Abstract
The ubiquitin-specific protease 9X (USP9X) is a conserved deubiquitinase that has been investigated in several types of human cancer. However, the clinical significance and the biological roles of USP9X in prostate cancer remain unexplored. In the present study, an investigation into the expression and clinical significance of USP9X in prostate cancer revealed that USP9X expression was downregulated in prostate cancer tissues compared with that in healthy tissues. In addition, decreased USP9X expression was associated with a higher Gleason score and local invasion. Depletion of USP9X in prostate cancer LNCaP and PC-3 cells by small interfering RNA promoted cell invasion and migration. Furthermore, USP9X depletion upregulated matrix metalloproteinase 9 (MMP9) and the phosphorylation of dynamin-related protein 1 (DRP1). Notably, a significant increase in phosphorylated extracellular signal-regulated kinase (ERK), an upstream activator of MMP9 and DRP1, was observed. To investigate whether ERK activation was able to increase MMP9 protein levels and induce DRP1 phosphorylation, an ERK inhibitor was used, demonstrating that ERK-mediated MMP9 production and change in mitochondrial function was critical for the biological function of USP9X in prostate cancer cells. In conclusion, the present study demonstrated that USP9X is downregulated in prostate cancer and functions as an inhibitor of tumor cell invasion, possibly through the regulation of the ERK signaling pathway.
Keywords: ubiquitin-specific protease 9X, invasion, epithelial-mesenchymal transition, mitochondrial dynamics, prostate cancer
Introduction
Prostate cancer is the second most frequently diagnosed cancer in men, and the second largest cause of cancer-associated mortalities globally in 2018 (1,2). Despite improvements in therapy, the 5-year survival rate of prostate cancer remained at 66.4% for patients diagnosed with prostate cancer in 2012 in China (3,4). Metastasis and invasion are the main causes of prostate cancer-associated mortality (5). Thus, deciphering the mechanism underlying invasive and metastatic behavior is of great importance for the early diagnosis and treatment of prostate cancer.
Ubiquitination and deubiquitination are core regulatory functions in the process of post-translational protein modification. Deubiquitinating enzymes serve an important role in the regulation of multiple biological processes, including cell cycle control, DNA repair, chromatin remodeling, the epithelial-mesenchymal transition (EMT) and several signaling pathways that are frequently dysregulated during tumor development (6).
Ubiquitin-specific protease (USP)9X is a deubiquitinase member of the USP family (7). USP9X has been demonstrated to remove ubiquitin moieties, principally directing target proteins toward proteasomal degradation, and to be involved a variety of diseases, including malignant tumors (8). A previous study identified this protein as a tumor suppressor with prognostic and therapeutic relevance in pancreatic ductal adenocarcinoma (PDA), as its depletion significantly increased colony formation in soft agar and suppressed anoikis in metastatic PDA (9). In clear cell renal cell carcinoma, USP9X expression was reported to be downregulated and associated with a poor prognosis (10). USP9X also ameliorates the oncogenic activity of transcriptional coactivator YAP and tafazzin, which are core components of the Hippo pathway, by deubiquitinating their inhibitor, angiomotin (10). The aforementioned evidence indicates that USP9X may be a potent tumor suppressor. However, whether USP9X exerts a similar effect on human prostate cancer remains unclear.
In the present study, the clinical significance of USP9X in prostate cancer was investigated, and its involvement in the biological features of prostate cancer cells and the underlying mechanisms were explored.
Materials and methods
Samples
A total of 102 cases of prostate adenocarcinoma tissue with no prior adjuvant treatment and 25 cases of normal tissue were obtained from male patients attended at the Second Affiliated Hospital of Kunming Medical University (Kunming, China) between January 2013 and November 2016, with informed consent. The samples were fixed in 10% neutral buffered formalin for 24 h and embedded in paraffin. The mean age ± standard deviation was 68.1±8.67 years (range, 40–87 years). The normal tissues were obtained from patients with benign prostate diseases. An additional set of8 fresh specimens of tumor tissue and adjacent normal tissue were stored at −70°C immediately following resection for subsequent protein expression analysis. This study was conducted with the approval of the Ethics Committee of the Second Affiliated Hospital of Kunming Medical University.
Immunohistochemistry
The sections were deparaffinized in xylene, rehydrated with graded alcohol, and then boiled in 0.01 M citrate buffer (pH 6.0) for 2 min in an autoclave. Hydrogen peroxide (0.3%) was applied to block endogenous peroxide activity, and the sections were incubated with normal goat serum (Fuzhou Maixin Biotech Co., Ltd., Fuzhou, China) at 37°C for 15 min to reduce non-specific binding. The tissue sections (4 µm thickness) were incubated with anti-USP9X antibody (1:400; cat. no. 55054-1-AP; ProteinTech Group, Inc., Chicago, IL, USA) overnight at 4°C. Biotinylated goat anti-mouse or anti-rabbit serum IgG was used as a secondary antibody and the incubation was performed at 37°C for 25 min. Following washing, the sections were incubated at 37°C for 25 min with streptavidin-biotin conjugated with horseradish peroxidase (HRP) (all supplied in the Elivision™ Super HRP IHC Kit-9922; Fuzhou Maixin Biotech Co., Ltd.), and the peroxidase reaction was developed with 3,3′-diaminobenzidine tetrahydrochloride. Counterstaining with hematoxylin was performed at room temperature for 2 min and the sections were dehydrated in ethanol prior to mounting.
Two independent blinded investigators examined all tumor slides randomly and a third pathologist was consulted in the case of any discrepancies. Five views were examined per slide, and 100 cells were observed per view at ×400 magnification using a light microscope. Immunostaining of USP9X was scored on a semiquantitative scale by evaluating the percentage of immunoreactive tumor cells and the staining intensity. Based on previous studies (11,12), the staining intensity was categorized as follows: 0, negative or weak; 1, moderate; and 2, strong. The percentage of stained tumor cells was scored as: 0, 0%; 1 1–5%; 2, 6–25%; 3, 26–75%; and 4, 76–100%. The scores of each tumor sample were multiplied to give a final score of 0–8, and those with a final score of 4–8 were classified as having high USP9X expression. Tumor samples with a score <4 were considered as having low USP9X expression.
Cell culture and reagents
All cell lines were obtained from the Cell Bank of Type Culture Collection of Chinese Academy of Sciences (Shanghai, China). The prostate cancer cell lines, LNCaP, DU145 and PC-3, were maintained in RPMI-1640 medium, minimum essential medium and F-12K medium (Thermo Fisher Scientific, Inc., Waltham, MA, USA), supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Inc.) at 37°C in 5% CO2. The human prostate epithelial RWPE-1 cell line was maintained in K-SFM medium (Thermo Fisher Scientific, Inc.). The extracellular signal-regulated kinase (ERK) inhibitor PD98059 was purchased from Selleck Chemicals (Houston, TX, USA) and the cells were treated at the concentration of 10 µM.
Transfection
ON-TARGETplus small interfering (si)RNAs and control siRNA (siControl) were obtained from GE Healthcare Dharmacon, Inc. (Lafayette, CO, USA). siRNAs were transfected into cells (50 nM) using DharmaFECT1 transfection reagent (GE Healthcare Dharmacon, Inc.) according to the manufacturer's protocol. The sequence of the siControl was 5′-GCGCGATAGCGCGAATATA-3′. The target sequences for USP9X siRNA were: siRNA1, 5′-AGAAAUCGCUGGUAUAAAU-3′; siRNA2, 5′-GUACGACGAUGUAUUCUCA-3′; and siRNA3, 5′-GAAAUAACUUCCUACCGAA-3′. siRNA3 exhibited the best efficiency, as determined by the mRNA and protein measurements described below, and was chosen for the knockdown experiments. Subsequent experiments were performed 55–72 h after transfection.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from cells using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.). For qPCR, cDNA was synthesized using the iScript™ Reverse Transcription SuperMix (Bio-Rad Laboratories, Inc., Hercules, CA, USA) using the following temperature protocol: 5 min at 25°C, 20 min at 46°C and 1 min at 95°C. The relative mRNA expression levels were determined from the cDNA using SYBR Master mix on an ABI 7500 Real-Time PCR system (both Applied Biosystems; Thermo Fisher Scientific, Inc.) using the following thermocycling conditions: 50°C for 2 min, 95°C for 10 min and 40 cycles of 95°C for 15 sec and 60°C for 60 sec. The primer sequences were as follows: USP9X forward, 5′GGACTCCTGGCAAACTCACAGA3′ and reverse, 5′-TCAAACAGCCCATCTCGGTC-3′; and GAPDH forward, 5′-CTGGGCTACACTGAGCACC-3′ and reverse, 5′-AAGTGGTCGTTGAGGGCAATG-3′. Relative expression was normalized against GAPDH and calculated using the comparative Cq method (2−ΔΔCq) (13). All experiments were performed in triplicate.
Western blot analysis
Total protein was extracted using Pierce lysis buffer (Thermo Fisher Scientific, Inc.). Protein quantification was performed using the Bradford method. A total of 50 µg sample protein was separated by 7–15% SDS-PAGE, which was transferred to polyvinylidene fluoride (PVDF) membranes (Merck KGaA, Darmstadt, Germany). The incubation of primary antibodies was performed overnight at 4°C. The antibodies used were against the following proteins: USP9X (1:1,000, cat. no. 55054-1-AP; ProteinTech Group, Inc.), phosphorylated (p-)ERK (cat. no. 4037), ERK (cat. no. 4695), p-protein kinase B (p-AKT; cat. no. 4060), AKT (cat. no. 4685), cyclin D1 (cat. no. 2978), vimentin (cat. no. 5741), p-transcription factor p65 (cat. no. 3033), p-dynamin-related protein 1 (p-DRP1; cat. no. 3455), DRP1 (cat. no. 8570), p65 (cat. no. 4282) (all 1:1,000; Cell Signaling Technology, Inc., Danvers, MA, USA), matrix metalloproteinase 9 (MMP9; 1:1,000; cat. no. ab38898; Abcam, Cambridge, UK), E-cadherin (1:1,500, cat. no. 610181; Becton, Dickinson and Company, Franklin Lakes, NJ, USA) and β-actin (1:2,000; cat. no. sc-47778; Santa Cruz Biotechnology, Inc., Dallas, TX, USA). Following incubation with HRP-coupled anti-mouse or rabbit IgG antibody (1:1,000 dilution; cat. nos. 7076 and 7074, respectively; Cell Signaling Technology, Inc.) at 37°C for 2 h. The target proteins on the PVDF membrane were visualized using the Pierce ECL kit (Thermo Fisher Scientific, Inc.) and captured using the MicroChemi imaging system (DNR Bio-Imaging Systems, Ltd., Neve Yamin, Israel). The western blot intensity was analyzed using ImageJ software version 1.48 (National Institutes of Health, Bethesda, MD, USA).
Cell invasion assay
A cell invasion assay was performed using a 24-well Transwell chamber with a pore size of 8 µm (Costar; Corning, Inc., Corning, NY, USA). The inserts were coated with Matrigel (Becton, Dickinson and Company). At 48 h post-transfection, the cells were trypsinized and 3×105 cells in 100 µl serum-free medium were transferred to the upper Matrigel chamber and incubated for 16 h. Medium supplemented with 10% FBS was added to the lower chamber as the chemoattractant. Following incubation, the non-invaded cells on the upper membrane surface were removed with a cotton tip, and the cells that passed through the filter were fixed with 4% paraformaldehyde and stained with hematoxylin for 2 min at room temperature. Images were captured using alight microscope (Olympus Corporation, Tokyo, Japan) at ×400 magnification. All experiments were performed in triplicate.
Scratch wound-healing assay
To observe cell migration, cells were seeded at a density of ~80% confluence as a monolayer, and scratched with a 1-ml pipette tip. The detached cells were washed away and the remaining cells were cultured for 24 h in corresponding medium (RPMI-1640 medium for the LNCaP cells and F-12K medium for the PC-3 cells) without FBS to slow down the growth rate. Images of the cell monolayer were taken using a light microscope (Olympus Corporation; ×200 magnification) and the width of the scratch wound was measured using ImageJ software version 1.48. All experiments were performed in triplicate.
Cell cycle analysis
For cell cycle analysis, cells were fixed in cold 70% ethanol for ≥1 h at 4°C. The cells were then washed with PBS, pelleted at 3,500 × g at 4°C for 10 min and the supernatant was discarded. The cells were subsequently resuspended in 500 µl PBS containing 10 µg/ml RNase A and 20 µg/ml propidium iodide (PI), and incubated at room temperature in the dark for 30 min. The cell cycle was analyzed by flow cytometry using the FACSCalibur™ flow cytometer and FloJo software version 10 (both Becton, Dickinson and Company).
Apoptosis analysis
Cell apoptosis detection was performed with Annexin V/PI double staining, using the FITC Annexin V Apoptosis Detection kit I (Becton, Dickinson and Company). At 48 h post-transfection, the cells were harvested, washed in chilled PBS and resuspended in 250 µl binding buffer. Annexin V-fluorescein isothiocyanate and PI solutions were added to the cell suspension and incubated for 30 min at room temperature. The cells were analyzed using the FACSCalibur flow cytometer and FlowJo software version 10.
Mitochondrial membrane potential
The mitochondrial membrane potential (Δψm) was detected using JC-1 staining. Briefly, cells were harvested, washed with PBS and incubated with 5 µM JC-1 (Cell Signaling Technology, Inc.) for 30 min at 37°C in the incubator. The cells were then washed and analyzed using the FACSCalibur flow cytometer and FlowJo software version 10.
MTT assay
To determine the proliferation rate of cells, 5,000 cells per well were plated in 96-well plates in medium containing 10% FBS. MTT solution (20 µl, 5 mg/ml) was added into each well and incubated for 1 hat 37°C. Dimethylsulfoxide was added into each well to dissolve the formazan and the dye was measured at 490 nm using a microplate reader (Thermo Fisher Scientific, Inc.).
Proliferation assay
Proliferation was assessed using the CyQUANT™ cell proliferation assay (Invitrogen; Thermo Fisher Scientific, Inc.). The cells were plated in 96-well black fluorescence microtitre plates, at a density of 5,000 cells/well. After 24 h of transfection at 37°C, the medium was discarded, and the plates frozen at −80°C until use. On the day of the analysis, the plates with the adherent cells were thawed and incubated with the CyQUANT dye for 5 min in the dark. The fluorescence was measured on a fluorescence microplate reader (BioTek Instruments, Inc., Winooski, VT, USA) with the excitation set at 480 nm and emission at 520 nm.
Mitochondrial function
To determine the subcellular distribution of mitochondria, the cells were treated with 50 nM MitoTracker™ Red (Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C for 30 min to stain the mitochondria and cell cytosol. The mean mitochondrial length was determined by measuring 10 individual mitochondria from cells obtained by fluorescence microscopy using the cellSens software version 1.16 (Olympus Corporation).
Statistical analysis
SPSS version 16.0 for Windows (SPSS Inc., Chicago, IL, USA) was used for all statistical analyses. The χ2 test was used to examine associations between USP9X expression levels and patient clinicopathological factors. Paired Student's t-test was used for the comparison of the intensity of USP9X in normal and cancer tissues. Analysis of variance, followed by a least-significant-difference post hoc test was performed for the comparison of USP9X expression among the cell lines and comparison of protein change in cells treated with USP9X siRNA and PD98059. Unpaired Student's t-test was used to perform the remaining comparisons. The P-values were based on a two-sided statistical analysis and P<0.05 was considered to indicate statistically significant differences.
Results
USP9X is downregulated in prostate cancer tissues
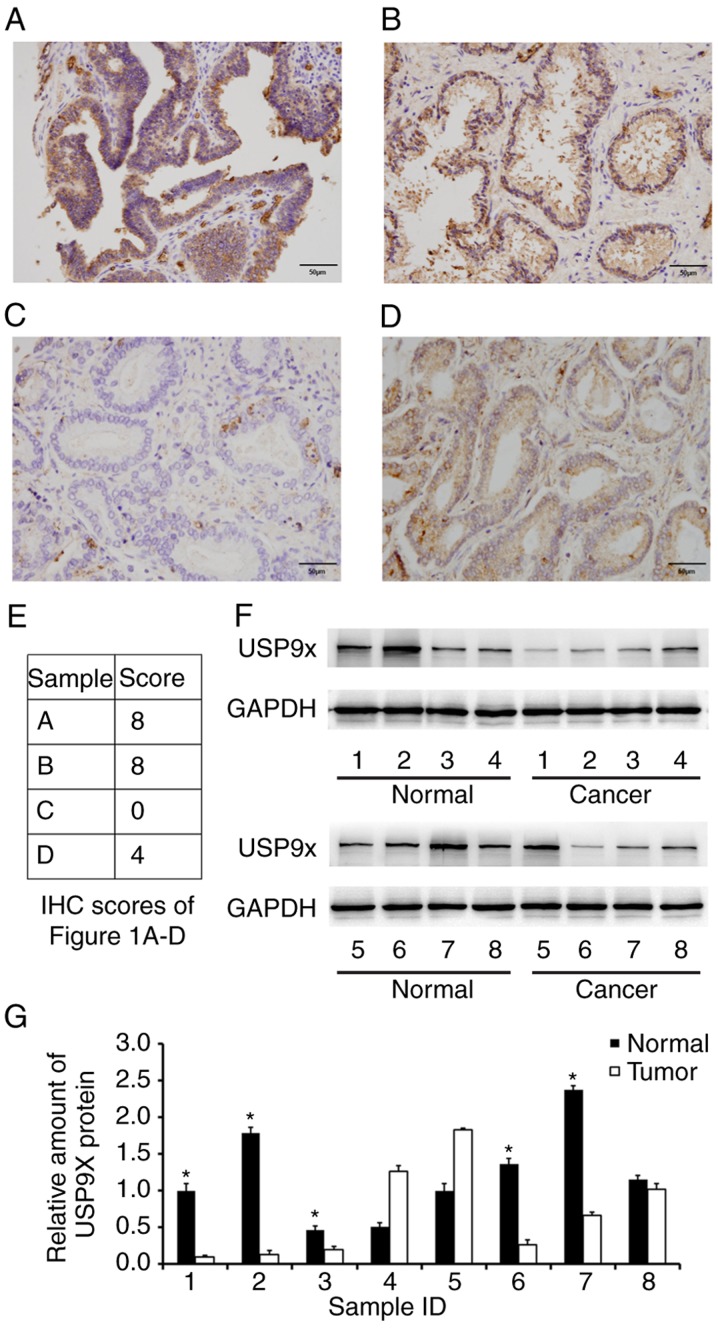
To assess the protein expression and distribution of USP9X in prostate cancer, immunohistochemistry was performed on 102 prostate adenocarcinoma tissue samples and 25 normal prostate tissue samples from different individuals. As displayed in Fig. 1A-D, USP9X was mainly localized in the cytoplasm with sporadic nuclear distribution. Low USP9X protein expression (downregulation) was demonstrated in 54.9% (56/102 samples) of the prostate cancer cases, whereas only 8% (2/25 samples) of the normal tissues exhibited low USP9X expression. The immunostaining scores of the images in Fig. 1A-D are indicated in Fig. 1E. Statistical analysis revealed that low USP9X expression was associated with higher Gleason scores (14) (>7; P=0.0046) and higher T stage (15) (T1-T2 vs. T3-T4, P=0.0138), indicating that downregulation of USP9X is linked with poor differentiation and local invasion. No statistical difference was found between USP9X expression and either age (P=0.7074), tumor-node-metastasis stage (16) (P=0.3103), distal metastasis (P=0.6715) or nodal status (16) (P=0.7045) (Table I). Additionally, USP9X protein expression levels were analyzed in 8 cases of paired prostate cancer/normal tissues using western blotting, and the overall downregulation of USP9X in cancer tissues compared with normal tissues at the protein level was confirmed in 5 out of 8 cases (Fig. 1F and G).
Figure 1.
Expression of USP9X is downregulated in prostate cancer tissues. (A) Strong cytoplasmic USP9X staining in a sample of normal prostate tissue. (B) Strong cytoplasmic and nuclear USP9X staining in a sample of normal prostate tissue. (C) USP9X downregulation (negative staining in most cells) in a sample of prostate cancer tissue. (D) Positive USP9X staining in a case of prostate cancer tissue; magnification, ×400. (E) IHC scores for the USP9X staining in the immunohistochemistry images of Fig. 1A-D. (F) Western blot analysis and (G) the relative quantification of USP9X in 8 paired prostate cancer/normal tissues. *P<0.05 vs. adjacent tumor tissue, using paired Student's t-test. The data are presented as the mean ± standard deviation. USP9X, ubiquitin-specific protease 9X; IHC, immunohistochemistry.
Table I.
Distribution of USP9X status in prostate cancer according to the clinicopathological characteristics of the patients.
| USP9X expression, n | ||||
|---|---|---|---|---|
| Characteristics | Patients, n | Negative/low | High | P-value |
| Age, years | 0.7074 | |||
| <65 | 33 | 19 | 14 | |
| ≥65 | 69 | 37 | 32 | |
| Tumor-node-metastasis stage | 0.3103 | |||
| I–II | 52 | 26 | 26 | |
| III–IV | 50 | 30 | 20 | |
| Gleason score | 0.0046a | |||
| ≤7 | 40 | 15 | 25 | |
| >7 | 62 | 41 | 21 | |
| T stage | 0.0138a | |||
| T1-T2 | 62 | 28 | 34 | |
| T3-T4 | 40 | 28 | 12 | |
| Lymph node metastasis | 0.7045 | |||
| Absent | 76 | 41 | 35 | |
| Present | 26 | 15 | 11 | |
| Metastasis | 0.6715 | |||
| Absent | 71 | 38 | 33 | |
| Present | 31 | 18 | 13 | |
P<0.05. USP9X, ubiquitin-specific protease 9X.
USP9X depletion facilitates invasion and migration in prostate cancer cells
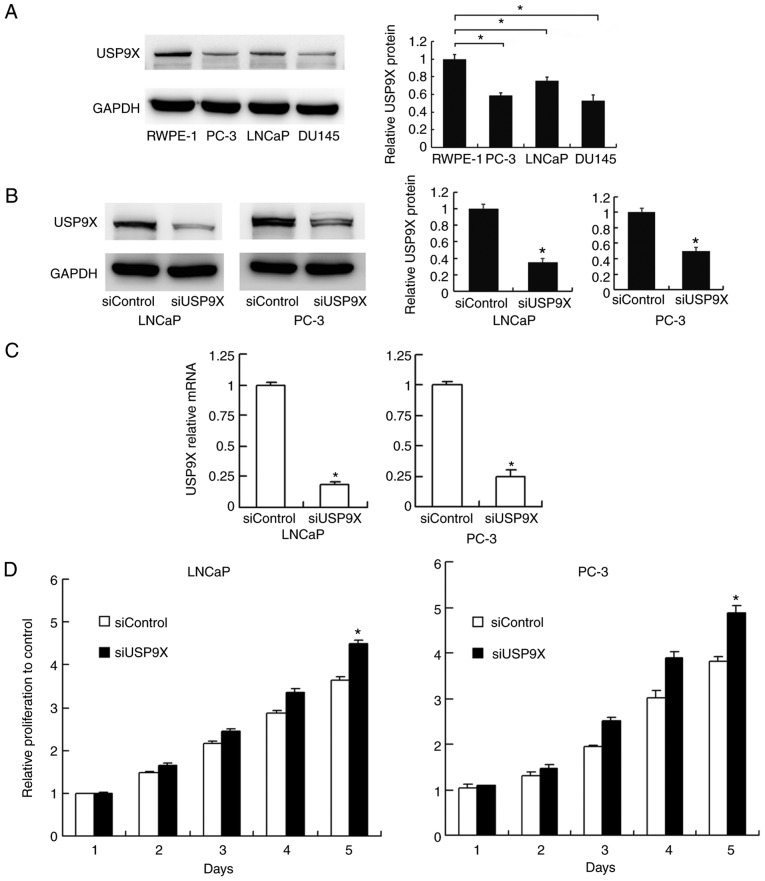
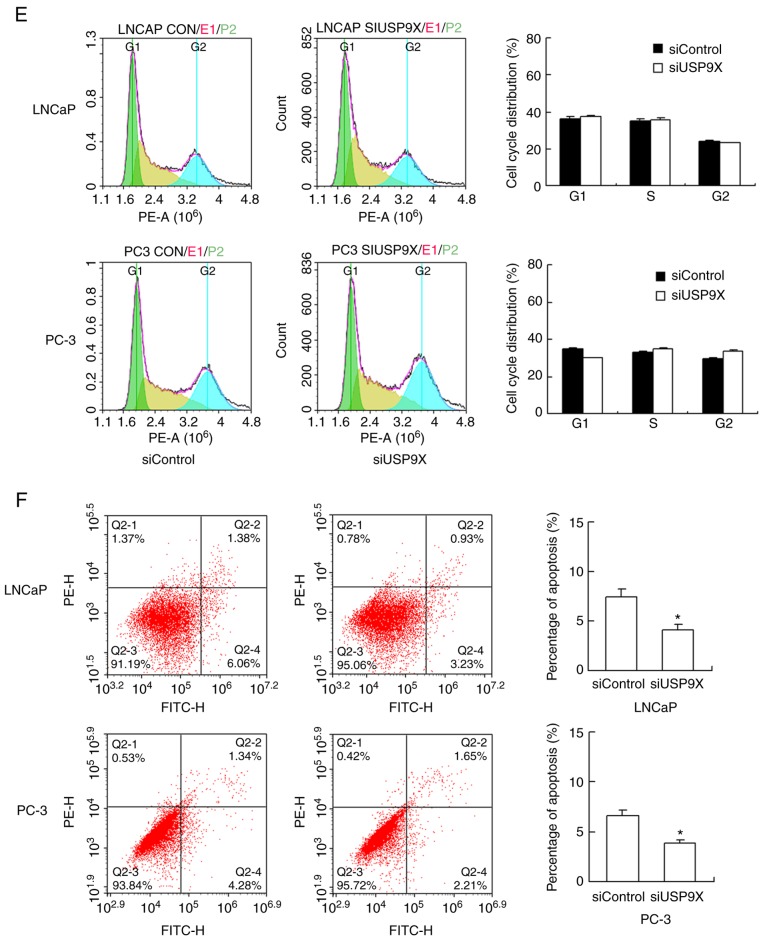
The levels of USP9X were examined by western blotting in normal prostate RWPE-1 cells and prostate cancer PC-3, LNCaP and DU145 cells. The expression of USP9X was higher in the normal RWPE-1 cells compared with that in the cancer cells (Fig. 2A). To characterize the impact of USP9X on the biological function of prostate cancer, USP9X silencing was performed in the prostate cancer PC-3 and LNCaP cells using siRNA. The siRNA knockdown efficiency was confirmed by western blotting (Fig. 2B) and RT-qPCR analysis (P<0.001 vs. siControl; n=3; Fig. 2C). Subsequently, an MTT assay and cell cycle analysis were performed to determine the involvement of USP9X in cell proliferation. The cell proliferation assay demonstrated that USP9X depletion increased the rate of cell proliferation in the prostate cancer cell lines only on day 5 (LNCaP, 21% increase; and PC-3, 28% increase; P<0.001 vs. siControl of the same day; n=3; Fig. 2D), which was not considered to be a notable overall effect. The cell cycle analysis demonstrated that USP9X knockdown did not cause any changes in the percentage distribution of cells in each phase (Fig. 2E). The change in the rate of cell apoptosis was then assessed. The Annexin V/PI staining revealed that USP9X silencing downregulated the apoptotic rate (P<0.001 vs. siControl; n=3; Fig. 2F). These results indicate that USP9X partially inhibits cell proliferation in prostate cancer cells.
Figure 2.
Effect of USP9X depletion on prostate cancer cell proliferation, the cell cycle and apoptosis. (A) USP9X protein expression in various cancer-derived and non-cancer-derived prostate cells (left panel) and its relative quantification (right panel. *P<0.05 vs. control prostate epithelial RWPE-1 cells. (B) siRNA efficiency tested by western blotting (left panel) and the relative quantification of USP9X (right panel). (C) siRNA efficiency tested by reverse transcription-quantitative polymerase chain reaction. (D) The MTT cell proliferation assay demonstrated that USP9X silencing increased the rate of cell proliferation in the LNCaP and PC-3 cells only on day 5 (21 and 28% increase, respectively, compared with siControl of the same day). The relative proliferation is presented as fold-change in cell count compared with the control on day 1. (E) The cell cycle analysis demonstrated no change in the percentage distribution of cells in each phase following USP9X silencing. (F) Annexin V/propidium iodide staining demonstrated that USP9X silencing decreased the rate of cell apoptosis. *P<0.05 vs. siControl, using Student's t-test. The data are presented as the mean ± standard deviation. USP9X, ubiquitin-specific protease 9X; siUSP9X, siRNA targeting the USP9X gene; siControl, control siRNA.
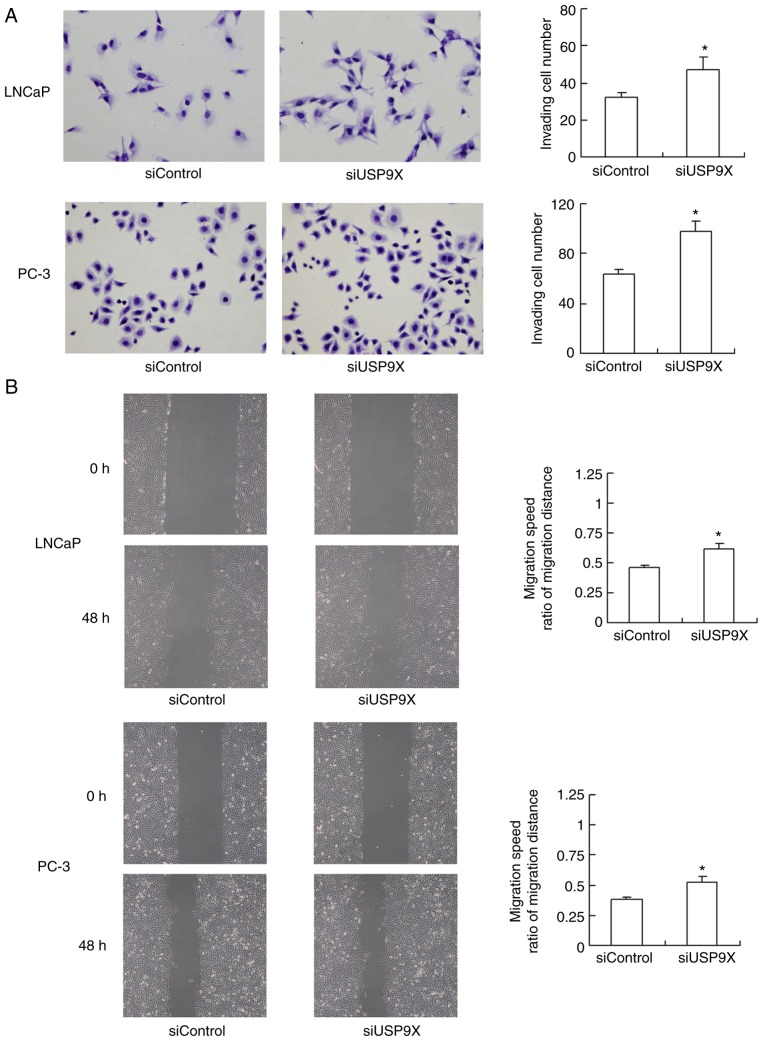
The effect of USP9X silencing on cell invasion and migration was also evaluated. As demonstrated in Fig. 3A, USP9X-knockdown significantly increased the invasive ability of the LNCaP and PC-3 cells (47 and 56% increase, respectively; P<0.05 vs. siControl). The migration of the cells was assessed using a wound-healing assay (Fig. 3B). USP9X-knockdown significantly upregulated the migratory ability of the LNCaP and PC-3 cell lines (37 and 42% increase; P<0.05 vs. siControl). The experiments were performed in triplicate. These results indicate that USP9X regulates the invasive ability of prostate cancer cells, and may therefore be a tumor suppressor in prostate cancer, which is in agreement with the immunohistochemistry results.
Figure 3.
Knockdown of USP9X in prostate cancer cells facilitates invasion and migration. (A) The invasion assay revealed that the USP9X-knockdown significantly increased the invading ability of LNCaP and PC-3 cells (47 and 56% increase, respectively). (B) The wound-healing assay revealed that USP9X silencing significantly upregulated the migratory ability of LNCaP and PC-3 cells (37 and 42% increase, respectively). The y-axis indicates the ratio of the wound gap distance closing over time. *P<0.05 vs. siControl, using Student's t-test. The data are presented as the mean ± standard deviation. USP9X, ubiquitin-specific protease 9X; siUSP9X, siRNA targeting the USP9X gene; siControl, control siRNA.
USP9X depletion upregulates MMP9 and induces mitochondrial fission in prostate cancer cells
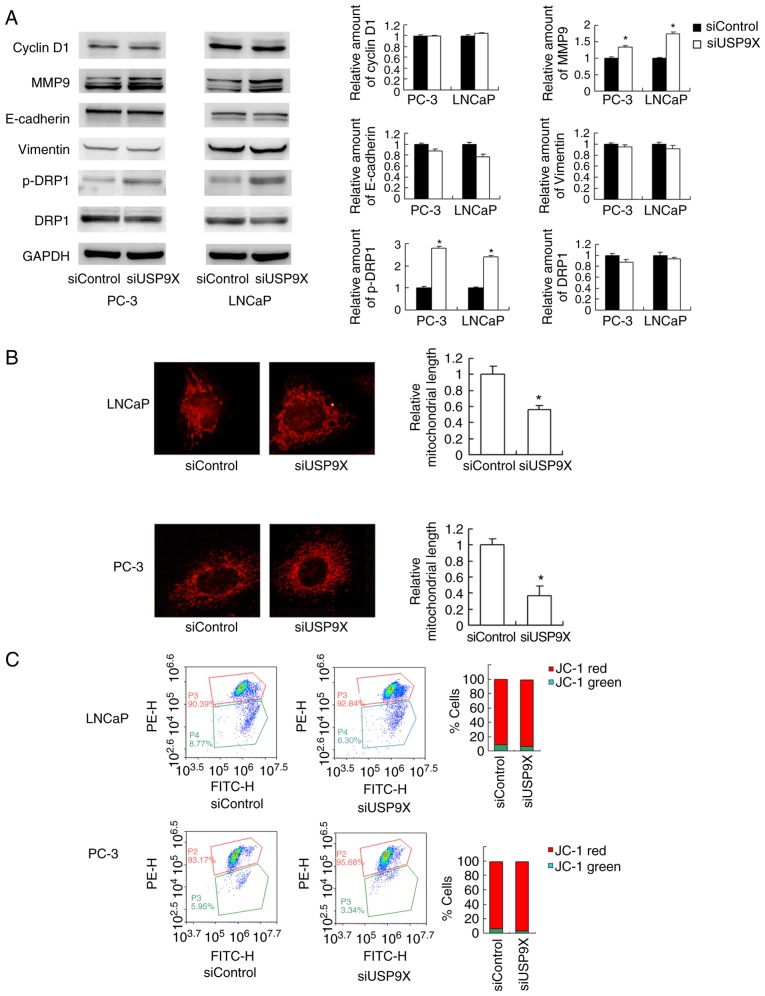
The aforementioned findings demonstrated that USP9X regulates prostate cancer cell invasion/migration more than proliferation. The mechanistic basis of USP9X-regulated invasion and migration was explored next. The MMP proteins serve pivotal roles during the initiation of cancer invasion (17). A western blot analysis revealed that the level of the MMP9 protein was increased in USP9X-knockdown LNCaP and PC-3 cells compared with that in the corresponding controls (Fig. 4A). Regarding cell cycle- and EMT-associated proteins, no significant differences were observed in the levels of cyclin D1, E-cadherin or vimentin (Fig. 4A). Cancer metabolism, particularly the mitochondrial status, has been reported to control cell migration (18). In the present study, changes in mitochondrial function were tested by examining the mitochondrial dynamics. As observed in Fig. 4B, USP9X-knockdown induced mitochondrial fission in the prostate cancer cell lines. In the USP9X-depleted LNCaP and PC-3 cells, the mitochondria displayed short tubules and spheres with an average length ~50% shorter than those in the control cells (P<0.001 vs. siControl; n=3; Fig. 4B). Accordingly, the level of p-DRP1, a member of the dynamin superfamily of GTPases that control mitochondrial fission/fusion balance (19), was increased. These results indicate that USP9X depletion induces prostate cancer cell invasion/migration, likely through the regulation of MMP9 and activation of DRP1.
Figure 4.
Silencing of USP9X in prostate cancer cell lines increases MMP9 protein level and mitochondrial fission. (A) A western blot analysis revealed that MMP9 and p-DRP1 protein levels were increased in USP9X-depleted LNCaP and PC-3 cells. No significant changes were observed in the levels of cyclin D1, E-cadherin and vimentin and total DRP1. (B) MitoTracker staining revealed that USP9X silencing induced mitochondrial fission in the prostate cancer cell lines. Relative mitochondrial length indicated the length of mitochondrial fragments in cells treated with siUSP9X relative to cells treated with siControl. The data are presented as the mean ± standard deviation (n=3). (C) JC-1 staining revealed no change in the mitochondrial membrane potential following USP9X silencing in prostate cancer cells. *P<0.05 vs. siControl, using Student's t-test. USP9X, ubiquitin-specific protease 9X; MMP9, matrix metalloproteinase 9; DRP1, dynamin-related protein 1; p-, phosphorylated; siUSP9X, siRNA targeting the USP9X gene; siControl, control siRNA.
JC-1 staining was used to examine the mitochondrial membrane potential, exhibiting red fluorescence under normal conditions that turns green when the Δψm is decreased. No significant change in the red/green ratio was observed (Fig. 4C), suggesting that USP9X-knockdown did not affect the mitochondrial membrane potential.
USP9X-induced effects on MMP9 and DRP1 in prostate cancer cells are dependent on the ERK signaling pathway
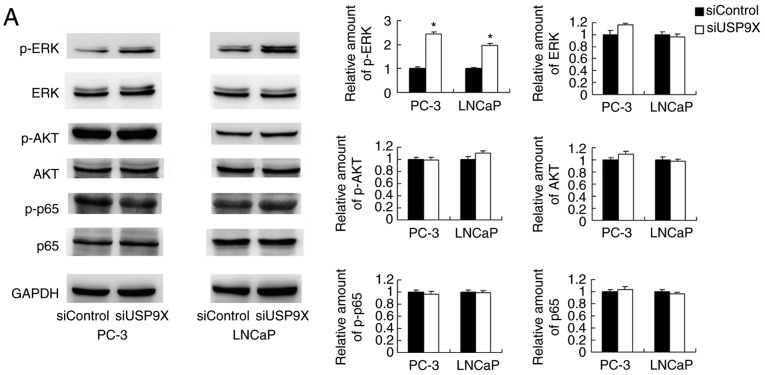
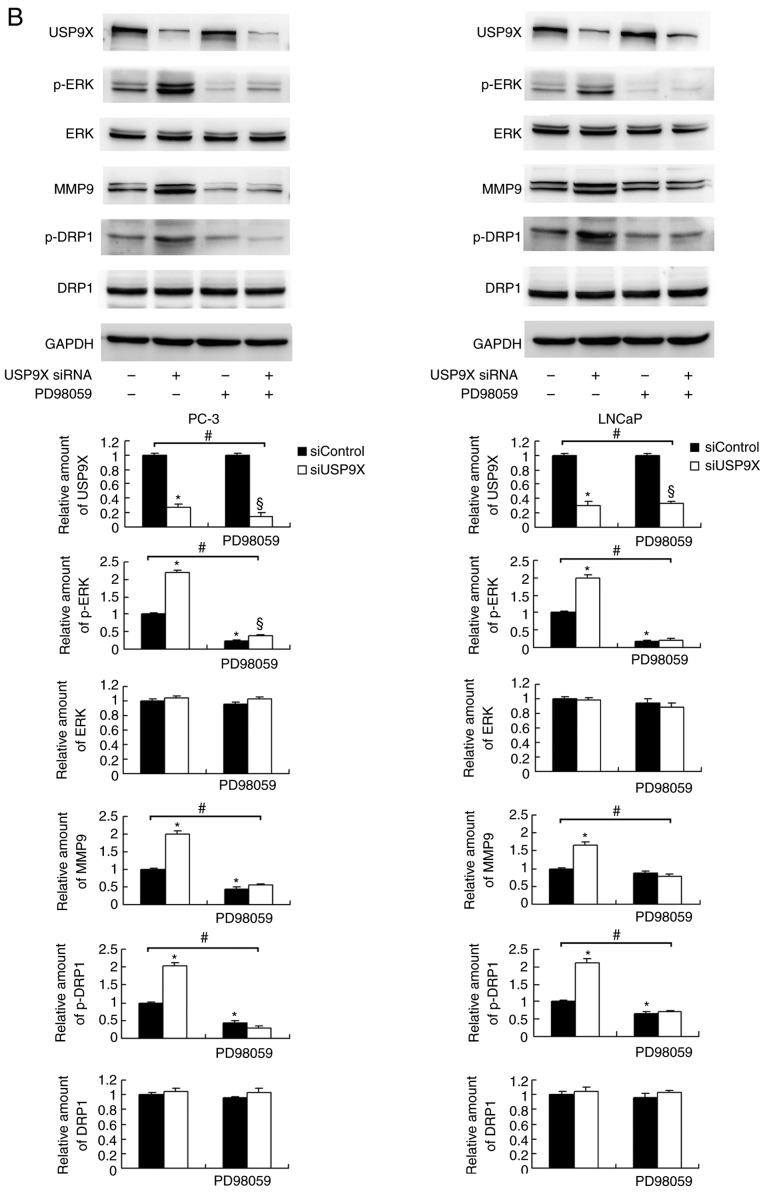
To further elucidate the mechanism of MMP9 and p-DRP1 regulation by USP9X, several signaling pathways were screened. USP9X-knockdown upregulated p-ERK, whereas no significant differences were observed in the levels of p-AKT or p-p65 (Fig. 5A). ERK signaling has been reported to induce transcriptional activation of MMP9 and DRP1 phosphorylation in cancer cells (20,21). To confirm the involvement of ERK signaling in the USP9X-induced regulation of MMP9 and phosphorylation of DRP-1, the ERK inhibitor PD98059 was used to block the ERK pathway, and protein expression levels were compared in the siUSP9X vs. siControl cells. The PD98059 treatment blocked ERK phosphorylation in the prostate cancer cells (Fig. 5B). ERK inhibition significantly decreased the levels of MMP9 and p-DRP1 (P<0.05). Notably, the inhibition of ERK abolished the effect of USP9X siRNA on MMP9 and p-DRP1 levels. These data support the conclusion that the effects of USP9X on MMP9 and p-DRP1 are dependent on ERK signaling (Fig. 5B).
Figure 5.
USP9X regulates MMP9 and p-DRP1 through the ERK signaling pathway. (A) Western blotting demonstrated that USP9X-knockdown upregulated the levels of p-ERK. No changes were observed for p-AKT and p-p65. (B) PD98059 treatment inhibited ERK phosphorylation in the prostate cancer cell lines, and decreased MMP9 and p-DRP1 protein levels. The inhibitor PD98059 abolished the effect of USP9X silencing on the levels of MMP9 and p-DRP1. #P<0.05, *P<0.05 vs. untreated siControl and §P<0.05 vs. PD98059-treated siControl using analysis of variance with a least-significant-difference post hoc test. The data are presented as the mean ± standard deviation. USP9X, ubiquitin-specific protease 9X; MMP9, matrix metalloproteinase 9; DRP1, dynamin-related protein 1; p-, phosphorylated; ERK, extracellular signal-regulated kinase; AKT, protein kinase B; p65, transcription factor p65; siUSP9X, siRNA targeting the USP9X gene; siControl, control siRNA.
Discussion
USP9X is a highly conserved member of the USP family of deubiquitinating enzymes (22) and serves an important role in development and disease (8,23). Previous studies have suggested that USP9X serves a context-specific role in the progression of cancer, acting either as a tumor suppressor or an oncoprotein (24–26). Elevated expression of USP9X has been linked to a poor prognosis in patients with non-small lung carcinoma (11), gastric cancer (12) and multiple myeloma (25). However, it was also reported that it suppresses tumor growth via serine/threonine-protein kinase LATS and the Hippo pathway (23). USP9X may also act as a tumor suppressor by maintaining genomic stability (23). These reports suggest that the role of USP9X in human cancer types is context-dependent. Therefore, it is necessary to characterize its biological function in different types of cancer. In the present study, the USP9X protein levels were demonstrated to be downregulated in prostate cancer tissues using immunohistochemistry and western blotting, and a statistical analysis revealed that its downregulation was associated with higher Gleason scores. Since tumors with higher Gleason scores tend to be more aggressive, the present results indicate that there is a possible link between USP9X downregulation and an aggressive phenotype in prostate cancer. USP9X downregulation was associated with a more advanced T stage, indicating its association with the local invasion of prostate cancer. In addition, USP9X protein levels were higher in the normal prostate RWPE-1 cells compared with those in various prostate cancer cells. These findings demonstrate that USP9X is downregulated in prostate cancer and acts as a potential tumor suppressor.
The biological functions of USP9X were investigated using siRNA treatment followed by a series of experiments, including proliferation, cell cycle, apoptosis, invasion and migration assays. USP9X silencing upregulated the proliferation rate only on day 5, with no significant effect on the cell cycle progression. However, the overall effect of USP9X silencing on proliferation promotion was not significant. The Annexin V/PI analysis revealed a downregulation of the apoptotic rate following USP9X silencing, suggesting that the effect of USP9X on prostate cancer cell growth may be due to its ability to inhibit apoptosis.
Compared with its effect on proliferation, USP9X siRNA led to a more notable effect on invasion and migration in the prostate cancer cells. To determine the underlying mechanism, proteins directly associated with invasion were investigated, and the results revealed that MMP9 levels were increased following the silencing of USP9X. MMP9 is a well-known collagenase involved in the degradation of the extracellular matrix (ECM) and prostate cancer cell invasion (27–30), explaining the increased invading ability induced by USP9X-knockdown.
The analysis of the clinical data of patients with prostate cancer revealed no significant association of USP9X expression with nodal or distal metastasis. Metastasis is a multi-step process, including the attachment and remodeling of the ECM, migration of the tumor cell body through the remodeled matrix, invasion through the lymph or blood vessels, survival within the circulation and colonization/proliferation at the secondary sites (31,32). In vitro invasion represents one of these steps. However, multiple genetic factors contribute to the metastasis in the clinical setting. Therefore, it is possible that a discrepancy exists between in vitro experiments and clinical data.
Cell migration is a complex cellular process influenced by numerous biological mechanisms, including the actin network, adhesion and energy metabolism. One key step in migration is the formation of lamellipodia at the leading edge, and this process consumes ATP produced by the mitochondria (33). Previous studies have suggested an association between mitochondrial function and cancer invasion/migration; for example, it was reported that increased mitochondrial fission induced cell migration (34,35). In the present study, USP9X silencing induced mitochondrial fission in prostate cancer cells, with a concomitant increase in DRP1 phosphorylation. The production of ATP by mitochondria is also important for cancer cell migration and invasion. During cell migration, the energy demands in different regions of the cell change. Under these circumstances, the mitochondria are cleaved by DRP1 into smaller segments due to the increased energy requirements (35,36). Mitochondrial fission directs the mitochondria to localize in neuronal areas that are predicted to have higher ATP consumption (37). It has also been reported that DRP1 is involved in cancer invasion and migration (38–40). Thus, based on the present findings, it is proposed that USP9X downregulation promotes invasion and migration through the induction of MMP9 and mitochondrial fission, which, to the best of our knowledge, has not been reported in other types of cancer.
To further elucidate how USP9X induces MMP9 and p-DRP1, several upstream signaling pathways were tested, and ERK signaling was revealed to be upregulated following the silencing of USP9X. The association between ERK and MMP9 has been demonstrated in various types of cells, including prostate cancer cell lines (41). ERK activation may also induce DRP1 phosphorylation and mitochondrial fission, which further promotes cancer cell invasion and drug resistance (40,42). The present findings further confirmed the association between ERK and MMP9/p-DRP1 using an inhibitor of the ERK pathway. The role of USP9X in cancer invasion/migration has scarcely been examined. To date, to the best of our knowledge, only a single study is available that suggests that miR-26b induces EMT through the downregulation of USP9X (43). In the present study, EMT markers, including E-cadherin and vimentin, were examined in prostate cancer cells, and no significant changes were observed in their levels. Therefore, EMT does not appear to serve a role in USP9X-regulated prostate cancer cell invasion and migration. These data suggest that USP9X inhibits prostate cancer invasion through the inhibition of ERK/MMP9/DRP1 signaling.
Two studies have reported on the role of USP9X inhibitors in cancer. In one study, the USP9X inhibitor WP1130 resulted in a decrease in the tumor growth in prostate cancer mouse xenograft models (44). Furthermore, USP9X inhibitor ABT-737 disrupted the interaction between USP9X and induced myeloid leukemia cell differentiation protein Mcl-1, and enhanced the antitumor activity of gemcitabine (45). However, the effects of WP1130 and ABT-737 on USP9X are not specific. WP1130 induces rapid proteasomal-dependent degradation of the c-Myc proto-oncogene protein. Additionally, it regulates the stability of tyrosine-protein kinase JAK2. The compound directly inhibits the deubiquitinating activity of USP9X, USP5, USP14, and ubiquitin carboxyl-terminal hydrolase isozymes L1 and L5. ABT-737 is a BH3 mimetic inhibitor of apoptosis regulator Bcl-2 and Bcl-2-like proteins 1 and 2. Furthermore, these reports mainly focused the role of USP9X on tumor growth. By contrast, the present results demonstrated that USP9X has a marked effect on invasion, and an involvement in cell proliferation, in prostate cancer cells.
In conclusion, the results of the present study suggest that USP9X is downregulated in prostate cancer and functions as an inhibitor of tumor cell invasion and migration, possibly through the regulation of the ERK/MMP9 and ERK/DRP1 signaling pathways.
Acknowledgements
Not applicable.
Funding
This study was supported by the Joint Project of Science and Technology, Department of Yunnan and Kunming Medical University, Kunming, China (grant no. 2017FE468), the National Natural Science Foundation of China (grant nos. 81660422 and 81660423) and the Doctor Newcomer Award Project (grant no. 60116090706).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
JZ, HW and BH performed the experiments, evaluated the data, wrote the manuscript and prepared the figures. JW designed the experiments. TL evaluated the data and wrote the manuscript. YZ and JC performed the surgery, evaluated the experiments and provided tissue samples. HZ and ZY performed and evaluated the experiments. All authors reviewed the manuscript. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
This study was approved by the Ethics Committee of the Second Affiliated Hospital of Kunming Medical University, Kunming, China. Informed written consent was obtained from all participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Karantanos T, Corn PG, Thompson TC. Prostate cancer progression after androgen deprivation therapy: Mechanisms of castrate resistance and novel therapeutic approaches. Oncogene. 2013;32:5501–5511. doi: 10.1038/onc.2013.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zeng H, Chen W, Zheng R, Zhang S, Ji JS, Zou X, Xia C, Sun K, Yang Z, Li H, et al. Changing cancer survival in China during 2003-15: A pooled analysis of 17 population-based cancer registries. Lancet Glob Health. 2018;6:e555–e567. doi: 10.1016/S2214-109X(18)30127-X. [DOI] [PubMed] [Google Scholar]
- 4.Hudson SV, O'Malley DM, Miller SM. Achieving optimal delivery of follow-up care for prostate cancer survivors: Improving patient outcomes. Patient Relat Outcome Meas. 2015;6:75–90. doi: 10.2147/PROM.S49588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goodwin JF, Kothari V, Drake JM, Zhao S, Dylgjeri E, Dean JL, Schiewer MJ, McNair C, Jones JK, Aytes A, et al. DNA-PKcs-mediated transcriptional regulation drives prostate cancer progression and metastasis. Cancer Cell. 2015;28:97–113. doi: 10.1016/j.ccell.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fraile JM, Quesada V, Rodriguez D, Freije JM, López-Otín C. Deubiquitinases in cancer: New functions and therapeutic options. Oncogene. 2012;31:2373–2388. doi: 10.1038/onc.2011.443. [DOI] [PubMed] [Google Scholar]
- 7.Salmena L, Pandolfi PP. Changing venues for tumour suppression: Balancing destruction and localization by monoubiquitylation. Nat Rev Cancer. 2007;7:409–413. doi: 10.1038/nrc2145. [DOI] [PubMed] [Google Scholar]
- 8.Oosterkamp HM, Hijmans EM, Brummelkamp TR, Canisius S, Wessels LF, Zwart W, Bernards R. USP9X downregulation renders breast cancer cells resistant to tamoxifen. Cancer Res. 2014;74:3810–3820. doi: 10.1158/0008-5472.CAN-13-1960. [DOI] [PubMed] [Google Scholar]
- 9.Pérez-Mancera PA, Rust AG, van der Weyden L, Kristiansen G, Li A, Sarver AL, Silverstein KA, Grützmann R, Aust D, Rümmele P, et al. The deubiquitinase USP9X suppresses pancreatic ductal adenocarcinoma. Nature. 2012;486:266–270. doi: 10.1038/nature11114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thanh Nguyen H, Andrejeva D, Gupta R, Choudhary C, Hong X, Eichhorn PJ, Loya AC, Cohen SM. Deubiquitylating enzyme USP9× regulates hippo pathway activity by controlling angiomotin protein turnover. Cell Discov. 2016;2:16001. doi: 10.1038/celldisc.2016.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, Liu Y, Yang B, Cao H, Yang CX, Ouyang W, Zhang SM, Yang GF, Zhou FX, Zhou YF, et al. Elevated expression of USP9X correlates with poor prognosis in human non-small cell lung cancer. J Thorac Dis. 2015;7:672–679. doi: 10.3978/j.issn.2072-1439.2015.04.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu X, Xie W, Song X, Wu K, Xiao L, Liu Y, Zhang L. Aberrant expression of deubiquitylating enzyme USP9X predicts poor prognosis in gastric cancer. Clin Res Hepatol Gastroenterol. 2017;41:687–692. doi: 10.1016/j.clinre.2017.01.008. [DOI] [PubMed] [Google Scholar]
- 13.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 14.Partin AW, Kattan MW, Subong EN, Walsh PC, Wojno KJ, Oesterling JE, Scardino PT, Pearson JD. Combination of prostate-specific antigen, clinical stage, and Gleason score to predict pathological stage of localized prostate cancer. A multi-institutional update. JAMA. 1997;277:1445–1451. doi: 10.1001/jama.1997.03540420041027. [DOI] [PubMed] [Google Scholar]
- 15.Fine SW. Evolution in prostate cancer staging: Pathology updates from AJCC 8th edition and opportunities that remain. Adv Anat Pathol. 2018;25:327–332. doi: 10.1097/PAP.0000000000000200. [DOI] [PubMed] [Google Scholar]
- 16.Braunhut BL, Punnen S, Kryvenko ON. Updates on grading and staging of prostate cancer. Surg Pathol Clin. 2018;11:759–774. doi: 10.1016/j.path.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 17.Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer. 2002;2:161–174. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- 18.Jia D, Park JH, Jung KH, Levine H, Kaipparettu BA. Elucidating the metabolic plasticity of cancer: Mitochondrial reprogramming and hybrid metabolic states. Cells. 2018;7 doi: 10.3390/cells7030021. doi: 10.3390/cells7030021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paupe V, Prudent J. New insights into the role of mitochondrial calcium homeostasis in cell migration. Biochem Biophys Res Commun. 2018;500:75–86. doi: 10.1016/j.bbrc.2017.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou C, Wang Y, Peng J, Li C, Liu P, Shen X. SNX10 plays a critical role in MMP9 secretion via JNK-p38-ERK signaling pathway. J Cell Biochem. 2017;118:4664–4671. doi: 10.1002/jcb.26132. [DOI] [PubMed] [Google Scholar]
- 21.Huang CY, Chiang SF, Chen WT, Ke TW, Chen TW, You YS, Lin CY, Chao KSC, Huang CY. HMGB1 promotes ERK-mediated mitochondrial Drp1 phosphorylation for chemoresistance through RAGE in colorectal cancer. Cell Death Dis. 2018;9:1004. doi: 10.1038/s41419-018-1019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wood SA, Pascoe WS, Ru K, Yamada T, Hirchenhain J, Kemler R, Mattick JS. Cloning and expression analysis of a novel mouse gene with sequence similarity to the Drosophila fat facets gene. Mech Dev. 1997;63:29–38. doi: 10.1016/S0925-4773(97)00672-2. [DOI] [PubMed] [Google Scholar]
- 23.Toloczko A, Guo F, Yuen HF, Wen Q, Wood SA, Ong YS, Chan PY, Shaik AA, Gunaratne J, Dunne MJ, et al. Deubiquitinating enzyme USP9X suppresses tumor growth via LATS kinase and core components of the hippo pathway. Cancer Res. 2017;77:4921–4933. doi: 10.1158/0008-5472.CAN-16-3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGarry E, Gaboriau D, Rainey MD, Restuccia U, Bachi A, Santocanale C. The deubiquitinase USP9X maintains DNA replication fork stability and DNA damage checkpoint responses by regulating CLASPIN during S-phase. Cancer Res. 2016;76:2384–2393. doi: 10.1158/0008-5472.CAN-15-2890. [DOI] [PubMed] [Google Scholar]
- 25.Wu Y, Yu X, Yi X, Wu K, Dwabe S, Atefi M, Elshimali Y, Kemp KT, II, Bhat K, Haro J, et al. Aberrant phosphorylation of SMAD4 Thr277-mediated USP9×-SMAD4 interaction by free fatty acids promotes breast cancer metastasis. Cancer Res. 2017;77:1383–1394. doi: 10.1158/0008-5472.CAN-16-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwickart M, Huang X, Lill JR, Liu J, Ferrando R, French DM, Maecker H, O'Rourke K, Bazan F, Eastham-Anderson J, et al. Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature. 2010;463:103–107. doi: 10.1038/nature08646. [DOI] [PubMed] [Google Scholar]
- 27.Ding X, Yang DR, Xia L, Chen B, Yu S, Niu Y, Wang M, Li G, Chang C. Targeting TR4 nuclear receptor suppresses prostate cancer invasion via reduction of infiltrating macrophages with alteration of the TIMP-1/MMP2/MMP9 signals. Mol Cancer. 2015;14:16. doi: 10.1186/s12943-014-0281-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moroz A, Delella FK, Almeida R, Lacorte LM, Fávaro WJ, Deffune E, Felisbino SL. Finasteride inhibits human prostate cancer cell invasion through MMP2 and MMP9 downregulation. PLoS One. 2013;8:e84757. doi: 10.1371/journal.pone.0084757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kato T, Fujita Y, Nakane K, Kojima T, Nozawa Y, Deguchi T, Ito M. ETS1 promotes chemoresistance and invasion of paclitaxel-resistant, hormone-refractory PC3 prostate cancer cells by up-regulating MDR1 and MMP9 expression. Biochem Biophys Res Commun. 2012;417:966–971. doi: 10.1016/j.bbrc.2011.12.047. [DOI] [PubMed] [Google Scholar]
- 30.London CA, Sekhon HS, Arora V, Stein DA, Iversen PL, Devi GR. A novel antisense inhibitor of MMP-9 attenuates angiogenesis, human prostate cancer cell invasion and tumorigenicity. Cancer Gene Ther. 2003;10:823–832. doi: 10.1038/sj.cgt.7700642. [DOI] [PubMed] [Google Scholar]
- 31.Zhang X, Yao X, Qin C, Luo P, Zhang J. Investigation of the molecular mechanisms underlying metastasis in prostate cancer by gene expression profiling. Exp Ther Med. 2016;12:925–932. doi: 10.3892/etm.2016.3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clarke NW, Hart CA, Brown MD. Molecular mechanisms of metastasis in prostate cancer. Asian J Androl. 2009;11:57–67. doi: 10.1038/aja.2008.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science. 2009;326:1208–1212. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Westermann B. Mitochondrial fusion and fission in cell life and death. Nat Rev Mol Cell Biol. 2010;11:872–884. doi: 10.1038/nrm3013. [DOI] [PubMed] [Google Scholar]
- 35.Zhao J, Zhang J, Yu M, Xie Y, Huang Y, Wolff DW, Abel PW, Tu Y. Mitochondrial dynamics regulates migration and invasion of breast cancer cells. Oncogene. 2013;32:4814–4824. doi: 10.1038/onc.2012.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pal AD, Basak NP, Banerjee AS, Banerjee S. Epstein-Barr virus latent membrane protein-2A alters mitochondrial dynamics promoting cellular migration mediated by Notch signaling pathway. Carcinogenesis. 2014;35:1592–1601. doi: 10.1093/carcin/bgu069. [DOI] [PubMed] [Google Scholar]
- 37.Hollenbeck PJ, Saxton WM. The axonal transport of mitochondria. J Cell Sci. 2005;118:5411–5419. doi: 10.1242/jcs.02745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yin M, Lu Q, Liu X, Wang T, Liu Y, Chen L. Silencing Drp1 inhibits glioma cells proliferation and invasion by RHOA/ROCK1 pathway. Biochem Biophys Res Commun. 2016;478:663–668. doi: 10.1016/j.bbrc.2016.08.003. [DOI] [PubMed] [Google Scholar]
- 39.Zhang J, Zhang Y, Wu W, Wang F, Liu X, Shui G, Nie C. Guanylate-binding protein 2 regulates Drp1-mediated mitochondrial fission to suppress breast cancer cell invasion. Cell Death Dis. 2017;8:e3151. doi: 10.1038/cddis.2017.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fu L, Dong Q, He J, Wang X, Xing J, Wang E, Qiu X, Li Q. SIRT4 inhibits malignancy progression of NSCLCs, through mitochondrial dynamics mediated by the ERK-Drp1 pathway. Oncogene. 2017;36:2724–2736. doi: 10.1038/onc.2016.425. [DOI] [PubMed] [Google Scholar]
- 41.Kato T, Fujita Y, Nakane K, Mizutani K, Terazawa R, Ehara H, Kanimoto Y, Kojima T, Nozawa Y, Deguchi T, Ito M. CCR1/CCL5 interaction promotes invasion of taxane-resistant PC3 prostate cancer cells by increasing secretion of MMPs 2/9 and by activating ERK and Rac signaling. Cytokine. 2013;64:251–257. doi: 10.1016/j.cyto.2013.06.313. [DOI] [PubMed] [Google Scholar]
- 42.Cai J, Wang J, Huang Y, Wu H, Xia T, Xiao J, Chen X, Li H, Qiu Y, Wang Y, et al. ERK/Drp1-dependent mitochondrial fission is involved in the MSC-induced drug resistance of T-cell acute lymphoblastic leukemia cells. Cell Death Dis. 2016;7:e2459. doi: 10.1038/cddis.2016.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shen G, Lin Y, Yang X, Zhang J, Xu Z, Jia H. MicroRNA-26b inhibits epithelial-mesenchymal transition in hepatocellular carcinoma by targeting USP9X. BMC Cancer. 2014;14:393. doi: 10.1186/1471-2407-14-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang S, Kollipara RK, Srivastava N, Li R, Ravindranathan P, Hernandez E, Freeman E, Humphries CG, Kapur P, Lotan Y, et al. Ablation of the oncogenic transcription factor ERG by deubiquitinase inhibition in prostate cancer. Proc Natl Acad Sci USA. 2014;111:4251–4256. doi: 10.1073/pnas.1322198111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang C, Cai TY, Zhu H, Yang LQ, Jiang H, Dong XW, Hu YZ, Lin NM, He QJ, Yang B. Synergistic antitumor activity of gemcitabine and ABT-737 in vitro and in vivo through disrupting the interaction of USP9X and Mcl-1. Mol Cancer Ther. 2011;10:1264–1275. doi: 10.1158/1535-7163.MCT-10-1091. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.