Abstract
Here we describe the design of a bioluminescent stem-loop probe for the sensitive detection of HIV-1 spliced RNA. In this study, we employed Gaussia luciferase (GLuc), a bioluminescent protein that has several advantages over other bioluminescent proteins, including smaller size, higher bioluminescent intensity, and chemical and thermal stability. GLuc was chemically conjugated to the DABCYL-modified stem-loop probe (SLP) and was purified with a 2-step process to remove unconjugated GLuc and SLP. The binding of the target RNA to the loop region of the SLP results in the open conformation separating the stem part of SLP. GLuc conjugated to the stem acts as a reporter that produces light by a chemical reaction upon adding its substrate, coelenterazine in the presence of the target, while DABCYL serves as a quencher of bioluminescence in the closed conformation of SLP in the absence of the target. The optimized GLuc based-SLP assay resulted in a signal-to-background ratio of 47, which is the highest reported with bioluminescent SLPs and is significantly higher compared to traditional fluorescence-based SLPs that yield low signal to background ratio. Moreover, the assay showed an excellent selectivity against a single and double mismatched nucleic acid target, low detection limit, and ability to detect spiked HIV-1 RNA in human serum matrix.
Introduction
Stem-loop probe (SLP), or molecular beacon (MB), is a self-complementary, single-stranded oligonucleotide with a hairpin-like structure that can undergo a structural change from closed to open conformation upon hybridization of complementary oligonucleotide target.1 The structural change can be monitored by conjugating a fluorophore–quencher pair to the SLP probe such that the binding of target nucleic acid to the loop region results in the separation of fluorophore– quencher pair resulting in the emission of fluorescence, which can be correlated with the concentration of nucleic acid target. This classical SLP and its various modifications have been employed in nucleic acid detection for a variety of applications including nucleic acid hybridization,1–3 real time polymerase chain reactions (RT-PCR),4,5 RNA hybridization in living cells,6,7 and DNA-protein interaction.8 Beside the classical design of SLP with fluorophore-quencher pair, different labels have been reported to improve sensitivity of SLP-based assays. As an alternative to the fluorophore, other signaling reporters such as quantum dots (QD)9 and metal complexes10,11 have been used. Likewise, enhancement of quenching capability has been demonstrated using gold nanoparticle (AuNP),12 silver nanoparticle (AgNP),13 graphene oxide,14 and carbon nanotubes.15
Our laboratory was the first to demonstrate the use of a protein reporter, specifically a bioluminescent protein, Renilla luciferase (Rluc), in the development of SLP.16 This bioluminescent SLP showed a higher signal-to-background (S/B) ratio and lower detection limit when directly compared to a fluorescent reporter-based SLP.16 The high sensitivity achieved was due to the fact that the bioluminescence emission was generated from a chemical reaction and did not require any excitation source, significantly reducing the background noise typically observed with fluorescence-based reporters. Low background observed with bioluminescent reporters also allows for direct target detection in complex matrices. In a recent study, a modular design of molecular beacon based on bioluminescence resonance energy transfer was developed using a bioluminescent protein Nanoluc as the donor and Cy3 as the acceptor yielding a pM detection limit.17
A prior report from our laboratory that employs bioluminescent protein showed promising results when bioluminescent reporter was used in the SLP design.16 However, there is still a room for further improvement in the bioluminescent SLP design in terms of size and activity of the bioluminescent protein. This prompted us to evaluate the use of other bioluminescent proteins that can further enhance the design of SLP to achieve better performance in terms of sensitivity and other analytical parameters. Specifically, in this report we evaluated Gaussia luciferase (GLuc) as a bioluminescent reporter in the development of a SLP for the detection of a spliced HIV-1 RNA. GLuc is the smallest coelenterazine-dependent luciferase and consists of 185 amino acids (19.9 kDa).18 It catalyzes the oxidation of coelenterazine to emit light at 480 nm and does not need ATP or other co-factors for bioluminescence activity.18 Apart from being small enough to potentially reduce steric hindrance for stem closure in the SLP design, GLuc has significantly higher bioluminescent activity than most other luciferases. For example, bioluminescence intensity is much stronger (over 200 fold) than that of RLuc18 and is comparable to the furimazine-dependent Nanoluc. GLuc is highly stable under a wide range of temperatures (up to 95 °C)19 and in an acidic environment (down to pH 1.5),20 whereas Rluc is stable up to 37 °C and pH (~7.4) and Nanoluc is stable up to 55 °C and pH (~7–9).21 These properties of GLuc make it an attractive reporter in terms of bioconjugation and bioanalytical applications. Therefore, it has been widely used for in vitro and in vivo imaging of tumor cells,22 gene expression analysis,18 protein–protein interaction study,23 immunoassay development,24 and DNA hybridization.25 We hypothesized that the small size of GLuc, high bioluminescence emission, and temperature and pH stability compared to other bioluminescent proteins would result in GLuc-based SLP with superior performance. We employed DABCYL as the quencher in this study since its absorption maximum overlaps well with the emission spectrum of GLuc.
In this manuscript, we demonstrate the design of Gluc- based SLP for the detection of HIV-1 RNA corresponding to the 3/5 splice junction in HIV-1 infected cells.26 Viral persistency has been a challenge with HIV,27–29 Herpes,30 Measles,31 Varicella,32 etc. With effective antiretroviral therapy, the level of plasma HIV-1 RNA falls below the detection limit of clinical assays (50 copies per ml).33 However, the virus cannot be completely eradicated, and a latent form of HIV-1 infection may persist in CD4+ cells that leads to HIV viral reactivation after interruption of the therapy.34 Typically, a number of pretreatment steps and cell isolation steps are performed prior to RNA detection,35 which prolongs the detection time and reduces the reliability of the method. The gold standard method for measuring HIV persistence is the quantitative viral outgrowth assay.36–38 This assay is performed on purified resting CD4+ T cells isolated from a patient, which are then activated to induce viral replication. After about two weeks the viral outgrowth is analyzed using ELISA assay or qPCR. In that respect, the use of bioluminescence detection provides a convenient alternative to ELISA or qPCR for sensitive detection of HIV following a viral outgrowth assay, as only minimal pretreatment is required and bioluminescence can be performed rapidly following cell lysis. Further, our strategy combines the enhanced sensitivity of bioluminescence with the specificity of stem-loop probes, which is key when developing methods for sample detection in physiological matrices.
Materials and methods
Materials
All purchased chemicals were used without further purification. Monobasic sodium phosphate anhydrous, dibasic sodium phosphate heptahydrate, imidazole, Tris-HCl, sodium chloride, Tween 20, 2-mercaptoethanol (β-ME), and nonyl phenoxypolyethoxylethanol (NP-40) were purchased from Sigma- Aldrich (St Louis, MO). Slide-A-Lyzer Dialysis G2 Cassettes, Pierce Centrifuge Column, Zeba desalt spin columns (7 kDa MWCO) and Bicinchoninic Acid (BCA) Protein Assay Kit were obtained from Thermo Fisher Scientific (Waltham, MA). Succinimidyl 4-hydrazinonicotinate acetone hydrazone (HyNic) was purchased from Solulink Biotechnologies (San Diego, CA). Isopropyl-1-thio-β-D-galactopyranoside (IPTG) and 1X ProBlock™ Gold Bacterial Protease Inhibitor Cocktail were purchased from Gold Biotechnology Inc. (St Louis, MO). Ni- NTA agarose was obtained from Qiagen (Valencia, CA). 4–20% Mini-PROTEAN® TGX™ precast polyacrylamide gels were supplied by Bio-Rad Laboratories Inc. (Hercules, California). Streptavidin resin was purchased from G-Biosciences, (St Louis, MO). 96-Well, black, non-binding microplate was purchased from Greiner Bio-One Inc. (Monroe, NC). Native coelenterazine was provided by NanoLight™ Technology (Prolume Ltd, Pinetop, AZ) and prepared in 1 mL of acidified methanol to make 1 mg mL−1. Stem-loop probe (SLP) with 5′-amino and 3′- DABCYL modification (Table 1) was purchased from TriLink Biotechnologies (San Diego, CA), all other synthetic oligonucleotides targets: fully matched (FM) HIV-1 DNA target, HIV-1 RNA target, DNA with single mismatch (SM), double mismatches (DM), and biotin modified target were purchased as lyophilized powder from Sigma-Aldrich (St Louis, MO).
Table 1.
Sequences of SLP (underlined bases are complementary to the targets), biotin-labeled DNA, HIV-1 DNA target, HIV-1 RNA target, single mismatched DNA, double mismatched DNA. Mismatched bases are shown in italic and underlined
| SLP | NH2-C6-CGTCACTCCGCTTCTTCCTTGTTATGTAGTGACG-DABCYL |
| Biotinylated oligo | ACATAACAAGGAAGAAGCGGAGTGACG-TEG Biotin |
| HIV-1 DNA target | AATATCAAGCAGGACATAACAAGGAAGAAGCGGAGACAGCGACGAA |
| HIV-1 RNA target | AAUAUCAAGCAGGACAUAACAAGGAAGAAGCGGAGACAGCGACGA |
| Single mismatch (SM) | AATATCAAGCAGGACATAACAATGAAGAAGCGGAGACAGCGACGAA |
| Double mismatches (DM) | AATATCAAGCAGGACATAACAATGAAGAAACGGAGACAGCGACGAA |
Methods
Expression and purification of Gaussia luciferase
The gene for the Gaussia luciferase (GLuc) was cloned into the pCold-I cold-shock expression vector and transformed into Origami 2 E. coli. For expression, a culture of 5 mL was grown overnight at 37 °C in terrific broth containing 0.1 mg mL−1 ampicillin. This culture was then used to inoculate 300 mL of the same broth and was grown at 37 °C to an OD600 nm of ~1.6. After that, the cells were centrifuged and resuspended in fresh media with antibiotic and put in an ice bath for 1 h to initiate cold-shock. Subsequently, expression was induced by adding 0.1 mM isopropyl-1-thio-β-D-galactopyranoside (IPTG) and incubated overnight at 15 °C. The cells were pelleted by centrifugation and resuspended in Lysis buffer (50 mM Tris-HCl pH 8.0, 150 mM sodium chloride, 10 mM imidazole, 1% (v/v) nonyl phenoxypolyethoxylethanol (NP-40), 0.2% (v/v) Tween 20, and 10 mM 2-mercaptoethanol (β-ME)). Before sonication, 1X ProBlock™ Gold Bacterial Protease Inhibitor Cocktail was added to the cell suspension then sonicated with microtip probe (Model 500 Sonic Dismembrator, Fisher Scientific, Pittsburgh, PA) on ice bath for 20 min with a 0.5 s on/off pulse sequence.
The lysate was centrifuged at 17 000g at 4 °C for 30 min to remove the insoluble material and the supernatant was filtered through 0.45 μm filter followed by 0.22 μm filter. The filtered crude protein was then incubated with Ni-NTA agarose at room temperature for 60 min, poured onto Pierce Centrifuge Column and unbound proteins were removed by a flow through using gravity flow. The beads were washed with 10 column volumes of lysis buffer followed by 20 column volumes of a wash buffer (50 mM Tris-HCl pH 8.0 containing 150 mM sodium chloride and 20 mM imidazole). The protein was eluted with an elution buffer of 50 mM Tris-HCl pH 8.0 containing 150 mM sodium chloride, and 150 mM imidazole in 6 tubes, each about 1 column volume. The eluted GLuc from each elution fraction was analyzed by SDS-PAGE using a 4–20% Tris-Glycine gel according to the manufacturer’s instructions.
Subsequently, eluted GLuc was dialyzed into a phosphate buffered saline (PBS, 100 mM sodium phosphate pH 7.2 and 150 mM sodium chloride) using Slide-A-Lyzer Dialysis G2 Cassettes with 3.5 kDa molecular weight cut-off (MWCO) then stored at 4 °C until further use. Protein concentration was measured using the Bicinchoninic Acid (BCA) Protein Assay Kit according to the manufacturer’s instructions.
Conjugation of the stem-loop probe (SLP) to GLuc
Conjugation of the SLP with GLuc was carried out using alde- hyde-amine bioconjugation chemistry by adopting a protocol used from Solulink Biotechnologies (http://www.solulink.com/). Briefly, SLP with 5′-amino modification was chemically converted to a benzaldehyde moiety by adding 50× excess of succinimidyl 4-formylbenzoate (SFB) in a modification buffer (100 mM sodium phosphate buffer pH 7.4, 150 mM sodium chloride). In parallel, amine residues of lysines on GLuc were modified with 20× excess of succinimidyl 4-hydrazinonicotinate acetone hydrazone (HyNic) in a modification buffer. Both reactions were incubated for 2 h at room temperature. Next, both reactions were desalted into a conjugation buffer (100 mM sodium phosphate, 150 mM sodium chloride, pH 6.0 buffer) using Zeba desalt spin columns, 0.5 mL, according to the manufacturer’s protocol. Subsequently, SFB-modified oligonucleotide was added in 4× excess to hydrazine-modified GLuc and conjugation reaction was performed for 2 h at room temperature followed by overnight at 4 °C. The conjugation of GLuc with SLP (G-SLP) was evaluated by SDS-PAGE using a 4–20% Tris-Glycine gel.
Purification of unconjugated GLuc and SLP
After conjugation, purification was necessary to remove any un-conjugated SLP and GLuc. Purification was performed in a two-step process: the first step involved removing unconjugated SLP using Ni-NTA agarose; while in the second step, unconjugated GLuc was removed using streptavidin resin. For the first step, conjugation mixture was incubated in 0.5 mL Ni-NTA agarose resin in Pierce Centrifuge Column for 1 h allowing for binding of His-tagged GLuc (unconjugated GLuc and G-SLP). The flow-through containing unconjugated SLP was removed followed by elution of unconjugated GLuc and G-SLP in an elution buffer. For the second step, first 20 μM of a short biotinylated oligonucleotide complementary to the SLP (Table 1) was added to 400 μL streptavidin resin in Pierce Centrifuge column. The eluted mixture of unconjugated GLuc and G-SLP was then added and incubated for >2 h at 37 °C to allow G-SLP to hybridize with complementary biotinylated oligonucleotide. The flow-through containing unconjugated GLuc was removed, and G-SLP was eluted by dehybridization in hot water. After purification, the conjugation of GLuc with SLP (G-SLP) was evaluated by SDS-PAGE and sliver staining using Thermo Scientific Pierce Silver Stain Kit according to the manufacturer’s instruction.
HIV-1 RNA assay development and optimization
Hybridization buffer optimization
A 20 nM of G-SLP and 50 nM HIV-1 synthetic DNA target in different hybridization buffers were added into a 96-well microplate in a total volume of 100 μL. Hybridization buffers employed for this study were: 100 mM sodium phosphate buffer pH 7.4, 1 mM EDTA; 100 mM sodium phosphate buffer pH 7.4, 75 mM sodium chloride buffer; 100 mM sodium phosphate buffer pH 7.4, 150 mM sodium chloride; 100 mM sodium phosphate buffer pH 7.4, 300 mM sodium chloride; 75 mM sodium chloride, 7.5 mM trisodium citrate buffer pH 7.4; 150 mM sodium chloride, 15 mM trisodium citrate buffer pH 7.4; 300 mM sodium chloride, 30 mM trisodium citrate buffer pH 7.4; 10 mM Tris buffer pH 7.4, 150 mM sodium chloride; and 10 mM Tris buffer pH 7.4, 500 mM sodium chloride. Hybridization reaction was performed for 60 min at 37 °C. Bioluminescence signal was measured using Clariostar Optima (BMG Labtech, Ortenberg, Germany) after injection of 50 μL of coelentrazine substrate (6 μM) prepared in 100 mM sodium phosphate buffer pH 7.4, 150 mM sodium chloride. Signal-to-background ratio was calculated by measuring the ratio of the emission intensity in the absence and presence of target (50 nM).
Hybridization time and temperature optimization
Hybridization assay was performed in a 96-well, black, non-binding microplate. Both G-SLP (20 nM) and HIV-1 synthetic target (50 nM) in a hybridization buffer (10 mM Tris buffer pH 7.4 with 500 mM NaCl) were added into microplate wells in a total volume of 100 μL. Then the microplate was sealed with adhesive tape to prevent evaporation and incubated for 15 and 60 min at 22, 37, and 45 °C in a temperature-controlled incubator. Bioluminescence signal was measured as described previously.
Assay sensitivity and specificity
To generate a dose-response curve, hybridization assay was performed in a 96-well, black, non-binding microplate. 20 nM of G-SLP and different amounts from 0.1 to 50 nM of HIV-1 DNA synthetic target in a hybridization buffer were added into microplate wells in a total volume of 100 μL and incubated for 60 min at 22, 37, and 45 °C. Bioluminescence signal was measured as described previously.
Assay specificity was evaluated by hybridizing 20 nM G-SLP with fully matched (FM), single mismatched (SM) and double mismatched (DM) synthetic HIV-1 target (Table 1) in various concentrations, and comparing the bioluminescence signal after hybridization for 1 h at 37 °C.
Detection of HIV-1 target RNA in a clinical sample matrix
To simulate clinical sample analysis, different known concentrations of the synthetic HIV-1 target RNA were spiked in 1 : 10 diluted pooled human serum (Sigma-Aldrich). G-SLP (20 nM) was added to serum samples, and hybridization was performed by incubation for 1 h at 37 °C. Bioluminescence signal was measured as described above.
Results and discussion
Stem-loop probe is a versatile design of hybridization probe in biosensing applications offering high specificity and sensitivity. In comparison to solid phase assays, which require multiple wash steps to decrease the non-specific adsorption and high background signal, SLP-based assay is a single-step, mix- and-measure assay. In this manuscript, we describe the design of a bioluminescent SLP that employs GLuc as the reporter protein and DABCYL as the quencher moiety. GLuc is the smallest known coelenterazine-dependent luciferase and is bright and stable at varied temperature and pH, which makes it an attractive reporter for use in the development of SLP assay.
Conjugation of GLuc with SLP
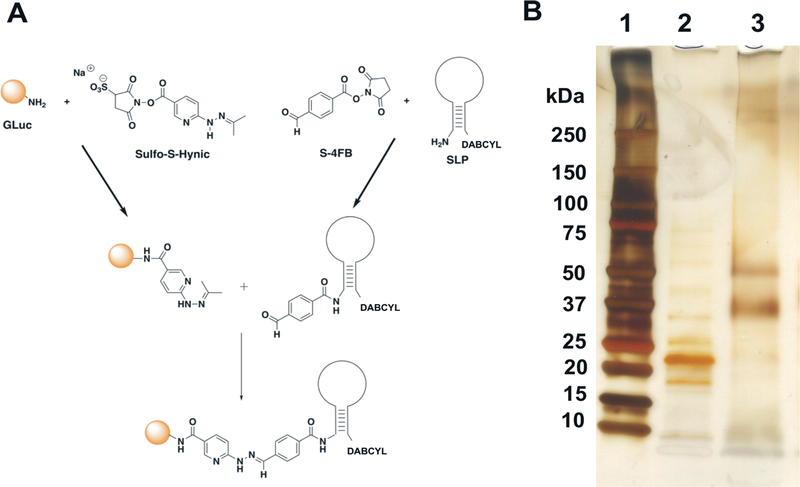
GLuc was expressed and purified following the protocol developed in our laboratory. SDS-PAGE was performed to verify the size and purity of GLuc protein. Purified GLuc was then conjugated to the DABCYL-modified SLP using SFB/HyNic chemistry as detailed in the methods section (Fig. 1A). SDS-PAGE analysis (ESI 1†) confirmed the successful conjugation between GLuc and SLP. Following conjugation, removal of un-conjugated GLuc and SLP was performed to reduce background stemming from the presence of unconjugated GLuc and signal depression resulting from unconjugated SLP. Assay sensitivity and signal-to-background (S/B) ratio depend significantly on the purification step. Purification of G-SLP was performed in a 2-step process as detailed in the methods section. Following purification, SDS-PAGE with silver stain was performed (Fig. 1B) and showed 2 distinct bands around 37 kDa and 50 kDa that corresponded to GLuc-SLP mono- and di-conjugates, respectively, as determined from the molecular weight of the stem-loop relative to the purified Gluc shown in the adjacent well. Based on the reproducibility of this analysis, the overwhelming majority of Gluc-SLP were represented as a ≤1: 2 conjugation ratio and provided a foundation for the excellent S/B ratio observed experimentally. This also provided evidence to suggest that higher-order conjugation was unfavorable, whether as a factor of appropriate molar ratios during conjugation or as steric effects that biased against proximal functionalization.
Fig. 1.
A: Schematic of chemical conjugation reaction between SLP and GLuc protein. B: SDS-PAGE showing analysis of GLuc before and after conjugation with SLP, visualized by silver staining. Lane 1; protein ladder, Lane 2; unconjugated GLuc, Lane 3; purified GLuc-SLP.
For attaching SLP to GLuc, we have chosen amine-reactive bioconjugation chemistry through lysine residues on GLuc because it offers an easy, simple method that doesn’t require protein mutation and capitalizes on the availability of commercial kits. The possibility of attaching multiple SLPs per bioluminescent protein offers a platform for increasing S/B ratio by decreasing the background. Over-conjugation of lysine residues may, indeed, affect the activity of a protein, although this was difficult to observe in our case due to the ever-present quenching effect of the DABCYL. If polyconjugates provided any artifacts within the assay system, the effect would be reproducible due to homogeneity and only apparent at extremely low target concentrations. Mitigation is as simple as shifting the dynamic range lower by reducing the total probe concentration. Conversely, conjugation through sulfhydryl or other groups often results in the loss of protein activity as these residues are typically essential for protein function. For example, GLuc has several Cys residues that are essential for its activity. This can also be mitigated by the introduction of a non-native conjugation site, as we demonstrated previously through a Tyr introduced at the N-terminus of Gluc to achieve site-specific conjugation since Gluc lacks Tyr residues.24
Assay condition optimization
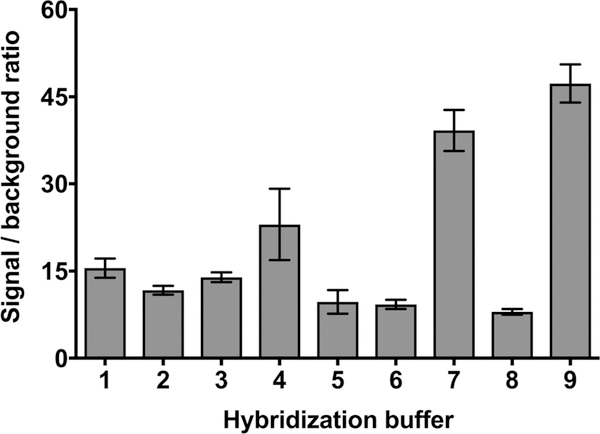
After the purification of GLuc-conjugated SLP, assay conditions such as hybridization buffer, temperature, and time were optimized in order to achieve the best S/B ratio. Buffer composition and salt concentration are known to play an important role in hybridization assay specificity and sensitivity. Therefore, we investigated the effect of several buffers with varying ionic strength on hybridization efficiency by monitoring the S/B ratio. Fig. 2 illustrates that the S/B ratio varied depending on the buffer composition. We found that the use of 10 mM Tris buffer pH 7.4 with 500 mM NaCl is the optimal hybridization buffer, resulting in an S/B ratio of about 47. This ratio is 5.2-fold higher compared to our previous study where Renilla luciferase (RLuc) was used as a reporter in the design of bioluminescent stem-loop probe (BSLP), where the S/B ratio was 9.16 As predicted, the GLuc S/B ratio increased relative to Rluc, and this was likely due primarily to the smaller size of GLuc. Besides presenting a less massive strain on the stem terminus that would reduce thermodynamic instability, the probability of the quencher being oriented at the outer limits of the Förster radius would be minimized. Stability to changes in pH and temperature also likely contributed to better performance of GLuc-based SLP.
Fig. 2.
Signal-to-background ratio (S/B) evaluation using different hybridization buffers in the presence of 50 nM of target. G-SLP probe and target were hybridized for 60 min at 37 °C. Buffers used for S/B ratio evaluation are: (1) 100 mM sodium phosphate buffer pH 7.4, 1 mM EDTA; (2) 100 mM sodium phosphate buffer pH 7.4, 75 mM sodium chloride; (3) 100 mM sodium phosphate buffer pH 7.4, 150 mM sodium chloride; (4) 100 mM sodium phosphate buffer pH 7.4, 300 mM sodium chloride; (5) 75 mM sodium chloride, 7.5 mM trisodium citrate buffer pH 7.4; (6) 150 mM sodium chloride, 15 mM trisodium citrate buffer pH 7.4; (7) 300 mM sodium chloride, 30 mM trisodium citrate buffer pH 7.4; (8) 10 mM Tris buffer pH 7.4, 150 mM sodium chloride; (9) 10 mM Tris buffer pH 7.4, 500 mM sodium chloride.
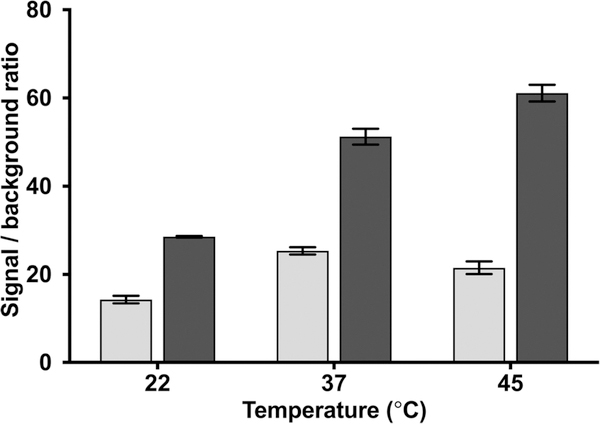
Following optimization of the hybridization buffer composition, hybridization time and temperature parameters were assessed. It has been reported that a SLP with a short stem has a lower melting temperature (Tm), which can easily open even in the absence of the complementary target. This results in a high background and low S/B ratio.39 While SLP with a long stem will give stable SLP (high Tm) with a low background and high S/B ratio, it is not favorable due to its slow hybridization kinetics in the presence of the target. Therefore, SLP should be carefully designed with optimum assay stringency to obtain the highest S/B ratio as well as an ultrasensitive and specific assay. Melting temperature and secondary structure of the SLP and SLP-target duplex were analyzed using the OligoAnalyzer 3.1 tool of IDT (https://www.idtdna.com/calc/analyzer Integrated DNA Technologies, Inc., USA). Our SLP consists of 34 nucleotides and was designed with 20 nucleotides at the loop region and stem with 7 bp length. Theoretical melting temperature (Tm) of the SLP was found to be 61.6 °C using Na+ concentration of 0.5 M. Theoretical Tm of the SLP-target duplex was found to be 74.8 °C (Na+ = 0.5 M). To test our designed SLP, a 50 nM of the target was hybridized with G-SLP for 15 and 60 min at room temperature (22 °C), physiologic temperature (37 °C), and at 45 °C. The bioluminescence result was then compared with control without the target added. As predicted, the S/B ratio was higher in the case of 60 min hybridization in comparison to 15 min (Fig. 3). Furthermore, with 60 min hybridization time we observed that the S/B ratio increased gradually with increase in assay temperature above 22 °C reaching more than two-fold at 45 °C. Temperature is an important factor in all hybridization assays, as temperature closer to the melting temperature of the stem facilitates opening of the self-complementary stem in the presence of the target.
Fig. 3.
Signal/background ratio calculated for SLP assay performed with 50 nM target at different temperatures (22, 37 and 45 °C) for 15 ( ) and 60 (∎) min and compared to the control without the target.
) and 60 (∎) min and compared to the control without the target.
Assay sensitivity and specificity
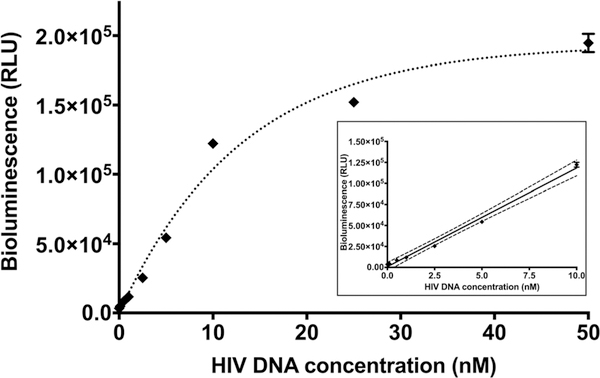
One of the important parameters in the assessment of bioassay performance is the assay sensitivity. To check for this, calibration curves were generated with different concentrations of synthetic HIV-1 DNA target from 0.1 to 50 nM at 22, 37 and 45 °C for 15 min and 60 min (data not shown). The lowest limit of detection (LOD) was achieved using a 60 min hybridization time at 37 °C. LOD was estimated to be 0.045 nM with a linear dynamic range between 0 and 10 nM (Fig. 4). Our study shows 9 times lower LOD using Gluc-SLP compared to using RLuc-SLP previously reported by our group,16 attesting to the advantage of using GLuc as a reporter over RLuc in the development of sensitive SLP assays. Moreover, our assay time (60 min) is shorter than in the previous report (180 min) where RLuc was employed in the bioluminescent SLP design.
Fig. 4.
Calibration curve obtained using G-SLP in the presence of various concentrations of HIV-1 DNA target. Assay was carried out for 60 min at 37 °C. Inset shows the linear range of the calibration curve, dotted lines represent linear fitting with 95% confidence interval. Each data point represents the average and standard deviation of three individual measurements. Some error bars that are not visible are obstructed by the symbols of the points.
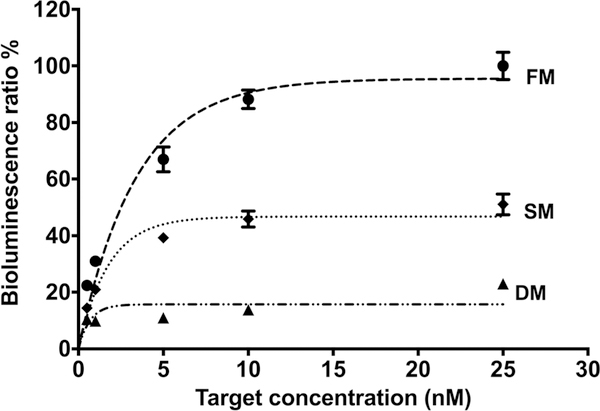
Another important parameter for nucleic acid bioassays is the assay specificity, which is the ability to discriminate between base mismatches. In that regard, the specificity of our assay was evaluated with 20 nM of G-SLP hybridized with 0.5, 1, 5, 10 and 25 nM of fully matched (FM), single mismatched (SM), and double mismatched (DM) DNA (Table 1) for 60 min at 37 °C. An excellent selectivity was seen for the assay, as the recorded bioluminescence signals reveal that our G-SLP assay was able to distinguish the FM target with high selectivity in the presence of interfering targets with one or two nucleotide mismatches at different target concentrations (Fig. 5).
Fig. 5.
Assay specificity, showing the bioluminescence signals for fully matched (- ● -), single mismatched (‧ ◆ ‧) and double mismatched (– ▲ ‧) DNA targets. Assay was performed for 60 min at 37 °C, in the presence of 0.5, 1, 5, 10 and 25 nM targets concentration. Each data point represents the average and standard deviation of three individual measurements.
Spiked samples in serum
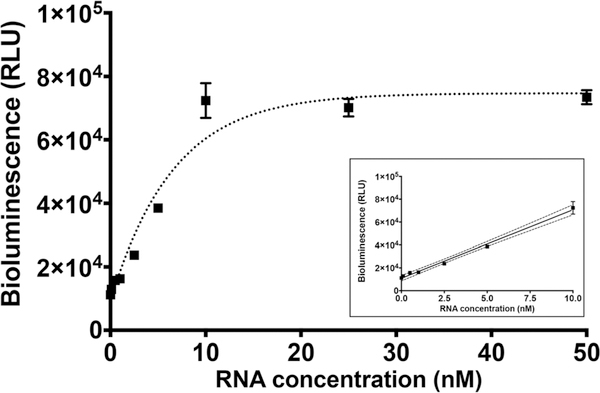
We performed a study to demonstrate the use of G-SLP assay for direct detection of HIV-1 RNA in a clinical sample matrix. For this, we spiked different amounts of the synthetic HIV-1 RNA target in human serum diluted to 10% in 10 mM Tris buffer pH 7.4, 500 mM NaCl, followed by incubation with G-SLP for 1 h at 37 °C. A calibration curve was generated as shown in Fig. 6. This curve resulted in a linear range between 0 and 10 nM with LOD 0.1 nM, which is only about two-fold higher than the LOD obtained with synthetic HIV-1 DNA in a buffer matrix. Although the LOD was decreased in 10% serum, it is sufficient for direct detection of HIV-1 RNA in a patient sample following cell-line expansion. If more sensitive detection is required, HIV-1 can be concentrated from a larger blood volume using ultracentrifugation; then the RNA assay could be performed in buffer following HIV particle lysis or RNA extraction with commercially available kits.
Fig. 6.
Calibration curve of G-SLP in the presence of various concentrations of RNA target in 10% serum. Assay was carried out for 1 h at 37 °C. Inset shows the linear range of the calibration curve, dots represent linear fitting with 95% confidence interval. Each data point represents the average and standard deviation of three individual measurements.
In conclusion, we have demonstrated that GLuc-based bioluminescent SLP results in a high S/B ratio yielding excellent sensitivity for the detection for target nucleic acid. GLuc, being smallest in size and brightest among the colenterazine- dependent bioluminescent proteins, contributes to the design of a superior SLP. The signal to background ratio obtained was the highest amongst the BSLPs reported to date. Additionally, GLuc is stable at high temperature, which should enhance the application of this construct for generic nucleic acid detection assays.
Supplementary Material
Acknowledgements
The work was supported through NIGMS funding (R01GM114321), the National Science Foundation (CHE-1506740), and the State of Florida Department of Health (7ZK01/7ZK11). Sylvia Daunert thanks the Miller School of Medicine of the University of Miami for the Lucille P. Markey Chair in Biochemistry and Molecular Biology.
Footnotes
Conflicts of interest
There are no conflicts of interest to declare.
Electronic supplementary information (ESI) available. See DOI: 10.1039/c8an00047f
References
- 1.Tyagi S and Kramer FR, Nat. Biotechnol, 1996, 14, 303–308. [DOI] [PubMed] [Google Scholar]
- 2.Yao G and Tan W, Anal. Biochem, 2004, 331, 216–223. [DOI] [PubMed] [Google Scholar]
- 3.Beni V, Zewdu T, Joda H, Katakis I and O’Sullivan C, Anal. Bioanal. Chem, 2012, 402, 1001–1009. [DOI] [PubMed] [Google Scholar]
- 4.Summerer D and Marx A, Angew. Chem., Int. Ed, 2002, 41, 3620–3622. [DOI] [PubMed] [Google Scholar]
- 5.Tyagi S, Bratu DP and Kramer FR, Nat. Biotechnol, 1998, 16, 49–53. [DOI] [PubMed] [Google Scholar]
- 6.Jayagopal A, Halfpenny KC, Perez JW and Wright DW, J. Am. Chem. Soc, 2010, 132, 9789–9796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sokol DL, Zhang X, Lu P and Gewirtz AM, Proc. Natl. Acad. Sci. U. S. A, 1998, 95, 11538–11543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li JJ, Fang X, Schuster SM and Tan W, Angew. Chem., Int. Ed, 2000, 39, 1049–1052. [DOI] [PubMed] [Google Scholar]
- 9.Chen AK, Behlke MA and Tsourkas A, Nucleic Acids Res, 2007, 35, e105–e105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Joshi HS and Tor Y, Chem. Commun, 2001, 549–550. [Google Scholar]
- 11.Li J, Zhou W, Ouyang X, Yu H, Yang R, Tan W and Yuan J, Anal. Chem, 2011, 83, 1356–1362. [DOI] [PubMed] [Google Scholar]
- 12.Dubertret B, Calame M and Libchaber AJ, Nat. Biotechnol, 2001, 19, 365–370. [DOI] [PubMed] [Google Scholar]
- 13.Peng H-I, Strohsahl CM, Leach KE, Krauss TD and Miller BL, ACS Nano, 2009, 3, 2265–2273. [DOI] [PubMed] [Google Scholar]
- 14.Lu C-H, Li J, Liu J-J, Yang H-H, Chen X and Chen G-N, Chem. - Eur.J, 2010, 16, 4889–4894. [DOI] [PubMed] [Google Scholar]
- 15.Yang R, Jin J, Chen Y, Shao N, Kang H, Xiao Z, Tang Z, Wu Y, Zhu Z and Tan W, J. Am. Chem. Soc, 2008, 130, 8351–8358. [DOI] [PubMed] [Google Scholar]
- 16.Hunt EA and Deo SK, Chem. Commun, 2011, 47, 9393–9395. [DOI] [PubMed] [Google Scholar]
- 17.Engelen W, van de Wiel KM, Meijer LHH, Saha B and Merkx M, Chem. Commun, 2017, 53, 2862–2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tannous BA, Kim D-E, Fernandez JL, Weissleder R and Breakefield XO, Mol. Ther, 2005, 11, 435–443. [DOI] [PubMed] [Google Scholar]
- 19.Rathnayaka T, Tawa M, Sohya S, Yohda M and Kuroda Y, Biochim. Biophys. Acta, Proteins Proteomics, 2010, 1814, 1912–1917. [Google Scholar]
- 20.Wiles S, Ferguson K, Stefanidou M, Young DB and Robertson BD, Appl. Environ. Microbiol, 2005, 71, 3427–3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hall MP, Unch J, Binkowski BF, Valley MP, Butler BL, Wood MG, Otto P, Zimmerman K, Vidugiris G, Machleidt T, Robers MB, Benink HA, Eggers CT, Slater MR, Meisenheimer PL, Klaubert DH, Fan F, Encell LP and Wood KV, ACS Chem. Biol, 2012, 7, 1848–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Venisnik KM, Olafsen T, Gambhir SS and Wu AM, Mol. Imaging Biol, 2007, 9, 267–277. [DOI] [PubMed] [Google Scholar]
- 23.Remy I and Michnick SW, Nat. Methods, 2006, 3, 977–979. [DOI] [PubMed] [Google Scholar]
- 24.Moutsiopoulou A, Hunt E, Broyles D, Pereira CA, Woodward K, Dikici E, Kaifer A, Daunert S and Deo SK, Bioconjugate Chem, 2017, 28, 1749–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Verhaegen M and Christopoulos TK, Anal. Chem, 2002, 74, 4378–4385. [DOI] [PubMed] [Google Scholar]
- 26.Purcell DF and Martin MA, J. Virol, 1993, 67, 6365–6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Finzi D, Blankson J, Siliciano JD, Margolick JB, Chadwick K, Pierson T, Smith K, Lisziewicz J, Lori F, Flexner C, Quinn TC, Chaisson RE, Rosenberg E, Walker B, Gange S, Gallant J and Siliciano RF, Nat. Med, 1999, 5, 512–517. [DOI] [PubMed] [Google Scholar]
- 28.Wong JK, Hezareh M, Günthard HF, Havlir DV, Ignacio CC, Spina CA and Richman DD, Science, 1997, 278, 1291–1295. [DOI] [PubMed] [Google Scholar]
- 29.Siliciano JD, Kajdas J, Finzi D, Quinn TC, Chadwick K, Margolick JB, Kovacs C, Gange SJ and Siliciano RF, Nat. Med, 2003, 9, 727–728. [DOI] [PubMed] [Google Scholar]
- 30.Grinde B, J. Oral Microbiol, 2013, 5, 22766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sever JL, Rev. Infect. Dis, 1983, 5, 467–473. [DOI] [PubMed] [Google Scholar]
- 32.Eshleman E, Shahzad A and Cohrs RJ, Future Virol, 2011, 6, 341–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perelson AS, Essunger P, Cao Y, Vesanen M, Hurley A, Saksela K, Markowitz M and Ho DD, Nature, 1997, 387, 188–191. [DOI] [PubMed] [Google Scholar]
- 34.Davey RT, Bhat N, Yoder C, Chun T-W, Metcalf JA, Dewar R, Natarajan V, Lempicki RA, Adelsberger JW, Miller KD, Kovacs JA, Polis MA, Walker RE, Falloon J, Masur H, Gee D, Baseler M, Dimitrov DS, Fauci AS and Lane HC, Proc. Natl. Acad. Sci. U. S. A, 1999, 96,15109–15114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richman DD, McCutchan JA and Spector SA, J. Infect. Dis, 1987, 156, 823–827. [DOI] [PubMed] [Google Scholar]
- 36.Chun T-W, Carruth L, Finzi D, Shen X, DiGiuseppe JA, Taylor H, Hermankova M, Chadwick K, Margolick J, Quinn TC, Kuo Y-H, Brookmeyer R, Zeiger MA, Barditch-Crovo P and Siliciano RF, Nature, 1997, 387, 183–188. [DOI] [PubMed] [Google Scholar]
- 37.Siliciano JD and Siliciano RF, in Human Retrovirus Protocols: Virology and Molecular Biology, ed. Zhu T, Humana Press, Totowa, NJ, 2005, pp. 3–15, DOI: 10.1385/1-59259-907-9:003. [DOI] [Google Scholar]
- 38.Fun A, Mok HP, Wills MR and Lever AM, Sci. Rep, 2017, 7, 43231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsourkas A, Behlke MA, Rose SD and Bao G, Nucleic Acids Res, 2003, 31, 1319–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.