miR-153 expression was associated with tumor progression and metastasis as well as poor prognosis in patients with breast cancer. A previous study demonstrated that miR-153 was downregulated in breast cancer (9). Downregulation of miR-153 expression may therefore serve a role during breast cancer progression. In the current study, overexpression of miR-153 led to a significant reduction in breast cancer cell proliferation, migration and invasion. Epithelial cancer cells acquire molecular alternations that facilitate the loss of epithelial features and gain of mesenchymal phenotype during EMT, promoting tumor cell migration and invasion (24). The present study demonstrated that overexpression of miR-153 inhibited EMT in breast cancer cells, which may have contributed to a reduction in tumor cell migration and invasion. Therefore, miR-153 may induce suppressive effects on the growth and metastasis of breast cancer.
The current study investigated the regulatory mechanism of miR-153-induced malignant phenotype in breast cancer cell lines. This study identified RUNX2 to be a novel target gene of miR-153 in breast cancer cells. RUNX2 is involved in osteogenesis and breast cancer bone metastases (16). The skeleton is one of the most common metastatic sites in breast cancer (16). The present study demonstrated that RUNX2 expression levels were significantly increased in breast cancer tissues and cell lines. High expression level of RUNX2 was associated with breast cancer progression including distant and lymph node metastasis. Similarly, Chang et al (25) demonstrated that higher expression of RUNX2 was associated with adverse outcomes in patients with breast cancer, including poor prognosis and recurrence (25). RUNX2 acts as an oncogene in breast cancer, activating the PI3K/AKT signaling, Indian Hedgehog signaling and a downstream bone metastatic pathway in breast cancer cells (26,27). The present study demonstrated that miR-153 may negatively regulate RUNX2 expression in breast cancer cells. An inverse correlation was identified between RUNX2 and miR-153 expression levels in breast cancer tissue samples. In addition, overexpression of RUNX2 impaired the suppressive effects of miR-153 on the malignant phenotype in breast cancer cells. Therefore, it was hypothesized that RUNX2 may be involved in the miR-153-induced malignant phenotype in breast cancer cell lines.
In conclusion, miR-153 inhibits cell proliferation, migration, invasion and EMT in breast cancer through direct targeting of RUNX2. miR-153 and RUNX2 may be potential molecular targets in the treatment of breast cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
YG designed the experiments. FY and YG performed the experiments. ZL analyzed the data. ZZ and JH prepared the manuscript.
Ethics approval and consent to participate
The current study was approved by the Ethics Committee of Second Xiangya Hospital of Central South University (Changsha, China). Written informed consent was obtained from patients involved in this study.
Patient consent for publication
Patient consent for publication has been obtained.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Xu X, Zhang Y, Jasper J, Lykken E, Alexander PB, Markowitz GJ, McDonnell DP, Li QJ, Wang XF. MiR-148a functions to suppress metastasis and serves as a prognostic indicator in triple-negative breast cancer. Oncotarget. 2016;7:20381–20394. doi: 10.18632/oncotarget.7953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng Y, Lv X, Wang X, Wang B, Shao X, Huang Y, Shi L, Chen Z, Huang J, Huang P. MiR-181b promotes chemoresistance in breast cancer by regulating Bim expression. Oncol Rep. 2016;35:683–690. doi: 10.3892/or.2015.4417. [DOI] [PubMed] [Google Scholar]
- 7.Yao Y, Hu J, Shen Z, Yao R, Liu S, Li Y, Cong H, Wang X, Qiu W, Yue L. MiR-200b expression in breast cancer: A prognostic marker and act on cell proliferation and apoptosis by targeting Sp1. J Cell Mol Med. 2015;19:760–769. doi: 10.1111/jcmm.12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu X, Li L, Li Y, Liu Z. MiR-153 promotes breast cancer cell apoptosis by targeting HECTD3. Am J Cancer Res. 2016;6:1563–1571. [PMC free article] [PubMed] [Google Scholar]
- 9.Li W, Zhai L, Zhao C, Lv S. MiR-153 inhibits epithelial-mesenchymal transition by targeting metadherin in human breast cancer. Breast Cancer Res Treat. 2015;150:501–509. doi: 10.1007/s10549-015-3346-y. [DOI] [PubMed] [Google Scholar]
- 10.Fkih M'hamed I, Privat M, Ponelle F, Penault-Llorca F, Kenani A, Bignon YJ. Identification of miR-10b, miR-26a, miR-146a and miR-153 as potential triple-negative breast cancer biomarkers. Cell Oncol (Dordr) 2015;38:433–442. doi: 10.1007/s13402-015-0239-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anaya-Ruiz M, Cebada J, Delgado-López G, Sánchez-Vázquez ML, Pérez-Santos JL. miR-153 silencing induces apoptosis in the MDA-MB-231 breast cancer cell line. Asian Pac J Cancer Prev. 2013;14:2983–2986. doi: 10.7314/APJCP.2013.14.5.2983. [DOI] [PubMed] [Google Scholar]
- 12.Wang B, Teng Y, Liu Q. MicroRNA-153 regulates NRF2 expression and is associated with breast carcinogenesis. Clin Lab. 2016;62:39–47. doi: 10.7754/Clin.Lab.2015.150518. [DOI] [PubMed] [Google Scholar]
- 13.Rashid H, Ma C, Chen H, Wang H, Hassan MQ, Sinha K, de Crombrugghe B, Javed A. Sp7 and Runx2 molecular complex synergistically regulate expression of target genes. Connect Tissue Res. 2014;55(Suppl 1):S83–S87. doi: 10.3109/03008207.2014.923872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McGee-Lawrence ME, Carpio LR, Bradley EW, Dudakovic A, Lian JB, van Wijnen AJ, Kakar S, Hsu W, Westendorf JJ. Runx2 is required for early stages of endochondral bone formation but delays final stages of bone repair in Axin2-deficient mice. Bone. 2014;66:277–286. doi: 10.1016/j.bone.2014.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen H, Ghori-Javed FY, Rashid H, Adhami MD, Serra R, Gutierrez SE, Javed A. Runx2 regulates endochondral ossification through control of chondrocyte proliferation and differentiation. J Bone Miner Res. 2014;29:2653–2665. doi: 10.1002/jbmr.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vishal M, Swetha R, Thejaswini G, Arumugam B, Selvamurugan N. Role of Runx2 in breast cancer-mediated bone metastasis. Int J Biol Macromol. 2017;99:608–614. doi: 10.1016/j.ijbiomac.2017.03.021. [DOI] [PubMed] [Google Scholar]
- 17.Tandon M, Othman AH, Ashok V, Stein GS, Pratap J. The role of Runx2 in facilitating autophagy in metastatic breast cancer cells. J Cell Physiol. 2018;233:559–571. doi: 10.1002/jcp.25916. [DOI] [PubMed] [Google Scholar]
- 18.Taipaleenmäki H, Browne G, Akech J, Zustin J, van Wijnen AJ, Stein JL, Hesse E, Stein GS, Lian JB. Targeting of Runx2 by miR-135 and miR-203 impairs progression of breast cancer and metastatic bone disease. Cancer Res. 2015;75:1433–1444. doi: 10.1158/0008-5472.CAN-14-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Wu Z, He B, He J, Mao X. Upregulation of miR-153 promotes cell proliferation via downregulation of the PTEN tumor suppressor gene in human prostate cancer. Prostate. 2013;73:596–604. doi: 10.1002/pros.22600. [DOI] [PubMed] [Google Scholar]
- 21.Liu JY, Lu JB, Xu Y. MicroRNA-153 inhibits the proliferation and invasion of human laryngeal squamous cell carcinoma by targeting KLF5. Exp Ther Med. 2016;11:2503–2508. doi: 10.3892/etm.2016.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Z, Liu C. MiR-153 regulates metastases of gastric cancer through Snail. Tumour Biol. 2015 doi: 10.1007/s13277-015-3846-8. [DOI] [PubMed] [Google Scholar]
- 23.Ghasemi A, Fallah S, Ansari M. MiR-153 as a tumor suppressor in glioblastoma multiforme is downregulated by DNA methylation. Clin Lab. 2016;62:573–580. doi: 10.7754/Clin.Lab.2015.150738. [DOI] [PubMed] [Google Scholar]
- 24.Wu Y, Sarkissyan M, Vadgama JV. Epithelial-mesenchymal transition and breast cancer. J Clin Med. 2016;5:E13. doi: 10.3390/jcm5020013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang CH, Fan TC, Yu JC, Liao GS, Lin YC, Shih AC, Li WH, Yu AL. The prognostic significance of RUNX2 and miR-10a/10b and their inter-relationship in breast cancer. J Transl Med. 2014;12:257. doi: 10.1186/s12967-014-0257-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pratap J, Wixted JJ, Gaur T, Zaidi SK, Dobson J, Gokul KD, Hussain S, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Runx2 transcriptional activation of Indian Hedgehog and a downstream bone metastatic pathway in breast cancer cells. Cancer Res. 2008;68:7795–7802. doi: 10.1158/0008-5472.CAN-08-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tandon M, Chen Z, Pratap J. Runx2 activates PI3K/Akt signaling via mTORC2 regulation in invasive breast cancer cells. Breast Cancer Res. 2014;16:R16. doi: 10.1186/bcr3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.