Abstract
Human leukocyte antigen-G (HLA-G) is highly expressed in numerous solid tumor cell types and has important roles in protecting tumor cells from host immune recognition and destruction. DNA methylation modification, which may regulate gene expression, is aberrant in numerous tumor cell types. However, whether the high expression of HLA-G in tumor cells is induced by aberrant DNA methylation has remained elusive. In the present study, HLA-G, DNA methyltransferase (DNMT) and ten-eleven translocation (TET) expression, as well as the DNA methylation level of HLA-G, were assessed in the HBL-100 breast cell line and the MCF-7 breast cancer cell line. The influence of TET on the expression and DNA methylation levels of HLA-G in MCF-7 was assessed through treatment with the TET inhibitor dimethyloxallyl glycine (DMOG). The results indicated that HLA-G expression was significantly greater in MCF-7 than that in HBL-100 cells; however, the DNA methylation level of HLA-G was lower in MCF-7 than that in HBL-100 cells. Furthermore, in MCF-7 cells, DNMT1 and DNMT3a were expressed at lower levels and TET2 was expressed at higher levels than in HBL-100 cells. Treatment with DMOG significantly decreased HLA-G expression, while increasing the DNA methylation level of HLA-G in MCF-7. In conclusion, the results indicated that overexpression of HLA-G in MCF-7 cells was induced by DNA methylation modification. The lower DNMT1 and DNMT3a and higher TET2 expression levels may be responsible for the abnormal DNA methylation of HLA-G in MCF-7. Treatment with TET inhibitor prevented aberrant HLA-G expression and DNA methylation in MCF-7. The present study may provide potential targets for novel anti-cancer drugs.
Keywords: human leukocyte antigen-G, DNA methylation, ten-eleven translocation, dimethyloxallyl glycine, MCF-7
Introduction
Human leukocyte antigen-G (HLA-G) is a molecule with immunomodulatory activity that belongs to the non-classical HLA class I family and its encoding gene is located at chromosome 6p21 (1,2). HLA-G may lead to immune tolerance by interacting with receptors that are expressed in immune regulatory cells, including immunoglobulin (Ig)-like transcript (ILT)2, ILT4, killer cell Ig-like receptor, two Ig domains and long cytoplasmic tail 4 and CD160 (3–5). Overexpression of HLA-G has been identified in numerous types of human solid tumor and hematological cancer (6), and high expression of HLA-G was reported to be associated with primary carcinogenesis and the metastatic capacity of breast invasive ductal carcinoma (7,8). Overexpression of HLA-G may be a means of tumor cells to avoid regulation by the immune system, by inhibiting natural killer and T cell-mediated lysis (3).
Aberrant DNA methylation is one of the characteristics of cancer cells (9,10). DNA methylation is a covalent modification of DNA and is performed by the DNA methyltransferase (DNMT) family, which mainly consists of three members, DNMT1, DNMT3a and DNMT3b (11). The removal of methyl groups from DNA is termed DNA demethylation. The ten-eleven translocation (TET) family, which includes TET1, TET2 and TET3, has been indicated to have important roles in DNA demethylation (12). DNA methylation has important roles in a number of key genomic functions, including gene imprinting, X chromosome inactivation, genome stability, retrotransposon silencing and gene inactivation in cancer (13–15). Dimethyloxallyl glycine (DMOG) is a small-molecule inhibitor of the TET protein. In mice, treatment of embryos with 1 mM DMOG from the germinal vesicle to the blastocyst stage effectively blocks the activity of TET enzymes in vitro (16).
In general, the evasion of immune surveillance is considered one of the emerging characteristics of cancer (17). High expression of HLA-G is essential for tumor cells to avoid immune recognition and destruction (18). DNA methylation modification has important roles in regulating gene expression, and DNMTs and TETs are responsible for the dynamic changes in DNA methylation (19,20). However, whether the high expression of HLA-G in tumor cells is induced by aberrant DNA methylation has remained elusive. Therefore, in the present study, the expression of HLA-G, DNMTs and TETs, as well as the DNA methylation levels of HLA-G, were assessed in the HBL-100 breast cell line and the MCF-7 breast cancer cell line. The effects of TET activity on the expression and DNA methylation levels of HLA-G in the MCF-7 cell line were also assessed by treating the cells with DMOG.
Materials and methods
Cell culture
The HBL-100 and MCF-7 cell lines were purchased from the Cell Bank of the Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences. The HBL-100 and MCF-7 cell lines were cultured in Dulbecco's modified Eagle's medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal calf serum (Bioind; Biological Industries), 100 IU/ml penicillin and 100 µg/ml streptomycin (HyClone; GE Healthcare). The cells were cultured in a humidified atmosphere with 5% CO2 at 37°C.
RNA isolation and reverse transcription quantitative (RT-q) PCR
Cells were treated with 100, 200 or 400 µM DMOG for 48 h and untreated cells were used as controls. Trypsin/EDTA was used to harvest the cultured cells, and RNA was isolated with TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. cDNA was synthesized using TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen Biotech Co., Ltd.). In brief, first-strand cDNA was synthesized in 20-µl reactions from 2 µl total RNA using SuperMix and gDNA Remover, and RT reaction steps consisted of 42°C for 15 min and 85°C for 5 sec. qPCR was performed using SYBR Premix Ex Taq (Takara Bio, Inc.) on a LightCycler 96 real-time PCR system (Roche). The 20-µl reaction mixtures consisted of 8 µl water, 1 µl cDNA, 1 µl (10 µM) primers (HLA-G, TET1, TET2, TET3, DNMT1, DNMT3a, DNMT3b and GAPDH) and 10 µl SYBR Premix Ex Taq. GAPDH was used as an internal control. The thermocycling program consisted of 95°C for 180 sec, followed by 50 cycles at 95°C for 10 sec, 60°C for 30 sec, then 95°C for 30 sec, 65°C for 60 sec and 97°C for 1 sec, and a final step at 37°C for 30 sec. Relative gene expression was quantified by using the 2−ΔΔCq method (21). The primer sequences for HLA-G, DNMT1, DNMT3a (22), DNMT3b (22), TET1, TET2 (23), TET3 (23) and GAPDH are listed in Table I.
Table I.
Primer sequences designed for PCR.
| Gene | Primer pair sequences (5′ to 3′) | Product size (bp) | Reference |
|---|---|---|---|
| RT-qPCR | |||
| HLA-G | F: AGAGGAGACACGGAACACCAAGG | 127 | NC_000006.12 |
| R: CAGGTCGCAGCCAATCATCCAC | |||
| DNMT1 | F: CCTCCAAAAACCCAGCCAAC | 101 | NC_000019.10 |
| R: TCCAGGACCCTGGGGATTTC | |||
| DNMT3a | F: CCAACATCGAATCCATGAAA | 140 | (22) |
| R: CTTGCGCTTGCTGATGTAGT | |||
| DNMT3b | F: CGAATTTTACCACCTGCTGAATT | 59 | (22) |
| R: AGAACGGCCGGTCATCAC | |||
| TET1 | F: ACCTATTCCCCGAATCAAGC | 100 | NC_000010.11 |
| R: TTGCACGGTCTCAGTGTTACTC | |||
| TET2 | F: AGCCCCATCACGTACAAAAC | 129 | (23) |
| R: TGTGGTGGCTGCTTCTGTAG | |||
| TET3 | F: CAGCAGCCGAGAAGAAGAAG | 125 | (23) |
| R: GGACAATCCACCCTTCAGAG | |||
| GAPDH | F: CAGGAGGCATTGCTGATGAT | 138 | NC_000012.12 |
| R: GAAGGCTGGGGCTCATTTT | |||
| Bisulfite-sequencing PCR | |||
| HLA-G | F: TGGGTTAAGATTTAGGGAGATA | 249 | (25) |
| R: TAACTTCTCTAAAAACCTATCACCTAA | |||
HLA-G, human leukocyte antigen-G; DNMT, DNA methyltransferase; TET, ten-eleven translocation; F, forward; R, reverse.
Bisulfite genomic sequencing
DNA was isolated using a TIANamp Genomic DNA kit [Tiangen Biotech (Beijing) Co., Ltd.] after the cells had been treated with DMOG or left untreated (control) for 48 h, followed by analysis using bisulfite sequencing. In brief, DNA was extracted from breast cancer cells treated as mentioned above and boiled in a water bath for 5 min, followed by chilling on ice. Subsequently, 4 µl of 2 M NaOH (final concentration, 0.3 M NaOH) was added to the DNA, and the mixture was incubated for 15 min at 50°C. Next, the solution was mixed with two volumes of 2% low-melting-point agarose (Sigma-Aldrich, Merck KGaA) and seven 10 µl-aliquotes of the DNA-agarose mixture were pipetted into ice-cold mineral oil to form beads. Then, at least seven beads were immersed in the fresh bisulfite solution (2.5 M sodium metabisulfite and 125 mM hydroquinone, pH 5) (24). These beads were incubated for 3–5 h in the dark and covered with mineral oil at 50°C. Next, the supernatant was discarded and the seven beads were washed four times in 1 ml Tris-EDTA buffer (pH 8.0) 15 min each time. After desulfonation in 0.5 ml of 0.2 M NaOH 2 times for 15 min each, the beads were washed with 1 ml Tris-EDTA buffer 3 times for 10 min each and with H2O 2 times for 15 min each, and then used as the input for PCR. The PCR primer sequence for HLA-G (25) is listed in Table I.
Statistical analysis
All experiments were performed at least three times. Statistical analysis was performed by one-way analysis of variance using SPSS 19.0 software (SPSS, Inc., Chicago, IL, USA) followed the LSD method to assess the differences between more than two groups. Differences were considered statistically significant at P<0.05.
Results
HLA-G expression and promoter DNA methylation levels in HBL-100 and MCF-7 cells
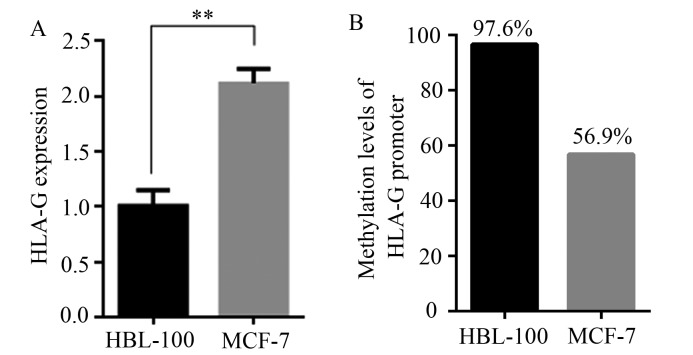
HLA-G expression was analyzed in HBL-100 and MCF-7 cells by RT-qPCR analysis. As presented in Fig. 1A, the expression of HLA-G was significantly greater in MCF-7 cells than in HBL-100 cells (P<0.01). Subsequently, the DNA methylation level of the HLA-G promoter region was compared between the two cell lines. As presented in Fig. 1B, the DNA methylation level of the HLA-G promoter region was 96.7% in HBL-100 cells, but only 56.9% in MCF-7 cells.
Figure 1.
Expression and promoter DNA methylation levels of HLA-G in HBL-100 and MCF-7 cells. (A) HLA-G expression was analyzed by reverse transcription-quantitative PCR in HBL-100 and MCF-7 cells. GAPDH was used for normalization. (B) Promoter DNA methylation level of HLA-G in HBL-100 and MCF-7 cells. Values are expressed as the mean ± standard deviation of three independent experiments. **P<0.01. HLA-G, human leukocyte antigen-G.
Expression of DNMTs in HBL-100 and MCF-7 cells
DNA methylation is catalyzed by DNMTs. The expression of various DNMTs was investigated by RT-qPCR. The expression of DNMT1, DNMT3a and DNMT3b was compared between HBL-100 and MCF-7 cells. The expression levels of DNMT1 and DNMT3a in HBL-100 cells were significantly greater than those in MCF-7 cells (DNMT1: P<0.05; DNMT3a: P<0.01; Fig. 2), but the expression levels of DNMT3b in HBL-100 cells were lower than those in MCF-7 cells (P<0.01).
Figure 2.
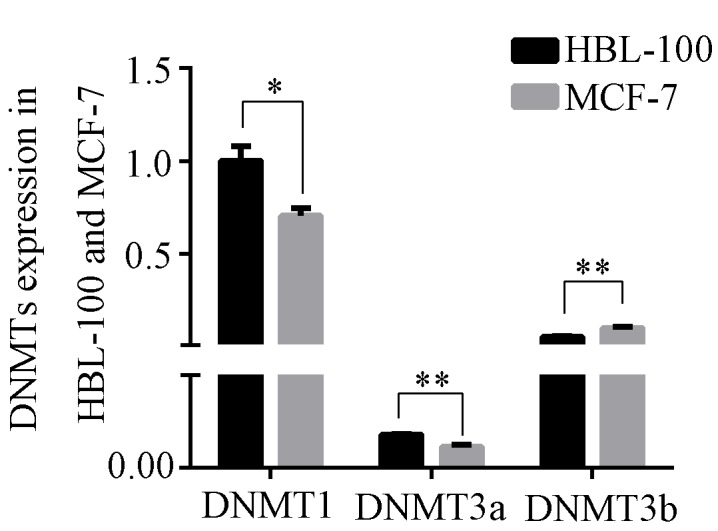
Expression of DNMTs in HBL-100 and MCF-7 cells. Expression of various DNMTs in HBL-100 and MCF-7 cells was analyzed by reverse transcription-quantitative PCR. GAPDH was used for normalization. The expression of DNTM1 in HBL-100 cells was used for calibration (expression was set to 1). Values are expressed as the mean ± standard deviation of three independent experiments. *P<0.05 and **P<0.01. DNMT, DNA methyltransferase.
Expression of TETs in HBL-100 and MCF-7 cells
The ten-eleven tanslocation (TET) family, which includes TET1, TET2 and TET3, is generally thought to be responsible for DNA demethylation. As presented in Fig. 3, the expression levels of TET1 and TET3 in MCF-7 cells were significantly lower than those in HBL-100 cells (P<0.01); however, TET2 expression in MCF-7 cells was greater than that in HBL-100 cells (P<0.01).
Figure 3.
Expression of TETs in HBL-100 and MCF-7 cells. Expression of various TETs in HBL-100 and MCF-7 cells was analyzed by reverse transcription-quantitative PCR. GAPDH was used for normalization. The expression of TET1 in HBL-100 cells was used for calibration (expression was set to 1). Values are expressed as the mean ± standard deviation of three independent experiments. **P<0.01. TET, ten-eleven translocation.
Effects of inhibition of TET on the expression and promoter DNA methylation of HLA-G in MCF-7 cells
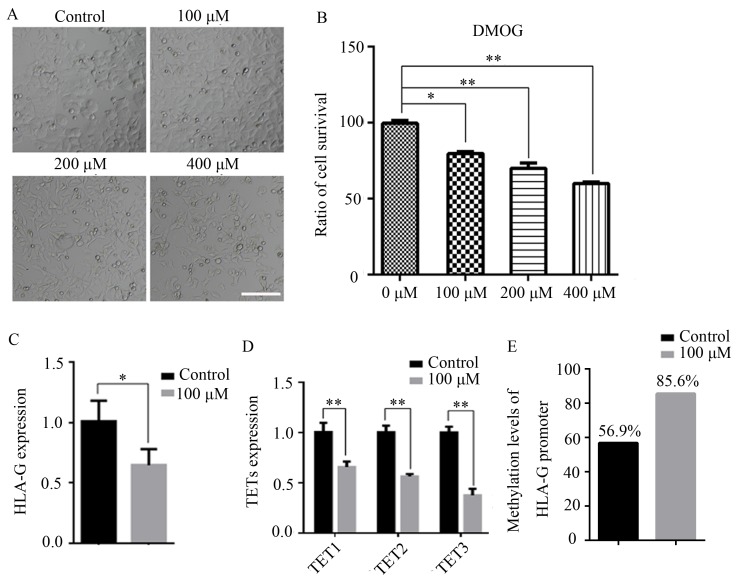
MCF-7 cells were treated with 100, 200 or 400 µM DMOG for 48 h, and untreated cells were used as controls. As presented in Fig. 4A, treatment with 200 and 400 µM DMOG changed the morphology of MCF-7 cells, indicating that the 200 and 400 µM DMOG treatments were cytotoxic. An MTT assay also indicated that DMOG had inhibitory effects on MCF-7 cells (Fig. 4B). Therefore, 100 µM DMOG was used as the final concentration for the subsequent experiments. It was indicated that treatment with DMOG significantly decreased the expression of HLA-G in MCF-7 cells (P<0.05; Fig. 4C). Furthermore, the expression levels of TET1, TET2 and TET3 were all significantly decreased (P<0.01; Fig. 4D). The promoter DNA methylation level of HLA-G in MCF-7 cells after DMOG treatment was then assessed, revealing that DMOG treatment significantly increased the promoter DNA methylation level of HLA-G in MCF-7 cells compared with that in the control cells (Fig. 4E).
Figure 4.
Effects of DMOG on the expression and promoter DNA methylation of HLA-G in MCF-7 cells. (A) Microscopy images of MCF-7 cells that were untreated or treated with 100, 200 or 400 µM DMOG for 48 h (scale bar, 100 µm). (B) The cell survival ratio of MCF-7 cells treated with DMOG was analyzed using an MTT assay. The untreated group was used for calibration (cell survival ratio was set to 100). (C) HLA-G expression was analyzed by reverse transcription-quantitative PCR in MCF-7 cells after treatment with 100 µM DMOG. GAPDH was used for normalization. HLA-G expression in MCF-7 cells untreated with DMOG was used for calibration (expression was set to 1). (D) Expression of TETs after treatment with DMOG. GAPDH was used for normalization. TET expression in MCF-7 cells untreated with DMOG was used for calibration (expression set to 1). (E) The promoter DNA methylation of HLA-G was analyzed by BSP-PCR in MCF-7 cells after treatment with DMOG. The black and white circles indicate methylated locus and unmethylated locus respectively. Values are expressed as the mean ± standard deviation of three independent experiments. *P<0.05 and **P<0.01. HLA-G, human leukocyte antigen-G; TET, ten-eleven translocation; DMOG, dimethyloxallyl glycine.
Discussion
HLA-G was first reported to allow tumors to avoid immunosurveillance in 1998 (26). Since then, numerous studies have been performed to support this hypothesis. HLA-G-induced suppression of T-cell responses has indicated the presence of an immune escape pathway in human glioblastoma (27). HLA-G has been reported to be overexpressed in a number of cancer types, including melanoma (28), primary cutaneous lymphomas (29), lung cancer (30) and breast cancer (31). However, the molecular mechanisms of the induction of HLA-G overexpression in cancer remain to be fully elucidated. Therefore, in the present study, the possible association between the expression of HLA-G and DNA methylation of its promoter region was determined in the MCF-7 breast cancer cell line as an underlying molecular mechanism that induces high HLA-G expression in cancer.
The present results indicated that HLA-G expression in MCF-7 cells was significantly greater than that in HBL-100 cells, which was in line with previously reported results in a variety of cancer types (28–31). High HLA-G expression may be responsible for the avoidance of immunosurveillance by MCF-7 cells. However, the mechanisms that cause high expression of HLA-G in MCF-7 cells remain elusive. DNA methylation is a means of gene expression regulation, and an aberrant DNA methylation pattern is one of the characteristics of cancer cells (9,10). Furthermore, previous studies have indicated the activation of HLA-G transcription after incubation with a DNMT inhibitor (5-aza-2′-deoxycytidine) in a broad panel of human leukemia cell lines (32), suggesting that DNA methylation regulates HLA-G expression in those cells (33). Therefore, it may be inferred that the differing DNA methylation in the HLA-G promoter region was responsible for the differences in HLA-G expression between HBL-100 and MCF-7 cells. In most cases, the DNA methylation level is negatively correlated with gene expression, e.g. the expression of the NANOG gene (34). The present results indicated that the DNA methylation level of the promoter region of the HLA-G gene in MCF-7 cells was lower than that in HBL-100 cells. Thus, similar to other genes, DNA methylation of the promoter region of the HLA-G gene negatively regulates its expression. Aberrant DNA methylation modification causes abnormally high expression of HLA-G in MCF-7 cells. DNMT and TET activities have important roles in dynamic changes in DNA methylation. The present results indicated that DNMT1 and DNMT3a were expressed at lower levels and that TET2 was expressed at higher levels in MCF-7 cells than in HBL-100 cells. However, DNMT3b expression was greater and TET1 and TET3 expression was lower in MCF-7 cells than in HBL-100 cells. The different downstream targets of DNMT1, DNMT3a and DNMT3b may be the reason for the different expression patterns of the members of the DNMT or TET families between HBL-100 and MCF-7 cells. Accumulating evidence suggests that somatic mutations in DNA methyltransferases and 5mC-modifying enzymes, including TET proteins, are associated with oncogenic transformation (12). Therefore, it may be inferred that the lower DNMT1 and DNMT3a expression levels and greater TET2 expressions levels were the reason for abnormal DNA methylation modification of the HLA-G gene in MCF-7 cells.
Treatment with small-molecule inhibitors of DNMT or TET may change the extent of DNA methylation and thereby gene expression (35,36). DMOG is a non-specific 2-OG-dependent dioxygenase inhibitor (37). In cows, treatment of parthenogenetic embryos with 1 mM DMOG effectively blocked the activity of TET enzymes and impeded parthenogenetic embryo development in vitro by disturbing the DNA demethylation progress (38). Therefore, it was assessed whether treatment with the TET inhibitor DMOG increases the DNA methylation level of HLA-G, and whether this decreases HLA-G expression in MCF-7 cells. The results suggested that treatment with 100 µM DMOG for 48 h significantly increased the DNA methylation level of the HLA-G promoter region in MCF-7 cells. More importantly, a negative association between the promoter region DNA methylation level and gene expression was observed, as treatment with DMOG significantly decreased HLA-G expression in MCF-7 cells. Unexpectedly, DMOG also significantly decreased TETs expression in MCF-7 cells, which indicated that TETs may regulate self-expression in MCF-7 cells. Overall, the results indicated that TETs are, at least in part, responsible for the lower DNA methylation level of the HLA-G promoter in MCF-7 cells.
In conclusion, the present study indicated that HLA-G was highly expressed and that the DNA methylation level of its gene promoter region was low in the MCF-7 breast cancer cell line. DNMTs and TETs were aberrantly expressed in MCF-7 cells, which may be the reason for the low DNA methylation level of the HLA-G promoter region. Inhibition of TET activity increased HLA-G promoter region DNA methylation levels and decreased the expression of HLA-G in MCF-7 cells. These results indicated that TETs are, at least in part, responsible for the lower DNA methylation level of the HLA-G promoter and overexpression of HLA-G in MCF-7 cells, which may provide potential targets for novel anti-cancer drugs. The exact upstream mechanisms that regulate the overexpression of HLA-G in cancers may require further investigation.
Acknowledgements
Not applicable.
Funding
This study was supported by the National Key R&D Program of China (grant no. 2017YFA0104400) and the Program for Changjiang Scholars and Innovative Research Team in University (grant no. IRT_16R32).
Availability of data and materials
The datasets used during the present study are available from the corresponding author on reasonable request.
Authors' contributions
DZ and XA mainly performed the experiments. ZL and SZ conceived the project, supervised the experiments and revised the manuscript. All authors agreed to be accountable for the content of the work. All authors read and approved the final manuscript.
Ethical approval and informed consent
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors have no competing interests to declare.
References
- 1.Carosella ED, Moreau P, Lemaoult J, Rouas-Freiss N. HLA-G: From biology to clinical benefits. Trends Immunol. 2008;29:125–132. doi: 10.1016/j.it.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 2.Jeong S, Park S, Park BW, Park Y, Kwon OJ, Kim HS. Human leukocyte antigen-G (HLA-G) polymorphism and expression in breast cancer patients. PLoS One. 2014;9:e98284. doi: 10.1371/journal.pone.0098284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pistoia V, Morandi F, Wang X, Ferrone S. Soluble HLA-G: Are they clinically relevant? Semin Cancer Biol. 2007;17:469–479. doi: 10.1016/j.semcancer.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baudhuin J, Migraine J, Faivre V, Loumagne L, Lukaszewicz AC, Payen D, Favier B. Exocytosis acts as a modulator of the ILT4-mediated inhibition of neutrophil functions. Proc Natl Acad Sci USA. 2013;110:17957–17962. doi: 10.1073/pnas.1221535110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Le Page ME, Goodridge JP, John E, Christiansen FT, Witt CS. Killer Ig-like receptor 2DL4 does not mediate NK cell IFN-gamma responses to soluble HLA-G preparations. J Immunol. 2014;192:732–740. doi: 10.4049/jimmunol.1301748. [DOI] [PubMed] [Google Scholar]
- 6.Morandi F, Rizzo R, Fainardi E, Rouas-Freiss N, Pistoia V. Recent advances in our understanding of HLA-G biology: Lessons from a wide spectrum of human diseases. J Immunol Res. 2016;2016:4326495. doi: 10.1155/2016/4326495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.da Silva GB, Silva TG, Duarte RA, Neto NL, Carrara HH, Donadi EA, Gonçalves MA, Soares EG, Soares CP. Expression of the classical and Nonclassical HLA molecules in breast cancer. Int J Breast Cancer. 2013;2013:250435. doi: 10.1155/2013/250435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elliott RL, Jiang XP, Phillips JT, Barnett BG, Head JF. Human leukocyte antigen G expression in breast cancer: Role in immunosuppression. Cancer Biother Radiopharm. 2011;26:153–157. doi: 10.1089/cbr.2010.0924. [DOI] [PubMed] [Google Scholar]
- 9.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 10.Gal-Yam EN, Saito Y, Egger G, Jones PA. Cancer epigenetics: Modifications, screening, and therapy. Annu Rev Med. 2008;59:267–280. doi: 10.1146/annurev.med.59.061606.095816. [DOI] [PubMed] [Google Scholar]
- 11.Zhang S, Chen X, Wang F, An X, Tang B, Zhang X, Sun L, Li Z. Aberrant DNA methylation reprogramming in bovine SCNT preimplantation embryos. Sci Rep. 2016;6:30345. doi: 10.1038/srep30345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu H, Zhang Y. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev. 2011;25:2436–2452. doi: 10.1101/gad.179184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bird AP, Wolffe AP. Methylation-induced repression-belts, braces, and chromatin. Cell. 1999;99:451–454. doi: 10.1016/S0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 14.Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet. 2000;9:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 15.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 16.Amouroux R, Nashun B, Shirane K, Nakagawa S, Hill PW, D'Souza Z, Nakayama M, Matsuda M, Turp A, Ndjetehe E, et al. De novo DNA methylation drives 5hmC accumulation in mouse zygotes. Nat Cell Biol. 2016;18:225–233. doi: 10.1038/ncb3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: Integrating immunity's roles in cancer suppression and promotion. Science. 2011;331:1565–1570. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- 18.de Kruijf EM, Sajet A, van Nes JG, Natanov R, Putter H, Smit VT, Liefers GJ, van den Elsen PJ, van de Velde CJ, Kuppen PJ. HLA-E and HLA-G expression in classical HLA class I-negative tumors is of prognostic value for clinical outcome of early breast cancer patients. J Immunol. 2010;185:7452–7459. doi: 10.4049/jimmunol.1002629. [DOI] [PubMed] [Google Scholar]
- 19.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/S0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 20.Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, Xie ZG, Shi L, He X, Jin SG, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Deivendran S, Marzook H, Santhoshkumar TR, Kumar R, Pillai MR. Metastasis-associated protein 1 is an upstream regulator of DNMT3a and stimulator of insulin-growth factor binding protein-3 in breast cancer. Sci Rep. 2017;7:44225. doi: 10.1038/srep44225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Collignon E, Canale A, Al Wardi C, Bizet M, Calonne E, Dedeurwaerder S, Garaud S, Naveaux C, Barham W, Wilson A, et al. Immunity drives TET1 regulation in cancer through NF-κB. Sci Adv. 2018;4:eaap7309. doi: 10.1126/sciadv.aap7309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu C, Zhang YL, Pan WW, Li XM, Wang ZW, Ge ZJ, Zhou JJ, Cang Y, Tong C, Sun QY, Fan HY. CRL4 complex regulates mammalian oocyte survival and reprogramming by activation of TET proteins. Science. 2013;342:1518–1521. doi: 10.1126/science.1244587. [DOI] [PubMed] [Google Scholar]
- 25.Verloes A, Spits C, Vercammen M, Geens M, LeMaoult J, Sermon K, Coucke W, Van de Velde H. The role of methylation, DNA polymorphisms and microRNAs on HLA-G expression in human embryonic stem cells. Stem Cell Res. 2017;19:118–127. doi: 10.1016/j.scr.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 26.Paul P, Rouas-Freiss N, Khalil-Daher I, Moreau P, Riteau B, Le Gal FA, Avril MF, Dausset J, Guillet JG, Carosella ED. HLA-G expression in melanoma: A way for tumor cells to escape from immunosurveillance. Proc Natl Acad Sci USA. 1998;95:4510–4515. doi: 10.1073/pnas.95.8.4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wiendl H, Mitsdoerffer M, Hofmeister V, Wischhusen J, Bornemann A, Meyermann R, Weiss EH, Melms A, Weller M. A functional role of HLA-G expression in human gliomas: An alternative strategy of immune escape. J Immunol. 2002;168:4772–4780. doi: 10.4049/jimmunol.168.9.4772. [DOI] [PubMed] [Google Scholar]
- 28.Ugurel S, Rebmann V, Ferrone S, Tilgen W, Grosse-Wilde H, Reinhold U. Soluble human leukocyte antigen-G serum level is elevated in melanoma patients and is further increased by interferon-alpha immunotherapy. Cancer. 2001;92:369–376. doi: 10.1002/1097-0142(20010715)92:2<369::AID-CNCR1332>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 29.Urosevic M, Willers J, Mueller B, Kempf W, Burg G, Dummer R. HLA-G protein up-regulation in primary cutaneous lymphomas is associated with interleukin-10 expression in large cell T-cell lymphomas and indolent B-cell lymphomas. Blood. 2002;99:609–617. doi: 10.1182/blood.V99.2.609. [DOI] [PubMed] [Google Scholar]
- 30.Urosevic M, Kurrer MO, Kamarashev J, Mueller B, Weder W, Burg G, Stahel RA, Dummer R, Trojan A. Human leukocyte antigen G up-regulation in lung cancer associates with high-grade histology, human leukocyte antigen class I loss and interleukin-10 production. Am J Pathol. 2001;159:817–824. doi: 10.1016/S0002-9440(10)61756-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lefebvre S, Antoine M, Uzan S, McMaster M, Dausset J, Carosella ED, Paul P. Specific activation of the non-classical class I histocompatibility HLA-G antigen and expression of the ILT2 inhibitory receptor in human breast cancer. J Pathol. 2002;196:266–274. doi: 10.1002/path.1039. [DOI] [PubMed] [Google Scholar]
- 32.Poláková K, Bandzuchová E, Kuba D, Russ G. Demethylating agent 5-aza-2′-deoxycytidine activates HLA-G expression in human leukemia cell lines. Leuk Res. 2009;33:518–524. doi: 10.1016/j.leukres.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 33.Moreau P, Mouillot G, Rousseau P, Marcou C, Dausset J, Carosella ED. HLA-G gene repression is reversed by demethylation. Proc Natl Acad Sci USA. 2003;100:1191–1196. doi: 10.1073/pnas.0337539100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang S, Tang B, Fan C, Shi L, Zhang X, Sun L, Li Z. Effect of DNMT inhibitor on bovine parthenogenetic embryo development. Biochem Biophys Res Commun. 2015;466:505–511. doi: 10.1016/j.bbrc.2015.09.060. [DOI] [PubMed] [Google Scholar]
- 35.Brueckner B, Garcia Boy R, Siedlecki P, Musch T, Kliem HC, Zielenkiewicz P, Suhai S, Wiessler M, Lyko F. Epigenetic reactivation of tumor suppressor genes by a novel small-molecule inhibitor of human DNA methyltransferases. Cancer Res. 2005;65:6305–6311. doi: 10.1158/0008-5472.CAN-04-2957. [DOI] [PubMed] [Google Scholar]
- 36.Stresemann C, Brueckner B, Musch T, Stopper H, Lyko F. Functional diversity of DNA methyltransferase inhibitors in human cancer cell lines. Cancer Res. 2006;66:2794–2800. doi: 10.1158/0008-5472.CAN-05-2821. [DOI] [PubMed] [Google Scholar]
- 37.Elvidge GP, Glenny L, Appelhoff RJ, Ratcliffe PJ, Ragoussis J, Gleadle JM. Concordant regulation of gene expression by hypoxia and 2-oxoglutarate-dependent dioxygenase inhibition: The role of HIF-1alpha, HIF-2alpha, and other pathways. J Biol Chem. 2006;281:15215–15226. doi: 10.1074/jbc.M511408200. [DOI] [PubMed] [Google Scholar]
- 38.Zhang J, Zhang S, Wang Y, Cheng H, Hao L, Zhai Y, Zhang Z, An X, Ma X, Zhang X, et al. Effect of TET inhibitor on bovine parthenogenetic embryo development. PLoS One. 2017;12:e0189542. doi: 10.1371/journal.pone.0189542. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used during the present study are available from the corresponding author on reasonable request.