Abstract
Genome-wide association studies (GWAS) have greatly expanded our understanding of the genetic architecture of cardiovascular diseases in the past decade. They have revealed hundreds of suggestive genetic loci that replicate known biological candidate genes and indicate the existence of a previously unsuspected new biology relevant to cardiovascular disorders. These data have been used successfully to create genetic risk scores that may improve risk prediction and the identification of susceptive individuals. Furthermore, these GWAS-identified novel pathways may herald a new era of novel drug development and stratification of patients. In this review, we will briefly summarize the literature on the candidate genes and signals discovered by GWAS on hypertension and coronary artery disease and discuss their implications on clinical medicine.
Keywords: Genome-wide association study, Genetic risk score, Hypertension, Coronary artery disease
Introduction
A large majority of heart diseases are polygenetic, that is, the result of a combination of multiple common genetic variants and environmental factors. Unravelling the genetic basis of heart diseases has proceeded slowly through linkage studies, because of the much smaller effect size attributable to common modifying variants in complex disorders [1]. Chip-based microarray technology for assaying over 1 million interindividual genetic variants provides the foundation of genome-wide association studies (GWAS), which are defined as any studies of common genetic variation across the entire human genome designed to identify genetic associations with observable traits [2]. The common variants usually refer to those with a prevalence of at least 5% in a population. A stringent genome-wide significance threshold of p < 5E–8 is routinely used as a correction for multiple testing, which is based on the estimation of approximately 1 million independent SNPs in a population [3].
The past decade has witnessed substantial advances in understanding the genetic basis of heart diseases through GWAS. Furthermore, HapMap- and 1000 Genomes Project-based meta-analyses including hundreds of thousands of subjects have expanded our understanding of the genetic architecture of heart diseases to a great extent. In the GWAS catalog (www.ebi.ac.uk/gwas/), hundreds of loci have been reported that show an association with more than 10 heart diseases or traits; the major achievements are listed in Table 1.
Table 1.
Genome-wide association studies of hypertension and several other cardiovascular diseases and abnormalities
| Study [Ref.], year | Initial sample size, n | Replication sample size, n | Major ethnic group | Top locus |
|---|---|---|---|---|
| Blood pressure | ||||
| Levyet al.[8], 2009 | 29,136 | 34,433 | European | SH2B3 |
| Newton-Chehet al.[7], 2009 | 34,433 | 113,236 | European | CNNM2 |
| Kato et al. [12], 2011 | 19,608 | 30,765 | Asian | HECTD4 |
| Wainet al. [10], 2011 | 74,064 | 48,607 | European | ATXN2 |
| Ehret et al. [9], 2011 | 69,395 | 133,361 | European | CSK |
| Warren et al. [11], 2017 | 140,886 | 190,318 | European | JAG1 |
| Coronary artery disease | ||||
| Samani et al. [17], 2007 | 4,864 | 2,519 | European | CDKN2B-AS1 - DMRTA1 |
| Helgadottir et al. [42], 2007 | 8,335 | 9,289 | European | CDKN2B-AS1 - DMRTA1 |
| Trégouët et al. [43], 2009 | 4,864 | 14,398 | European | LPA |
| Erdmannet al. [44], 2009 | 2,520 | 38,253 | European | MRAS |
| Reillyet al. [45], 2011 | 2,723 | 17,053 | European | ADAMTS7 |
| C4DGeneticsConsortium et al. [46], 2011 | 30,482 | 40,593 | European, Asian | PHACTR1 |
| Schunkert et al. [47], 2011 | 86,995 | 56,682 | European | CDKN2B-AS1 |
| Lu et al. [48], 2012 | 6,534 | 26,932 | Asian | CDKN2B-AS1 |
| Nikpay et al. [19], 2015 | 187,599 | None | European, Asian | CDKN2B-AS1 |
| Nelson et al. [20], 2017 | 63,731 | 276,868 | European, | CDKN2B-AS1 |
| Congestive heart failure | ||||
| Smith et al. [49], 2010 | 23,821 | None | European, African | LRIG3 |
| Dilated cardiomyopathy | ||||
| Villard et al. [50], 2011 | 2,287 | 2,467 | European | TSPB7 |
| Atrial fibrillation | ||||
| Gudbjartsson et al. [51], 2009 | 36,137 | 8,855 | European | ZFHX3 |
| Ellinor et al. [52], 2010 | 14,179 | 4,771 | European | KCNN3 |
| Ellinor et al. [53], 2012 | 59,133 | 5,381 | European | PITX2 - MIR297 |
| Lowet al. [54], 2017 | 36,792 | 257,896 | Asian, European | PITX2 - MIR297 |
| Electrocardiographic abnormalities in the PR interval | ||||
| Pfeufer et al. [55], 2010 | 28,517 | None | European | SCN10A |
| Chambers et al. [56], 2010 | 6,543 | 6,243 | Asian | SCN10A |
| QRS duration | ||||
| Sotoodehnia et al. [57], 2010 | 40,407 | 7,170 | European | SCN10A |
| QT interval | ||||
| Arking et al. [58], 2006 | 200 | 4,451 | European | NOS1AP, LOC105371475 |
| Newton-Cheh et al. [59], 2009 | 13,685 | 15,854 | European | LOC107985450 - NOS1AP |
| Arking et al. [60], 2014 | 70,389 | 33,316 | European | LOC107985450 – NOS1AP |
| Heart rate | ||||
| den Hoed et al. [61], 2013 | 92,355 | 88,823 | European | MYH6 |
| Nolte et al. [62], 2017 | 26,523 | 38,558 | European, Hispanic, African American | NDUFA11 |
| Dyslipidemia | ||||
| Saxena et al. [63], 2007 | 5,217 | None | European | APOC1 – APOC1P1 |
| Kooner et al. [64], 2008 | 2,011 | 10,536 | European, Mexican, Asian | MLXIPL |
| Kathiresan et al. [35], 2008 | 19,840 | 20,623 | European | HERPUD1 – CETP |
| Sandhu et al. [65], 2008 | 11,685 | 4,979 | European | CELSR2 - PSRC1 |
| Teslovich et al. [16], 2010 | 96,598 | None | European | ZPR1 |
| Kim et al. [66], 2011 | 12,545 | 30,395 | Asian | ZPR1 |
| Willer et al. [67], 2013 | 94,595 | 93,982 | European | LDLR |
| Ko et al. [68], 2014 | 3,323 | 6,017 | Mexican | ZPR1 |
| Surakka et al. [69], 2015 | 62,166 | None | European | APOE |
In this review, we will give a brief summary of the achievements of GWAS regarding blood pressure (BP) and coronary artery disease (CAD), the progress in risk prediction at individual level, the novel pathophysiological pathways, and drug discoveries.
GWAS-Significant Loci
Blood Pressure
BP is a quantitative trait, normally distributed in the general population, whereas 30–50% of the variation in BP is determined by inherited genetic factors [4, 5]. Hypertension ranks as the leading cause of morbidity and mortality worldwide, contributing to CAD, atrial fibrillation, and heart failure, so that the knowledge about the genetics of hypertension or BP is an important factor in the diagnosis, control, and treatment of all of these heart diseases.
Until the middle of 2017, GWAS have identified and replicated genetic variants of modest or weak effect on BP over 200 loci; the strongest SNPs associated with different BP traits are summarized in Table 2a and b. However, the attempts to identify genetic variants associated with BP have been challenging and of relatively low yield in the early phase. In 2007, the first GWAS (by the Wellcome Trust Case Control Consortium [WTCCC]) [6] adopted a case-control study design using 3,000 shared controls and 14,000 cases (2,000 for hypertension) of European ancestry to study 7 complex diseases simultaneously. Hypertension was the only disease without any significant results. The first GWAS of quantitative BP phenotypes was conducted in the Framingham Heart Study, which included 1,400 family subjects and found no significant results either [5]. These two studies let researchers realize the complexity of the genetic mechanisms underlying BP regulation and the need for much larger sample sizes when looking for genes associated with BP/hypertension.
Table 2.
Significant genetic loci for blood pressure and hypertension reported in genome-wide association studies in Europeans (a) and Asians and Africans (b)
| Chr | Strongest SNP |
Position | EA | EAF | OR or BETA | p value | N | Closest gene | Trait | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|
|
a 1 |
Europeans rs1886773 |
1319056 | A | 0.03 | −0.63 | 1.90E–10 | 233,789 | NADK, CPSF3L | PP | [11] |
| 1 | rs14057 | 6623180 | A | 0.35 | −0.3 | 2.50E–10 | 329,584 | RNF207 | SBP | [11] |
| 1 | rs9662255 | 9381890 | A | 0.43 | −0.21 | 1.90E–10 | 310,618 | chr1mb9 | PP | [11] |
| 1 | rs880315 | 10736809 | C | 0.35 | 0.64 | 9.10E–16 | 133,125 | CASZ1 | SBP | [70] |
| 1 | rs17367504 | 11802721 | G | 0.15 | −0.53 | 2.68E–18 | 149,714 | MTHFR, NPPB | DBP | [70] |
| 1 | rs12744757 | 11846764 | T | 0.06 | −0.7 | 7.54E–23 | 359,786 | NPPA-AS1, NPPA | SBP | [70] |
| 1 | rs1042010 | 15467418 | A | 0.19 | −0.41 | 3.03E–13 | 345,219 | CELA2A | SBP | [70] |
| 1 | rs1048238 | 16015154 | T | 0.57 | 0.37 | 8.09E–07 | 140,299 | HSPB7 | SBP | [70] |
| 1 | rs6686889 | 24703979 | T | 0.25 | 0.19 | 3.60E–09 | 322,575 | chr1mb25 | DBP | [11] |
| 1 | rs871524 | 37945773 | A | 0.33 | 0.23 | 1.27E–11 | 304,987 | INPP5B | PP | [70] |
| 1 | rs4360494 | 37990219 | C | 0.55 | 0.28 | 3.70E–16 | 282,851 | SF3A3 | PP | [11] |
| 1 | rs112557609 | 56111252 | A | 0.35 | 0.23 | 6.80E–12 | 325,952 | RP4–710M16.1, PPAP2B | PP | [11] |
| 1 | rs3889199 | 59188070 | A | 0.71 | 0.35 | 1.80E–24 | 329,486 | FGGY | PP | [11] |
| 1 | rs61767086 | 88600899 | G | 0.14 | −0.41 | 1.90E–09 | 136,914 | PKN2-AS1 | PP | [70] |
| 1 | rs10922502 | 88894475 | A | 0.62 | −0.38 | 2.20E–15 | 323,666 | GTF2B | SBP | [11] |
| 1 | rs12129649 | 112688881 | T | 0.06 | 0.55 | 5.93E–19 | 343,172 | MOV10 | DBP | [70] |
| 1 | rs11466111 | 115286557 | T | 0.02 | 0.65 | 5.08E–10 | 319,490 | chr1mb115 | DBP | [11] |
| 1 | rs12405515 | 172388301 | T | 0.56 | −0.17 | 1.40E–09 | 328,543 | DNM3 | DBP | [11] |
| 1 | rs12408022 | 217545447 | T | 0.26 | 0.2 | 2.40E–10 | 320,983 | GPATCH2 | DBP | [11] |
| 1 | rs10916082 | 227064925 | A | 0.73 | −0.18 | 8.40E–09 | 327,636 | CDC42BPA | DBP | [11] |
| 1 | rs2760061 | 228003374 | A | 0.47 | 0.23 | 2.10E–16 | 312,761 | WNT3A | DBP | [11] |
| 1 | rs953492 | 243307890 | A | 0.46 | 0.22 | 7.40E–16 | 325,253 | SDCCAG8 | DBP | [11] |
| 2 | rs2289081 | 20682080 | C | 0.36 | −0.22 | 5.50E–12 | 329,140 | C2orf43 | PP | [11] |
| 2 | rs55701159 | 24916727 | T | 0.89 | 0.29 | 7.20E–11 | 321,052 | ADCY3 | DBP | [11] |
| 2 | rs1275988 | 26691496 | T | 0.61 | −0.58 | 2.35E–15 | 149,285 | KCNK3 | SBP | [70] |
| 2 | rs7562 | 28412873 | T | 0.52 | 0.26 | 1.90E–08 | 319,942 | FOSL2 | SBP | [11] |
| 2 | rs13420463 | 37290423 | A | 0.77 | 0.36 | 7.00E–11 | 330,307 | PRKD3 | SBP | [11] |
| 2 | rs4952611 | 40340603 | T | 0.58 | −0.16 | 4.00E–08 | 309,395 | SLC8A1 | DBP | [11] |
| 2 | rs76326501 | 42940738 | A | 0.91 | 0.42 | 3.60E–18 | 318,127 | AC016735.1 | DBP | [11] |
| 2 | rs72876037 | 42967456 | T | 0.12 | −0.53 | 1.65E–14 | 319,468 | HAAO, ZFP36L2 | SBP | [70] |
| 2 | rs11690961 | 46136197 | A | 0.88 | 0.34 | 3.90E–12 | 327,847 | PRKCE | PP | [11] |
| 2 | rs74181299 | 65056838 | T | 0.62 | 0.23 | 9.60E–13 | 324,224 | CEP68 | PP | [11] |
| 2 | rs11689667 | 85264242 | T | 0.54 | 0.18 | 1.70E–08 | 330,634 | TCF7L1 | PP | [11] |
| 2 | rs2579519 | 96009418 | T | 0.63 | −0.2 | 4.80E–12 | 311,557 | GPAT2, FAHD2CP | DBP | [11] |
| 2 | rs62167177 | 145181135 | T | 0.14 | −0.26 | 1.04E–09 | 289,338 | TEX41 | DBP | [11] |
| 2 | rs1446468 | 164106976 | C | 0.55 | 0.54 | 2.98E–13 | 141,331 | FIGN, GRB14 | SBP | [70] |
| 2 | rs79146658 | 178921341 | T | 0.91 | −0.31 | 2.40E–10 | 321,318 | CCDC141 | DBP | [11] |
| 2 | rs13407401 | 179850979 | A | 0.29 | −0.43 | 4.30E–08 | 145,919 | ZNF385B | SBP | [70] |
| 2 | rs144073138 | 182410033 | G | 0.06 | 0.35 | 1.90E–08 | 295,411 | PDE1A | DBP | [11] |
| 2 | rs7592578 | 190574865 | T | 0.19 | −0.24 | 9.50E–12 | 304,672 | TMEM194B | DBP | [11] |
| 2 | rs55780018 | 207661416 | T | 0.54 | −0.39 | 5.90E–16 | 304,567 | METTL21A, AC079767.3 | SBP | [11] |
| 2 | rs1250259 | 215435759 | A | 0.74 | −0.31 | 8.70E–19 | 325,485 | FN1 | PP | [11] |
| 2 | rs10932679 | 216787868 | T | 0.19 | −0.23 | 1.70E–08 | 371,085 | LOC101928278 | PP | [70] |
| 2 | rs1063281 | 217804009 | T | 0.6 | −0.2 | 1.30E–12 | 315,354 | TNS1 | DBP | [11] |
| 3 | rs142892876 | 1110752 | T | 0.001 | 10.69 | 5.00E–08 | 19,546 | CNTN6 | DBP | [71] |
| 3 | rs11716531 | 27415717 | A | 0.24 | 0.21 | 5.72E–14 | 373,386 | SLC4A7 | DBP | [70] |
| 3 | rs13082711 | 27496418 | C | 0.23 | 0.33 | 2.72E–10 | 146,340 | SLC4A7 | DBP | [70] |
| 3 | rs36022378 | 49876272 | T | 0.8 | −0.2 | 4.70E–09 | 319,983 | CAMKV, ACTBP13 | DBP | [11] |
| 3 | rs743757 | 50438947 | C | 0.14 | 0.25 | 2.40E–10 | 328,836 | CACNA2D2 | DBP | [11] |
| 3 | rs9827472 | 56692618 | T | 0.37 | −0.18 | 4.30E–10 | 323,058 | FAM208A | DBP | [11] |
| 3 | rs9834975 | 122398816 | T | 0.44 | 0.15 | 1.14E–08 | 330,670 | chr3mb122 | DBP | [11] |
| 3 | rs62270945 | 128483046 | T | 0.03 | 0.61 | 1.80E–09 | 279,925 | GATA2 | PP | [11] |
| 3 | rs9859176 | 134281183 | T | 0.4 | 0.32 | 1.30E–11 | 322,428 | RYK | SBP | [11] |
| 3 | rs143112823 | 154990178 | A | 0.09 | −0.4 | 1.40E–14 | 297,343 | RP11–439C8.2 | DBP | [11] |
| 3 | rs419076 | 169383098 | T | 0.47 | 0.41 | 1.78E–13 | 193,725 | MECOM | SBP | [9] |
| 3 | rs12374077 | 185599886 | C | 0.35 | 0.16 | 9.20E–09 | 327,513 | SENP2 | DBP | [11] |
| 3 | rs528908640 | 193644588 | C | 0.0005 | 14.67 | 2.00E–08 | 19,546 | OPA1 | DBP | [71] |
| 3 | rs9815354 | 41912651 | A | 0.17 | 0.4 | 7.12E–11 | 135,381 | ULK4 | DBP | [70] |
| 4 | rs871606 | 53933078 | C | 0.1 | −0.5 | 1.53E–10 | 144,494 | CHIC2 | PP | [70] |
| 4 | rs17004869 | 80283879 | T | 0.05 | 0.72 | 1.02E–09 | 278,498 | FGF5 | SBP | [11] |
| 4 | rs2014912 | 85794517 | C | 0.85 | −0.42 | 3.86E–10 | 143,933 | ARHGAP24 | PP | [70] |
| 4 | rs13107325 | 102267552 | T | 0.05 | −0.68 | 2.28E–17 | 150,920 | SLC39A8 | DBP | [9] |
| 4 | rs13112725 | 105990585 | C | 0.77 | 0.41 | 3.11E–16 | 357,230 | NPNT, TBCK | SBP | [70] |
| 4 | rs33966350 | 110510288 | A | 0.01 | 1.66 | 2.10E–11 | 216,630 | ENPEP | SBP | [11] |
| 4 | rs66887589 | 119588124 | T | 0.52 | −0.22 | 3.40E–15 | 324,397 | PDE5A | DBP | [11] |
| 4 | rs78049276 | 147506351 | C | 0.13 | 0.27 | 1.04E–08 | 321,315 | chr4mb148 | PP | [11] |
| 4 | rs13139571 | 155724361 | C | 0.76 | 0.26 | 2.17E–10 | 185,393 | GUCY1A3, GUCY1B3 | DBP | [9] |
| 4 | rs1566497 | 168795997 | A | 0.42 | 0.24 | 1.90E–13 | 320,948 | PALLD | PP | [11] |
| 4 | rs17059668 | 173663512 | C | 0.92 | −0.33 | 2.80E–08 | 313,277 | chr4mb174 | PP | [11] |
| 5 | rs1173771 | 32814922 | G | 0.6 | 0.5 | 1.79E–16 | 158,664 | C5orf23, NPR3 | SBP | [9] |
| 5 | rs10078021 | 75742606 | T | 0.63 | −0.16 | 1.30E–08 | 314,172 | POC5 | DBP | [11] |
| 5 | rs10057188 | 78541966 | A | 0.46 | −0.21 | 6.70E–11 | 325,985 | LHFPL2 | PP | [11] |
| 5 | rs10059921 | 88218698 | T | 0.08 | −0.53 | 4.00E–09 | 298,543 | TMEM161B | SBP | [11] |
| 5 | rs337100 | 123210816 | A | 0.41 | −0.28 | 4.61E–09 | 149,200 | PRDM6 - SUMO1P5 | PP | [70] |
| 5 | rs6595838 | 128532506 | A | 0.3 | 0.34 | 7.60E–12 | 328,401 | FBN2 | SBP | [11] |
| 5 | rs31864 | 158793185 | A | 0.55 | 0.21 | 5.50E–11 | 326,557 | EBF1 | PP | [11] |
| 5 | rs72812846 | 173950633 | A | 0.28 | −0.21 | 2.20E–11 | 312,601 | CPEB4 | DBP | [11] |
| 6 | rs6911827 | 22130372 | T | 0.45 | 0.3 | 2.00E–10 | 326,471 | CASC15 | SBP | [11] |
| 6 | rs1800562 | 26092913 | A | 0.06 | 0.39 | 8.69E–17 | 373,770 | HFE | DBP | [70] |
| 6 | rs151168737 | 31638615 | A | 0.46 | 0.25 | 3.60E–20 | 315,660 | PRRC2A - BAG6 | DBP | [70] |
| 6 | rs805303 | 31648589 | G | 0.61 | 0.38 | 1.49E–11 | 201,745 | BAT2, BAT5 | SBP | [9] |
| 6 | rs185819 | 32082290 | C | 0.51 | 0.37 | 1.04E–17 | 364,144 | TNXB | SBP | [70] |
| 6 | rs78648104 | 50715296 | T | 0.92 | −0.48 | 1.30E–08 | 305,426 | TFAP2D | SBP | [11] |
| 6 | rs13205180 | 51967696 | T | 0.49 | 0.17 | 7.00E–10 | 325,419 | PKHD1 | DBP | [11] |
| 6 | rs9372498 | 118251323 | A | 0.08 | 0.33 | 1.80E–11 | 330,625 | SLC35F1 | DBP | [11] |
| 6 | rs11154027 | 121460244 | T | 0.47 | 0.21 | 1.10E–10 | 316,708 | GJA1 | PP | [11] |
| 6 | rs13209747 | 126794309 | T | 0.45 | 0.27 | 1.12E–09 | 146,105 | RSPO3 | DBP | [70] |
| 6 | rs17080102 | 150683634 | C | 0.07 | −0.57 | 1.04E–10 | 147,869 | PLEKHG1 | DBP | [70] |
| 6 | rs36083386 | 152076778 | I | 0.11 | 0.44 | 1.50E–18 | 323,303 | ESR1 | PP | [11] |
| 6 | rs449789 | 159278093 | C | 0.14 | 0.36 | 2.40E–15 | 325,584 | FNDC1 | PP | [11] |
| 6 | rs147212971 | 165764963 | T | 0.06 | −0.36 | 1.60E–09 | 296,010 | PDE10A | DBP | [11] |
| 6 | rs1322639 | 169187008 | A | 0.78 | 0.32 | 4.80E–17 | 319,866 | THBS2 | PP | [11] |
| 7 | rs6461992 | 27181212 | G | 0.93 | 0.91 | 8.49E–24 | 311,598 | HOXA3 | SBP | [11] |
| 7 | rs1859168 | 27202740 | C | 0.92 | 0.44 | 1.85E–20 | 373,643 | HOTTIP | DBP | [70] |
| 7 | rs76206723 | 40408372 | A | 0.1 | −0.35 | 7.40E–12 | 328,162 | SUGCT | PP | [11] |
| 7 | rs10260816 | 45970501 | G | 0.43 | −0.3 | 5.31E–10 | 143,371 | IGFBP3 | PP | [70] |
| 7 | rs17477177 | 106771412 | C | 0.21 | 0.55 | 4.14E–22 | 149,296 | PIK3CG | PP | [70] |
| 7 | rs13238550 | 131374297 | A | 0.4 | 0.33 | 1.90E–12 | 325,647 | MKLN1 | SBP | [11] |
| 7 | rs1011018 | 139763465 | A | 0.2 | −0.33 | 1.50E–08 | 325,110 | HIPK2 | SBP | [11] |
| 7 | rs3918226 | 150993088 | T | 0.08 | 0.67 | 2.27E–13 | 118,604 | NOS3 | DBP | [70] |
| 8 | rs9693857 | 9409607 | T | 0.45 | −0.34 | 2.40E–15 | 374,178 | LOC105379231 | SBP | [70] |
| 8 | rs1821002 | 10782555 | G | 0.6 | −0.42 | 4.26E–19 | 323,712 | BLK, GATA4 | SBP | [11] |
| 8 | rs10107145 | 10900703 | G | 0.53 | −0.36 | 1.44E–17 | 374,413 | XKR6 | SBP | [70] |
| 8 | rs6557876 | 26043159 | T | 0.25 | −0.37 | 2.85E–14 | 369,457 | EBF2 | SBP | [70] |
| 8 | rs2978456 | 42467247 | T | 0.55 | −0.19 | 1.20E–08 | 304,964 | SLC20A2 | PP | [11] |
| 8 | rs2978098 | 100664447 | A | 0.54 | 0.17 | 1.50E–09 | 324,424 | SNX31 | DBP | [11] |
| 8 | rs35783704 | 104954030 | A | 0.11 | −0.41 | 7.08E–09 | 349,452 | LRP12, ZFPM2 | SBP | [70] |
| 8 | rs2071518 | 119423572 | T | 0.17 | 0.3 | 1.56E–08 | 149,046 | NOV | PP | [10] |
| 8 | rs894344 | 134600502 | A | 0.6 | −0.26 | 3.20E–08 | 329,834 | ZFAT | SBP | [11] |
| 8 | rs4454254 | 140049929 | A | 0.63 | −0.26 | 5.10E–16 | 330,022 | TRAPPC9 | PP | [11] |
| 8 | rs62524579 | 142979538 | A | 0.53 | −0.18 | 3.80E–09 | 268,645 | RP11–273G15.2 | DBP | [11] |
| 8 | rs4076877 | 143820544 | T | 0.05 | −0.42 | 3.57E–08 | 252,983 | chr8mb144 | DBP | [11] |
| 9 | rs872256 | 2496480 | NA | NA | 0.1 | 9.00E–09 | 8,423 | SMARCA2, VLDLR | SBP | [72] |
| 9 | rs4364717 | 21801531 | A | 0.55 | −0.18 | 1.30E–10 | 327,173 | MTAP | DBP | [11] |
| 9 | rs568998724 | 107040156 | A | 0.0007 | 13.49 | 3.00E–08 | 19,546 | LOC340512 | DBP | [71] |
| 9 | rs72765298 | 125138717 | T | 0.87 | −0.37 | 2.70E–14 | 316,271 | SCAI | PP | [11] |
| 10 | rs9337951 | 30028144 | A | 0.34 | 0.28 | 2.50E–15 | 299,646 | KIAA1462 | PP | [11] |
| 10 | rs10826995 | 31793730 | T | 0.71 | −0.21 | 1.10E–09 | 327,373 | ARHGAP12, ZEB1 | PP | [11] |
| 10 | rs4590817 | 61707795 | G | 0.83 | −0.44 | 3.52E–13 | 146,522 | C10orf107 | MAP | [10] |
| 10 | rs932764 | 94136183 | G | 0.44 | 0.48 | 7.10E–16 | 160,885 | PLCE1 | SBP | [9] |
| 10 | rs112184198 | 100844757 | A | 0.1 | −0.66 | 3.60E–18 | 323,791 | PAX2 | SBP | [11] |
| 10 | rs11191548 | 103086421 | T | 0.91 | 1.1 | 6.90E–26 | 161,709 | NT5C2, CYP17A1 | SBP | [9] |
| 10 | rs10787517 | 114055047 | A | 0.62 | 0.44 | 3.31E–18 | 272,852 | ADRB1 - RNU6–709P | SBP | [70] |
| 11 | rs2649044 | 9742422 | T | 0.55 | 0.2 | 1.12E–14 | 343,652 | SWAP70 | DBP | [70] |
| 11 | rs7129220 | 10328991 | G | 0.89 | −0.62 | 2.97E–12 | 182,871 | ADM | SBP | [9] |
| 11 | rs177542 | 16901107 | A | 0.5 | 0.24 | 4.18E–08 | 143,257 | PLEKHA7 | DBP | [70] |
| 11 | rs11030119 | 27706555 | A | 0.31 | −0.16 | 2.90E–08 | 330,002 | BDNF | DBP | [11] |
| 11 | rs11442819 | 45186591 | I | 0.11 | −0.28 | 7.10E–09 | 326,483 | PRDM11 | PP | [11] |
| 11 | rs751984 | 61510774 | C | 0.12 | −0.41 | 1.98E–09 | 140,821 | LRRC10B | DBP | [70] |
| 11 | rs4980532 | 63913247 | T | 0.56 | 0.3 | 1.53E–10 | 147,416 | RCOR2 | PP | [70] |
| 11 | rs67330701 | 69312240 | T | 0.09 | −0.37 | 2.10E–12 | 276,760 | MYEOV | DBP | [11] |
| 11 | rs2289125 | 89491285 | A | 0.21 | −0.38 | 9.10E–22 | 307,682 | NOX4 | PP | [11] |
| 11 | rs633185 | 100722807 | G | 0.28 | −0.56 | 1.21E–17 | 160,461 | TMEM133, FLJ32810 | SBP | [9] |
| 11 | rs8258 | 117412960 | T | 0.38 | 0.24 | 2.90E–13 | 327,038 | CEP164 | PP | [11] |
| 11 | rs11222084 | 130403335 | T | 0.35 | 0.35 | 2.26E–12 | 139,748 | ADAMTS8 | PP | [70] |
| 12 | rs10770612 | 20077705 | A | 0.8 | 0.31 | 6.90E–15 | 311,586 | PDE3A | PP | [11] |
| 12 | rs73099903 | 53046995 | T | 0.07 | 0.51 | 1.95E–10 | 343,318 | LOC283335 | SBP | [70] |
| 12 | rs7312464 | 65980467 | G | 0.52 | 0.21 | 5.35E–10 | 289,978 | chr12mb66 | PP | [11] |
| 12 | rs17249754 | 89666809 | A | 0.16 | −0.9 | 6.59E–20 | 146,304 | ATP2B1 | SBP | [70] |
| 12 | rs139236208 | 94486966 | A | 0.1 | −0.36 | 1.60E–10 | 291,244 | CCDC41 | PP | [11] |
| 12 | rs3184504 | 111446804 | T | 0.47 | 0.45 | 3.59E–25 | 120,633 | SH2B3 | DBP | [9] |
| 12 | rs10850519 | 115490635 | C | 0.3 | −0.21 | 5.10E–13 | 327,837 | TBX5, TBX3 | DBP | [11] |
| 13 | rs9549328 | 112981842 | T | 0.23 | 0.32 | 1.50E–08 | 313,787 | MCF2L | SBP | [11] |
| 14 | rs12050260 | 23291885 | T | 0.35 | 0.19 | 2.60E–08 | 304,390 | MYH6 | PP | [11] |
| 14 | rs8904 | 35402011 | A | 0.38 | 0.31 | 1.31E–12 | 365,195 | NFKBIA | SBP | [70] |
| 14 | rs9888615 | 52910822 | T | 0.29 | −0.32 | 3.50E–10 | 326,235 | FERMT2 | SBP | [11] |
| 14 | rs8016306 | 63461828 | A | 0.8 | 0.34 | 3.70E–09 | 329,869 | PPP2R5E | SBP | [11] |
| 14 | rs12434998 | 93989208 | C | 0.37 | −0.19 | 1.64E–08 | 315,683 | chr14mb94 | PP | [11] |
| 14 | rs9323988 | 98121293 | T | 0.63 | −0.21 | 4.10E–11 | 327,551 | RP11–61O1.1 | PP | [11] |
| 15 | rs1036477 | 48622729 | G | 0.1 | −0.42 | 3.81E–08 | 149,967 | FBN1 | PP | [70] |
| 15 | rs7178615 | 66576734 | A | 0.37 | −0.18 | 2.60E–10 | 318,076 | RP11–321F6.1 | DBP | [11] |
| 15 | rs1563894 | 68343437 | NA | NA | 0.09 | 3.00E–08 | 10,090 | ITGA11 | SBP | [72] |
| 15 | rs117539635 | 69390577 | G | 0.03 | −0.65 | 2.15E–08 | 196,789 | chr15mb69 | DBP | [11] |
| 15 | rs1378942 | 74785026 | C | 0.35 | 0.42 | 2.69E–26 | 163,115 | CYP1A1, ULK3 | DBP | [9] |
| 15 | rs6495122 | 74833304 | A | 0.42 | 0.4 | 1.84E–10 | NA | CSK, ULK3 | DBP | [8] |
| 15 | rs187680191 | 76002970 | T | 0.0006 | 18.58 | 3.00E–09 | 19,546 | NRG4 | DBP | [71] |
| 15 | rs62012628 | 78777658 | T | 0.29 | −0.24 | 5.10E–12 | 244,143 | ADAMTS7 | DBP | [11] |
| 15 | rs11634851 | 80736624 | G | 0.46 | 0.32 | 5.38E–14 | 374,208 | FAM108C1 | SBP | [70] |
| 15 | rs2521501 | 90894158 | T | 0.31 | 0.65 | 5.20E–19 | 127,208 | FURIN, FES | SBP | [9] |
| 15 | rs12906962 | 94768842 | T | 0.68 | −0.22 | 5.60E–14 | 319,952 | chr15mb95 | DBP | [11] |
| 15 | rs4984496 | 96092669 | G | 0.67 | −0.18 | 2.86E–09 | 307,660 | chr15mb96 | DBP | [11] |
| 16 | rs12921187 | 4893018 | T | 0.43 | −0.17 | 2.50E–10 | 326,469 | PPL | DBP | [11] |
| 16 | rs13333226 | 20354332 | A | 0.81 | 0.85 | 1.50E–13 | 39,706 | UMOD | HT | [25] |
| 16 | rs72799341 | 30925422 | A | 0.24 | 0.19 | 5.80E–09 | 324,502 | FBXL19 | DBP | [11] |
| 16 | rs56249585 | 65231799 | T | 0.53 | 0.18 | 8.98E–09 | 318,082 | chr16mb65 | PP | [11] |
| 16 | rs141767645 | 69869262 | T | 0.02 | 1.3 | 1.90E–08 | 174,313 | NFAT5 | PP | [11] |
| 16 | rs200337503 | 70084312 | I | 0.03 | 1.31 | 6.88E–13 | 193,318 | NFAT5 | PP | [11] |
| 16 | rs117006983 | 70721707 | A | 0.01 | 0.99 | 4.10E–12 | 250,766 | VAC14 | PP | [11] |
| 16 | rs11643209 | 75297146 | T | 0.42 | −0.34 | 1.80E–12 | 309,242 | CFDP1 | SBP | [11] |
| 16 | rs8059962 | 81540592 | T | 0.42 | −0.17 | 1.30E–09 | 319,839 | CMIP | DBP | [11] |
| 16 | rs7500448 | 83012185 | A | 0.75 | 0.33 | 1.10E–19 | 321,958 | CDH13 | PP | [11] |
| 17 | rs12941318 | 1430304 | T | 0.49 | −0.27 | 2.50E–08 | 299,739 | CRK | SBP | [11] |
| 17 | rs7226020 | 6570508 | T | 0.56 | −0.26 | 2.30E–14 | 303,389 | KIAA0753 | PP | [11] |
| 17 | rs5417 | 7281743 | A | 0.57 | 0.21 | 1.10E–13 | 319,299 | TP53, SLC2A4 | DBP | [11] |
| 17 | rs138643143 | 42557849 | A | 0.07 | 0.5 | 3.30E–09 | 229,161 | KCNH4, HSD17B1 | PP | [11] |
| 17 | rs62080325 | 43983263 | A | 0.66 | −0.19 | 4.00E–08 | 315,689 | PYY | PP | [11] |
| 17 | rs12946454 | 45130754 | T | 0.28 | 0.57 | 1.00E–08 | 77,690 | PLCD3 | SBP | [7] |
| 17 | rs551011992 | 45851264 | G | 0.24 | −0.33 | 3.32E–08 | 275,778 | GOSR2 | SBP | [11] |
| 17 | rs17608766 | 46935905 | C | 0.14 | 0.55 | 1.97E–15 | 137,066 | GOSR2 | PP | [70] |
| 17 | rs79917357 | 48747312 | A | 0.17 | 0.34 | 5.33E–09 | 336,863 | LOC105371811 –LOC105371812 | SBP | [70] |
| 17 | rs585736 | 48796910 | A | 0.03 | 0.61 | 2.50E–11 | 301,845 | HOXB7 | PP | [11] |
| 17 | rs12940887 | 49325445 | T | 0.37 | 0.26 | 1.18E–08 | 144,603 | ZNF652 | DBP | [70] |
| 17 | rs16948048 | 49363104 | G | 0.39 | 0.31 | 8.24E+04 | 82,441 | PHB, ZNF652 | DBP | [7] |
| 17 | rs740698 | 62689790 | T | 0.56 | −0.23 | 3.10E–12 | 311,450 | MRC2 | PP | [11] |
| 17 | rs4459609 | 63471587 | A | 0.61 | −0.2 | 2.03E–14 | 361,581 | CYB561, ACE | DBP | [70] |
| 17 | rs4308 | 63482264 | A | 0.37 | 0.21 | 6.80E–14 | 319,394 | ACE | DBP | [11] |
| 17 | rs2467099 | 75952964 | T | 0.22 | −0.31 | 3.30E–08 | 326,401 | ACOX1 | SBP | [11] |
| 17 | rs57927100 | 77321218 | G | 0.26 | −0.31 | 4.04E–10 | 347,188 | SPET9 | SBP | [70] |
| 18 | rs7236548 | 45517785 | A | 0.18 | 0.35 | 2.00E–18 | 330,075 | SLC14A2 | PP | [11] |
| 18 | rs745821 | 50616484 | T | 0.76 | 0.19 | 1.40E–09 | 330,954 | MAPK4 | DBP | [11] |
| 19 | rs36047283 | 7255690 | G | 0.11 | −0.8 | 3.25E–26 | 281,588 | INSR | SBP | [70] |
| 19 | rs2116941 | 10223767 | C | 0.81 | −0.22 | 2.77E–08 | 321,960 | chr19mb10 | PP | [11] |
| 19 | rs77279095 | 11415506 | A | 0.04 | 0.86 | 7.26E–09 | 224,215 | RGL3 | SBP | [11] |
| 19 | rs62104477 | 29804084 | T | 0.33 | 0.18 | 1.20E–09 | 320,347 | CCNE1 | DBP | [11] |
| 19 | rs9710247 | 40254542 | G | 0.45 | 0.16 | 1.61E–09 | 308,028 | AKT2 | DBP | [70] |
| 20 | rs6108168 | 8645624 | A | 0.25 | −0.21 | 1.10E–11 | 327,368 | PLCB1 | DBP | [11] |
| 20 | rs1327235 | 10988382 | G | 0.46 | 0.3 | 1.41E–15 | 158,454 | JAG1 | DBP | [9] |
| 20 | rs6081613 | 19485263 | A | 0.28 | 0.26 | 1.60E–13 | 315,546 | SLC24A3 | PP | [11] |
| 20 | rs80346118 | 48794612 | A | 0.15 | −0.27 | 1.10E–12 | 327,614 | PREX1 | DBP | [11] |
| 20 | rs6015450 | 59176062 | G | 0.12 | 0.9 | 3.87E–23 | 159,190 | EDN3, GNAS | SBP | [9] |
| 21 | rs12627651 | 43340723 | A | 0.29 | 0.32 | 2.40E–09 | 121,169 | CRYAA, SIK1 | DBP | [70] |
| 22 | rs4819852 | 20000644 | A | 0.29 | 0.26 | 1.43E–13 | 367,460 | ARVCF | PP | [70] |
| 22 | rs73161324 | 41642782 | T | 0.05 | 0.5 | 2.80E–11 | 267,722 | XRCC6 | PP | [11] |
| b Asians and Africans | ||||||||||
| Asians | ||||||||||
| 1 rs880315 | 10736809 | C | 0.65 | 0.56 | 3.05E–10 | 32,611 | CASZ1 | DBP | [12] | |
| 1 | rs10745332 | 112646431 | A | 0.82 | 0.96 | 2.52E–09 | 46,269 | MOV10 | SBP | [13] |
| 2 | rs1344653 | 19531084 | A | 0.53 | −0.27 | 7.79E–12 | 220,853 | OSR1 | PP | [73] |
| 2 | rs1275988 | 26691496 | T | 0.53 | −0.37 | 4.95E–21 | 236,311 | KCNK3 | OH | [73] |
| 2 | rs7604423 | 43155602 | C | 0.66 | −0.21 | 2.40E–08 | 217,072 | NR | DBP | [73] |
| 2 | rs6736587 | 81628601 | C | 0.16 | −1.38 | 5.3E–08 | NA | CTNNA2 | OH | [74] |
| 2 | rs16849225 | 164050310 | C | 0.61 | 0.75 | 3.45E–11 | 49,511 | GRB14, FIGN | SBP | [12] |
| 3 | rs820430 | 27507409 | A | 0.32 | 0.76 | 1.36E–12 | 79,318 | SLC4A7 | SBP | [13] |
| 3 | rs9810888 | 53601568 | G | 0.39 | 0.39 | 4.0E–12 | 77,555 | CACNA1D | DBP | [13] |
| 4 | rs1902859 | 80236549 | C | 0.41 | 1.34 | 1.76E–22 | 45,856 | FGF5 | SBP | [13] |
| 4 | rs2014912 | 85794517 | T | 0.16 | 0.62 | 5.37E–17 | 242,456 | ARHGAP24 | SBP | [73] |
| 4 | rs6825911 | 110460482 | C | 0.51 | 0.39 | 8.96E–09 | 49,511 | ENPEP | DBP | [12] |
| 4 | rs13143871 | 155698052 | T | 0.80 | 0.96 | 5.16E–08 | 45,737 | GUCY1A3 | SBP | [13] |
| 5 | rs1173766 | 32804422 | C | 0.60 | 0.63 | 1.95E–08 | 49,970 | NPR3 | SBP | [12] |
| 5 | rs13359291 | 123140763 | A | 0.31 | 0.53 | 8.88E–16 | 229,584 | PRDM6 | SBP | [73] |
| 5 | rs9687065 | 149011577 | A | 0.78 | 0.26 | 7.36E–11 | 259,216 | ABLIM3, SH3TC2 | DBP | [73] |
| 6 | rs2021783 | 32077074 | C | 0.79 | 0.49 | 2.18E–12 | 78,911 | CYP21A2 | DBP | [13] |
| 6 | rs1563788 | 43340625 | T | 0.31 | 0.51 | 2.22E–16 | 220,757 | ZNF318 | SBP | [73] |
| 6 | rs1474698 | 56199399 | T | 0.62 | −0.20 | 4.52E–09 | 274,981 | NR | PP | [73] |
| 7 | rs2107595 | 19009765 | A | 0.23 | 0.31 | 3.91E–11 | 209,305 | HDAC9 | PP | [73] |
| 7 | rs10260816 | 45970501 | C | 0.61 | 0.31 | 1.51E–14 | 207,070 | IGFBP3 | PP | [73] |
| 7 | rs17477177 | 106771412 | T | 0.77 | −0.53 | 3.69E–12 | 99,344 | PIK3CG | PP | [73] |
| 10 | rs4919669 | 102712218 | A | 0.43 | −0.65 | 2.63E–08 | NA | ARL3 | SBP | [74] |
| 10 | rs284844 | 102794772 | A | 0.49 | −0.75 | 1.05E–11 | NA | WBP1L | SBP | [74] |
| 10 | rs4409766 | 102856906 | T | 0.71 | 1.24 | 6.08E–17 | 46,030 | CYP17A1 | SBP | [13] |
| 10 | rs11191548 | 103086421 | T | 0.74 | 1.18 | 3.94E–17 | 41,315 | CNNM2 | SBP | [12] |
| 10 | rs11191580 | 103146454 | T | 0.74 | 0.97 | 4.44E–15 | NA | NT5C2 | SBP | [74] |
| 11 | rs4757391 | 16281393 | C | 0.28 | 0.88 | 5.20E–9 | 46,336 | SOX6 | SBP | [13] |
| 11 | rs751984 | 61510774 | T | 0.79 | 0.33 | 7.66E–12 | 233,082 | LRRC10B, SYT7 | MAP | [73] |
| 12 | rs12579720 | 20020830 | C | 0.32 | −0.32 | 2.2E–16 | 218,606 | PDE3A | DBP | [73] |
| 12 | rs17249754 | 89666809 | G | 0.64 | 1.17 | 7.72E–20 | 40,719 | ATP2B1 | SBP | [12] |
| 12 | rs3184504 | 111446804 | T | 0.46 | 0.54 | 2.20E–10 | 35,342 | SH2B3 | DBP | [73] |
| 12 | rs653178 | 111569952 | T | 0.50 | −0.55 | 1.20E–08 | 35,342 | ATXN2 | DBP | [73] |
| 12 | rs11066280 | 112379979 | T | 0.75 | 1.01 | 1.32E–35 | 46,957 | HECTD4 | DBP | [12] |
| 12 | rs35444 | 115114632 | A | 0.75 | 0.52 | 9.62E–08 | 29,746 | TBX3 | DBP | [12] |
| 12 | rs11067763 | 115760536 | A | 0.62 | 0.51 | 2E–18 | 79,651 | MED13L | DBP | [13] |
| 17 | rs2240736 | 61408032 | T | 0.66 | 0.35 | 2.20E–16 | 217,197 | C17orf82, TBX2 | MAP | [73] |
| 18 | rs403814 | 6282594 | A | NA | 1.15 | 6.13E–09 | NA | L3MBTL4 | HT | [75] |
| 19 | rs740406 | 2232222 | A | 0.87 | −0.55 | 3.10E–15 | 193,219 | PLEKHJ1, DOT1L | PP | [73] |
| 20 | rs1887320 | 10985350 | A | 0.53 | 0.78 | 1.48E–08 | 46,123 | JAG1 | SBP | [13] |
|
Africans 3 |
rs1717027 | 41946428 | T | 0.64 | 0.49 | 4.90E–13 | 29,322 | ULK4 | HT | [76] |
| 6 | rs13209747 | 126794309 | T | 0.19 | 0.56 | 2.40E–11 | 28,708 | RSPO3 | DBP | [76] |
| 6 | rs76987554 | 133759717 | C | 0.09 | −1.69 | 2.28E–10 | 62,297 | TARID, TCF21 | SBP | [15] |
| 6 | rs17080102 | 150683634 | C | 0.1 | −0.74 | 1.90E–11 | 29,323 | PLEKHG1 | DBP | [76] |
| 7 | rs6969780 | 27119517 | C | 0.3 | 0.46 | 3.27E–10 | 85,397 | HOXA | DBP | [15] |
| 7 | rs17428471 | 27298248 | T | 0.14 | 1.20 | 2.00E–12 | 29,326 | HOXA, EVX1 | SBP | [76] |
| 7 | rs11563582 | 27312031 | A | 0.13 | 0.67 | 1.71E–11 | 84,816 | EVX1, HOXA | DBP | [15] |
| 7 | rs7801190 | 100860471 | C | 0.72 | 1.31 | 3.40E–08 | 10,771 | SLC12A9 | HT | [77] |
| 8 | rs7006531 | 94098516 | G | 0.85 | −1.02 | 5.96E–12 | 72,887 | CDH17 | PP | [15] |
| 8 | rs78192203 | 141364973 | T | 0.2 | 0.84 | 4.00E–11 | 62,681 | GPR20 | DBP | [15] |
| 9 | rs115795127 | 83378986 | T | 0.89 | 0.17 | 1.13E–08 | 32,149 | FRMD3 | HT | [15] |
Chr, chromosome; EA, effect allele; EAF, effect allele frequency; SBP, systolic blood pressure; DBP, diastolic blood pressure; PP, pulse pressure; MAP, mean arterial pressure; HT, hypertension; OH, orthostatic hypotension; NA, not available.
To enhance the statistical power, international collaborations were established between studies and organized in consortia. Furthermore, detecting associations with BP as a continuous variable rather than in case-control studies also was successful. In 2009, two large-scale meta-analyses of GWAS from the Global Blood Pressure Genetics (Global BPgen) [7] and Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) [8] consortia identified associations withstanding correction for multiple testing (genome-wide significance). Each of the study contained nearly 30,000 subjects at the discovery phase and found 8 genomic loci, among which 3 loci overlapped. In 2011, the International Consortium for Blood Pressure (ICBP) [9] used a multistage design with 200,000 individuals of European descent, replicated the previous 13 loci effectively and discovered 16 new loci significant at the genome-wide level. Then, the ICBP analyzed two derived BP phenotypes: mean arterial pressure and pulse pressure (PP) [10]. This study discovered 4 novel loci for PP and 2 novel loci for mean arterial pressure. The latest GWAS among UK Biobank participants of European ancestry included ∼140,000 subjects from a single source at the discovery phase. With dense 1000 Genomes Project and UK10K imputation, they yielded a data set with ∼9.8 million variants for the meta-analysis. With the help of major international consortia for parallel replication, they included GWAS data from 330,956 individuals in total and reported 107 significant loci, among which 24 were associated with systolic BP (SBP), 41 with diastolic BP (DBP), and 42 with PP [11].
The first large meta-analysis of GWAS on BP traits among East Asians was conducted by the Asian Genetic Epidemiology Network (AGEN) consortium [12]. Our group was one of the 8 groups participating in stage 1 meta-analysis, which included 19,608 individuals totally. After de novo genotyping in two stages of replication involving 10,518 and 20,247 East Asian samples, we confirmed 7 loci previously identified in the European population reported by Global BPgen and CHARGE, and additionally identified 6 novel loci: ST7L-CAPZA1, FIGN-GRB14, ENPEP, NPR3, TBX3, and ALDH2. In another meta-analysis of a Han Chinese population, which involved 11,816 subjects in the discovery stage and 69,146 subjects for replication [13], the authors replicated 8 previously reported loci and 4 novel loci: CASZ1, MOV10, FGF5, CYP17A1, SOX6, ATP2B1, ALDH2, JAG1, CACNA1D, CYP21A2, MED13L, and SLC4A7. These findings suggest a possible presence of allelic heterogeneity in BP regulation between Europeans and Asians and provide new mechanistic insight into hypertension.
African-descent populations are the most ancestral and have smaller regions of linkage disequilibrium due to the accumulation of more recombination events in that group, so that GWAS performed on Africans need more SNPs with better overall genomic coverage [14]. There are fewer loci reaching GWAS significance in African populations than in Europeans or Asians. The largest GWAS performed in an African-origin population analyzed 21 GWAS comprising 31,968 individuals of African ancestry and validated their results with an additional 54,395 individuals from multiethnic studies [15]. The authors found 9 loci with 11 independent variants for either SBP, DBP, hypertension, or combined traits.
Coronary Artery Disease
Heritability of CAD has been estimated between 40 and 60% based on family and twin studies [16]. The first robust locus associated with CAD identified by GWAS was reported in 2007 [6, 17, 18], when three independent groups reported common variants at the 9p21 locus. It was associated with a ∼30% increased risk of CAD per copy of the risk allele. Since then, progressively larger-sample-size meta-analyses of additional GWAS, mainly on subjects of European descent, have identified additional loci of smaller effect size. In 2015, the CARDIoGRAMplusC4D Consortium published a GWAS meta-analysis of 185,000 CAD cases and controls, interrogating 6.7 million common variants as well as 2.7 million low-frequency variants [19]. In this study, the authors confirmed most CAD loci known at that time and identified 10 novel loci. The majority of the significant loci had a frequency of more than 5%, which indicated that the genetics of CAD was largely determined by the cumulative effect of multiple common risks of small effect size and strongly supported the common disease/common variant hypothesis. In 2017, Nelson et al. [20] further meta-analyzed the results from 10,801 cases in the UK Biobank against 137,914 controls in combination with the CARDIoGRAMplusC4D 1000 Genomes and the MIGen/CARDIoGRAM Exome chip studies. They found 12 new signals reaching genome-wide significance. They also reported that 304 independent variants meeting the 5% false discovery rate threshold explained 21.2% of CAD heritability. This finding highlighted the importance of the false discovery rate approach in the expansion of associated variants. The strongest SNPs associated with CAD which had reached genome-wide significance are listed in Table 3a and b.
Table 3.
Significant loci for coronary artery disease reported by genome-wide association studies in Europeans (a) and Asians (b)
| Chr | Strongest SNP | Position | EA | EAF | OR or BETA | p value | n | Closest gene | Ref. |
|---|---|---|---|---|---|---|---|---|---|
|
a 1 |
Europeans rs11206510 |
55030366 | T | 0.85 | 1.08 | 2.34E–08 | 188,372 | PCSK9 | [19] |
| 1 | rs17114036 | 56497149 | A | 0.91 | 1.17 | 3.81E–19 | 133,226 | PPAP2B | [47] |
| 1 | rs629301 | 109275684 | NA | NA | NA | 1.00E–49 | NA | CELSR2 | [78] |
| 1 | rs11810571 | 151789832 | G | 0.79 | 1.06 | 4.34E–08 | 345,035 | TDRKH | [20] |
| 1 | rs6689306 | 154423470 | A | 0.44 | 1.05 | 1.50E–09 | NA | IL6R | [20] |
| 2 | rs16986953 | 19742712 | A | 0.07 | 1.11 | 4.80E–10 | NA | AK097927 | [20] |
| 2 | rs7568458 | 85561052 | A | 0.45 | 1.06 | 2.40E–13 | NA | VAMP5, VAMP8, GGCX | [20] |
| 2 | rs17678683 | 144528992 | G | 0.09 | 1.10 | 3.00E–09 | 179,585 | ZEB2, ACO74093.1 | [19] |
| 2 | rs1250229 | 215439661 | T | 0.26 | 1.07 | 2.88E–13 | 270,189 | FN1 | [20] |
| 3 | rs7623687 | 49411133 | A | 0.86 | 1.08 | 3.54E–10 | 333,836 | RHOA | [20] |
| 3 | rs142695226 | 124756354 | G | 0.14 | 1.07 | 1.58E–09 | 345,188 | UMPS, ITGB5 | [20] |
| 3 | rs9818870 | 138403280 | T | 0.15 | 1.15 | 7.44E–13 | 40,773 | MRAS | [44] |
| 3 | rs12493885 | 154122077 | C | 0.87 | 1.07 | 3.23E–08 | 251,805 | ARHGEF26 | [20] |
| 4 | rs17087335 | 56972417 | T | 0.21 | 1.06 | 4.60E–08 | NA | NOA1, REST | [19] |
| 4 | rs10857147 | 80259918 | T | 0.27 | 1.05 | 5.81E–09 | 345,133 | PRDM8, FGF5 | [20] |
| 4 | rs7678555 | 119988346 | C | 0.28 | 1.05 | 1.35E–08 | 345,126 | PDE5A, MAD2L1 | [20] |
| 4 | rs6841581 | 147480038 | A | 0.15 | 1.07 | 4.60E–10 | NA | EDNAA | [20] |
| 6 | rs6909752 | 22612400 | A | 0.33 | 1.05 | 2.26E–09 | 343,171 | HDGFL1 | [20] |
| 6 | rs17609940 | 35067023 | G | 0.75 | 1.07 | 1.36E–08 | 137,412 | ANKS1A | [47] |
| 6 | rs56336142 | 39166323 | T | 0.81 | 1.07 | 1.85E–08 | 188,349 | KCNK5 | [19] |
| 6 | rs12190287 | 133893387 | C | 0.62 | 1.08 | 1.70E–12 | 130,888 | TCF21 | [47] |
| 6 | rs10455872 | 160589086 | G | 0.07 | 1.31 | 1.70E–49 | NA | LPA | [20] |
| 6 | rs4252185 | 160702419 | C | 0.06 | 1.34 | 1.64E–32 | NA | PLG | [19] |
| 7 | rs2107595 | 19009765 | A | 0.18 | 1.08 | 3.40E–13 | NA | HDAC9 | [20] |
| 7 | rs11556924 | 130023656 | C | 0.62 | 1.09 | 9.18E–18 | 134,200 | ZC3HC1 | [47] |
| 7 | rs3918226 | 150993088 | T | 0.07 | 1.13 | 1.60E–12 | NA | NOS3 | [20] |
| 8 | rs2954029 | 125478730 | A | 0.54 | 1.06 | 5.20E–13 | NA | TRIB1 | [20] |
| 9 | rs579459 | 133278724 | C | 0.21 | 1.10 | 4.08E–14 | 123,978 | ABO | [47] |
| 10 | rs2505083 | 30046193 | C | 0.39 | 1.06 | 6.85E–09 | 169,421 | KIAA1462 | [19] |
| 10 | rs501120 | 44258419 | T | 0.87 | 1.33 | 9.46E–08 | 7,181 | CXCL12 | [17] |
| 10 | rs2246942 | 89245129 | G | 0.35 | 1.08 | 3.50E–16 | NA | LIPA | [20] |
| 10 | rs12413409 | 102959339 | G | 0.89 | 1.12 | 1.03E–09 | 129,741 | NT5C2, CNNM2, CYP17A1 | [47] |
| 11 | rs10840293 | 9729649 | A | 0.55 | 1.05 | NA | SWAP70 | [20] | |
| 11 | rs3993105 | 13281524 | T | 0.68 | 1.05 | 4.88E–08 | 340,699 | ARNTL | [20] |
| 11 | rs974819 | 103789839 | T | 0.32 | 1.07 | 2.41E–09 | 64,881 | PDGFD | [46] |
| 11 | rs964184 | 116778201 | NA | NA | NA | 2.00E–108 | NA | ZPR1 | [78] |
| 12 | rs3184504 | 111446804 | T | 0.42 | 1.07 | 1.03E–09 | NA | SH2B3 | [19] |
| 12 | rs11830157 | 117827636 | G | 0.36 | 1.12 | 2.12E–09 | NA | KSR2 | [19] |
| 12 | rs2244608 | 120979185 | G | 0.35 | 1.05 | 7.90E–10 | 345,106 | HFN1A | [20] |
| 13 | rs1924981 | 28448508 | T | 0.33 | 1.05 | 1.90E–07 | NA | FLT1 | [20] |
| 14 | rs2895811 | 99667605 | C | 0.43 | 1.07 | 1.14E–09 | 114,238 | HHIPL1 | [47] |
| 15 | rs56062135 | 67163292 | C | 0.79 | 1.07 | 4.52E–09 | 184,037 | SMAD3 | [19] |
| 15 | rs7164479 | 78830712 | T | 0.58 | 1.07 | 6.40E–18 | NA | ADAMTS7 | [20] |
| 15 | rs8042271 | 89030987 | G | 0.90 | 1.10 | 3.68E–08 | 162,206 | MFGE8, ABHD2 | [19] |
| 15 | rs17514846 | 90873320 | A | 0.44 | 1.05 | 3.10E–07 | NA | FURIN, FES | [19] |
| 16 | rs7500448 | 83012185 | A | 0.77 | 1.06 | 4.90E–10 | 337,399 | CDH13 | [20] |
| 17 | rs216172 | 2223210 | C | 0.37 | 1.07 | 1.15E–09 | 111,538 | SMG6, SRR | [47] |
| 17 | rs46522 | 48911235 | T | 0.53 | 1.06 | 1.81E–08 | 137,633 | UBE2Z | [47] |
| 17 | rs7212798 | 60936127 | C | 0.15 | 1.08 | 1.88E–08 | 188,377 | BCAS3 | [19] |
| 18 | rs8082812 | 8522684 | NA | NA | NA | 5.00E–67 | NA | THEM1S3P - AKR1B1P6 | [78] |
| 18 | rs663129 | 60171168 | A | 0.26 | 1.06 | 3.20E–08 | NA | PMA1P1, MC4R | [19] |
| 19 | rs6511720 | 11091630 | G | 0.88 | 1.14 | 7.90E–22 | NA | LDLR | [20] |
| 19 | rs12976411 | 32391114 | A | 0.09 | 1.49 | 1.18E–14 | NA | ZNF507, LOC400684 | [19] |
| 19 | rs8108632 | 41348629 | T | 0.48 | 1.05 | 4.13E–08 | 345,058 | TGFB1 | [20] |
| 19 | rs4420638 | 44919689 | NA | NA | NA | 1.00E–22 | NA | APOC1 – APOC1P1 | [78] |
| 19 | rs1964272 | 45687010 | G | 0.52 | 1.05 | 2.52E–08 | 343,548 | SNAPD2 | [20] |
| 21 | rs28451064 | 34221526 | A | 0.12 | 1.14 | 2.60E–23 | NA | KCNE2 | [20] |
| 21 | rs460976 | 41463567 | NA | NA | NA | 1.00E–08 | NA | MX1 - TMPRSS2 | [78] |
| 22 | rs180803 | 24262890 | G | 0.97 | 1.20 | 1.64E–10 | 174,076 | POM121L9P, ADORA2A | [19] |
| bAsians | |||||||||
| 2 | rs2123536 | 19745816 | T | 0.39 | 1.12 | 6.83E–11 | 33,466 | TTC32, WDR35 | [48] |
| 4 | rs1842896 | 155590307 | T | 0.76 | 1.14 | 1.26E–11 | 33,466 | GUCY1A3 | [48] |
| 6 | rs9349379 | 12903725 | G | 0.349 | 1.34 | 8.02E–10 | 4,496 | PHACTR1 | [79] |
| 6 | rs9268402 | 32373576 | G | 0.59 | 1.16 | 2.77E–15 | 33,466 | BTNL2, C6orf10 | [48] |
| 6 | rs12524865 | 133875536 | C | 0.61 | 1.11 | 1.87E–07 | 24,724 | TCF21 | [48] |
| 9 | rs10738607 | 22088095 | G | 0.64 | 0.78 | 2.37E–08 | 1,237 | CDKN2A, CDKN2B | [80] |
| 12 | rs7136259 | 89687411 | T | 0.39 | 1.11 | 5.68E–10 | 33,466 | ATP2B1 | [48] |
| 12 | rs3782889 | 110912851 | C | 0.21 | 1.26 | 3.95E–14 | 13,742 | MYL2 | [81] |
| 12 | rs671 | 111803962 | A | 0.23 | 1.43 | 1.6E–34 | 12,041 | ALDH2 | [82] |
| 12 | rs11066280 | 112379979 | A | 0.17 | 1.19 | 1.69E–11 | 24,741 | C12orf51 | [48] |
Chr, chromosome; EA, effect allele; EAF, effect allele frequency; NA, not available.
Although the majority of GWAS for CAD has been carried out on European populations, there were still important studies performed on Asian and Black populations with smaller sample sizes [19]. Unlike the similar effect sizes of CAD risk alleles in East Asian and European populations, the effect size was apparently attenuated in South Asian and Black populations. The lower level of linkage disequilibrium in the African genome might lead to failure to tag potentially shared causal variants [21].
Clinical Implications
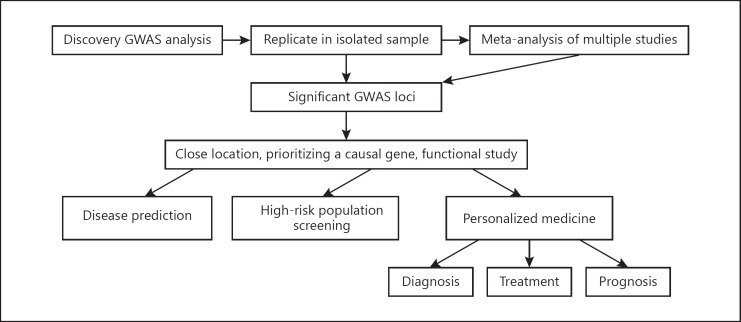
It is undeniable that GWAS have achieved considerable success in exploring the genetic architecture of heart diseases. However, it is important to point out that the significant variant is often merely tagged. The gene indicated by the GWAS signal would not be considered as the causal variant until the functional mechanism can be found. The ultimate goals of the study of the genetics of CAD or hypertension are to understand the pathophysiological mechanism and subsequently to establish risk prediction methods and develop effective therapeutic strategies (Fig. 1).
Fig. 1.
Clinical implications from genome-wide association studies (GWAS).
The effect of the susceptibility variants identified by GWAS is small individually, but their effects are independent and additive, which can be calculated as a genetic risk score (GRS) in an unweighted manner by adding risk allele numbers or with a weighted mean [22]. As DNA is stable over the course of the lifetime, a genetic risk can be ascertained from birth; therefore, GRS may be particularly useful in risk prediction among younger patients in whom the cumulative impact of lifestyle factors is less pronounced.
The interpretation of GWAS often is complicated by the nature of significant loci, which are mostly located in the intergenic region. Bioinformatic approaches could narrow down the list, prioritizing a gene for subsequent functional study according to expression quantitative trait locus [23], genome-wide chromatin profiles of histone modifications, data on transcription factor-binding sites, chromatin immunoprecipitation sequencing [24], etc. Below, we describe some examples of clinical implications of GWAS results for risk prediction and mechanism interpretation.
Hypertension
With the expansion of significant BP loci by GWAS, robust associations with some biological candidate genes previously suspected to influence BP were detected. The latest GWAS by Warren et al. [11] identified multiple loci involved in BP regulation, including angiotensin-converting enzyme, voltage-dependent calcium channel auxiliary subunit, metalloendopeptidase/neutral endopeptidase, adrenergic β 2B receptor, and phosphodiesterase 5a. Moreover, in the pathway analysis, they also showed an enrichment of pathways associated with cardiovascular disease, including the α-adrenergic pathway, CXCR4 chemokine signaling pathway, endothelin system, and angiotensin-receptor pathways.
In the ICBP GWAS [10], the difference in SBP and DBP between the top and the bottom quintile of the GRS was 4.6 and 3.0 mm Hg, respectively. Furthermore, the GRS generated by combination of 107 loci by Warren et al. [11] showed that the group with GRS in the lowest quintile had an SBP approximately 9–10 mm Hg lower than those with GRS in the highest quintile. Reductions in BP of such a magnitude might lead to significantly lower cardiovascular morbidity and mortality among hypertensive patients. The GRS could therefore be useful in early life to assess the risk of hypertension and direct dietary or lifestyle modifications.
The GWAS findings had also provided clues for personalized prevention and treatment of hypertension. One good example is the uromodulin gene (UMOD), which was identified in an extreme case-control design [25]: rs13333226-G in UMOD showed an association with lower risk of hypertension and reduced urinary UMOD excretion. Uromodulin is mainly expressed in the thick portion of the ascending limb of the loop of Henle, which indicates that it may participate in BP regulation through an effect on sodium hemostasis. Trudu et al. [26] modeled the effect of UMOD in transgenic mice and demonstrated that uromodulin influenced BP through activation of the renal sodium cotransporter NKCC2. The subjects with the UMOD risk variant had increased UMOD excretion, greater salt sensitivity, hypertension, and a greater BP response to loop diuretics. This finding presents an opportunity for hypertension precision medicine and new drug development.
The AGEN study [12] found an ethnicity-specific locus on 12q24.13, where the acetaldehyde dehydrogenase (ALDH2) gene is located. ALDH2 is a key enzyme in the major pathway of alcohol metabolism, and glutamate-lysine substitution (rs671, E504K) produces an inactive subunit of ALDH2, resulting in an inability to metabolize acetaldehyde and subsequent accumulation of acetaldehyde after alcohol intake [27]. The SNP rs671 is not polymorphic in Europeans, but it is close to rs3184504 at the SH2B3 locus, which is significantly associated with BP in Europeans and has no polymorphism in East Asians. This phenomenon indicates the natural selection that has occurred. Furthermore, rs671 displays pleiotropic effects both on risk factors for cardiovascular disease and on CAD susceptibility. Interestingly, the locus exhibits a deleterious effect on BP but has protective effects on HDL cholesterol and LDL cholesterol, which results in a net reduction in CAD risk. Moreover, most of the associations between rs671 and each of the cardiovascular risk factors are influenced by alcohol intake.
Coronary Artery Disease
Since 2007, nearly 70 distinct genetic loci for CAD have been found due to the progressively larger sample sizes. A minority of all these risk variants appears to modulate CAD risk by influencing classic risk factors such as plasma lipids, diabetes, and hypertension, re inforcing the key role for these pathways in the development of CAD. The rest of the risk vari ants identified by GWAS are located in regulatory regions of unclear function.
In a large prospective cohort study with a median of 10.7 years of follow-up, Ripatti et al. [28] found that individuals with a GRS in the top quintile derived from 13 multilocus SNPs of CAD exhibited a 1.66-fold increased risk adjusting for traditional risk factors. However, the GRS did not improve the C-index over the traditional risk factors and family history or the net reclassification of risk categories. A recent survey on 55,685 subjects from three prospective studies and one cross-sectional study confirmed the association between GRS and incidence of CAD [29], with subjects in the top GRS quintiles having a 91% higher relative risk than those in the bottom quintiles. They also reported that a favorable lifestyle was associated with a relative risk of CAD nearly 50% lower than with an unfavorable lifestyle.
The 9p21.3 locus was the first one identified by GWAS, consisting of a cluster of 59 linked SNPs in a 53,000-bp region [30]. Among the CAD risk alleles, rs10811656 and rs10757278 are located in an enhancer element and disrupt a binding site for ATAT1 [31]. This enhancer locus interacts with CDKN2A/B and IFNA21, which encodes interferon-γ in human vascular endothelial cells and participates in the inflammatory process. It is also suggested that this region probably encodes for a long noncoding RNA designated as ANRIL (antisense noncoding RNA in the INK4 locus). ANRIL influences the expression of CDKN2A/B, which is involved in regulating the cell cycle and cellular proliferation [32]. A meta-analysis also confirmed that the 9p21.3 locus increases the burden of atherosclerosis but not the risk of myocardial infarction, which indicated the stimulatory effect of this locus in atherosclerosis [33].
The 1p13 locus has been independently associated with CAD in several GWAS. Among a number of genes located in this region, SORT1 was found as the target one by expression quantitative trait locus and expression studies. SORT1 encodes sortilin, which is a multiligand type 1 receptor and is expressed in many cell and tissue types [34]. Overexpression of Sort1 might decrease the very-low-density lipoprotein secretion rate and increase plasma low-density lipoprotein turnover [35]. However, the knockout or knockdown of Sort1 in different studies resulted in either increased or decreased very-low-density lipoprotein secretion [36, 37]. Whether Sort1 was the gene causing CAD or just a signal found in the GWAS still needs to be clarified. The concrete molecular mechanism by which sortilin influences lipid metabolism and CAD risk has not yet been elucidated.
Perspectives
The GWAS findings have identified novel biological signals involved in human heart diseases. However, there are still certain challenges. The heritability of BP as derived from family studies varies from 30 to 50%, but the collective effect of all BP loci identified through GWAS only explains ∼3.5% of BP heritability. The missing heritability is not unique to BP genetics but is universally observed in almost all common heart diseases. Heritability reflects the degree of phenotypic resemblance between relatives, which not only depends on the genetic architecture contributing to the trait but also is the result of environmental factors and interactions within the genome. In GWAS on hypertension-related phenotypes, significant gene-environment interaction has been identified for alcohol consumption [38], body mass index, smoking [39], educational level, and other modifiable lifestyle factors. These findings may help identify the causal genetic loci that contribute to missing heritability. In addition, gene-gene interaction [40], rare variants [41], and epigenetic mechanisms likely explain a certain fraction of complex heart diseases. Future research will focus on the application of variants identified in GWAS for the identification of individuals at risk of disease, guidance of clinical management decisions, and prediction of prognosis.
Disclosure Statement
No potential conflict of interest was reported by the authors.
References
- 1.Watkins H, Farrall M. Genetic susceptibility to coronary artery disease: from promise to progress. Nat Rev Genet. 2006 Mar;7((3)):163–73. doi: 10.1038/nrg1805. [DOI] [PubMed] [Google Scholar]
- 2.Pearson TA, Manolio TA. How to interpret a genome-wide association study. JAMA. 2008 Mar;299((11)):1335–44. doi: 10.1001/jama.299.11.1335. [DOI] [PubMed] [Google Scholar]
- 3.Pe'er I, Yelensky R, Altshuler D, Daly MJ. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol. 2008 May;32((4)):381–5. doi: 10.1002/gepi.20303. [DOI] [PubMed] [Google Scholar]
- 4.Miall WE, Oldham PD. The hereditary factor in arterial blood-pressure. BMJ. 1963 Jan;1((5323)):75–80. doi: 10.1136/bmj.1.5323.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levy D, Larson MG, Benjamin EJ, Newton-Cheh C, Wang TJ, Hwang SJ, et al. Framingham Heart Study 100K Project: genome-wide associations for blood pressure and arterial stiffness. BMC Med Genet. 2007 Sep;8(Suppl 1):S3. doi: 10.1186/1471-2350-8-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wellcome Trust Case Control C, Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007 Jun;447((7145)):661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Newton-Cheh C, Johnson T, Gateva V, Tobin MD, Bochud M, Coin L, et al. Wellcome Trust Case Control Consortium Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet. 2009 Jun;41((6)):666–76. doi: 10.1038/ng.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levy D, Ehret GB, Rice K, Verwoert GC, Launer LJ, Dehghan A, et al. Genome-wide association study of blood pressure and hypertension. Nat Genet. 2009 Jun;41((6)):677–87. doi: 10.1038/ng.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, Chasman DI, et al. CHARGE-HF consortium Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011 Sep;478((7367)):103–9. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wain LV, Verwoert GC, O'Reilly PF, Shi G, Johnson T, Johnson AD, et al. LifeLines Cohort Study. EchoGen consortium. AortaGen Consortium. CHARGE Consortium Heart Failure Working Group. KidneyGen consortium. CKDGen consortium. Cardiogenics consortium. CardioGram Genome-wide association study identifies six new loci influencing pulse pressure and mean arterial pressure. Nat Genet. 2011 Sep;43((10)):1005–11. doi: 10.1038/ng.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warren HR, Evangelou E, Cabrera CP, Gao H, Ren M, Mifsud B, et al. International Consortium of Blood Pressure (ICBP) 1000G Analyses. BIOS Consortium. Lifelines Cohort Study. Understanding Society Scientific group. CHD Exome+ Consortium. ExomeBP Consortium. T2D-GENES Consortium. GoT2DGenes Consortium. Cohorts for Heart and Ageing Research in Genome Epidemiology (CHARGE) BP Exome Consortium. International Genomics of Blood Pressure (iGEN-BP) Consortium. UK Biobank CardioMetabolic Consortium BP working group Genome-wide association analysis identifies novel blood pressure loci and offers biological insights into cardiovascular risk. Nat Genet. 2017 Mar;49((3)):403–15. [Google Scholar]
- 12.Kato N, Takeuchi F, Tabara Y, Kelly TN, Go MJ, Sim X, et al. Meta-analysis of genome-wide association studies identifies common variants associated with blood pressure variation in east Asians. Nat Genet. 2011 Jun;43((6)):531–8. doi: 10.1038/ng.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu X, Wang L, Lin X, Huang J, Charles Gu C, He M, et al. Genome-wide association study in Chinese identifies novel loci for blood pressure and hypertension. Hum Mol Genet. 2015 Feb;24((3)):865–74. doi: 10.1093/hmg/ddu478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bush WS, Moore JH. Chapter 11: genome-wide association studies. PLOS Comput Biol. 2012;8((12)):e1002822. doi: 10.1371/journal.pcbi.1002822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang J, Le TH, Edwards DR, Tayo BO, Gaulton KJ, Smith JA, et al. Single-trait and multi-trait genome-wide association analyses identify novel loci for blood pressure in African-ancestry populations. PLoS Genet. 2017 May;13((5)):e1006728. doi: 10.1371/journal.pgen.1006728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010 Aug;466((7307)):707–13. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, et al. WTCCC and the Cardiogenics Consortium Genomewide association analysis of coronary artery disease. N Engl J Med. 2007 Aug;357((5)):443–53. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McPherson R, Pertsemlidis A, Kavaslar N, Stewart A, Roberts R, Cox DR, et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007 Jun;316((5830)):1488–91. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nikpay M, Goel A, Won HH, Hall LM, Willenborg C, Kanoni S, et al. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet. 2015 Oct;47((10)):1121–30. doi: 10.1038/ng.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nelson CP, Goel A, Butterworth AS, Kanoni S, Webb TR, Marouli E, et al. EPIC-CVD Consortium. CARDIoGRAMplusC4D. UK Biobank CardioMetabolic Consortium CHD working group Association analyses based on false discovery rate implicate new loci for coronary artery disease. Nat Genet. 2017 Sep;49((9)):1385–91. doi: 10.1038/ng.3913. [DOI] [PubMed] [Google Scholar]
- 21.Marigorta UM, Navarro A. High trans-ethnic replicability of GWAS results implies common causal variants. PLoS Genet. 2013 Jun;9((6)):e1003566. doi: 10.1371/journal.pgen.1003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McPherson R, Tybjaerg-Hansen A. Genetics of Coronary Artery Disease. Circ Res. 2016 Feb;118((4)):564–78. doi: 10.1161/CIRCRESAHA.115.306566. [DOI] [PubMed] [Google Scholar]
- 23.Consortium EP, ENCODE Project Consortium A user's guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011 Apr;9((4)):e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park PJ. ChIP-seq: advantages and challenges of a maturing technology. Nat Rev Genet. 2009 Oct;10((10)):669–80. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Padmanabhan S, Melander O, Johnson T, Di Blasio AM, Lee WK, Gentilini D, et al. Global BPgen Consortium Genome-wide association study of blood pressure extremes identifies variant near UMOD associated with hypertension. PLoS Genet. 2010 Oct;6((10)):e1001177. doi: 10.1371/journal.pgen.1001177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trudu M, Janas S, Lanzani C, Debaix H, Schaeffer C, Ikehata M, et al. SKIPOGH team Common noncoding UMOD gene variants induce salt-sensitive hypertension and kidney damage by increasing uromodulin expression. Nat Med. 2013 Dec;19((12)):1655–60. doi: 10.1038/nm.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takeshita T, Morimoto K, Mao XQ, Hashimoto T, Furuyama J. Phenotypic differences in low Km aldehyde dehydrogenase in Japanese workers. Lancet. 1993 Mar;341((8848)):837–8. [PubMed] [Google Scholar]
- 28.Ripatti S, Tikkanen E, Orho-Melander M, Havulinna AS, Silander K, Sharma A, et al. A multilocus genetic risk score for coronary heart disease: case-control and prospective cohort analyses. Lancet. 2010 Oct;376((9750)):1393–400. doi: 10.1016/S0140-6736(10)61267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khera AV, Emdin CA, Drake I, Natarajan P, Bick AG, Cook NR, et al. Genetic Risk, Adherence to a Healthy Lifestyle, and Coronary Disease. N Engl J Med. 2016 Dec;375((24)):2349–58. doi: 10.1056/NEJMoa1605086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen HH, Almontashiri NA, Antoine D, Stewart AF. Functional genomics of the 9p21.3 locus for atherosclerosis: clarity or confusion? Curr Cardiol Rep. 2014 Jul;16((7)):502. doi: 10.1007/s11886-014-0502-7. [DOI] [PubMed] [Google Scholar]
- 31.Harismendy O, Notani D, Song X, Rahim NG, Tanasa B, Heintzman N, et al. 9p21 DNA variants associated with coronary artery disease impair interferon-γ signalling response. Nature. 2011 Feb;470((7333)):264–8. doi: 10.1038/nature09753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holdt LM, Hoffmann S, Sass K, Langenberger D, Scholz M, Krohn K, et al. Alu elements in ANRIL non-coding RNA at chromosome 9p21 modulate atherogenic cell functions through trans-regulation of gene networks. PLoS Genet. 2013;9((7)):e1003588. doi: 10.1371/journal.pgen.1003588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chan K, Patel RS, Newcombe P, Nelson CP, Qasim A, Epstein SE, et al. Association between the chromosome 9p21 locus and angiographic coronary artery disease burden: a collaborative meta-analysis. J Am Coll Cardiol. 2013 Mar;61((9)):957–70. doi: 10.1016/j.jacc.2012.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petersen CM, Nielsen MS, Nykjaer A, Jacobsen L, Tommerup N, Rasmussen HH, et al. Molecular identification of a novel candidate sorting receptor purified from human brain by receptor-associated protein affinity chromatography. J Biol Chem. 1997 Feb;272((6)):3599–605. doi: 10.1074/jbc.272.6.3599. [DOI] [PubMed] [Google Scholar]
- 35.Kathiresan S, Melander O, Guiducci C, Surti A, Burtt NP, Rieder MJ, et al. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet. 2008 Feb;40((2)):189–97. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kjolby M, Andersen OM, Breiderhoff T, Fjorback AW, Pedersen KM, Madsen P, et al. Sort1, encoded by the cardiovascular risk locus 1p13.3, is a regulator of hepatic lipoprotein export. Cell Metab. 2010 Sep;12((3)):213–23. doi: 10.1016/j.cmet.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 37.Musunuru K, Strong A, Frank-Kamenetsky M, Lee NE, Ahfeldt T, Sachs KV, et al. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature. 2010 Aug;466((7307)):714–9. doi: 10.1038/nature09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simino J, Sung YJ, Kume R, Schwander K, Rao DC. Gene-alcohol interactions identify several novel blood pressure loci including a promising locus near SLC16A9. Front Genet. 2013 Dec;4:277. doi: 10.3389/fgene.2013.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sung YJ, de Las Fuentes L, Schwander KL, Simino J, Rao DC. Gene-smoking interactions identify several novel blood pressure loci in the Framingham Heart Study. Am J Hypertens. 2015 Mar;28((3)):343–54. doi: 10.1093/ajh/hpu149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Slavin TP, Feng T, Schnell A, Zhu X, Elston RC. Two-marker association tests yield new disease associations for coronary artery disease and hypertension. Hum Genet. 2011 Dec;130((6)):725–33. doi: 10.1007/s00439-011-1009-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu X, Peloso GM, Liu DJ, Wu Y, Zhang H, Zhou W, et al. GLGC Consortium Exome chip meta-analysis identifies novel loci and East Asian-specific coding variants that contribute to lipid levels and coronary artery disease. Nat Genet. 2017 Dec;49((12)):1722–30. doi: 10.1038/ng.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Helgadottir A, Thorleifsson G, Manolescu A, Gretarsdottir S, Blondal T, Jonasdottir A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007 Jun;316((5830)):1491–3. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 43.Trégouët DA, König IR, Erdmann J, Munteanu A, Braund PS, Hall AS, et al. Wellcome Trust Case Control Consortium. Cardiogenics Consortium Genome-wide haplotype association study identifies the SLC22A3-LPAL2-LPA gene cluster as a risk locus for coronary artery disease. Nat Genet. 2009 Mar;41((3)):283–5. doi: 10.1038/ng.314. [DOI] [PubMed] [Google Scholar]
- 44.Erdmann J, Grosshennig A, Braund PS, König IR, Hengstenberg C, Hall AS, et al. Italian Atherosclerosis, Thrombosis, and Vascular Biology Working Group. Myocardial Infarction Genetics Consortium. Wellcome Trust Case Control Consortium. Cardiogenics Consortium New susceptibility locus for coronary artery disease on chromosome 3q22.3. Nat Genet. 2009 Mar;41((3)):280–2. doi: 10.1038/ng.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reilly MP, Li M, He J, Ferguson JF, Stylianou IM, Mehta NN, et al. Myocardial Infarction Genetics Consortium. Wellcome Trust Case Control Consortium Identification of ADAMTS7 as a novel locus for coronary atherosclerosis and association of ABO with myocardial infarction in the presence of coronary atherosclerosis: two genome-wide association studies. Lancet. 2011 Jan;377((9763)):383–92. doi: 10.1016/S0140-6736(10)61996-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peden JF, Hopewell JC, Saleheen D, Chambers JC, Hager J, Soranzo N, et al. Coronary Artery Disease (C4D) Genetics Consortium A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nat Genet. 2011 Mar;43((4)):339–44. doi: 10.1038/ng.782. [DOI] [PubMed] [Google Scholar]
- 47.Schunkert H, König IR, Kathiresan S, Reilly MP, Assimes TL, Holm H, et al. Cardiogenics. CARDIoGRAM Consortium Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011 Mar;43((4)):333–8. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu X, Wang L, Chen S, He L, Yang X, Shi Y, et al. Coronary ARtery DIsease Genome-Wide Replication And Meta-Analysis (CARDIoGRAM) Consortium Genome-wide association study in Han Chinese identifies four new susceptibility loci for coronary artery disease. Nat Genet. 2012 Jul;44((8)):890–4. doi: 10.1038/ng.2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith NL, Felix JF, Morrison AC, Demissie S, Glazer NL, Loehr LR, et al. Association of genome-wide variation with the risk of incident heart failure in adults of European and African ancestry: a prospective meta-analysis from the cohorts for heart and aging research in genomic epidemiology (CHARGE) consortium. Circ Cardiovasc Genet. 2010 Jun;3((3)):256–66. doi: 10.1161/CIRCGENETICS.109.895763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Villard E, Perret C, Gary F, Proust C, Dilanian G, Hengstenberg C, et al. Cardiogenics Consortium A genome-wide association study identifies two loci associated with heart failure due to dilated cardiomyopathy. Eur Heart J. 2011 May;32((9)):1065–76. doi: 10.1093/eurheartj/ehr105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gudbjartsson DF, Holm H, Gretarsdottir S, Thorleifsson G, Walters GB, Thorgeirsson G, et al. A sequence variant in ZFHX3 on 16q22 associates with atrial fibrillation and ischemic stroke. Nat Genet. 2009 Aug;41((8)):876–8. doi: 10.1038/ng.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ellinor PT, Lunetta KL, Glazer NL, Pfeufer A, Alonso A, Chung MK, et al. Common variants in KCNN3 are associated with lone atrial fibrillation. Nat Genet. 2010 Mar;42((3)):240–4. doi: 10.1038/ng.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ellinor PT, Lunetta KL, Albert CM, Glazer NL, Ritchie MD, Smith AV, et al. Meta-analysis identifies six new susceptibility loci for atrial fibrillation. Nat Genet. 2012 Apr;44((6)):670–5. doi: 10.1038/ng.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Low SK, Takahashi A, Ebana Y, Ozaki K, Christophersen IE, Ellinor PT, et al. AFGen Consortium Identification of six new genetic loci associated with atrial fibrillation in the Japanese population. Nat Genet. 2017 Jun;49((6)):953–8. doi: 10.1038/ng.3842. [DOI] [PubMed] [Google Scholar]
- 55.Pfeufer A, van Noord C, Marciante KD, Arking DE, Larson MG, Smith AV, et al. Genome-wide association study of PR interval. Nat Genet. 2010 Feb;42((2)):153–9. doi: 10.1038/ng.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chambers JC, Zhao J, Terracciano CM, Bezzina CR, Zhang W, Kaba R, et al. Genetic variation in SCN10A influences cardiac conduction. Nat Genet. 2010 Feb;42((2)):149–52. doi: 10.1038/ng.516. [DOI] [PubMed] [Google Scholar]
- 57.Sotoodehnia N, Isaacs A, de Bakker PI, Dörr M, Newton-Cheh C, Nolte IM, et al. Common variants in 22 loci are associated with QRS duration and cardiac ventricular conduction. Nat Genet. 2010 Dec;42((12)):1068–76. doi: 10.1038/ng.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Arking DE, Pfeufer A, Post W, Kao WH, Newton-Cheh C, Ikeda M, et al. A common genetic variant in the NOS1 regulator NOS1AP modulates cardiac repolarization. Nat Genet. 2006 Jun;38((6)):644–51. doi: 10.1038/ng1790. [DOI] [PubMed] [Google Scholar]
- 59.Newton-Cheh C, Eijgelsheim M, Rice KM, de Bakker PI, Yin X, Estrada K, et al. Common variants at ten loci influence QT interval duration in the QTGEN Study. Nat Genet. 2009 Apr;41((4)):399–406. doi: 10.1038/ng.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Arking DE, Pulit SL, Crotti L, van der Harst P, Munroe PB, Koopmann TT, et al. CARe Consortium. COGENT Consortium. DCCT/EDIC. eMERGE Consortium. HRGEN Consortium Genetic association study of QT interval highlights role for calcium signaling pathways in myocardial repolarization. Nat Genet. 2014 Aug;46((8)):826–36. doi: 10.1038/ng.3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.den Hoed M, Eijgelsheim M, Esko T, Brundel BJ, Peal DS, Evans DM, et al. CHARGE-AF Consortium Identification of heart rate-associated loci and their effects on cardiac conduction and rhythm disorders. Nat Genet. 2013 Jun;45((6)):621–31. doi: 10.1038/ng.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nolte IM, Munoz ML, Tragante V, Amare AT, Jansen R, Vaez A, et al. Genetic loci associated with heart rate variability and their effects on cardiac disease risk. Nat Commun. 2017 Jun;8:15805. doi: 10.1038/ncomms15805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, et al. Diabetes Genetics Initiative of Broad Institute of Harvard and MIT, Lund University, and Novartis Institutes of BioMedical Research Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007 Jun;316((5829)):1331–6. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 64.Kooner JS, Chambers JC, Aguilar-Salinas CA, Hinds DA, Hyde CL, Warnes GR, et al. Genome-wide scan identifies variation in MLXIPL associated with plasma triglycerides. Nat Genet. 2008 Feb;40((2)):149–51. doi: 10.1038/ng.2007.61. [DOI] [PubMed] [Google Scholar]
- 65.Sandhu MS, Waterworth DM, Debenham SL, Wheeler E, Papadakis K, Zhao JH, et al. Wellcome Trust Case Control Consortium LDL-cholesterol concentrations: a genome-wide association study. Lancet. 2008 Feb;371((9611)):483–91. doi: 10.1016/S0140-6736(08)60208-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim YJ, Go MJ, Hu C, Hong CB, Kim YK, Lee JY, et al. MAGIC consortium Large-scale genome-wide association studies in East Asians identify new genetic loci influencing metabolic traits. Nat Genet. 2011 Sep;43((10)):990–5. doi: 10.1038/ng.939. [DOI] [PubMed] [Google Scholar]
- 67.Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S, et al. Global Lipids Genetics Consortium Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013 Nov;45((11)):1274–83. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ko A, Cantor RM, Weissglas-Volkov D, Nikkola E, Reddy PM, Sinsheimer JS, et al. Amerindian-specific regions under positive selection harbour new lipid variants in Latinos. Nat Commun. 2014 Jun;5((1)):3983. doi: 10.1038/ncomms4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Surakka I, Horikoshi M, Mägi R, Sarin AP, Mahajan A, Lagou V, et al. ENGAGE Consortium The impact of low-frequency and rare variants on lipid levels. Nat Genet. 2015 Jun;47((6)):589–97. doi: 10.1038/ng.3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wain LV, Vaez A, Jansen R, Joehanes R, van der Most PJ, Erzurumluoglu AM, et al. Novel Blood Pressure Locus and Gene Discovery Using Genome-Wide Association Study and Expression Data Sets From Blood and the Kidney. Hypertension. 2017 Jul;70((3)):HYPERTENSIONAHA.117.09438. doi: 10.1161/HYPERTENSIONAHA.117.09438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nagy R, Boutin TS, Marten J, Huffman JE, Kerr SM, Campbell A, et al. Exploration of haplotype research consortium imputation for genome-wide association studies in 20,032 Generation Scotland participants. Genome Med. 2017 Mar;9((1)):23. doi: 10.1186/s13073-017-0414-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Parmar PG, Taal HR, Timpson NJ, Thiering E, Lehtimäki T, Marinelli M, et al. Early Genetics and Lifecourse Epidemiology Consortium International Genome-Wide Association Study Consortium Identifies Novel Loci Associated With Blood Pressure in Children and Adolescents. Circ Cardiovasc Genet. 2016 Jun;9((3)):266–78. doi: 10.1161/CIRCGENETICS.115.001190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kato N, Loh M, Takeuchi F, Verweij N, Wang X, Zhang W, et al. BIOS-consortium. CARDIo GRAMplusCD. LifeLines Cohort Study. InterAct Consortium Trans-ancestry genome-wide association study identifies 12 genetic loci influencing blood pressure and implicates a role for DNA methylation. Nat Genet. 2015 Nov;47((11)):1282–93. doi: 10.1038/ng.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li C, Kim YK, Dorajoo R, Li H, Lee IT, Cheng CY, et al. Genome-Wide Association Study Meta-Analysis of Long-Term Average Blood Pressure in East Asians. Circ Cardiovasc Genet. 2017 Apr;10((2)):e001527. doi: 10.1161/CIRCGENETICS.116.001527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu X, Hu C, Bao M, Li J, Liu X, Tan X, et al. Genome Wide Association Study Identifies L3MBTL4 as a Novel Susceptibility Gene for Hypertension. Sci Rep. 2016 Aug;6((1)):30811. doi: 10.1038/srep30811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Franceschini N, Fox E, Zhang Z, Edwards TL, Nalls MA, Sung YJ, et al. Asian Genetic Epidemiology Network Consortium Genome-wide association analysis of blood-pressure traits in African-ancestry individuals reveals common associated genes in African and non-African populations. Am J Hum Genet. 2013 Sep;93((3)):545–54. doi: 10.1016/j.ajhg.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lettre G, Palmer CD, Young T, Ejebe KG, Allayee H, Benjamin EJ, et al. Genome-wide association study of coronary heart disease and its risk factors in 8,090 African Americans: the NHLBI CARe Project. PLoS Genet. 2011 Feb;7((2)):e1001300. doi: 10.1371/journal.pgen.1001300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.He L, Kernogitski Y, Kulminskaya I, Loika Y, Arbeev KG, Loiko E, et al. Pleiotropic Meta-Analyses of Longitudinal Studies Discover Novel Genetic Variants Associated with Age-Related Diseases. Front Genet. 2016 Oct;7:179. doi: 10.3389/fgene.2016.00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hager J, Kamatani Y, Cazier JB, Youhanna S, Ghassibe-Sabbagh M, Platt DE, et al. FGENTCARD Consortium Genome-wide association study in a Lebanese cohort confirms PHACTR1 as a major determinant of coronary artery stenosis. PLoS One. 2012;7((6)):e38663. doi: 10.1371/journal.pone.0038663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wakil SM, Ram R, Muiya NP, Mehta M, Andres E, Mazhar N, et al. A genome-wide association study reveals susceptibility loci for myocardial infarction/coronary artery disease in Saudi Arabs. Atherosclerosis. 2016 Feb;245:62–70. doi: 10.1016/j.atherosclerosis.2015.11.019. [DOI] [PubMed] [Google Scholar]
- 81.Lee JY, Lee BS, Shin DJ, Woo Park K, Shin YA, Joong Kim K, et al. A genome-wide association study of a coronary artery disease risk variant. J Hum Genet. 2013 Mar;58((3)):120–6. doi: 10.1038/jhg.2012.124. [DOI] [PubMed] [Google Scholar]
- 82.Takeuchi F, Yokota M, Yamamoto K, Nakashima E, Katsuya T, Asano H, et al. Genome-wide association study of coronary artery disease in the Japanese. Eur J Hum Genet. 2012 Mar;20((3)):333–40. doi: 10.1038/ejhg.2011.184. [DOI] [PMC free article] [PubMed] [Google Scholar]