Abstract
The high mortality of colorectal cancer (CRC) is likely caused by early invasion and metastasis. The chemoresistance of tumor cells is the critical reason for treatment failure. The present study aimed to develop targeted solutions to overcome chemotherapy drug resistance in CRC. CCK-8 assay was used to examine SW480 cell viability. SW480 cell apoptosis was examined using flow cytometry. The present study demonstrated that the expression of miR-1271 was significantly decreased in CRC tumors and cell lines compared with control tissues. Furthermore, the expression of microRNA (miR)-1271 was increased and decreased following the transfection of miR-1271 mimics and an inhibitor, respectively. Furthermore, miR-1271 regulated mammalian target of rapamycin (mTOR) expression by directly binding to the mTOR 3′-untranslated region and the relative luciferase activity of mTOR was decreased following miR-1271 overexpression. The results of the present study indicate that miR-1271 may be a potential target for anti-CRC therapy, particularly in the sensitivity of chemotherapeutic drugs. miR-1271 may therefore enhance the sensitivity of CRC cells to chemotherapy drugs and provide a novel approach for the gene therapy of CRC.
Keywords: colorectal cancer, microRNA, microRNA-1271, mechanistic target of rapamycin
Introduction
Colorectal cancer (CRC) has one of the highest morbidity and mortality rates of tumors worldwide. There are ~1,000,000 newly diagnosed cases, at an increasing rate of >20% per year (1). The 5-year survival rate of CRC is <15% due to the poor sensitivity of radiotherapy and chemotherapy (2). In the past decades, progress has been achieved in the CRC field, particularly in molecular carcinogenesis and novel targeted therapies (3–5). However, the overall survival rate of CRC remains low (6,7). The resistance of tumor cells to anticancer drugs is the primary factor affecting the curative effect of CRC chemotherapy (8).
At present, the standard therapy for CRC is surgical intervention followed by combination chemotherapy, consisting of cisplatin and platinum-based drugs (9). Cisplatin, one of the most effective non-specific cell cycle drugs, serves a pivotal role in the treatment of CRC (10,11). Cisplatin is an effective primary chemotherapeutic agent for the treatment of CRC. The development of drug resistance has resulted in poor survival rates in CRC and has become an increasing clinical challenge for patients with cancer (12). Therefore, there is an urgent requirement to develop targeted solutions to overcome drug resistance in the chemotherapeutic treatment of CRC based on the distinct molecular background of CRC and the mechanism of resistance to platinum-based drugs.
MicroRNAs (miRNA) (13,14) are a type of short single small non-coding RNA that are induced via the specific combination with target gene mRNA (mRNA). This interaction induces the degradation or inhibits the translation of mRNA to change the expression of target proteins and regulate their signaling pathways, leading to the occurrence and development of tumors (15). miRNA can regulate the expression of hundreds of target genes, the expression profiles of which differ significantly between tumor and normal tissues (16). It is also associated with the early diagnosis and prognostic prediction of tumors, alongside individualized treatment with novel molecular markers that exhibit tumor specificity (17). miRNAs are abnormally expressed or mutated in a variety of tumors (18). Therefore, it has been hypothesized that miRNAs serve the role of an oncogene or tumor suppressor gene in tumor development (13,19). A widened understanding of the association between miRNA and CRC may provide novel agents for its diagnosis, treatment and prognosis.
The change in the expression of miRNAs serves an important role in the resistance to anticancer drugs. Thus, determining the association between miRNA expression and drug resistance will provide a novel mechanism to reverse resistance to anticancer drugs. The primary mechanism used to achieve this, is the regulation of target gene expression by directly targeting the 3′-untranslated region (UTR) of mRNA (20). Recently, numerous studies have revealed that miRNAs are involved in the regulation of cancer occurrence and development (21–24). Specifically, miR-1271 serves an important role in the regulation of CRC cell apoptosis, proliferation and metastasis (25). The present study revealed that miR-1271 was abnormally expressed in CRC tissues; however, the mechanism of miR-1271 function in CRC remains unclear. Therefore, the aim of the present study was to assess the influence of miR-1271 on the sensitivity of CRC to chemotherapeutic drugs and the possible mechanisms of molecular regulation in CRC.
Materials and methods
Clinical sample collection
Tumor tissues and adjacent normal tissues (>2 cm away from the tumor tissue) were extracted from 30 patients with CRC (male, 17; female, 13; age range, 45–63 years; mean age, 56 years) who had undergone surgical resection at Shanghai Jiao Tong University Affiliated Sixth People's Hospital (Shanghai, China). All enrolled patients had not received chemotherapy or radiotherapy prior to the study. Tissue samples were stored in liquid nitrogen at −80°C until further use. Written informed consent was obtained from each patient or their relatives prior to surgery. The present study was approved by the Ethics Committee of Shanghai Jiao Tong University Affiliated Sixth People's Hospital (Shanghai, China).
Cell culture
The CRC SW480 cell line was obtained from the American Type Culture Collection (Manassas, VA, USA). Cell lines were cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA), supplemented with 10% fetal bovine serum (Hyclone; GE Healthcare Life Sciences, Logan, UT, USA) and 100 U/ml penicillin and 100 µg/ml streptomycin (Invitrogen; Thermo Fisher Scientific, Inc.) in an incubator with a humidified atmosphere at 5% CO2 and 37°C. Cell propagation was performed for 2 days. Cells in the logarithmic phase of growth were used for subsequent experimentation.
Drug treatment and cell transfection
Cells were treated with 10 µg/ml of cisplatin (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). SW480 cells in the logarithmic stage were were transfected with 200 nmol/l miR-1271 mimic, miR-NC mimic, 200 nmol/l miR-1271 inhibitor or miR-NC inhibitor (all Thermo Fisher Scientific, Inc.) using Lipofectamine RNAiMAX (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Following 72-h transfection, the cisplatin-miR-1271 mimic, cisplatin-miR-1271 inhibitor and the cisplatin group were treated with cisplatin (10 µg/ml) for 48 h at 37°C. The cells of the control group were untreated. The sequences of miR-1271 mimics and miR-NC mimics were as follows: 5′-CUUGGCACCUAGCAAGCACUCA-3′ and 3′-UUCUCCGAACGUGUCACGUTT-5′. The sequences of miR-1271 inhibitor and miR-NC inhibitor were 5′-UGAGUGCUUGCUAGGUGCCAAG-3′ and 5′-UUGUACUACACAAAAGUACUG-3′, respectively.
Drug sensitivity assay
Cell proliferation was assessed in vitro using the cell counting kit-8 (CCK-8) assay (Dojindo Molecular Technologies, Inc., Kumamoto, Japan). Briefly, SW480 cells were transfected with miR-1271 mimic, inhibitor or negative control for 72 h and incubated with cisplatin for 48 h (as described previously). Cell were seeded into 96-well plates at a density of 5.0×103 cells/well and incubated in 100 µl RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.) at 37°C. Following 0, 12, 24 or 48 h incubation, 10 µl CCK-8 was added to each well and incubated for a further 2 h at 37°C. The absorbance of each sample was measured at a wavelength of 450 nm using a microplate reader (Bio-Rad Laboratories Inc., Hercules, CA, USA).
Flow cytometry
Cell apoptosis was analyzed using the Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide apoptosis detection kit (cat. no. 556570; BD Biosciences, San Jose, CA, USA). Briefly, cells were collected and resuspended at ~5×105 cells/ml in binding solution. Cells were transferred into the flow tube (100 µl/tube) and incubated for 5 min at room temperature with 5 µl Annexin V/FITC and PI. Following incubation, cells were washed thrice with ice-cold PBS and apoptotic cells were analyzed using a FACS Calibur flow cytometer and CellQuest software (version 6.0; BD Biosciences).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cells and tissues using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). Total RNA was reverse transcribed into cDNA using the Primer-Script™ One Step RT-PCR kit (Takara Biotechnology Co., Ltd., Dalian, China). qPCR was subsequently performed using SYBR® Premix Ex Taq (Takara Biotechnology Co., Ltd.), according to the manufacturer's protocol. The following thermocycling conditions were used: 95°C for 5 min; 40 cycles of 95°C for 10 sec, 60°C for 20 sec and 72°C for 20 sec. Differences in the expression of Bcl-2-associated X (Bax), B-cell lymphoma-2 (Bcl-2), caspase-3 and mammalian target of rapamycin (mTOR) in the four groups were compared. The relative mRNA expression levels were quantified using the 2−ΔΔCq method (26) and normalized to the internal control GAPDH.
For the detection of miRNA, total RNA was isolated using the miRNA Extraction kit (Qiagen GmbH, Hilden, Germany). Total RNA was reverse transcribed into cDNA using the miRNA cDNA Synthesis kit (Qiagen GmbH). qPCR was subsequently performed using the SYBR® Green (Takara Biotechnology Co., Ltd.), according to the manufacturer's protocol. The experiment was performed as described above. U6 served as an internal control. The sequences of the primers utilized are listed in Table I.
Table I.
Oligonucleotide primer pairs used for quantitative polymerase chain reaction.
| Gene | Primer sequence (5′-3′) |
|---|---|
| miR-1271 | F: CAGCACTTGGCACCTAGCA |
| R: TATGGTTGTTCTCCTCTCTGTCTC | |
| U6 | F: CTCGCTTCGGCAGCACA |
| R: AACGCTTCACGAATTTGCGT | |
| Bax | F: CACCAGCTCTGAACAGATCATGA |
| R: TCAGCCCATCTTCTTCCAGATGT | |
| Bcl-2 | F: CACCCCTGGCATCTTCTCCTT |
| R: AGCGTCTTCAGAGACAGCCAG | |
| caspase-3 | F: AACTGGACTGTGGCATTGAG |
| R: ACAAAGCGACTGGATGAACC | |
| mTOR | F: AACAACGGCTTTCCACCAGG |
| R: CACCCTAAGTGAGCCCTTGGA | |
| GAPDH | F: GGAGCGAGATCCCTCCAAAAT |
| R: GGCTGTTGTCATACTTCTCATGG |
miR, microRNA; Bax, Bcl-2-associated X; Bcl-2, B cell lymphoma-2; mTOR, mammalian target of rapamycin.
Western blot analysis
Following transfection, SW480 cells from each group were collected and lysed using radioimmunoprecipitation assay buffer (Beyotime Institute of Biotechnology, Haimen, China) on ice. Total protein was quantified using a bicinchoninic acid assay kit (Beyotime Institute of Biotechnology). The supernatant was used to detect its protein concentration following high-speed centrifugation at 11,180 × g for 15 min at 4°C. Total protein was quantified using a bicinchoninic acid assay (Beyotime Institute of Biotechnology) and 50 µg protein/lane was separated via SDS-PAGE on a 10% gel. The separated proteins were transferred onto polyvinylidene fluoride membranes and blocked for 2 h at 37°C with non-fat milk. The membranes were incubated with primary antibodies against Bax (cat. no. B3428; 1:1,000), Bcl-2 (cat. no. SAB3500343; 1:1,000), caspase-3 (cat. no. C9598; 1:1,000), mTOR (cat. no. SAB2701843; 1:1,000) and anti-GAPDH (cat. no. G9545; 1:100; all Sigma-Aldrich; Merck KGaA) overnight at 4°C. The membranes were subsequently incubated with horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (cat. no. ab205718; 1:5,000; Abcam, Cambridge, UK) for 1 h at room temperature. Protein bands were visualised using an the ECL Plus immunoblotting detection system (Thermo Fisher Scientific, Inc.) and quantified using Gel-Pro analyzer software (version 6.0; Media Cybernetics, Inc., Rockville, MD, USA). The results are expressed as the ratio of objective protein to GAPDH.
Bioinformatics analysis and dual-luciferase reporter assay
TargetScan bioinformatics software (www.targetscan.org/vert_71) was used to predicted mTOR was a target of miR-1271. The potential miR-1271 binding site in the 3′UTR of mTOR and the corresponding mutant were synthesized and cloned into the pGL3 vector (Switchgear Genomics, Menlo Park, CA, USA). The cDNA fragment corresponding to the wild-type 3′-UTR of mTOR were inserted downstream of the luciferase in the psiCHECK-2 reporter plasmid (cat. no. C8021; Promega Corporation, Madison, WI, USA). A deletion mutant lacking the seed sequence of the miR-1271 binding site in the 3′-UTR of mTOR was created using the QuikChange Site-directed Mutagenesis kit (Agilent Technologies, Inc., Santa Clara, CA, USA). For the reporter assay, SW480 cells were co-transfected with miR-1271 mimic, inhibitor, or negative control, and WT/MUT-3′UTR of mTOR. Following 72-h transfection, luciferase activities were detected using the Dual-Luciferase Reporter assay system (Promega Corporation), according to manufacturer's protocol. Firefly luciferase activity was normalized to Renilla luciferase activity.
Statistical analysis
SPSS 20.2 software (IBM Corp., Armonk, NY, USA) was used to analyze the data. Data are presented as the mean ± standard deviation of ≥3 independent experiments. A two-tailed Student's t-test was used to analyze the difference between two groups. One-way analysis of variance followed by a Newman-Keuls post-hoc test was employed to analyze data among multiple groups. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-1271 expression decreases in CRC tissue samples
Tumor and matched non-tumor tissue samples were profiled using RT-qPCR to identify the expression of miR-1271 in CRC. The results revealed that miR-1271 expression was significantly decreased in tumor tissues compared with non-tumor tissues, indicating that miR-1271 was abnormally expressed at low levels in CRC tissues (Fig. 1).
Figure 1.
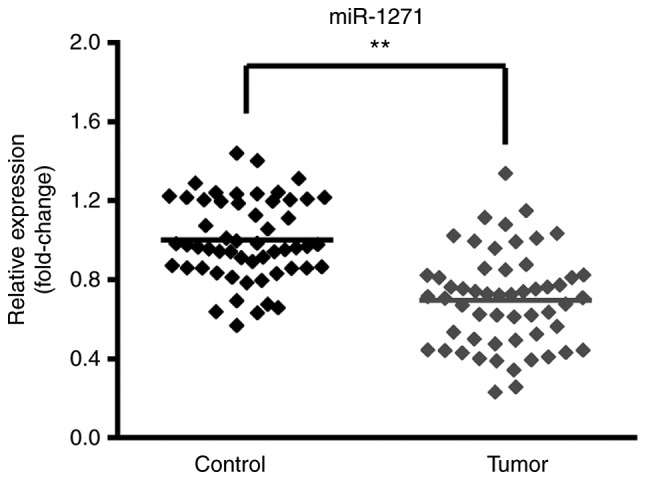
miR-1271 expression in CRC tissues. Reverse transcription-quantitative polymerase chain reaction confirmed that miR-1271 was downregulated in CRC tissues compared with adjacent normal tissues (control). **P<0.01 vs. Control. miR, microRNA; CRC, colorectal cancer.
Expression of miR-1271
SW480 cells were transfected with miR-1271 mimics, miR-NC mimics, a miR-1271 inhibitor or a miR-NC inhibitor. The expression of miR-1271 was significantly increased and decreased following transfection with miR-1271 mimics and the miR-1271 inhibitor, respectively (Fig. 2A and B).
Figure 2.
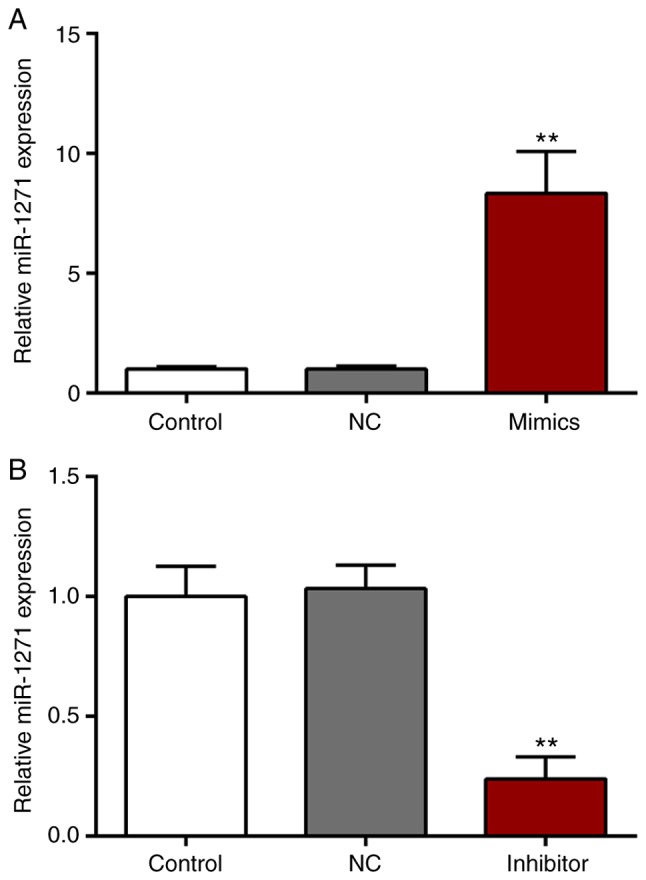
miR-1271 expression in CRC cell line. (A) SW480 cells were transfected with miR-1271 mimics or a miR-NC inhibitor. No significance was observed between the NC and control group. Compared with NC group, the expression of miR-1271 was significantly upregulated. **P<0.01 vs. miR-NC mimics. (B) Following transfection with a miR-1271 inhibitor, the expression of miR-1271 was significantly decreased compared with the miR-NC inhibitor. **P<0.01 vs. miR-NC inhibitor. miR, microRNA; CRC, colorectal cancer; NC, negative control.
miR-1271 inhibits SW480 cell viability
A CCK-8 was performed to assess the effect of SW480 cell activity and proliferation (Fig. 3). The results revealed that, compared with the cisplatin group, the cisplatin + miR-1271 mimics group significantly decreased the cell viability of SW480 cells at 48 h. However, when compared with the cisplatin group, the transient downregulation of miR-1271 increased the cell viability of cisplatin-treated SW480 cells.
Figure 3.
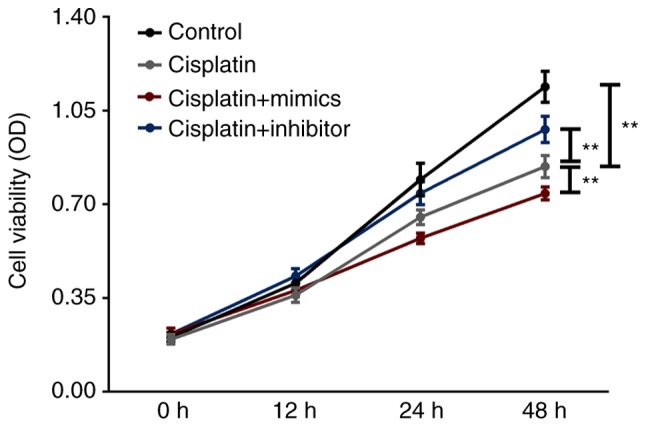
miR-1271 upregulation inhibits SW480 cell viability. Cell proliferation was examined was assessed in the cisplatin, cisplatin+mimics, cisplatin+inhibitor and the control group via a cell counting kit-8 assay. **P<0.01 vs. Cisplatin group. miR, microRNA.
miR-1271 promotes SW480 cell apoptosis
Flow cytometry was performed to assess the effect of miR-1271 on the apoptosis of cisplatin-treated SW480 cells (Fig. 4). The results demonstrated that transfection of miR-1271 mimics induced a significant increase in SW480 cell apoptosis compared with the cisplatin group. Furthermore, transfection with the miR-1271 inhibitor significantly decreased the rate of apoptosis induced by cisplatin treatment.
Figure 4.

miR-1271 upregulation promotes SW480 cell apoptosis. (A) SW480 cell apoptosis was assessed in the cisplatin, cisplatin+mimics, cisplatin+inhibitor and the control group via flow cytometric analysis. (B) Quantitative analysis of A. **P<0.01 vs. Cisplatin group. miR, microRNA; PI, propidium iodide; FITC, fluorescein isothiocyanate.
mTOR expression
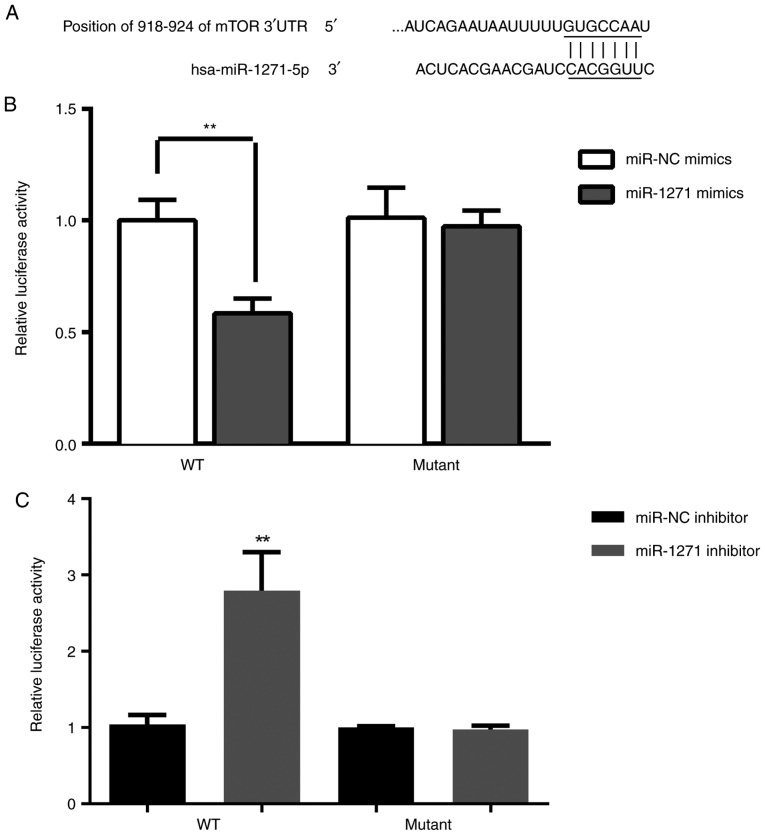
To further gain insight into the potential underlying mechanism of miR-1271 in chemoresistance, TargetScan (www.targetscan.org/vert_71) was used to predict the targets of miR-1271. It was revealed that mTOR can be directly regulated by miR-1271 and is therefore a potential candidate target gene for further study (Fig. 5A). Dual-luciferase reporter assay demonstrated that the luciferase activity in SW480 cells was significantly decreased following co-transfection with miR-1271 mimic and WT 3′-UTR of mTOR (Fig. 5B). Furthermore, the luciferase activity was significantly increased following co-transfection with miR-1271 inhibitor and WT 3′-UTR of mTOR (Fig. 5C). The results of RT-qPCR and western blotting were used to determine the effect of miR-1271 expression on the regulation of mTOR. miR-1271 was revealed to be overexpressed in CRC tissues (Fig. 6A-C).
Figure 5.
mTOR is a direct a target of miR-1271. (A) mTOR 3′-UTR contains two putative miR-1271 binding sites. A luciferase assay was performed to measure relative luciferase activity of mTOR following treatment with (B) miR-1271 mimics and (C) a miR-1271 inhibitor. **P<0.01 vs. miR-NC mimics or miR-NC inhibitor. mTOR, mammalian target of rapamycin; UTR, untranslated region; miR, microRNA; WT, wild-type; NC, negative control.
Figure 6.
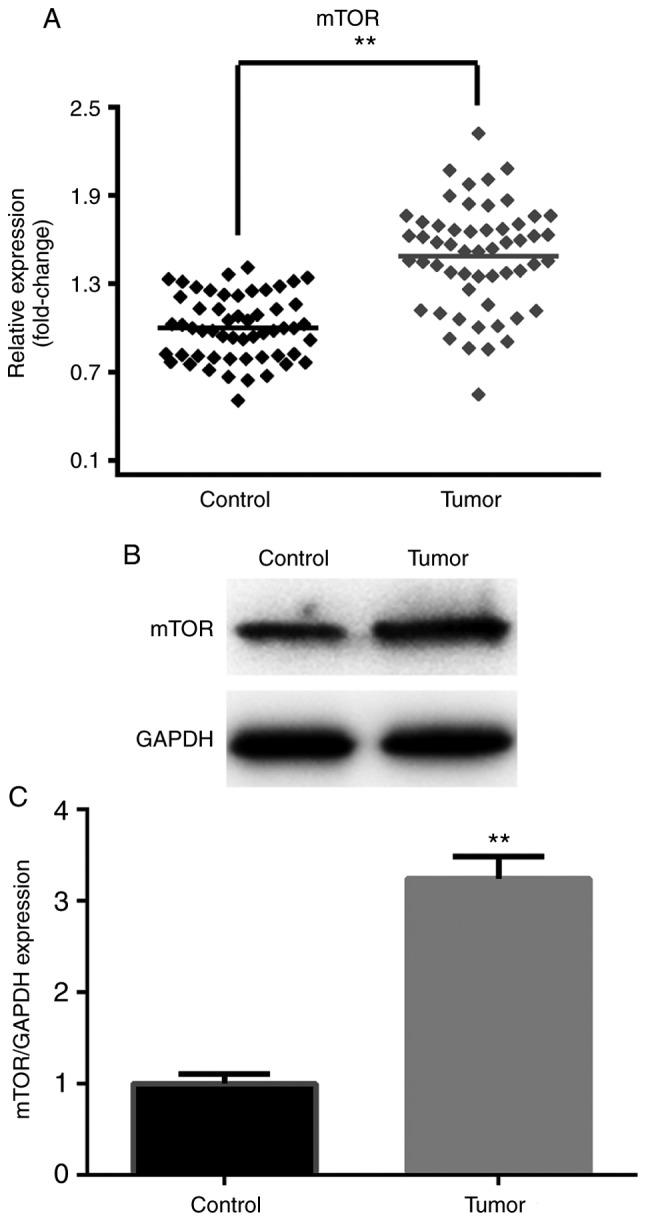
mTOR is upregulated in CRC tissues. (A) Reverse transcription-quantitative polymerase chain reaction, (B) western blotting and (C) densitometry were performed to confirm that mTOR was overexpressed in CRC tissues. **P<0.01 vs. Control. mTOR, mammalian target of rapamycin; CRC, colorectal cancer.
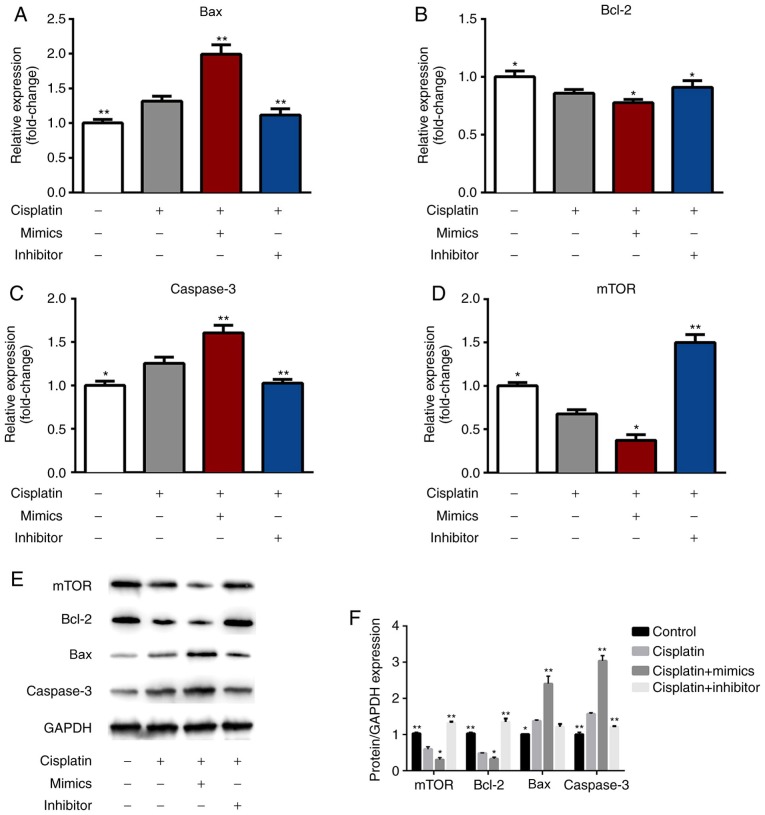
miR-1271 regulated the expression of Bcl-2, mTOR, Bax and Caspase-3
RT-qPCR results revealed that the pro-apoptosis genes (Bax and caspase-3) were upregulated and that the anti-apoptotic gene (Bcl-2) and mTOR were downregulated in the cisplatin + mimics group compared with the cisplatin treatment group (Fig. 7A-D). However, transfection with miR-1271 inhibitor exhibited the opposite effect. The protein expression of Bax and caspase-3 in the cisplatin + mimics group were significantly increased, while the expression of Bcl-2 and mTOR was significantly decreased in the cisplatin + mimics group compared with the cisplatin treatment group (Fig. 7E and F). However, transfection with miR-1271 inhibitor exhibited the opposite effect.
Figure 7.
miR-1271 regulates Bax, Bcl-2, caspase-3 and mTOR expression. Reverse transcription-quantitative polymerase chain reaction was performed to detect the transfection efficiency of control, cisplatin, cisplatin+mimics and cisplatin+inhibitor in SW480 cells. (A) Bax, (B) Bcl-2, (C) caspase-3 and (D) mTOR expression. (E) Western blotting and (F) densitometry were performed to assess the protein levels of Bax, Bcl-2, caspase-3 and mTOR. *P<0.05 vs. Cisplatin group; **P<0.01 vs. Cisplatin group. miR, microRNA; Bax, Bcl-2-associated X; Bcl-2, B cell lymphoma-2; mTOR, mammalian target of rapamycin.
Discussion
Recently, a number of studies have reported that abnormally expressed miRNAs are associated with cancer due to their affect on cell proliferation, apoptosis and invasion (27–29). miRNAs may thus serve as biomarkers for the diagnosis and prognosis of different types of cancer, including CRC. It is therefore necessary to identify specific miRNAs and their targets as they may provide valuable insight for the diagnosis and treatment of patients with CRC (30). The present study revealed that the expression of miR-1271 was significantly decreased in CRC tumors and cell lines compared with control tissues. Furthermore, the expression of miR-1271 was increased following treatment with miR-1271 mimics and decreased following transfection with a miR-1271 inhibitor.
To the best of our knowledge, it is necessary to identify specific miRNA target genes to elucidate the mechanism involved in the progression and development of cancer. Previous studies have demonstrated that miRNAs regulate their target genes by binding to the mRNA 3′-UTR (31–33), which is consistent with the results of the present study. Furthermore, the relative luciferase activity of mTOR was decreased following miR-1271 overexpression and increased following the suppression of miR-1271 expression.
Cisplatin is the most widely used drug in the chemotherapeutic treatment of CRC and other types of cancer, including ovarian (8), lung (20) and gastric cancer (31). However, the effect of chemotherapy is not satisfactory due to drug resistance (8,34). The resistance of tumor cells is involved in a variety of biochemical cellular changes, including the reduced accumulation of drugs, elevated levels of metallothionein in cells, a higher expression of drug resistance-associated genes and enhanced DNA repair ability (35). At present, no drug has been able to effectively reverse cisplatin resistance in the body (36). The drug sensitivities of miR-1271 mimics and miR-1271 inhibitor were measured via cell proliferation and apoptosis assays. The results indicated that miR-1271 mimics could decrease the proliferation and increase the apoptosis of cisplatin-treated cells, while miR-1271 inhibitor exhibit the opposite results. In addition, the pro-apoptosis genes, Bax and caspase-3, were upregulated, however the anti-apoptotic gene Bcl-2, and mTOR were downregulated in miR-1271-mimic-treated cells. The proportion of Bax and Bcl-2 can be a predictor of apoptosis (37). In the present study, miR-1271 served an anti-tumor role in CRC. Cells transfected with miR-1271 mimics upregulated the pro-apoptotic genes Bax and caspase-3 and downregulated the anti-apoptotic gene Bcl-2. In addition, the miR-1271 inhibitor exhibited adverse effects. The results indicated that the inhibitor reduced cisplatin sensitivity. mTOR has been demonstrated to be involved in the progression and metastasis of various types of cancer (38,39), acting as an oncogene via a variety of mechanisms. mTOR expression was associated with lung and prostate cancer, as well as osteosarcoma (40–42). The present study revealed that miR-1271 may regulate the chemosensitivity of CRC cells by regulating the expression of mTOR.
In conclusion, miR-1271 may be a potential target for antitumor therapy, particularly in the sensitivity of cells to chemotherapy drugs. We hypothesized that miR-1271 regulates chemotherapeutic sensitivity in CRC by targeting mTOR. In order to further clarify the mechanism of chemotherapeutic drug sensitivity, in vivo assays are required to verify the sensitivity of miR-1271 to chemotherapy drugs and to lay a foundation for the application of miR-1271 in future studies.
However, there is a limitation in our study. Since the level of Bax in the cisplatin + mimics group was significantly increased compared with the cisplatin group, it is possible that there may be another mechanism mediated by Bax in these conditions, which should be investigated in future study.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
HY was responsible for drafting the manuscript, as well as the acquisition, analysis and interpretation of data. JZ collected, analyzed and interpreted the data. QS contributed to the conception and design of the current study. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Shanghai Jiao Tong University Affiliated Sixth People's Hospital (Shanghai, China). Written informed consent was obtained from each patient or their relatives prior to surgery.
Patient consent for publication
All patients agreed with the publication of this study.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Li S, Li C, Fang Z. MicroRNA 214 inhibits adipocyte enhancer-binding protein 1 activity and increases the sensitivity of chemotherapy in colorectal cancer. Oncol Lett. 2019;17:55–62. doi: 10.3892/ol.2018.9623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tie Y, Chen C, Yang Y, Qian Z, Yuan H, Wang H, Tang H, Peng Y, Du X, Liu B. Upregulation of let-7f-5p promotes chemotherapeutic resistance in colorectal cancer by directly repressing repressing several pro-apoptotic proteins. Oncol Lett. 2018;15:8695–8702. doi: 10.3892/ol.2018.8410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lo Russo G, Proto C, Garassino MC. Afatinib in the treatment of squamous non-small cell CRC: A new frontier or an old mistake? Transl Lung Cancer Res. 2016;5:110–114. doi: 10.3978/j.issn.2218-6751.2015.12.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kazandjian D, Suzman DL, Blumenthal G, Mushti S, He K, Libeg M, Keegan P, Pazdur R. FDA approval summary: Nivolumab for the treatment of metastatic non-small cell CRC with progression on or after platinum-based chemotherapy. Oncologist. 2016;21:634–642. doi: 10.1634/theoncologist.2015-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Antonia S, Goldberg SB, Balmanoukian A, Chaft JE, Sanborn RE, Gupta A, Narwal R, Steele K, Gu Y, Karakunnel JJ, Rizvi NA. Safety and antitumour activity of durvalumab plus tremelimumab in non-small cell CRC: A multicentre, phase 1b study. Lancet Oncol. 2016;17:299–308. doi: 10.1016/S1470-2045(15)00544-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou YY, Hu ZG, Zeng FJ, Han J. Clinical profile of cyclooxygenase-2 inhibitors in treating non-small cell CRC: A meta-analysis of nine randomized clinical trials. PLoS One. 2016;11:e0151939. doi: 10.1371/journal.pone.0151939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fenchel K, Sellmann L, Dempke WC. Overall survival in non-small cell CRC-what is clinically meaningful? Transl Lung Cancer Res. 2016;5:115–119. doi: 10.3978/j.issn.2218-6751.2016.01.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi X, Xiao L, Mao X, He J, Ding Y, Huang J, Peng C, Xu Z. miR-205-5p mediated downregulation of PTEN contributes to cisplatin resistance in C13K human ovarian cancer cells. Front Genet. 2018;9:555. doi: 10.3389/fgene.2018.00555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu D, Lu P, Mi X, Miao J. Downregulation of miR-503 contributes to the development of drug resistance in ovarian cancer by targeting pi3K p85. Arch Gynecol Obstet. 2018;297:699–707. doi: 10.1007/s00404-018-4649-0. [DOI] [PubMed] [Google Scholar]
- 10.Siddik ZH. Cisplatin: Mode of cytotoxic action and molecular basis of resistance. Oncogene. 2003;22:7265–7279. doi: 10.1038/sj.onc.1206933. [DOI] [PubMed] [Google Scholar]
- 11.Dasari S, Tchounwou PB. Cisplatin in cancer therapy: Molecular mechanisms of action. Eur J Pharmacol. 2014;740:364–378. doi: 10.1016/j.ejphar.2014.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang R, Lin JY, Chi YJ. miR-519d reduces the 5-fluorouracil resistance in colorectal cancer cells by down-regulating the expression of CCND1. Eur Rev Med Pharmacol Sci. 2018;22:2869–2875. doi: 10.26355/eurrev_201805_14989. [DOI] [PubMed] [Google Scholar]
- 13.Paliouras AR, Monteverde T, Garofalo M. Oncogene-induced regulation of microrna expression: Implications for cancer initiation, progression and therapy. Cancer Lett. 2018;421:152–160. doi: 10.1016/j.canlet.2018.02.029. [DOI] [PubMed] [Google Scholar]
- 14.Zhang C, Qian D, Zhao H, Lv N, Yu P, Sun Z. miR17 improves insulin sensitivity through inhibiting expression of ask1 and anti-inflammation of macrophages. Biomed Pharmacother. 2018;100:448–454. doi: 10.1016/j.biopha.2018.02.012. [DOI] [PubMed] [Google Scholar]
- 15.Rupaimoole R, Calin GA, Lopez-Berestein G, Sood AK. miRNA deregulation in cancer cells and the tumor microenvironment. Cancer Discov. 2016;6:235–246. doi: 10.1158/2159-8290.CD-15-0893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yin W, Shi L, Mao Y. miR-194 regulates nasopharyngeal carcinoma progression by modulating MAP3K3 expression. FEBS Open Bio. 2018;9:43–52. doi: 10.1002/2211-5463.12545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rupaimoole R, Slack FJ. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 18.Kaalund SS, Venø MT, Bak M, Møller RS, Laursen H, Madsen F, Broholm H, Quistorff B, Uldall P, Tommerup N, et al. Aberrant expression of miR-218 and miR-204 in human mesial temporal lobe epilepsy and hippocampal sclerosis-convergence on axonal guidance. Epilepsia. 2014;55:2017–2027. doi: 10.1111/epi.12839. [DOI] [PubMed] [Google Scholar]
- 19.Shen J, Li M. Microrna-744 inhibits cellular proliferation and invasion of colorectal cancer by directly targeting oncogene notch1. Oncol Res. 2018 doi: 10.3727/096504018X15188747585738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Q, Zhang B, Sun L, Yan Q, Zhang Y, Zhang Z, Su Y, Wang C. MicroRNA-130b targets PTEN to induce resistance to cisplatin in lung cancer cells by activating Wnt/β-catenin pathway. Cell Biochem Funct. 2018;36:194–202. doi: 10.1002/cbf.3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao Y, Ma H, Gao C, Lv Y, Chen X, Xu R, Sun M, Liu X, Lu X, Pei X, Li P. Tumor-promoting properties of miR-8084 in breast cancer through enhancing proliferation, suppressing apoptosis and inducing epithelial-mesenchymal transition. J Transl Med. 2018;16:38. doi: 10.1186/s12967-018-1419-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu Y, Guo X, Wang J, Liu Y, Gao H, Fan H, Nong X, Yang X, Liu M, Li S, Tang H. A novel microrna identified in hepatocellular carcinomas is responsive to lef1 and facilitates proliferation and epithelial-mesenchymal transition via targeting of nfix. Oncogenesis. 2018;7:22. doi: 10.1038/s41389-017-0010-x. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Xia D, Tian S, Chen Z, Qin W, Liu Q. miR302a inhibits the proliferation of esophageal cancer cells through the mapk and PI3K/Akt signaling pathways. Oncol Lett. 2018;15:3937–3943. doi: 10.3892/ol.2018.7782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu Z, Xu N, Yang W, Liu Y, Yan F. Microrna-411 promoted the osteosarcoma progression by suppressing MTSS1 expression. Environ Sci Pollut Res Int. 2018;25:12064–12071. doi: 10.1007/s11356-018-1331-9. [DOI] [PubMed] [Google Scholar]
- 25.Sun X, Zhai H, Chen X, Kong R, Zhang X. MicroRNA-1271 suppresses the proliferation and invasion of colorectal cancer cells by regulating metadherin/Wnt signaling. J Biochem Mol Toxicol. 2018;32 doi: 10.1002/jbt.22028. Doi: 10.1002/jbt.22028. [DOI] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Xing F, Wang S, Zhou J. The expression of microRNA-598 inhibits ovarian cancer cell proliferation and metastasis by targeting URI. Mol Ther Oncolytics. 2018;12:9–15. doi: 10.1016/j.omto.2018.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, Qin H. miR-338-3p targets RAB23 and suppresses tumorigenicity of prostate cancer cells. Am J Cancer Res. 2018;8:2564–2574. [PMC free article] [PubMed] [Google Scholar]
- 29.Weng L, Ma J, Jia YP, Wu SQ, Liu BY, Cao Y, Yin X, Shang MY, Mao AW. miR-4262 promotes cell apoptosis and inhibits proliferation of colon cancer cells: Involvement of GALNT4. Am J Transl Res. 2018;10:3969–3977. [PMC free article] [PubMed] [Google Scholar]
- 30.Toiyama Y, Okugawa Y, Fleshman J, Richard Boland C, Goel A. MicroRNAs as potential liquid biopsy biomarkers in colorectal cancer: A systematic review. Biochim Biophys Acta Rev Cancer. 2018;1870:274–282. doi: 10.1016/j.bbcan.2018.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang M, Shan X, Zhou X, Qiu T, Zhu W, Ding Y, Shu Y, Liu P. miR-1271 regulates cisplatin resistance of human gastric cancer cell lines by targeting IGF1R, IRS1, mTOR, and BCL2. Anticancer Agents Med Chem. 2014;14:884–891. doi: 10.2174/1871520614666140528161318. [DOI] [PubMed] [Google Scholar]
- 32.Shao Q, Zhang P, Ma Y, Lu Z, Meng J, Li H, Wang X, Chen D, Zhang M, Han Y, et al. Microrna-139-5p affects cisplatin sensitivity in human nasopharyngeal carcinoma cells by regulating the epithelial-to-mesenchymal transition. Gene. 2018;652:48–58. doi: 10.1016/j.gene.2018.02.003. [DOI] [PubMed] [Google Scholar]
- 33.Yang Y, Liu L, Zhang Y, Guan H, Wu J, Zhu X, Yuan J, Li M. miR-503 targets pi3K p85 and IKK-beta and suppresses progression of non-small cell CRC. Int J Cancer. 2014;135:1531–42. doi: 10.1002/ijc.28799. [DOI] [PubMed] [Google Scholar]
- 34.Chen Z, Gao YJ, Hou RZ, Ding DY, Song DF, Wang DY, Feng Y. MicroRNA-206 facilitates gastric cancer cell apoptosis and suppresses cisplatin resistance by targeting MAPK2 signaling pathway. Eur Rev Med Pharmacol Sci. 2019;23:171–180. doi: 10.26355/eurrev_201901_16761. [DOI] [PubMed] [Google Scholar]
- 35.Baguley BC. Multiple drug resistance mechanisms in cancer. Mol Biotechnol. 2010;46:308–316. doi: 10.1007/s12033-010-9321-2. [DOI] [PubMed] [Google Scholar]
- 36.Li J, Sun H, Liu T, Kong J. MicroRNA-423 promotes proliferation, migration and invasion and induces chemoresistance of endometrial cancer cells. Exp Ther Med. 2018;16:4213–4224. doi: 10.3892/etm.2018.6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wei Y, Wu S, Xu W, Liang Y, Li Y, Zhao W, Wu J. Depleted aldehyde dehydrogenase 1A1 (ALDH1A1) reverses cisplatin resistance of human lung adenocarcinoma cell A549/DDP. Thorac Cancer. 2017;8:26–32. doi: 10.1111/1759-7714.12400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li L, Sun JX, Wang XQ, Liu XK, Chen XX, Zhang B, He ZD, Liu DZ, Chen LX, Wang LW, Huang Z. Notoginsenoside R7 suppresses cervical cancer via PI3K/PTEN/Akt/mTOR signaling. Oncotarget. 2017;8:109487–109496. doi: 10.18632/oncotarget.22721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fujishita T, Kojima Y, Kajino-Sakamoto R, Taketo MM, Aoki M. Tumor microenvironment confers mTOR inhibitor resistance in invasive intestinal adenocarcinoma. Oncogene. 2017;36:6480–6489. doi: 10.1038/onc.2017.242. [DOI] [PubMed] [Google Scholar]
- 40.Wang C, Liu E, Li W, Cui J, Li T. miR-3188 inhibits non-small cell lung cancer cell proliferation through FOXO1-mediated mTOR-p-PI3K/AKT-c-JUN signaling pathway. Front Pharmacol Dec. 2018;9:1362. doi: 10.3389/fphar.2018.01362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao S, Zhao Z, Wu R, Wu L, Tian X, Zhang Z. miR-146b inhibits autophagy in prostate cancer through affecting PTEN/AKT/mTOR signaling pathway. Aging (Albany NY) 2019;11:284. doi: 10.18632/aging.101780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Song L, Zhou Z, Gan Y, Li P, Xu Y, Zhang Z, Luo F, Xu J, Zhou Q, Dai F. Long noncoding RNA OIP5-AS1 causes cisplatin resistance in osteosarcoma through inducing the LPAATβ/PI3K/AKT/mTOR signaling pathway by sponging the miR-340-5p. J Cell Biochem. 2018 Dec 11; doi: 10.1002/jcb.28244. (Epub ahead of print). doi: 10.1002/jcb.28244. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.