Abstract
The recent technological advancement in digital pathology has enabled 3D tissue-based investigation of human diseases at extremely high resolutions. Discovering and verifying spatial patterns among massive 3D micro-anatomic biological objects such as blood vessels and cells derived from 3D pathology image volumes plays a pivotal role in understanding diseases. However, the exponential increase of available 3D data and the complex structures of biological objects make it extremely difficult to support spatial queries due to high I/O, communication and computational cost for 3D spatial queries. In this demonstration, we present our scalable in-memory based spatial query system iSPEED for large-scale 3D data with complex structures. Low latency is managed by storing in memory with progressive compression including successive levels of detail on object level. On the other hand, low computational cost is achieved by pre-generation of global spatial indexes in memory and additional on-demand generation of indexing at run-time. Furthermore, iSPEED applies structural indexing on complex structured objects in multiple query types to gain performance advantage. During query processing, the memory footprint of iSPEED is minimal due to its indexing structure and progressive decompression on-demand. We demonstrate iSPEED query capability with three representative queries: 3D spatial joins, nearest neighbor and spatial proximity estimation on multiple datasets using a web based RESTful interface. Users can furthermore explore the input data structure, manage and adjust query pipeline parameters on the interface.
PVLDB Reference Format:
Hoang Vo, Yanhui Liang, Jun Kong, and Fusheng Wang. iSPEED: a Scalable and Distributed In-Memory Based Spatial Query System for Large and Structurally Complex 3D Data.
1. INTRODUCTION
3D spatial data has presence in a wide range of industrial applications, for instance, urban planning, 3D mapping and navigation, terrain modeling, environmental assessments. Managing and analyzing large amount of 3D spatial data to derive values and guide decision making have become essential to business success and scientific discoveries. The emergence of 3D spatial data is driven by not only applications but also scientific applications that are becoming increasingly data- and compute-intensive.
1 In the past decade, digital pathology has grown to high popularity in advancing biomedical research. The very first step in quantitative 3D pathology imaging is to extract micro-anatomic objects such as blood vessels, cells and nuclei, together with their associated features using 3D registration, segmentation and reconstruction [5, 6]. The following step is to explore spatial relationships among a massive number of such 3D objects, to discover spatial patterns and their correlations with disease progression. For instance, in brain tumor studies, we would like to measure the distances from cells to their nearest neighboring tumor vessels. As another example, we would like to use a containment query to identify only cells of interest contained in a blood vessel or within certain distance from the vessel. Therefore to support efficient spatial queries and analytics on the 3D objects, we need to address four major challenges: 1) the “big data” challenge due to explosion of 3D data, 2) the complex 3D object representation, and 3) the high geometric computation complexity.
Explosion of 3D Data.
Current digital microscopy scanners generate images with extremely high resolution. While a typical 2D microscopy image may contain 100,000 × 100,000 pixels, with a million micro-anatomic objects. A typical 3D tissue volume may generate hundreds of slices, and contain tens of millions of 3D biological objects, with each object represented with hundreds to thousands of mesh facets. A typical study may involve hundreds of patients and contain hundreds of tissue volumes. This unprecedented scale of 3D objects poses significant challenges on data processing, leading to tremendous I/O, communication, and computational cost.
Complex Structures.
3D objects frequently come with complex structures. For example, blood vessels are reconstructed to capture the 3D structural variations such as bifurcations where one main vessel grows into two or more branches at multiple points. While minimal bounding boxes (MBBs) have been successfully used in traditional spatial indexing, MBBs are not effective to represent such complex 3D objects for distance based spatial queries.
Multiple Object Resolution Levels.
In practice, 3D objects are commonly represented with multiple resolutions with different Level of Detail (LOD). In digital pathology, one frequently wants to quickly visualize rough 3D shapes of blood vessels to explore spatial relationships, and another user may want to explore 3D structure details for fine calculation of vessel features. LOD in higher resolution provides more accurate results for spatial computations, but could significantly increase data volume and computation cost. Thus, a 3D spatial querying system has to balance between accuracy and computational cost.
High Computation Complexity.
3D spatial queries involve computationally intensive geometric operations for quantitative measurements and identifications of topology relationships. For spatial joins on large datasets, MBBs are most often useful only to identify potential intersection of convex objects. However, for actual spatial refinement, and especially queries requiring quantitative spatial measurements, such as computation of intersected volumes of objects with LOD at high resolution, geometric computation clearly dominate the majority of query cost.
To tackle the above challenges, we introduce iSPEED, an efficient and scalable in-memory based spatial query processing system for large scale 3D data. To achieve low latency, iSPEEED stores data in memory in a highly compressed form using an effective progressive compression approach that compresses each 3D object individually with successive levels of detail. To minimize search space and computation cost, iSPEED provides global spatial indexing in memory through partitioning at subspace level and partitioned cuboid level. iSPEED provides an in-memory 3D spatial query engine INTENSE, which can be invoked on-demand for running many instances in parallel.
At run time, iSPEED dynamically decompresses only required 3D objects at the specified level of detail, and generates necessary spatial indexes in-memory to accelerate query processing, such as on-demand object-level indexing and structural indexing on complex structured objects. Multiple spatial queries are supported, including spatial joins, nearest neighbor, and spatial proximity estimation. iSPEED further allows user to model 3D objects with multiple levels of detail for spatial queries and provides options to select parameters for faster queries or higher accuracy to meet application specific requirements.
Lastly, iSPEED provides a user friendly interface allowing customization of parameters and visualization of intermediate results. In Section 2.2 we describe our indexing approach and in Section 3 we discuss the in-memory spatial query engine. Finally, we describe the demonstration settings in Sec. 4
2. BACKGROUND AND OVERVIEW
Pathology Image Properties.
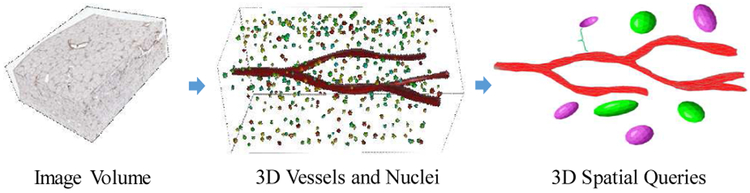
3D image analysis of whole slide image volumes produces large amount of quantifications such as 3D spatial objects and features [5]. In a typical 3D analytical pathology imaging pipeline, selected biopsies are sectioned into thin slices and mounted on physical glasses. These slides are then scanned into digital images to form 3D image volumes. With the image volume, micro-anatomic objects of interest such as blood vessels and nuclei are reconstructed in 3D models, as shown in Fig. 1. Finally, the 3D objects as well as their extracted features are managed and queried by a spatial data management system.
Figure 1:
The workflow of 3D pathology imaging with spatial queries.
Common biological objects obtained from pathology images include nuclei or cells, fats, blood vessels and ducts. While nuclei have relatively simple shapes, blood vessels and ducts could have complex structures such as bifurcations. The spatial relationships and distribution patterns among these objects play a critical role for understanding of tumor micro-environment and investigations of disease progression [2].
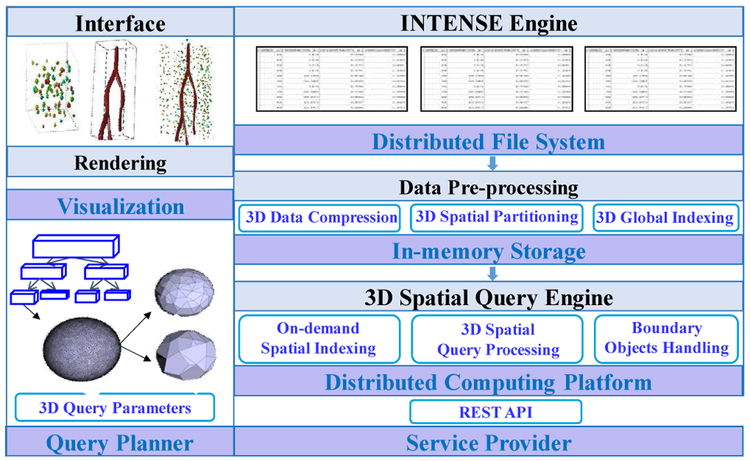
One major objective of iSPEED (in-memory spatial query system for three dimensional spatial data) is to reduce I/O and communication cost, exploit indexing techniques for complex objects to accelerate queries, and provide high scalability to run on large computer clusters or computing clouds. The architecture overview of iSPEED is shown in Fig. 2. 3D data is first staged in a distributed file system and pre-processed for compression and indexing. After effective compression, each individually compressed 3D geometric object will be stored in memory, and a master object index will be generated to track the MBB and in-memory location of each object. Pre-processing will also provide spatial data partitioning to generate partitioned cuboids which form the unit of parallel query tasks. Partitioning creates two level global spatial indexes: partitioned cuboid index to represent the MBB of all cuboids, and subspace index to group neighboring cuboids into large subspaces to form a higher level spatial index.
Figure 2:
Architecture of iSPEED.
iSPEED provides an on-demand in-memory three dimensional spatial query engine INTENSE to run query tasks. INTENSE can be invoked on-demand to run many instances in parallel. For each query task, the IDs and MBBs of the 3D objects contained in the partitioned cuboid will be identified with the master object index, and INTENSE will create an in-memory index such as R*-tree for query processing. Only at the refinement or spatial measurement step, the original geometries will be needed for geometric computations such as computing if two polyhedrons intersect or intersecting volume. INTENSE will read the compressed 3D objects from the memory and decompress the objects based on specified level of detail to feed them to the query engine.
2.1. Compression and In-Memory Storage
They key advantage factor in iSPEED is the usage of progressive compression approach to compress each 3D object individually. This is performed before actual queries, while extracting the MBB of each object and create a master object index to record the MBB and the location of each object. This serves as an additional layer of index for objects. This one time process guarantees the speed up of all subsequent queries.
3D Mesh Compression.
iSPEED uses a 3D mesh compression approach to progressively compress and decompress individual objects [7, 4]. The steps include decimation of mesh LOD, removing vertices and adding new edges, modifying entropy and maintain compressed symbol lists. Since the data compression algorithm is not lossless, high compression rate could incur potential structure distortion. We observed that on average the error rate is about 0.21% for join query. As spatial data is generated from image analysis algorithms which themselves come with errors [10], and spatial queries involve massive number of 3D objects for statistical purpose, such precision loss is negligible in practice.
In-memory Data Storage.
High effective compression enables data storage in memory, and the compression ratio depends on the structure complexity of the mesh object. Our experiments indicate that the size of the base mesh L0 is less than 1% of the total file size of the raw data, and the size of the whole compressed file with multiple LODs is only about 3% of the raw file size. For a typical 3D volume in digital pathology of 1TB in size, the final size after compression will be about 30GB, which could well fit into the memory of a compute cluster node.
While each 3D object is compressed individually, a master object index is created to track the MBB and the in-memory location of each object. Each record in the master object index is in the format of ⟨object_id, dataset_id, object_class, MBB, offset, length⟩, where object_id is a unique ID for each object within the dataset with dataset_id; object_class indicates object class such as cell or different vessel types; offset and length indicate the byte offset and length of the object in the memory respectively.
2.2. Spatial Partitioning and Global Indexing
To achieve scalability, we provide spatial partition level parallelism by spatial partitioning. We have performed extensive studies on spatial partitioning for 2D space [9]. Similarly, for 3D space, by partitioning the input data into partitioned cuboids, we can take the objects contained in each cuboid to form the processing unit for basic parallelism. In digital pathology, as the distributions of biological objects are relatively homogenous compared to geospatial data, we take multiple partitioning approaches, including fixed grid, oc-tree, sort-tile-recursive, which is most suitable for such distributions based on our experiences. The partitions can be used to form two levels of global spatial indexes: partitioned cuboid indexing based on the partitioned cuboids, and subspace indexing based on aggregation of neighboring cuboids.
iSPEED uses R*-tree for the two-level global spatial indexing. Besides the partitioned cuboid and subspace indexes, object-level spatial index is created for objects contained in each cuboid, and structural index is created for individual complex objects (Section 3). All indexes have insignificant size and are maintained in memory at pre-processing phase (partitioned cuboid and subspace indexes) or on-demand (object-level and structural indexes). The master object index is non-spatial and keeps the MBB and in-memory location of each 3D object (compressed 3D geometry).
3. IN-MEMORY SPATIAL QUERY ENGINE
INTENSE (in-memory three dimensional spatial query engine) is a standalone in-memory based query engine for iSPEED. INTENSE is generic and can be extended (Fig. 2). It leverages multiple types of indexing structures(pre-generated in memory or on-demand) to accelerate spatial queries. First, INTENSE offers parallelism with decoupled spatial query processing on individual partitions to support multiple querying pipelines with optimal spatial access methods, and provides result normalization to handle boundary crossing objects. This is critical in distributed system, as there always exists boundary issues affecting query results. Second, INTENSE performs on-demand in-memory indexing for object-level index (many objects contained in a partitioned cuboid; R*-Tree provides a relative good performance) and structural index (individual complex object such as a blood vessel).
3.1. Indexing levels
Object-level indexing.
Object-level spatial indexing provides access support objects in each partitioned cuboid to support indexed based spatial queries. For instance, joining objects from two cuboids can be supported through R-Tree based indexing. We apply an on-demand based indexing approach by creating suitable indexes for the current query at runtime. This provides much flexibility and reduces storage, with very small overhead. Our extensive profiling shows that, for data and computation intensive spatial queries such as spatial join, the overhead for index building on modern hardware is very small.
Structural Indexing.
Traditional approaches for distance based queries simplify spatial objects with points or MBBs. However, these simplifications are not applicable to complex structured 3D objects such as vessels. INTENSE provides two in-memory on-demand structural indexing to accelerate spatial queries on complex structures: topological skeleton based indexing, and hierarchical Axis-Aligned Bounding Box (AABB) tree based indexing. They are discussed in details in our previous work. [3, 4]
In spatial proximity estimation query, the accurate distance between objects (e.g., cell and vessel) needs to be computed accurately. Rather than iterating on every primitive of a vessel structure that is computationally expensive, iSPEED uses a hierarchy on the AABB tree of its primitives (facets) to minimize the traversal search space on the complex structured vessel [8].
3.2. Sample Join Workflow
Spatial join is one of the most commonly used spatial queries. For digital pathology, spatial queries could be used to determine relationships of different types of biological objects such as containment relationship. One particular query type is spatial cross-matching, to compare or consolidate results of segmented and reconstructed 3D objects from different algorithms.
We illustrate the general workflow of such two-way spatial join. Here we take a filter-and-refine strategy to reduce the computational cost of spatial predicate on 3D geometries. After identifying the objects with the same cuboid_id from two datasets, iSPEED builds a spatial index in bulk on one dataset (here dataset2) to generate an object-level index, and here we use 3D R*-tree [1]. Following a distributed refinement strategy, for every 3D object in dataset1, we first query its MBB on the R*-tree as a rough filtering step to eliminate objects pairs with no MBB intersection. For those candidates with MBB intersection, we load their compressed data from main memory and perform geometry decompression at specified level of detail to obtain polyhedrons. Then we perform spatial refinement step on the polyhedron pairs through 3D geometric operations. Other spatial operators such as overlaps and touches can also be processed similarly and can be found in our previous works[3].
3.3. Query Task Parallelization
iSPEED provides a streamlined and parallel querying pipelines for efficiency and scalability. With partitioned cuboids, iSPEED can run multiple query tasks through distributed computing paradigms such as Hadoop or Spark on commodity clusters. In our implementation and demonstration, we use MapReduce for the query task parallelization. All other components are written exclusively to support the best performance and eliminate redundancy.
4. INTERFACE AND DEMONSTRATION
Interface.
While the engine INTENSE is implemented with C++, using open source libraries, including the Computational Geometry Algorithms Library (CGAL), and SpatialIndex extended for 3D R*-tree support, the front-end interface that user will be experiencing is client based and built purely on JavaScript with OpenGL support for 3D. The visualization is rendered on the client side with data pulled from the server via REST API requests. The server hosts a database and is also a node in the INTENSE Hadoop Cluster. The server forwards 3D processing requests to INTENSE, while it serves static raw geometry from HDFS and non-spatial attribute filter with assistance from the database. Non-geometric or non-spatial attributes of objects are stored on the server Post-greSQL database. The schema is hierarchical containing tables for data sets, images and individual objects. Attributes are indexed by column, providing very fast support when querying.
Demo Queries.
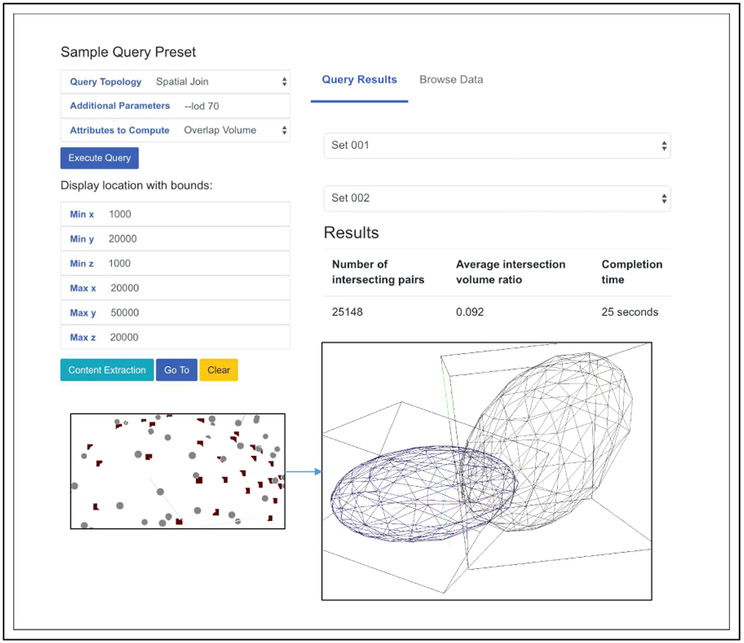
In our demo, we provide users with options to browse, explore data visualization or perform analytics to obtain query statistics. The data exploration interface contains an interacting canvas. Users can view various 3D micro-anatomic at different levels of details to visualize the complexity. For analytics users can select an option from a set of pre-defined queries, including spatial join and nearest neighbor queries on different datasets. Additional query predicate and parameters can be entered, such as LOD value to speed up query processing. The results will be returned and displayed to users when the query finishes. Currently we provide a limited support to result browsing due to the large volume of data that needs to be displayed (Fig. 4); Users have the ability to select a spatial region and explore objects within them. A typical query is comparing intersection volume ratio between two sets of machine-segmented nuclei; another is obtaining an average 3-nearest distance as a metric for density of nuclei along blood vessels.
Figure 4:
iSPEED Interface.
Demonstration Setup.
Three datasets from analytical pathology imaging are used for system performance evaluation. The 3D objects including nuclei (cells) and blood vessels are derived from 3D image volumes with different number of slides, and have been validated and represented in OFF format. We have multiple dataset sizes at 10GB, 40GB, 80GB and 500GB.
The performance of iSPEED is demonstrated on a remote cluster environment. The cluster has five nodes with 124 cores (Intel(R) Xeon(R) CPU E5-2650 v3 at 2.30GHz). Each node comes with 5TB hard drive at 7200rmp and 128GB memory. Cluster nodes are connected via a 1Gb network. The 3D datasets are uploaded in Hadoop Distributed File System.
Scenarios.
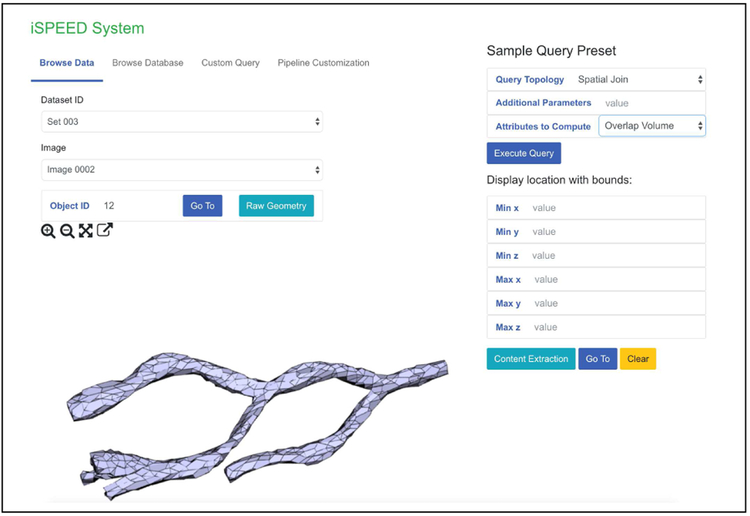
For demonstration, we explore three representative data- and compute- intensive 3D spatial queries: spatial joins, nearest neighbor, and spatial proximity estimation. We also provide user with the ability to explore and visualize the structural complexity of the original input data as in Fig. 3. The canvas like environment allows examination of individual object as well as specific image region. More complex query can also be generated and customized via the web base interface.
Figure 3:
iSPEED Interface.
5. ACKNOWLEDGEMENTS
The research is supported in part by grants from National Science Foundation ACI 1443054 and IIS 1350885, National Institute of Health K25CA181503, and the Emory University Research Committee.
Footnotes
The work was conducted while Dr. Liang was at Stony Brook. Her current affiliation is Google Inc.
Contributor Information
Hoang Vo, Stony Brook University, Stony Brook, NY, USA.
Yanhui Liang, Stony Brook University, Stony Brook, NY, USA.
Jun Kong, Emory University, Atlanta, GA, USA.
Fusheng Wang, Stony Brook University, Stony Brook, NY, USA.
6. REFERENCES
- [1].Arge L, Procopiuc O, Ramaswamy S, Suel T, Vahrenhold J, and Vitter JS. A unified approach for indexed and non-indexed spatial joins In International Conference on Extending Database Technology, pages 413–429. Springer, 2000. [Google Scholar]
- [2].Charles NA, Holland EC, Gilbertson R, Glass R, and Kettenmann H. The brain tumor microenvironment. Glia, 60(3):502–514, 2012. [DOI] [PubMed] [Google Scholar]
- [3].Liang Y, Vo H, Aji A, Kong J, and Wang F. Scalable 3d spatial queries for analytical pathology imaging with mapreduce In The 24th ACM SIGSPATIAL GIS, page 52 ACM, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Liang Y, Vo H, Kong J, and Wang F. ispeed: an efficient in-memory based spatial query system for large-scale 3d data with complex structures In Proceedings of the 25th ACM SIGSPATIAL International Conference on Advances in Geographic Information Systems, page 17 ACM, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Liang Y, Wang F, Treanor D, Magee D, Teodoro G, Zhu Y, and Kong J. A 3d primary vessel reconstruction framework with serial microscopy images In MICCAI 2015, pages 251–259. Springer, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Liang Y, Wang F, Treanor D, Magee D, Teodoro G, Zhu Y, and Kong J. Liver whole slide image analysis for 3d vessel reconstruction In ISBI 2015, pages 182–185. IEEE, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Maglo A, Courbet C, Alliez P, and Hudelot C. Progressive compression of manifold polygon meshes. Computers & Graphics, 36(5):349–359, 2012. [Google Scholar]
- [8].Terdiman P. OPCODE 3D collision detection library, 2005. [Google Scholar]
- [9].Vo H, Aji A, and Wang F. Sato: a spatial data partitioning framework for scalable query processing In The 22nd ACM SIGSPATIAL GIS, pages 545–548. ACM, 2014. [Google Scholar]
- [10].Wang F, Kong J, Gao J, Cooper LA, Kurc T, Zhou Z, Adler D, Vergara-Niedermayr C, Katigbak B, Brat DJ, et al. A high-performance spatial database based approach for pathology imaging algorithm evaluation. Journal of pathology informatics, 4(1):5, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]