Abstract
Actin-related protein 2/3 complex (ARPC2) is an actin-binding component involved in the regulation of actin polymerization. It mediates the formation of branched actin networks and contacts the mother actin filament. Migration and invasion are key processes which enable tumor cells to infiltrate blood vessels or lymphatic vessels, and the actin pathway plays a very important role. Given that ARPC2 is critical to this progression, the present study focused on ARPC2 activity in breast cancer (BrCa) cell invasion and migration. Limited data are available on the expression and role of ARPC2 proteins in breast carcinomas. We screened the Oncomine database for messenger RNAs (mRNAs) that are upregulated in BrCa and found that ARPC2 was one of the most consistently involved mRNAs in BrCa. The analysis of immunohistochemical data revealed that ARPC2 expression was higher in breast cancerous tissues than in adjacent non-cancerous tissues. In addition, ARPC2 was highly associated with the tumor stage, nodal metastasis, and overall survival of patients with BrCa. We performed siRNA-ARPC2 transfection to investigate the effect of ARPC2 on the proliferation, migration, invasion and arrest of BrCa cells. It was revealed that ectopic ARPC2 expression significantly upregulated N-cadherin, vimentin, ZEB1, MMP-9 and MMP-3 expression and also activated the TGF-β pathway to contribute to epithelial-mesenchymal transition (EMT). These results collectively indicated that ARPC2 promoted the tumorigenesis of breast carcinoma and the initiation of EMT. Therefore, ARPC2 was revealed to be a potential therapeutic target in patients with BrCa.
Keywords: breast carcinoma, ARPC2 protein, proliferation, metastasis, molecular mechanism
Introduction
Breast cancer (BrCa) is the most prevalent malignancy among women. Approximately 252,710 new cases of invasive BrCa and 40,610 cancer-related deaths are expected to occur among American women in 2017 (1). BrCa is a heterogametic disease that can be categorized into luminal A, luminal B, human epithelial growth factor receptor-2 (HER-2) overexpressing and basal/triple negative (TN) molecular subtypes on the basis of gene expression profiling (2–5). Given the high morbidity and poor prognosis of patients with BrCa, additional research is required to understand the potential molecular mechanism that mediates BrCa tumorigenesis. To date, a variety of biological factors have been identified to be involved in the development of this disease. Therefore, it is imperative to identify potential therapeutic targets such as progesterone receptor (PR) and estrogen receptor (ER), human epidermal growth factor receptor 2 (HER2) among the various biological factors involved in BrCa development (6–8).
The Arp2/3 protein complex consists of 7 subunits: The actin-related proteins ARP2 (ACTR2) and ARP3 (ACTR3), ARPC1B (ARC41), ARPC2 (ARC34), ARPC3 (ARC21), ARPC4 (ARC20), and ARPC5 (ARC16) (9,10). Among these subunits, ARPC2 (subunit 2, 34 kDa) has a fundamental role in the nucleation of actin filaments (AFs) and participates in the polarization of the Golgi apparatus (11). Al Ghouleh et al demonstrated that ARPC2 participates in smooth muscle cell migration (12), Zhang et al also reported that ARPC2 may be involved in human gastric cancer cell proliferation or migration (13). Thus ARPC2 is critical to cell migration and invasion (14). Activation of epithelial-mesenchymal transition (EMT) has been implicated in the metastasis of human tumors in previous studies (15–17).
EMT is a specific physiological or pathophysiological phenomenon that is characterized by the loss of the epithelial phenotype and gain of mesenchymal characteristics by epithelial cells. These processes are evidenced by the decrease in E-cadherin expression and increase in vimentin/N-cadherin expression (18). The activation of EMT has been implicated in the metastasis of human tumors. In cancer, the frequent occurrence of EMT is associated with poor prognosis and distant metasis. TGF-β is a pathway involved in EMT regulation (19,20). The downstream factors of the TGF-β pathway include N-cadherin, vimentin, ZEB1, MMP-9 and MMP-3, and these factors have important roles in EMT regulation (21,22). However, whether the TGF-β/EMT pathway is involved in ARPC2-mediated carcinogenesis has never been reported.
The present study was aimed to determine whether ARPC2 is a BrCa oncogene and to further understand the mechanisms that underlie its in vitro and in vivo effect. By searching the Oncomine database and performing immunohistochemistry (IHC), we found that ARPC2 mRNA and protein expression levels were upregulated in BrCa. The results of the cell cycle, wound-healing, migration and invasion assays revealed that ARPC2 significantly promoted the proliferation, migration and invasion of BrCa cells. Clinical data, revealed that ARPC2 was associated with lymph node invasion and tumor stage. High ARPC2 expression levels were associated with a low overall survival (OS) rate. Lastly, we performed western blot analysis to further explore the potential mechanism of ARPC2 in BrCa. We found that ARPC2 increased the expression of EMT-inducing transcription factors (EMT-TFs), such as N-cadherin, vimentin, ZEB1, MMP-9 and MMP-3. Notably, ARPC2 may contribute to the malignant progression of BrCa.
Materials and methods
Patients and tissue collection
We obtained 172 paraffin-embedded surgical BrCa tissue specimens from the Department of Pathology at the First Affiliated Hospital of Anhui Medical University (Hefei, Anhui, China). We recruited 68 female patients with benign breast diseases from the same hospital. The 68 female patients were used as the control group. No patients had received preoperative adjuvant therapy. Two pathologists confirmed the diagnoses of breast tumors in all the samples used. All cases were followed-up for 60 months. Overall survival (OS) was calculated on the basis of data. Patient samples were categorized into two groups in accordance with median expression (high vs. low expression) (Table I) and assessed using a Kaplan-Meier survival plot. The protocol of the present study was approved by the Institutional Review Board of Anhui Medical University. Detailed information of patients including ER/PR expression, lymph node metastasis and age are listed in Table II. The agreement on the use of these tissue samples was approved by the Biomedical Ethics Committee of Anhui Medical University and the confidentiality of patient information was maintained. We obtained informed consent from all patients before enrolling participants in the present study.
Table I.
Expression of ARPC2 in breast cancer and normal tissues.
| ARPC2 expression | |||
|---|---|---|---|
| Group | n | Negative/low positive, n (%) | High positive, n (%) |
| Cancer | 172 | 76 (44.0) | 96 (56.0)a |
| Normal | 68 | 48 (72.0) | 20 (28.0)a |
P<0.001. ARPC2, actin-related protein 2/3 complex.
Table II.
Association of ARPC2 expression with clinicopathological parameters from breast cancer patients.
| ARPC2 expression | ||||
|---|---|---|---|---|
| Parameter | n | High | Low or none | P-value |
| Age (years) | ||||
| ≤35 | 6 | 4 | 2 | 0.075 |
| 36–55 | 111 | 57 | 54 | |
| >60 | 55 | 28 | 27 | |
| Tumor size (cm) | ||||
| ≤2 | 41 | 16 | 25 | <0.05 |
| 3–5 | 110 | 51 | 59 | |
| >6 | 21 | 15 | 6 | |
| Lymph node involvement | ||||
| pN0 | 79 | 35 | 44 | <0.05 |
| pN1 | 45 | 24 | 21 | |
| pN2 | 48 | 35 | 13 | |
| Grade | ||||
| I | 79 | 38 | 41 | <0.05 |
| II | 45 | 25 | 20 | |
| III | 48 | 32 | 16 | |
| Stage | ||||
| I | 24 | 9 | 15 | 0.65 |
| II | 132 | 66 | 66 | |
| III–IV | 16 | 7 | 9 | |
| ER | ||||
| + | 106 | 37 | 69 | 0.864 |
| − | 66 | 24 | 42 | |
| PR | ||||
| + | 96 | 46 | 50 | 0.65 |
| − | 76 | 37 | 40 | |
| HER-2 | ||||
| + | 47 | 25 | 22 | 0.288 |
| − | 125 | 70 | 55 | |
ARPC2, actin-related protein 2/3 complex; ER, estrogen receptor; PR, progesterone receptor.
Oncomine, The cancer genome atlas data and cBioPortal analysis
The Oncomine microarray database (http://www.oncomine.org) was used for analysis. We screened for relevant entries on ARPC2 expression in BrCa tissues and normal breast tissues. With ‘Cancer vs. normal’ analysis as the primary filter. Data sets were used in the present study in accordance with Ma et al (23), Curtis et al (24), Perou et al (25) and Zhao et al (26). We screened for differentially expressed genes (Pearson's correlation >0.4) in several important node locations of ARPC2 in BrCa (Pearson's correlation >0.4) by using BioPortal for Cancer Genomics (http://www.cbioportal.org). All searches were performed according to the cBioPortal's online instructions.
Cell culture
Human BrCa cell lines (MCF-10A, MCF-7, T47D, BT474 and MDA-MB-231) were obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA) and cultured under ATCC-recommended conditions. Cells were routinely incubated at 37°C and 5% CO2 under a humidified atmosphere.
ARPC2 overexpression and downregulation
The full length of Homo sapiens ARPC2 was synthesized and subcloned into an entry vector by OriGene Technologies Technologies, Inc., (Beijing, China). siRNA-ARPC2 (siARPC2) was purchased from Shanghai GenePharma Co., Ltd. (Shanghai, China). MCF-7 and MDA-MB-231 cells were transfected using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) in accordance to the manufacturer's instructions. Transfection efficiency was confirmed by performing reverse transcription-quantitative real-time polymerase chain reaction (qRT-PCR) and western blotting.
Western blotting and RT-PCR
We used RT-PCR to evaluate the mRNA levels of ARPC2. Total RNA was isolated from cultured cells using TRIzol reagent. RNA was reverse transcribed into cDNA using the PrimeScript RT reagent kit (Takara Biotechnology Co., Ltd., Dalian, China). qRT-PCR was performed using the SYBR Prime Script kit (Takara Bio, Inc., Otsu, Japan). ARPC: Forward primer, 5′-TCCGACTCTACCAGCTGATGC-3′ and reverse primer, 5′-AAGCTGGACTCATCCCACAGC-3′. PCR was conducted at 95°C for 15 min, followed by 40 cycles of 94°C for 15 sec, 55°C for 30 sec, and 64°C for 30 sec. Experiment was repeated three times and the expression of ARPC2 was quantified using the 2−ΔΔCq method (27). Cells were lysed via a lysis buffer which consisted of RIPA (Beyotime Institute of Biotechnology, Haimen, China). The protein concentration was determined using BCA kit. Equal amounts of total protein (10 µg) were separated by 10% SDS polyacrylamide gels and transferred onto polyvinylidene fluoride (PVDF) membranes, followed by blocking with 5% non-fat milk for 1 h. The membranes were incubated with primary antibodies overnight at 4°C. Protein expression was quantified by densitometry (Quantity One software; Bio-Rad Laboratories, Hercules, CA, USA). The antibodies used for western blotting were: N-cadherin (cat. no. 4061; Cell Signaling Technology, Inc., Danvers, MA, USA), E-cadherin (cat. no. 5296; Cell Signaling Technology, Inc.), vimentin (cat. no. 5296; Cell Signaling Technology, Inc.), ZEB1 (cat. no. 3396; Cell Signaling Technology, Inc.) and MMP-9 (cat no. 3852; Cell Signaling Technology, Inc.), MMP-3 (cat. no. ab52915; Abcam, Cambridge, UK), β-actin (cat. no. ab5694; Abcam), ARPC2 (cat. no. ab11798; Abcam) and GAPDH (cat. no. g9545; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). The bands were displayed by ECL illumination and analyzed by the Tanon 5200 image acquisition system (Tanon Science and Technology Co., Ltd., Shanghai, China).
Colony formation assays
For the colony formation assay, transfected cells were grown in a fresh 6-well plate with RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum. For the cell colony formation assay, the transfected cells were seeded in 6-well plates (1×103/well) and cultured for 14 days at 37°C. After 14 days, all visible colonies per well were manually counted. Assays were performed in triplicate, and the results were calculated as the mean ± standard deviation (SD).
Transwell assay
The siRNA-control (siControl) and siRNA-ARPC2 (siARPC2) were transfected into MCF-7 and MDA-MB-231 cells for 48 h. For the migration assay, 5×104 cells were seeded with serum-free RPMI-1640 medium in the upper chamber of Transwell inserts (8-µm; Corning Incorporated, Corning, NY, USA). For the invasion assay, inserts were coated with Matrigel (Sigma-Aldrich; Merck KGaA). For the migration and invasion assays, the lower chamber of the Transwell unit was filled with culture medium supplemented with 10% serum. After 36–48 h, the cell filters were fixed using methanol and stained with 0.1% crystal violet. The number of cells was manually counted. Each experiment was assessed in triplicate.
Wound healing assay
Cells (5×106) were seeded in 6-well plates. Once the cells achieved 100% confluence, the culture plate was scratched along its central axis of with 200-µl sterile pipette tips. An inverted microscope was used to observe wound healing at three time-points (0, 24 and 48 h) after wounding. The wound healing rate was calculated and wounds were observed and images were captured. The assays were performed three times.
In vivo tumor formation
Five-week-old, female, and BALB/c nude mice (7 in each group) were used for the xenograft tumor formation assay. All animal testing was performed at the Model Animal Research Center of The University of Science and Technology of China (Hefei, China). All of the experiments were approved by the Institute Research Ethics Committee of Anhui Medical University (Hefei, China). The suspended cells (5×106) were injected into each mouse on the right side of the posterior flank. Seven days after injections, tumor nodules were checked every 5 days and tumor volumes were calculated by width × length × (width + length)/2. All tumors were removed from the nude mice after 40 days.
IHC staining
For IHC staining, 4-µm sections of the TMA blocks were incubated overnight with a pre-diluted goat antihuman ARPC2 monoclonal antibody (Fuzhou Maixin Biotech Co., Ltd., Fuzhou, China). Immunoreactivity was evaluated semi-quantitatively, and the result was calculated on the basis of the percentage of positive cells. Immunostaining was considered as low positive when the percentage of the stained tumor cells was <30%, and as high positive when the percentage of stained tumor cells exceeded 30%.
Cell cycle and apoptosis analyses
Cells (0.5×106−1×106) were washed twice with cold phosphate-buffered saline. Cells were stained with propidium iodide (PI; Cycletest™ Plus DNA reagent kit; BD Biosciences, Franklin Lakes, NJ, USA) in accordance with the manufacturer's protocol. Analyses and measurements were performed using FACSVerse flow cytometer (BD Biosciences). The percentage of cells in the S, G0/G1 and G2/M phases were counted and compared. Similarly, cell apoptosis was analyzed by flow cytometry using the Annexin V-FITC/PI Apoptosis Detection kit (BD Biosciences) 48 h after transfection.
Statistical analysis
Data were analyzed with SPSS 19.0 software (SPSS Inc., Chicago, IL, USA). The association between the ARPC2 expression level and the clinopathological characteristics of patients was analyzed by the Chi-square test. Student's t-test was used for comparing the differences between groups. Cumulative recurrence and survival rates were analyzed by the Kaplan-Meier method. P<0.05 was considered to indicate a statistically significant difference.
Results
ARPC2 expression levels in different BrCa cell lines
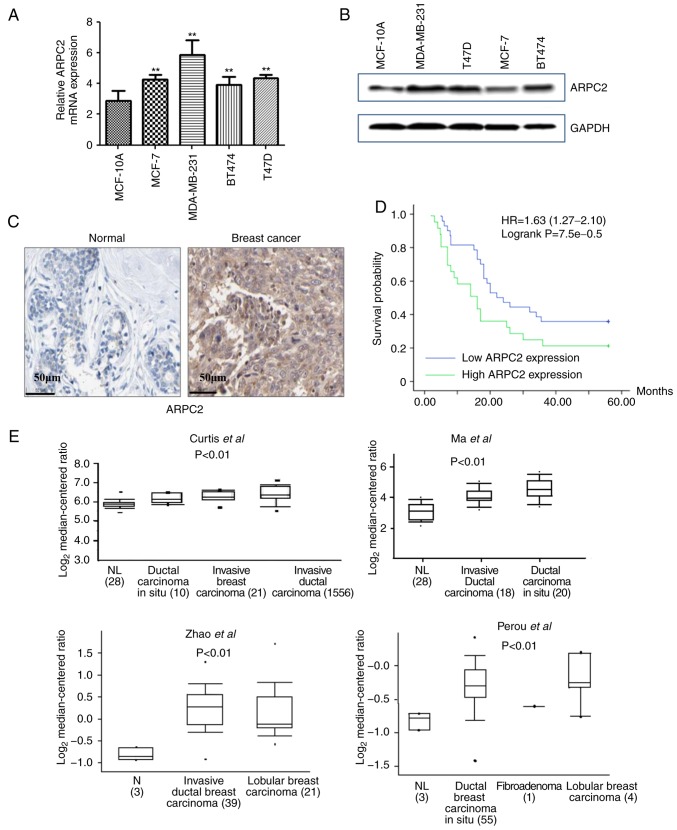
We quantified the expression of ARPC2 in different types of human BrCa cell lines (MCF-7, T47D, BT474 and MDA-MB-231) and human breast epithelial cell line (MCF-10A) by RT-PCR and western blotting. As revealed in Fig. 1A and B, we found that ARPC2 was highly expressed in BrCa cell lines. However, changes in ARPC2 expression in four types of BrCa cell lines did not exhibit a consistent pattern. Since MCF-7 and MDA-MB-231 cell lines had a good tumorigenic effect, we selected MCF-7 and MB-MDA-231 cell lines as experimental models.
Figure 1.
ARPC2 is upregulated in breast cancer cell lines and specimens from patients with breast cancer. (A) The level of ARPC2 mRNA was higher in breast cancer cell lines than MCF-10A non-malignant cells through qRT-PCR. **P<0.01. (B) ARPC2 protein expression in breast cancer cell lines was higher than in MCF-10A non-malignant cells. (C) Relative ARPC2 expression in breast cancer and normal breast tissues analyzed though IHC. Breast cancer cells feature strong-moderate cytoplasmic staining; normal tissues exhibit weak staining. (D) Kaplan-Meier overall survival curves generated in accordance to the ARPC2 protein levels of patients with breast cancer (low vs. high); ARPC2 was associated with worse overall survival. (E) Ratio of ARPC2 mRNA in different BrCa types was analyzed using TCGA database. ARPC2, actin-related protein 2/3 complex; TCGA, The Cancer Genome Atlas. The dots mean extreme outlier.
ARPC2 mRNA and protein expression in BrCa and normal tissues
We performed IHC to assess ARPC2 protein expression in different types of BrCa (n=172) and paired normal breast (n=68) tissues. As revealed in Fig. 1C, the stain for ARPC2 protein expression was predominantly located in the cytosol. Our results demonstrated that the expression level of ARPC2 protein in normal tissues was significantly lower than in the cancer tissues (Table I). The Kaplan-Meier curve and log-rank test analyses revealed that ARPC2 expression was associated with the OS of patients with BrCa. We evaluated the prognostic value of ARPC2 using the protein expression data and survival information of 172 patients with BrCa. We divided patient samples into two groups in accordance with the median expression (high vs. low expression). As revealed in Fig. 1D, high ARPC2 levels were associated with a low OS rate. Moreover, to illustrate the potential relationship between ARPC2 and BrCa progression, we used the Oncomine database (http://www.oncomine.org) for the analysis of ARPC2 mRNA expression. We found that ARPC2 mRNA expression was significantly upregulated in BrCa relative to that in normal breast tissues (Fig. 1E).
Relationship of ARPC2 with the clinicopathological characteristics and poor prognosis of patients with BrCa
We analyzed the relationship between ARPC2 protein expression and the clinicopathological parameters of patients with BrCa. As presented in Table II, ARPC2 protein expression was significantly and positively associated with lymph node metastasis, tumor size and grade of BrCa (P<0.05). The protein expression of ARPC2, however, was not significantly associated with other clinical parameters, including age, HER-2 expression, and estrogen and progesterone receptor status of patients with BrCa.
ARPC2 silencing inhibits the migration and invasion capabilities of BrCa cell lines
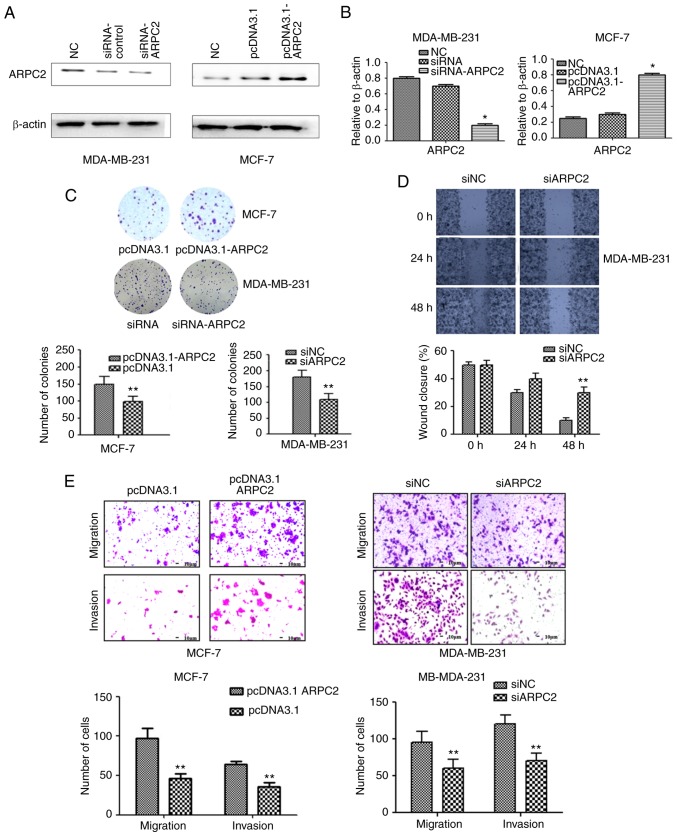
To delineate the role of ARPC2 in BrCa tumorigenesis, we knocked out ARPC2 in MDA-MB-231 cells and overexpressed ARPC2 expression in MCF-7 cell lines. We confirmed ARPC2 expression by performing western blotting and qRT-PCR (Fig. 2A and B). To further confirm the role of ARPC2 in BrCa migration and invasion, we used a colony formation and scratch wound healing assays (Fig. 2C and D). The colony forming ability of cells was decreased and the percentage of wound closure was increased by ARPC2 silencing in MDA-MB-231 cells. Similarly, the migration and the invasion of MDA-MB-231 was also inhibited by ARPC2 silencing as determined by Transwell assays. Thus, the inducive effect of ARPC2 in BrCa cell migration and invasion was revealed.
Figure 2.
Effect of ARPC2 on MCF-7 or MDA-MB-231 cells which were grown and transfected with siRNA-ARPC2 or pcDNA3.1-ARPC2. (A) ARPC2 expression level was confirmed at the transcription level by western blotting. Cells were transfected with siRNAs or plasmid targeting ARPC2 to knock down or induce ARPC2 expression. (B) ARPC2 mRNA expression level was confirmed by RT-PCR. *P<0.05. (C) Cell colony formation assay was performed in MDA-MB-231 or MCF-7 cells after transfection. **P<0.01. (D) A wound healing assay; the migrated cells were fixed and counted. Data represent the average of three experiments. **P<0.01. (E) Cell migration was examined by Boyden chamber assay and type I collagen-coated Transwell. **P<0.01. Cell invasion was examined using Matrigel-coated Transwell cell culture chambers. Ability of migration and invasion of cells were quantified by counting the number of cells that migrated or invaded the underside of the porous polycarbonate membrane under a phase-contrast microscope. ARPC2, actin-related protein 2/3 complex.
ARPC2 overexpression inhibits the growth of BrCa cells
We assessed the effects of pcDNA3.1-ARPC2 transfection on cellular proliferation and growth through colony formation assays. Our results demonstrated that pcDNA3.1-ARPC2 displayed a relatively rapid proliferation rate compared to control cells when MCF-7 cells were transfected with pcDNA3.1-ARPC2. The colony forming ability (Fig. 2C) as well as the migration and invasion abilities (Fig. 2E) were enhanced as determined by colony formation and Transwell assays.
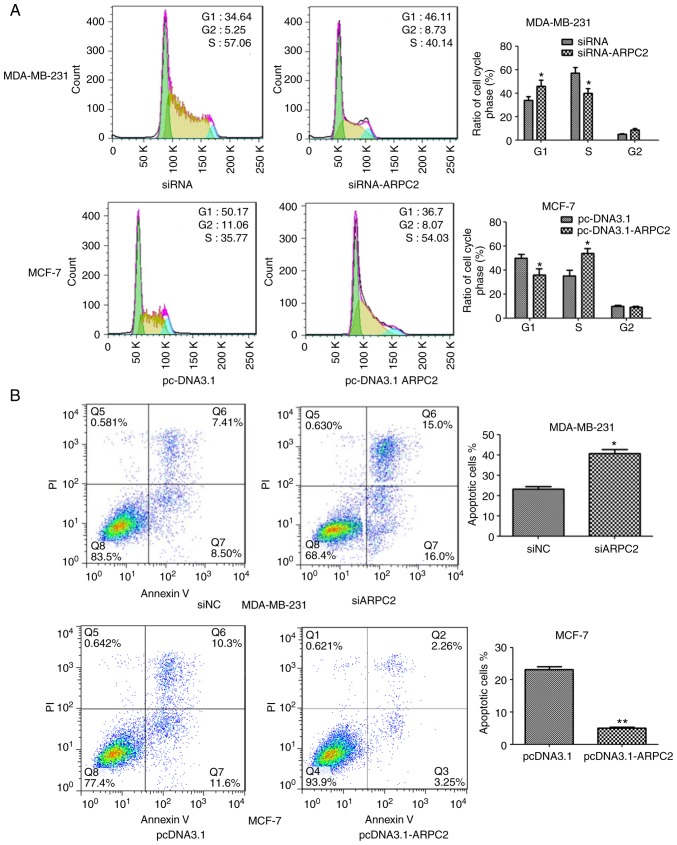
ARPC2 silencing inhibits BrCa cell proliferation and induces apoptosis
We downregulated ARPC2 expression in the MDA-MB-231 cell line through transfection with an ARPC2-specific siRNA or a non-specific siRNA control. As revealed in Fig. 3A, ARPC2 silencing altered the cell cycle of the MDA-MB-231 cell line. We found that a higher number of siARPC2 MDA-MB-231 cells were in the G1 phase compared with the control cells. In addition, the transfection of siARPC2 increased apoptosis of MDA-MB-231 cells, as determined by performing Annexin V-FITC/PI staining (Fig. 3B).
Figure 3.
(A) The cell cycle was analyzed by flow cytometry. *P<0.05 (B) Transfected cells were cultured for an apoptosis assay, and stained with Annexin V and PI. *P<0.05, **P<0.01. PI, propidium iodide.
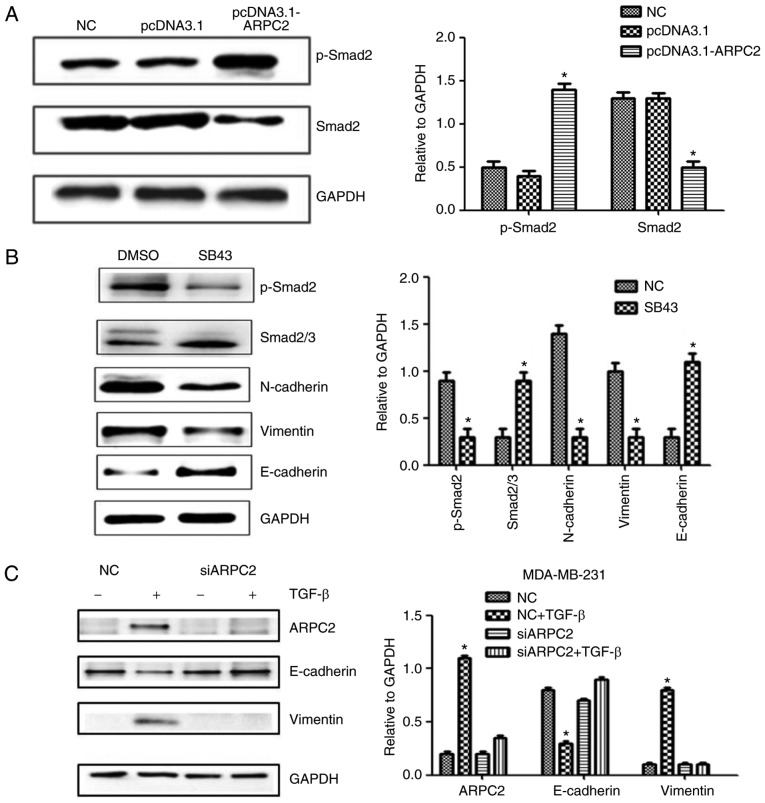
TGF-β signaling is activated in ARPC2-induced EMT
ARPC2 overexpression significantly increased the level of phosphorylated Smad2 (p-Smad2) protein, a downstream effector of the TGF-β pathway (Fig. 4A). Moreover, ARPC2 overexpression efficiently increased phosphorylated Smad2 levels. To further validate that TGF-β signaling is responsible for ARPC2-induced EMT, we used TGF-β receptor kinase inhibitor SB43 to block TGF-β signaling. The results revealed that suppression of TGF-β signaling reduced Smad2 phosphorylation levels and downregulated vimentin expression (Fig. 4B). To investigate whether ARPC2 is required for TGF-β-induced EMT, we transfected MDA-MB-231 cells with siARPC2 or non-target control siRNA and examined their responses to TGF-β treatments after 48 h. It was determined that TGF-β stimulation induced EMT transfer in MDA-MB-231-siCtrl cells, but not in MB-MDA-231-siARPC2 cells. TGF-β treatment inhibited E-cadherin expression and increased vimentin expression. However, under the same conditions E-cadherin and vimentin expression levels were not altered in MB-MDA-231-siARPC2 cells (Fig. 4C). These experiments suggested that ARPC2 is involved in TGF-β-induced EMT.
Figure 4.
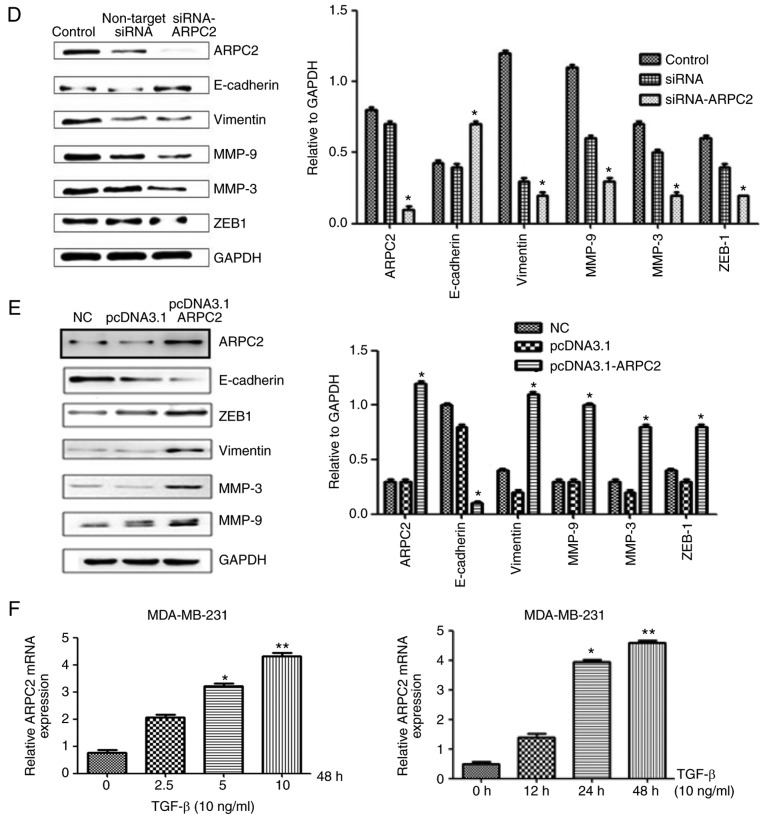
Western blotting detects EMT-related protein expression levels after ARPC2 knockdown or overexpression. (A) MCF-7 cells were stably transfected with NC or pcDNA3.1-ARPC2, and the levels of p-Smad2 and Smad2/3 were assessed by western blotting. *P<0.05. (B) MCF-7 cells were treated with 10 ng/l SB43 or DMSO for 48 h, and the protein expression levels of EMT markers, p-Smad2 and total Smad2/3 protein were assessed by western blotting. *P<0.05. (C) Western blotting was performed to examine ARPC2, E-cadherin, and vimentin expression after 48 h of treatment with TGF-β in siCtrl and siARPC2. *P<0.05. (D) Effect of ARPC2 downregulation, analysis of the expression of ARPC2, E-cadherin, vimentin, MMP-9, MMP-3 and ZEB-1 by western blotting. *P<0.05. (E) Effect of ARPC2 overexpression. Analysis of the level of ARPC2, E-cadherin, vimentin, MMP-9, MMP-3 and ZEB-1 by western blotting. *P<0.05. (F) ARPC2 mRNA expression levels in MDA-MB-231 cells treated with different concentrations of TGF-β at 48 h and (10 ng/ml) TGF-β at different time-points. EMT, epithelial-mesenchymal transition; ARPC2, actin-related protein 2/3 complex. *P<0.05, **P<0.01.
ARPC2 regulates EMT-related protein expression
Cellular adhesive ability is associated with EMT. To determine whether ARPC2 expression triggers EMT in BrCa cells, we examined MMP-3, MMP-9 (cell migration marker), E-cadherin (epithelial cell marker), N-cadherin, vimentin (mesenchymal marker) and ZEB-1 protein expression after transfection with siARPC2 and pcDNA3.1-ARPC2 (Fig. 4D and E). Compared to the control cell line, the expression levels of, vimentin, MMP-3, MMP-9 and ZEB-1 were upregulated by transfection with pcDNA3.1-ARPC2 and the level of E-cadherin was downregulated. These results demonstrated that ARPC2 stimulated the expression of EMT-related proteins and EMT-TFs. The transfection of pcDNA3.1-ARPC2 resulted in a markedly high invasiveness of BrCa. Knockdown of ARPC2 by siRNA resulted in the inhibition of invasion and metastasis of BrCa. To examine whether the TGF-β could stimulate the expression of ARPC2 in MDA-MB-231 cells, cells were incubated with the presence 10 ng/ml TGF-β for 12, 24 and 48 h. Subsequently, we used 2.5, 5, and 10 ng/ml TGF-β to culture cells for 48 h. qPCR analysis revealed that ARPC2 mRNA expression was time- and dose-dependent on TGF-β stimulation (Fig. 4F).
ARPC2 overexpression promotes mammary tumor growth in a xenograft model
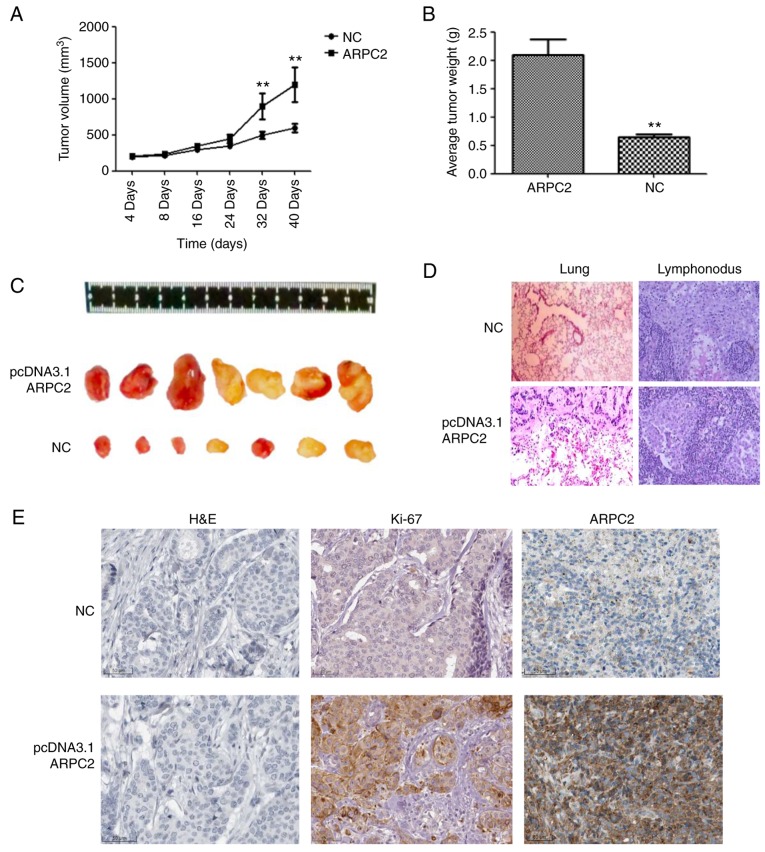
We demonstrated the biological role of ARPC2 in MDA-MB-231 cells through an in vivo experiment. We transfected the ARPC2 vector into MDA-MB-231 cells which were then subcutaneously injected into BALB/c nude mice. MDA-MB-231 and ARPC2-MDA-MB-231 groups were examined 40 days after implantation (Fig. 5). The ARPC2-MDA-MB-231 group of mice formed significantly larger tumor sizes than the MDA-MB-231 control group (Fig. 5C). As revealed in Fig 5, a high ARPC2 level increased tumor growth and weight (Fig. 5A and B), and generated lung or lymphonodus metastases in nude mice (Fig. 5D). We subsequently explored the effects of ARPC2 on tumor growth in vivo by IHC, the results of which revealed that tumour growth and tumor weight were significantly increased in the ARPC2-overexpression group compared with the negative control group and that the Ki-67 expression levels in the ARPC2-overexpression group were higher than those in the negative control group.
Figure 5.
Effect of ARPC2 on MCF-7 cells in vivo. (A) The subcutaneous tumor growth curves. **P<0.01. (B) The quantification of the tumor weight. **P<0.01. (C) The representative images of subcutaneous implanted tumor cells. (D) Lung and lymphonodus sections were counterstained with H&E. (E) Immunohistochemical index Ki-67 and ARPC2-positively stained cells in the tissue sections of a xenograft model compared to the control group. ARPC2, actin-related protein 2/3 complex; H&E, hematoxylin and eosin.
Discussion
Recurrent and metastatic BrCa gravely threatens the health of women worldwide. Thus, intensive studies on the potential candidate molecules involved in the metastasis of BrCa are urgently required for the development of BrCa treatment. Previous studies on microarray profiling have revealed that ARPC2 is aberrantly expressed during BrCa progression. In the present study, it was revealed that ARPC2 expression was increased in BrCa cell lines (MCF-7, BT474, MDA-MB-231 and T47D). Furthermore, ARPC2 expression was uniquely linked with the invasion, apoptosis and proliferation of mammary carcinoma cells. This positive association was further supported by evidence obtained through in vitro studies and a nude mouse model. The analysis of clinical specimens, IHC results, and clinicopathological and prognostic information indicated that ARPC2 was significantly associated with tumor size, lymph node metastasis, and tumor grade. Moreover, Kaplan-Meier analyses revealed that ARPC2 expression was linked with the poor OS of patients with BrCa. Therefore, we proposed that ARPC2 has regulatory functions in BrCa development and that it can be treated as an independent prognostic marker for BrCa.
The evolutionarily conserved Arp2/3 complex consists of five subunits (ARPC1-5) (28,29), and two actin-related proteins ARP2 and ARP3. ARPC2 can promote actin assembly in lamellipodia and participate in lamellipodial protrusions, which promote cell shape change and locomotion. ARPC2, along with some co-expressed genes, plays a crucial role in invasion and metastasis (30–32). To explore the regulatory network of ARPC2, we found that ARPC2 may be involved in EMT, a key early event in breast tumor invasion and metastasis. A growing body of evidence has revealed that EMT, a specific physiological or pathophysiological phenomenon, is characterized by the conversion of epithelial cells into mesenchymal-like cells through the loss of polarity and disruption of cell-cell connections (33,34). Although the significance of EMT-induced carcinogenesis has been defined, its molecular mechanism remains unclear.
EMT involves the dissolution of epithelial tight junctions, the modulation of adherent junctions, the remodeling of the cytoskeleton, and the loss of apical-basal polarity. Matrix metalloproteinases (MMPs) are EMT-related proteins (35) and ZEB1 is crucial for the transcriptional regulation of EMT (14). N-cadherin and vimentin are present in the microfilaments (actins) and microtubules (tubulins) (36,37). Arp2/3 subunits exhibit a tissue-specific expression pattern characterized by the increased expression of mesenchymal proteins. To directly test this hypothesis, we quantified the expression levels of ARPC2, E-cadherin, vimentin, ZEB1, MMP-9 and MMP-3 proteins. We found that in vitro ARPC2 increased breast tumor cell migration and invasion, as well as vimentin, ZEB1, MMP-9 and MMP-3 proteins. The results demonstrated that ARPC2 is an oncogene or has oncogenic effects on BrCa cell growth and metastasis by stimulating EMT. Recently, a study revealed that a TGF-β/Smad-dependent gene regulated fibronectin and smooth muscle actin (38) in human lung mesenchymal cells. In the present study, we found that TGF-β signaling was activated in ARPC2-induced EMT. In addition, TGF-β signaling induced ARPC2 expression in breast epithelial cells, and the knockdown of ARPC2 blocked TGF-β-induced EMT. We proposed that a feedback loop exists between ARPC2 and TGF-β expression. This feedback loop could regulate EMT and BrCa progression.
To the best of our knowledge, we are the first to report that ARPC2 plays a critical role in BrCa in vitro and in vivo. ARPC2 overexpression promoted BrCa cell proliferation, invasion, and metastasis through EMT. Our results indicated that ARPC2 is a potential biomarker or therapeutic target for BrCa.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Scientific Research Foundation of the Institute for Translational Medicine of Anhui Province, SRFITMAP (no. 2017zhyx36).
Availability of data and materials
All data generated or analyzed during the present study are included in this published article.
Authors' contributions
QW conceived and designed the study and the experiments and acquired the funding. ZC, WW and YH performed the experiments. JW selected the patients and collected the clinical samples; YS, YH and JW analyzed the data. ZW, XD and YS collected data. ZC and WW wrote the manuscript. ZW and XD reviewed and edited the manuscript. All authors have read and approved the final manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work re appropriately investigated and resolved.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Anhui Medical University (Hefei, China) and consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
There authors declare that they have no competing interests.
References
- 1.DeSantis CE, Ma J, Goding Sauer A, Newman LA, Jemal A. Breast cancer statistics, 2017, racial disparity in mortality by state. CA Cancer J Clin. 2017;67:439–448. doi: 10.3322/caac.21412. [DOI] [PubMed] [Google Scholar]
- 2.Ngamcherdtrakul W, Castro DJ, Gu S, Morry J, Reda M, Gray JW, Yantasee W. Current development of targeted oligonucleotide-based cancer therapies: Perspective on HER2-positive breast cancer treatment. Cancer Treat Rev. 2016;45:19–29. doi: 10.1016/j.ctrv.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maruti SS, Willett WC, Feskanich D, Rosner B, Colditz GA. A prospective study of age-specific physical activity and premenopausal breast cancer. J Natl Cancer Inst. 2008;100:728–737. doi: 10.1093/jnci/djn135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hiatt RA, Klabunde C, Breen N, Swan J, Ballard-Barbash R. Cancer screening practices from National Health Interview Surveys: Past, present, and future. J Natl Cancer Inst. 2002;94:1837–1846. doi: 10.1093/jnci/94.24.1837. [DOI] [PubMed] [Google Scholar]
- 5.Slatkin M. Linkage disequilibrium-understanding the evolutionary past and mapping the medical future. Nat Rev Genet. 2008;9:477–485. doi: 10.1038/nrg2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim IA, No M, Lee JM, Shin JH, Oh JS, Choi EJ, Kim IH, Atadja P, Bernhard EJ. epigenetic modulation of radiation response in human cancer cells with activated EGFR or HER-2 signaling: Potential role of histone deacetylase 6. Radiother Oncol. 2009;92:125–132. doi: 10.1016/j.radonc.2009.03.008. [DOI] [PubMed] [Google Scholar]
- 7.Lakhani SR, Van De Vijver MJ, Jacquemier J, Anderson TJ, Osin PP, McGuffog L, Easton DF. The pathology of familial breast cancer: Predictive value of immunohistochemical markers estrogen receptor, progesterone receptor, HER-2, and p53 in patients with mutations in BRCA1 and BRCA2. J Clin Oncol. 2002;20:2310–2318. doi: 10.1200/JCO.2002.09.023. [DOI] [PubMed] [Google Scholar]
- 8.Kyndi M, Sørensen FB, Knudsen H, Overgaard M, Nielsen HM, Overgaard J, Danish Breast Cancer Cooperative Group Estrogen receptor, progesterone receptor, HER-2, and response to postmastectomy radiotherapy in high-risk breast cancer: The Danish Breast Cancer Cooperative Group. J Clin Oncol. 2008;26:1419–1426. doi: 10.1200/JCO.2007.14.5565. [DOI] [PubMed] [Google Scholar]
- 9.Jeng RL, Goley ED, D'Alessio JA, Chaga OY, Svitkina TM, Borisy GG, Heinzen RA, Welch MD. A Rickettsia WASP-like protein activates the Arp2/3 complex and mediates actin-based motility. Cell Microbiol. 2004;6:761–769. doi: 10.1111/j.1462-5822.2004.00402.x. [DOI] [PubMed] [Google Scholar]
- 10.Goley ED, Welch MD. The ARP2/3 complex: An actin nucleator comes of age. Nat Rev Mol Cell Biol. 2006;7:713–726. doi: 10.1038/nrm2026. [DOI] [PubMed] [Google Scholar]
- 11.Frank DJ, Hopmann R, Lenartowska M, Miller KG. Capping protein and the Arp2/3 complex regulate nonbundle actin filament assembly to indirectly control actin bundle positioning during Drosophila melanogaster bristle development. Mol Biol Cell. 2006;17:3930–3939. doi: 10.1091/mbc.e06-06-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Al Ghouleh I, Rodríguez A, Pagano PJ, Csányi G. Proteomic analysis identifies an NADPH oxidase 1 (Nox1)-mediated role for actin-related protein 2/3 complex subunit 2 (ARPC2) in promoting smooth muscle cell migration. Int J Mol Sci. 2013;14:20220–20235. doi: 10.3390/ijms141020220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang J, Liu Y, Yu CJ, Dai F, Xiong J, Li HJ, Wu ZS, Ding R, Wang H. Role of ARPC2 in human gastric cancer. Mediators Inflamm. 2017;2017:5432818. doi: 10.1155/2017/5432818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moreno-Bueno G, Portillo F, Cano A. Transcriptional regulation of cell polarity in EMT and cancer. Oncogene. 2008;27:6958–6969. doi: 10.1038/onc.2008.346. [DOI] [PubMed] [Google Scholar]
- 15.Wright JA, Richer JK, Goodall GJ. microRNAs and EMT in mammary cells and breast cancer. J Mammary Gland Biol Neoplasia. 2010;15:213–223. doi: 10.1007/s10911-010-9183-z. [DOI] [PubMed] [Google Scholar]
- 16.Creighton CJ, Chang JC, Rosen JM. Epithelial-mesenchymal transition (EMT) in tumor-initiating cells and its clinical implications in breast cancer. J Mammary Gland Biol Neoplasia. 2010;15:253–260. doi: 10.1007/s10911-010-9173-1. [DOI] [PubMed] [Google Scholar]
- 17.Hardy KM, Booth BW, Hendrix MJ, Salomon DS, Strizzi L. ErbB/EGF signaling and EMT in mammary development and breast cancer. J Mammary Gland Biol Neoplasia. 2010;15:191–199. doi: 10.1007/s10911-010-9172-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doble BW, Woodgett JR. Role of glycogen synthase kinase-3 in cell fate and epithelial-mesenchymal transitions. Cells Tissues Organs. 2007;185:73–84. doi: 10.1159/000101306. [DOI] [PubMed] [Google Scholar]
- 19.Xu J, Lamouille S, Derynck R. TGF-beta-induced epithelial to mesenchymal transition. Cell Res. 2009;19:156–172. doi: 10.1038/cr.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kudo-Saito C, Shirako H, Takeuchi T, Kawakami Y. Cancer metastasis is accelerated through immunosuppression during snail-induced EMT of cancer cells. Cancer Cell. 2009;15:195–206. doi: 10.1016/j.ccr.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 21.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 22.Gavert N, Ben-Ze'ev A. Epithelial-mesenchymal transition and the invasive potential of tumors. Trends Mol Med. 2008;14:199–209. doi: 10.1016/j.molmed.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 23.Ma XJ, Dahiya S, Richardson E, Erlander M, Sgroi DC. Gene expression profiling of the tumor microenvironment during breast cancer progression. Breast Cancer Res. 2009;11:R7. doi: 10.1186/bcr2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 26.Zhao H, Langerod A, Ji Y, Nowels KW, Nesland JM, Tibshirani R, Bukholm IK, Kåresen R, Botstein D, Børresen-Dale AL, et al. Different gene expression patterns in invasive lobular and ductal carcinomas of the breast. Mol Biol Cell. 2004;15:2523–2536. doi: 10.1091/mbc.e03-11-0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Abella JV, Galloni C, Pernier J, Barry DJ, Kjær S, Carlier MF, Way M. Isoform diversity in the Arp2/3 complex determines actin filament dynamics. Nat Cell Biol. 2016;18:76–86. doi: 10.1038/ncb3286. [DOI] [PubMed] [Google Scholar]
- 29.Robinson R, Turbedsky K, Kaiser D, Marchand J, Higgs H, Choe S, Pollard T. Crystal structure of Arp2/3 complex. Science. 2001;294:1679–1684. doi: 10.1126/science.1066333. [DOI] [PubMed] [Google Scholar]
- 30.Daugherty KM, Goode BL. Functional surfaces on the p35/ARPC2 subunit of Arp2/3 complex required for cell growth, actin nucleation, and endocytosis. J Biol Chem. 2008;283:16950–16959. doi: 10.1074/jbc.M800783200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saedler R, Mathur N, Srinivas BP, Kernebeck B, Hülskamp M, Mathur J. Actin control over microtubules suggested by DISTORTED2 encoding the Arabidopsis ARPC2 subunit homolog. Plant Cell Physiol. 2004;45:813–822. doi: 10.1093/pcp/pch103. [DOI] [PubMed] [Google Scholar]
- 32.Franke A, Balschun T, Karlsen TH, Sventoraityte J, Nikolaus S, Mayr G, Domingues FS, Albrecht M, Nothnagel M, Ellinghaus D, et al. Sequence variants in IL10, ARPC2 and multiple other loci contribute to ulcerative colitis susceptibility. Nat Genet. 2008;40:1319–1323. doi: 10.1038/ng.221. [DOI] [PubMed] [Google Scholar]
- 33.Shapiro IM, Cheng AW, Flytzanis NC, Balsamo M, Condeelis JS, Oktay MH, Burge CB, Gertler FB. An EMT-driven alternative splicing program occurs in human breast cancer and modulates cellular phenotype. PLoS Genet. 2011;7:e1002218. doi: 10.1371/journal.pgen.1002218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cannito S, Novo E, di Bonzo LV, Busletta C, Colombatto S, Parola M. Epithelial-mesenchymal transition: From molecular mechanisms, redox regulation to implications in human health and disease. Antioxid Redox Signal. 2010;12:1383–1430. doi: 10.1089/ars.2009.2737. [DOI] [PubMed] [Google Scholar]
- 35.Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: Regulators of the tumor microenvironment. Cell. 2010;141:52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mendez MG, Kojima S, Goldman RD. Vimentin induces changes in cell shape, motility, and adhesion during the epithelial to mesenchymal transition. FASEB J. 2010;24:1838–1851. doi: 10.1096/fj.09-151639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prasain N, Stevens T. The actin cytoskeleton in endothelial cell phenotypes. Microvasc Res. 2009;77:53–63. doi: 10.1016/j.mvr.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hur W, Rhim H, Jung CK, Kim JD, Bae SH, Jang JW, Yang JM, Oh ST, Kim DG, Wang HJ, et al. SOX4 overexpression regulates the p53-mediated apoptosis in hepatocellular carcinoma: Clinical implication and functional analysis in vitro. Carcinogenesis. 2010;31:1298–1307. doi: 10.1093/carcin/bgq072. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during the present study are included in this published article.