Abstract
Post-transcriptional regulation of gene expression by RNA binding proteins (RBPs) and non-coding RNAs plays an important role in global gene expression. Many post-transcriptional regulators are misexpressed and misregulated in cancers, resulting in altered programs of protein biosynthesis that can drive tumor progression. While comparative studies of several RBPs and microRNAs expressed in various cancer types have been reported, a model system that can be used to quantify RBP regulation and functional outcomes during the initiation and early stages of tumorigenesis is lacking. It was previously demonstrated that oncogenic transformation of normal human cells can be induced by expressing hTERT, p53DD, cyclin D1, CDK4R24C, C-MYCT58A and H-RASG12V. Here we describe a user-friendly method for generating this genetically defined model of step-wise tumorigenesis beginning with normal donor-derived human cells. This method immortalizes a donor’s normal cells in about a week, reducing the chances of senescence. The entire stable system can be established in less than 12 weeks. We then demonstrate the utility of such a system in elucidating the expression of multiple RBPs at an early step of tumor formation. We identify significant changes in the expression levels of transcripts encoding RBPs prior to transformation, suggesting that our described donor-derived isogenic system can provide insight about post-transcriptional regulation during the earliest stages of tumorigenesis in the context of diverse genetic backgrounds.
Keywords: Post-transcriptional regulation, RNA binding proteins, RNA regulons, Tumorigenesis, Isogenic models
1. Introduction
Precise control of gene expression is achieved through regulation at both the transcriptional and post-transcriptional levels. While many studies focus on steady state mRNA levels to measure gene expression, it has become increasingly clear that this does not provide an accurate picture of the complex regulatory features of gene expression occurring within a cell. Post-transcriptional regulation (PTR) of mRNA expression is controlled and coordinated by trans-acting RNA binding proteins (RBPs), and also non-coding RNAs, that bind to cis regulatory elements contained in nascent transcripts [1]. RBPs coordinate functionally regulated mRNAs into RNA regulons, allowing for combinatorial changes in PTR expression in response to perturbations [1]. Tightly regulated PTR events include splicing, nuclear export, localization, mRNA stability and ultimately translation [1–4]. Large numbers of mRNAs undergo PTR, as evidenced by the fact that steady state protein levels do not always correlate with steady state mRNA levels [5–9]. RBPs in eukaryotes have been estimated to outnumber DNA binding proteins [10,11], are highly expressed when compared to any other class of genes [12], and are, as a class, more conserved than transcription factors in metazoans [13], highlighting the importance of PTR.
Many recent studies have shown that PTR is an important determinant of gene expression in both normal cellular processes and pathological states. It is now widely recognized that RBPs robustly influence cancer-related gene expression patterns, as they regulate many mRNAs encoding proto-oncogenes, growth factors, cytokines, and cell cycle regulators [14,15]. Cancer has traditionally been viewed as being driven by aberrant transcriptional regulation and signaling events, though, over the past several years, many RBPs have emerged as critical players in tumor development [14,16]. Global studies have identified many RBPs that are significantly misexpressed in tumors compared to normal tissues [12,17], and several studies have suggested that RBPs dynamically and differentially regulate target mRNAs in different states and contexts [18–22]. However, cancers are derived from normal cells that often evolve step-wise and progressively to a neoplastic state, and the involvement of PTR in this progression has not been looked at in the context of tumor initiation and step-wise progression. Thus, more studies are needed in order to fully understand the PTR regulators and downstream genetic programs activated by cancer driver mutations that coordinate tumor origins, evolution and progression.
Studies of tumorigenesis typically involve the use of cancer cell lines. While cell line models have been informative and have demonstrated the importance of transcriptional and post-transcriptional control of cancer [18,19,23], they typically involve cell lines that are already immortalized, and thus they may already be well along the path to becoming a cancer cell. For example, it is common to set up an isogenic system of late stage tumor formation by adding an oncogene, such as H-RASG12V, to an already immortal ized cell line, for example, MCF10A human mammary epithelial cell line. However, one major drawback of such an isogenic system is that it cannot be used to study the earliest stages of oncogenesis, the process by which a normal cell becomes progressively tumorigenic, since there are no normal isogenic primary cells to compare. Likewise, comparisons between patient tumor tissues and normal matched tissues do not provide information about the intermediate stages of transformation. In addition, certain cancer cell lines, such as the ever-popular MCF7 breast cancer cell line, have been demonstrated to evolve and adapt differently over time [24]. Thus, findings from one laboratory may not be replicable in the same cell line in another laboratory due to accumulated mutations in these lines. While mouse models can help to illuminate genetic alterations that lead to cancer, many alterations characteristic of human cancers do not yield the same cancers in mice [9,25,26].
Seminal work in Robert Weinberg’s laboratory demonstrated that normal human epithelial and fibroblast cells are converted to a tumorigenic state through the combined ectopic expression of hTERT, oncogenic HRASG12V, and the Simian Virus 40 (SV40) large and small t tumor antigens [27]. Expression of these transgenes systematically establishes: 1) telomere maintenance, 2) unlimited replication potential and, 3) complete oncogenic transformation. Importantly, this work identified minimal intracellular pathways whose disruption is sufficient for creation of a human tumor. Subsequent work in Chris Counter’s laboratory expanded on these findings, and identified a core set of proteins that together disrupt the same pathways as the Weinberg SV40 system to drive the process of tumorigenesis [8,28]. In this system, the introduction of hTERT, p53DD, cyclin D1, CDK4R24C and c-MYCT58A immortalizes the cell, while subsequent expression of HRASG12V converts the cell to a fully tumorigenic state. The elimination of proteins of viral origin better recapitulates what happens in human tumors, since most cancers are not caused by viruses, and introduction of viral proteins into human cells has been shown to significantly alter other physiological and biological properties [29]. Using this system, well-defined genetic models of tumorigenesis can be established for most normal human cells, allowing for the investigation of the earliest stages of transformation in a variety of tumor types.
An important aspect of pharmacogenomics concerns differences in genotype and gene expression among individual patients that vary in drug responses. To this end, we believe that a donor derived, genetically defined, model offers an advantage for taking into consideration individual genetic differences in preclinical cancer studies. Thus, rather than using standard cancer cell lines as pre-clinical models, establishing multiple donor derived isogenic lines in which to conduct initial studies could be highly beneficial. Despite this benefit, relatively few laboratories have taken advantage of the cell systems developed by the Weinberg and Counter laboratories. Unfortunately, these cell lines were not available to be shared between laboratories due in part to the fact that primary cells have extremely limited passage numbers and are in short supply. While the immortalized and transformed lines could be shared if available, the main advantage of this system, the comparison to primary cells, would be lost. In addition, if one is to make comparisons between different individuals of various genetic backgrounds, multiple primary cells from several donors must be obtained and step-wise transformed. Therefore, the cell lines must be engineered by the laboratory wishing to use them.
The process of engineering a stable isogenic system in the absence of viral genes has been described [8,9]. We have simplified this method, as detailed in this paper, with a few useful modifications to streamline the numerous steps of transfection and infection involved in establishing this system. Additionally, we have determined that for cell system expansion, which is necessary for the application of many methods of global analysis, modifications of the cell culture media are necessary. Furthermore, we have cultured this system in the absence of selective pressures for over 20 passages, and demonstrated that cells still robustly express the essential transgenes. These modifications allow expansion and application of various published methods to study post-transcriptional regulation during the earliest stages of tumorigenesis. We demonstrate large changes in expression of RBPs during the immortalization step of normal human primary cells, many of which have been previously identified to play a role in cancer formation. It is important to note that we did not observe additional changes in expression levels of transcripts encoding RBPs during subsequent RAS transformation. Therefore, this isogenic model system can provide insights into how RBPs can affect global RNA regulation that are otherwise lost using standard immortalized cell lines.
2. Materials and methods
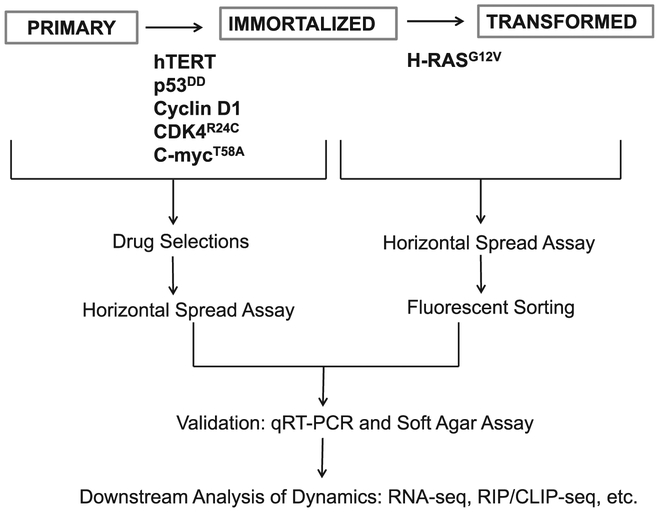
A workflow of the procedure for establishing an isogenic model is outlined in Fig. 1. Here we detail a user-friendly, step-by-step procedure for establishing and validating this model. This method makes a few minor modifications from the methods described by Chris Counter’s laboratory for the sake of simplification [8,9]. This protocol is optimized for human mammary epithelial cells (HMECs); however this method can be adapted to virtually any available normal human cell. It is critical to start with cells that are not yet immortalized.
Fig. 1.
Workflow to establish and validate a stepwise isogenic system of tumorigenesis. The procedure begins with expression of hTERT, p53DD, cyclin D1, CDK4R24C and c-MycT58A in normal human primary cells to generate immortalized, intermediary cells. Immortalized cells are subsequently transformed with HRASG12V. All transfections are done through generating amphotropic retroviruses in a 293T packaging cell line and infecting the cells of interest. Stable cells are selected through either antibiotic selections or fluorescent sorting. Cells are then checked with a horizontal spread assay to ensure they do not shed recombinant retrovirus. Transgene expression and cell phenotype is then validated with qRT-PCR and soft agar assays. Once verified, many global gene expression studies, drug tests, and post-transcriptional assays can be carried out in this system.
2.1. Materials
2.1.1. Cell lines and cell culture reagents
HEK 293T/17 cells (ATCC)
HEK 293 cells (ATCC); or any cell line that is not resistant to antibiotics listed
Low passage primary cells of choice; in this paper, we use HMECs obtained at p9 (Lonza)
DMEM (Thermo Fisher Scientific) +/− 10% FBS
Primary cell specific media (for HMECs, MEGM (Lonza))
Appropriate cell culture reagents (for HMECs, reagent pack (Lonza))
2× MEM (Thermo Fisher Scientific)
2.1.2. Plasmids
Plasmids to make this system are obtained from multiple published sources. As with all plasmids, we highly recommend sequence validating before use. Alternatively, all pBabe empty vector backbones with all appropriate selection markers are publicly available from Addgene, and one could easily clone vectors of interest
p0467/pCL-10A1 viral packaging plasmid (gift of Dr. Chris Counter; can be obtained from Novus Biologicals, catalog #NBP2–29542)
pBabe-hygro-hTERT (addgene plasmid #1773, gift of Dr. Bob Weinberg [30])
pBabe-puro-cyclinD1-HA (addgene plasmid #9050, gift of Dr. William Hahn)
pBabe-YFP-FLAG-HRASG12V (gift of Dr. Chris Counter [27])
pBabe-GFP control (addgene plasmid #10668, gift of Dr. William Hahn)
2.1.3. Primers
Sequences:
GapDH F 5′-CAT GTT CGT CAT GGG TGT GAA CCA-3′ R 5′-AGT GAT GGC ATG GAC TGT GGT CAT-3′
pBabe hTERT F 5′-GAG GTG CAG AGC GAC TAC-3′ R 5′-GCT GTT CAC CTG CAA ATC CA −3′ [8]
pBabe p53DD F 5′GCT CAC TCC AGC TAC CTG AAG A 3′ R 5′ TCC ACA CCC TAA CTG ACA CAC A 3′
pBabe cyclinD1 F 5′-CCC AGC AGA ACA TGG ACC C R 5′-TTC TGC CTG CTG GGG AG
pBabe CDK4R25C F 5′ GAC TGG CCT CGA GAT GTA T-3′ R 5′-TAC TTC TGC CTG CTG GGG-3′ [8]
pBabe C-MycT58A F 5′ ACT CGC TGC TGT CCT CCG A R 5′ GAG TGA GAC GTG GCA CCT CTG A
pBabe HRASG12V F 5′-GCA TCC CCT ACA TCG AGA-3′ R 5′-TAC TTC TGC CTG CTG GG-3′
2.1.4. Transfection reagents
FuGene-6 (Roche #1814443)
Polybrene (hexadimethrine bromide) (Sigma-Aldrich #H9268)
2.1.5. Antibiotics
Hygromycin
G418/Geneticin
Puromycin
Zeocin
Blasticidin
2.1.6. Other
0.45um acrodisc syringe filter with HT tuffryn membrane (VWR #28144–007)
10 ml syringe
10 cm tissue culture plates
1.5 ml tubes
6-well tissue culture plates
Real-time PCR reagents of choice (we use Sybr Green (Thermo Fisher Scientific))
1.8% agarose in sterile water, autoclaved Liquid Nitrogen
2.2. Immortalization of normal human primary cells (~9 days)
Day 1: Split 293T/17 cells into 6 10 cm plates so that they will be ~30–40% confluent the next day
Day 2: Set up 7 × 1.5 mL tubes and mix the following in order:
X μl serum free media (to 200 μl)
12 μl FuGENE-6
3 μg p0467/pcl-10A
3 μg pBabe plasmid containing transgene of interest (one tube for each transgene plus GFP control)
Tap gently to mix, Incubate at room temperature for 15–30 min, Drip slowly onto 293T/17 cells, and incubate overnight.
Day 3: Repeat transfection of 293T/17 cells from Day 2. Incubate at least 8 h, and then replace media with media specific to the cells to be infected (for example, for HMECs, add MEGM). Allow cells to incubate in this new media for 48 h.
NOTE: At this point, the 293T/17 cells produce Amphotropic retrovirus. Please follow your institute’s guidelines for working with amphotropic retroviruses.
Day 4: Make sure that primary cells to be transfected will be 25–30% confluent the next day.
Day 5: Collect media from 293T/17 cells and filter through a sterile 0.45um Acrodisc with an HT tufftyn membrane. Snap freeze all media, except the one containing pbabe-hTERT, in liquid nitrogen and store at −70 °C- −80 °C. Alternatively, one can set up the experiment to do one transfection at a time, and use the virus media fresh. To troubleshoot, use media fresh instead of freezing or measure viral titers.
To the pBabe-hTERT media, add polybrene to a final concentration of 4μg/ml and mix gently. Replace the medium from the cells to be infected with 6 ml of the filtered media plus 2 ml of fresh media and incubate. Add fresh MEGM (or appropriate media) to 293T/17 cells.
12 h later: Repeat Day 5 protocol. Discard 293T/17s.
Day 6: 12–24 h after the last infection, thaw media containing the pBabe-p53DD viruses at 37 °C. Use the media immediately to infect the cells as described for Day 5. Primary cells will be slightly more confluent than the previous day, but should not need to be split.
Alternatively, if not freezing the viral media, new 293T/17s should be transfected on days 3 and 4 so that they are ready for harvest and infections on Day 6. If using the media fresh and not freezing, continue with this pattern for the rest of the plasmids.
Days 7–9: Repeat infections for all other transgenes except HRASG12V.
By the last infection day, primary cells will have been in culture for about one week. Some primary cells may need to be split if they reach a high confluency within this time period. However, many are slow growing and will not need to be split during the week of infections. If desired, cells can be split and saved at any step during the infections if one is interested in generating more intermediary cells.
Note: Once cells are immortalized/ transformed, required media conditions may change. For example, while primary HMECs grow best in serum-free media, we’ve found that immortalized and transformed HMECs require a minimum of 5% serum to grow in a monolayer. We routinely culture these cells in 10% FBS. Immortalized and transformed cells grow in spheroids under serum-free conditions (data not shown).
2.3. Generation of stable cell lines (~60 days)
We use the following selection criteria for immortalized HMECs, but we suggest setting up drug response curves for antibiotic selection to determine proper concentrations for your cells of interest. Cells are typically allowed to recover for 5 days between drug selections.
Hygromycin: 80 μg/ml for 7 days
G418: 250 μg/ml for 10 days
Puromycin: 0.5 μg/ml for 5 days
Zeocin: 800 μg/ml for 8 days
Blasticidin: 4.5 μg/ml for 7 days
2.4. Horizontal spread assay (~8–13 days)
Cells must be tested to be sure they do not shed recombinant virus.
Day 1: Split 293 cells (or any cell line that does not contain antibiotic resistance markers) into 3 × 6-well plates at a sparse density (we usually use a 1:20 split for 293 cells). Split immortalized and transformed cells 1:4 into 2 × 10 cm plates each and 1:12 1 × 6-well plates each.
Day 2: Collect the media from the 10 cm plates of immortalized and transformed cells, filter through a 0.45um acrodisc filter and add 4 μg/μl polybrene. Wash one of the plates of 293 cells 2× with PBS, and then add 3 ml of immortalized and transformed conditioned media to 1×6-well plate each. Use the other plate of 293 cells as a negative control and the 6-well plates of immortalized and transformed cells as positive controls- to each of these plates, add fresh media supplemented with 4 μg/ml polybrene.
Day 3: 24 h later, replace all wells with fresh, cell specific media.
Day 4: 24 h later, replace all wells with fresh media supplemented with selection antibiotics. Use 1 well from each plate to select with hygro, neo, puro, bleo and blast at the same concentration used to make stable cell lines. Leave the 6th well antibiotic free.
Day 7: Replace with fresh selection media. Replace media every 3 days as needed until:
Positive Controls (Immortalized and Transformed) are confluent.
Negative Controls (293 cells not treated) are all dead.
Experimental Cells (293 treated) are all dead and well left antibiotic free is visually assessed and YFP-. If all cells are dead, then the immortalized and transformed cells do not produce virus. If cells are viable, then immortalized and transformed cells should be discarded, as they produce recombinant virus.
2.5. Generation of transformed cells (~7 days)
Repeat the infection protocol as described on Day 5, infecting the immortalized cells with the HRAS or GFP control viral media. Perform the horizontal spread assay, using fluorescence as readout. Sort cells by flow cytometry to enrich for fluorescent positive cells.
2.6. Validation of transgene overexpression (~1 day)
Harvest RNA from all cell lines and reverse transcribe to cDNA. Perform qRT-PCR with transgene specific primers (listed above) to test for transgene overexpression.
2.7. Soft agar Assay: Anchorage independent growth (~3 weeks)
For each cell line to be tested, combine 0.75 mL of 2× MEM with 0.25 mL sterile water, and incubate at 37 °C for 15–20 min. Add 0.5 mL of 1.8% agarose (to 0.6%) and plate in one well of a 6-well plate. Allow agarose to solidify at 37 °C for 30–60 min. For each cell line to be tested, resuspend 25,000–50,000 cells in 0.5 ml sterile water. Combine with 0.75 mL of 2× MEM and 0.25 mL melted 1.8% agarose (cooled to at least 42 °C before adding), and plate 1.5 mL on top of the bottom agarose layer. Incubate at room temperature for 30 min before moving to a 37 °C incubator. The next morning, add 1× media. Continue adding 1× media every few days as needed to prevent drying. Image wells after about 2–3 weeks.
3. Results
3.1. Isogenic system validation
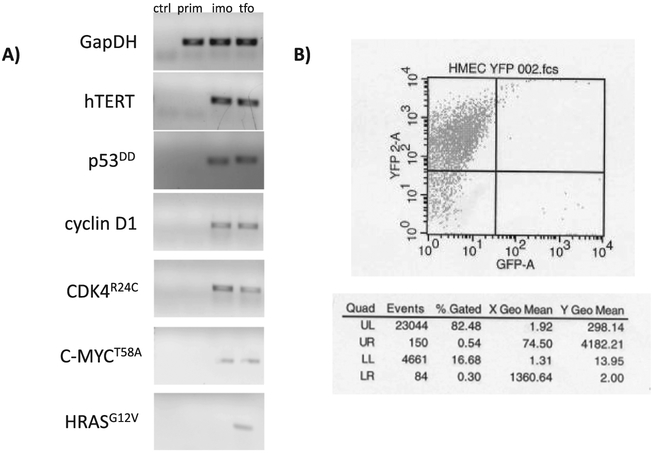
After generating this system from normal HMECs as described in section 2, we confirmed expression of all transgenes with quantitative real-time PCR. Cells were retested for transgene expression after being maintained in selection-free media for 20–25 additional passages post-selection, and all transgenes were still expressed (Fig. 2A). As an additional verification, transformed cells are over 80% fluorescent at p46 (Fig. 2B). This confirms that, once stable cells are made, cells continue to express the transgenes of interest when selective media is removed. Therefore, one does not need to maintain these cells in selective antibiotics once proper selection is carried out. This is likely due to the fact that all trans-genes confer a growth and survival advantage to cells.
Fig. 2.
Confirming expression of transgenes. A) All transfections were validated by transgene confirmation using quantitative RT-PCR for the prim, imo, and tfo cell lines, and a “no template” control (ctrl). The imo and tfo cells were maintained in antibiotic-free media for 20–25 passages before reconfirming transgene expression. The final PCR products are shown. B) Transformed cells were sorted using FACS at p46. As seen in the table, the selected cells maintained over 80% fluorescence.
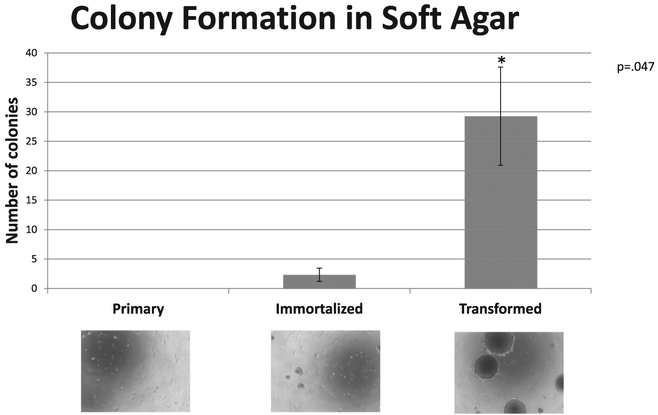
We used soft agar assays, an in vitro proxy for tumor forming potential, to confirm that only transformed cells exhibit anchorage independent growth, (Fig. 3). Likewise, this assay was performed initially and then repeated at 20–25 additional passages. Results were replicated, indicating that the phenotype is stable over time.
Fig. 3.
Confirmation of tumorigenic phenotype in an isogenic system. Soft agar assays were used to test for anchorage independent growth. Representative images of cell fields are shown to scale for each of the three cell lines. The data were quantified by photographing 4 fields of 4 independent experiments. The mean and standard deviations of the 4 fields were calculated as number of colonies per field as shown in the graph. These images confirm that the RAS-transformed cells form large spheroidal foci, the expected phenotypic property.
3.2. Evidence that PTR is important during tumor initiation
In order to determine if this system could be useful as a model for studying PTR during the early stages of tumorigenesis, we assessed the expression dynamics of RNA binding proteins in this system using RNA-sequencing. Total RNA was extracted from each cell type, and sequencing libraries were made using Epicentre’s Scriptseq v2 kit with 16 amplification cycles. Samples were sequenced on an Illumina Hi-Seq 2000 (one replicate; 125 bp PE) and an Illumina Hi-Seq 4000 (two replicates; 150 bp PE) (Duke University Genomics Core). There were approximately 60–80 million reads per library. Reads were mapped to the human genome (hg19) using TopHat2, and significant changes in gene expression patterns were determined using Cufflinks/Cuffdiff.
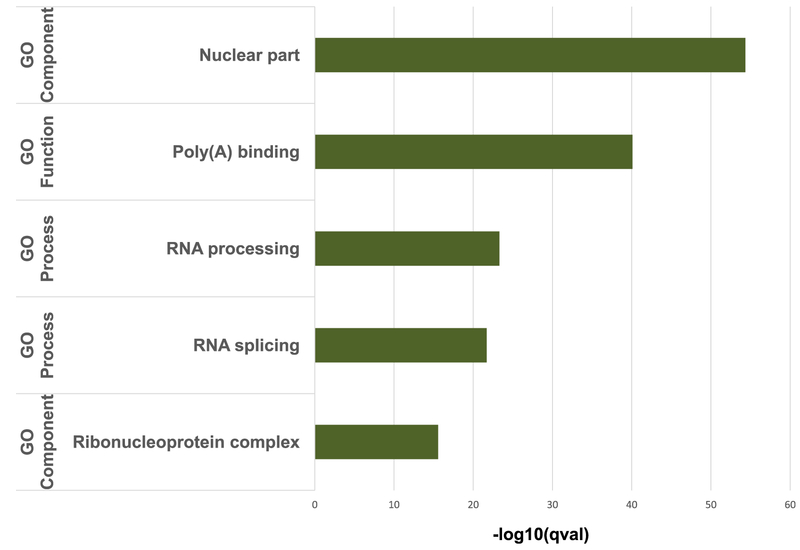
GO category analysis revealed significant enrichment in upregulated genes for post-transcriptional processes, including nuclear part, polyA binding, RNA processing, RNA splicing and Ribonucleo-protein complex during immortalization (Fig. 4). These changes were significant during immortalization but not during transformation.
Fig. 4.
HMEC immortalization increases RNA expression of RBPs. GO category analysis of RNA-seq data shows significant changes in RNA regulatory processes during immortalization. Horizontal bars indicate statistically significant increases during the transition from primary HMECs to immortalized HMECs. These changes in expression were not observed during RAS transformation.
To see if our findings could be have potential clinical significance, we looked for expression levels of RBPs identified by two recent in silico studies as over-expressed in a variety of patient tumors compared to normal tissues [12,17]. We combined published lists of RBPs that were strongly upregulated across 9 different cancers profiled in the TCGA database compared to normal tissues sequenced as part of the Human BodyMap 2.0 project [12], or that were significantly upregulated specifically in breast tumors compared to normal breast cells as determined using SAGE data and ONCOMINE microarray datasets [17]. The sum of these two data sets was 58 significantly upregulated RBPs. We then filtered this dataset for those RBPs that had a mean FPKM value greater than 1 for all three conditions across all of our RNA-seq replicates. This list consisted of 46 expressed RBPs, 26 of which had significant changes in mRNA expression during immortalization and none of which changed significantly during transformation (Fig. 5).
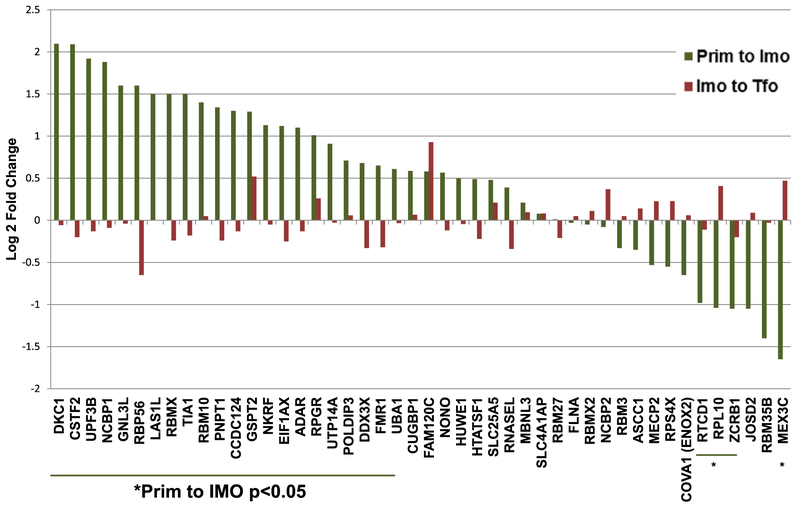
Fig. 5.
RBPs previously demonstrated to be upregulated in patient tumors show statistically significant expression changes during immortalization, but not transformation. Log 2-fold change mRNA levels during immortalization (Prim to Imo) and transformation (Imo to Tfo) for a subset of RBPs found in previous studies to have significant RNA expression changes in tumor samples compared to normal tissue [12,17].
Many of the RBPs identified as being alternatively expressed in tumors versus normal tissues have previously demonstrated roles in cancer. For example, TIA1 promotes stress granule assembly and has several putative cancer-relevant targets [33,34]. CSTF2, an alternative polyadenylation factor, is upregulated, and it has been suggested that cancer cells exhibit high incidences of alternative polyadenylation [35]. Interestingly, mRNA encoding the ADAR RNA editing enzyme is upregulated as well. A-to-I editing is highly prevalent in tumor cells compared to normal cells, and ADAR silencing reduces proliferation and increases apoptosis [36].
In addition, we identified many other RBPs that have altered RNA expression in immortalized cells compared to primary cells (Fig. 4). Notably, we found ESRP1, an epithelial splicing factor known to be suppressed in metastatic cells [37], to be significantly downregulated. In contrast, Sam68/KHDRBS1, an RBP that regulates a cancer-promoting alternative splicing program [16], is significantly upregulated. Other examples of RBPs previously demonstrated to be cancer-promoting that are upregulated in immortalized cells include ELAVL1 [14], LARP1 [38], and hnRNPM [39].
Our data suggest that RBP expression differences that are important in human tumors can occur early in the process, before transformation. These RBPs are therefore high priority candidates for future studies of tumor initiation. It is likely that previous studies that used model systems that begin with immortalized cell lines rather than primary cells for comparison could fail to identify important changes in RBP dynamics that regulate tumor initiation and progression.
4. Discussion
The earliest stages of oncogenesis that unleash the processes that eventually convert a normal cell to become tumorigenic are the most difficult to study. Here we describe a streamlined proto col for establishing and validating a donor derived isogenic model of the early stages of tumor initiation using HMECs. This model should be adaptable to most types of human cells, allowing for the study of the early stages of many different tumor types for which normal primary cells are available. When selecting primary cells of origin, the amount of material that will be needed in downstream applications should be taken into consideration. Primary cells must be able to divide several times in cell culture if one is to produce enough material for downstream methods such as RNA-seq and RIP/CLIP-seq methods. We used commercially available HMECs that had undergone post-stasis selection and had spontaneously lost expression of p16INK4 [40], allowing them to undergo several additional population doublings. When adapting this method to other primary cells, one should work out optimal cell culturing conditions and drug selections. When selecting stable cell lines, we recommend performing a dose response curve for antibiotic selection to determine optimal concentrations for the cells of interest; a paper by O’Hayer and Counter can serve as a useful starting point [9].
We intentionally do not use any viral genes to immortalize the cells, as was originally done by the Weinberg laboratory [27]. SV40 is not believed to be causative in the formation of human tumors [41,42]. Additionally, expression of SV40 has been demonstrated to induce DNA damage response pathways [43,44] and to target several host pathways in addition to the minimal perturbations needed for immortalization [45,46]. Therefore, while SV40-containing isogenic systems, such as the common HMLE lines, may be attractive to researchers due to the fact that they are readily available and relatively easy to culture, they may not accurately recapitulate in vivo changes during the onset of human tumorigenesis.
Several published studies have also utilized isogenic models without introducing viral oncoproteins to study tumorigenesis. The immortalized MCF10A cell line has been used in many labs to study changes in PTR after perturbations such as TGFb induced epithelial to mesenchymal transition (EMT) or RAS transformation. These studies have been useful for revealing important mechanisms of PTR. For example, Joel Neilson’s laboratory recently uncovered a novel role for CELF1 in EMT. CELF1 mediates the translation of a subset of mRNAs that are necessary and sufficient for EMT [23]. However, these studies are limited to understanding PTR during later stages of tumor progression, as these cell line models do not allow direct comparison with the normal primary source. Our finding that the most significant changes in genes encoding RBPs are observed during the transition step from normal to immortalized, not during the step to transformation, suggests that PTR plays an important role in the very early stages of tumor formation; i.e. before a cell has the ability to form a tumor. These changes could be missed in many other standard cell line models.
Numerous diseases, including cancers, have been associated with altered expression and functions of RBPs [15,16,47]. We previously estimated there are approximately 1500 RBPs in mammalian cells [11]. Recently, there has been significant progress made towards mass discovery of RBPs, and over 800 have been identified to date[47]. Additionally, efforts have been made to understand and quantify dynamic changes in RBP expression, targeting and regulation in dynamic settings, including disease states, stress conditions and drug responses [20,22,47–51]. The donor-derived isogenic system described in this paper, when combined with next-generation sequencing, is a valuable approach for identifying pertinent RBP expression changes occurring during biologically dynamic states that are otherwise difficult to identify. Many RBPs experimentally determined to be involved in tumorigenesis were initially identified due to aberrant expression levels in cancers [17]. Therefore, the RBPs identified as being dynamic in this system (Figs. 4 and 5) open the door for future studies of PTR in tumor initiation and progression. In addition, parallel comparisons of RBP dynamics in a cohort of donor-derived isogenic cancer systems could provide insights into how patients with diverse genetic backgrounds respond differently to drugs targeting PTR.
While mRNA targets of many RBPs have been demonstrated to be dynamic under various perturbations [18,20–23], precision is of the essence. For example, detection of subtle changes in cellular expression of RBPs or RBP-mRNA interactions is challenging and does not always indicate precise functional outcomes. Therefore, comparative studies using this donor-derived isogenic system can apply methods that precisely quantify subtle changes in RBP binding sites, protein binding regions and RNA-protein interactions [48–53]. Indeed, our lab has gone to great lengths to develop quantifiable methods to identify and quantify multiple binding sites of RBPs in order to study dynamic changes in PTR [52,53]. Moreover, these approaches can be integrated with other methods to measure changes in global mRNA stability [54] and translation [55]. The combination of methods that quantify RNA-protein interactions and link them to functional outcome can provide a comprehensive picture of overall gene coordination during tumor initiation and early stage tumorigenesis. In sum, these combined approaches using the donor derived isogenic system have the potential to identify novel cancer targets and drug candidates with great precision.
Acknowledgements
We thank Matt Friedersdorf for his insights and critical reviewing of the manuscript. We also thank Kyle Mansfield, Jeff Blackinton and Cindo Nicholson for helpful discussions. This work was funded by grants from the NIH: R01CA157268, F31CA185892, and NSF: 0842621.
References
- [1].Keene JD, RNA regulons: coordination of post-transcriptional events, Nat. Rev. Genet 8 (7) (2007) 533–543. [DOI] [PubMed] [Google Scholar]
- [2].Maniatis T, Reed R, An extensive network of coupling among gene expression machines, Nature 416 (6880) (2002) 499–506. [DOI] [PubMed] [Google Scholar]
- [3].Morris AR, Mukherjee N, Keene JD, Systematic analysis of posttranscriptional gene expression, Wiley Interdiscip. Rev. Syst. Biol. Med 2 (2) (2010) 162–180. [DOI] [PubMed] [Google Scholar]
- [4].Orphanides G, Reinberg D, A unified theory of gene expression, Cell 108 (4) (2002) 439–451. [DOI] [PubMed] [Google Scholar]
- [5].Mansfield KD, Keene JD, The ribonome: a dominant force in co-ordinating gene expression, Biol. Cell 101 (3) (2009) 169–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Vogel C, Marcotte EM, Insights into the regulation of protein abundance from proteomic and transcriptomic analyses, Nat. Rev. Genet 13 (4) (2012) 227–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ideker T et al. , Integrated genomic and proteomic analyses of a systematically perturbed metabolic network, Science 292 (5518) (2001) 929–934. [DOI] [PubMed] [Google Scholar]
- [8].Kendall SD et al. , A network of genetic events sufficient to convert normal human cells to a tumorigenic state, Cancer Res. 65 (21) (2005) 9824–9828. [DOI] [PubMed] [Google Scholar]
- [9].O’Hayer KM, Counter CM, A genetically defined normal human somatic cell system to study ras oncogenesis in vivo and in vitro, Methods Enzymol. 407 (2006) 637–647. [DOI] [PubMed] [Google Scholar]
- [10].Gerber AP, Herschlag D, Brown PO, Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast, PLoS Biol. 2 (3) (2004) E79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Keene JD, Ribonucleoprotein infrastructure regulating the flow of genetic information between the genome and the proteome, Proc. Natl. Acad. Sci. USA 98 (13) (2001) 7018–7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kechavarzi B, Janga SC, Dissecting the expression landscape of RNA-binding proteins in human cancers, Genome Biol. 15 (1) (2014) R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Alie A et al. , The ancestral gene repertoire of animal stem cells, Proc. Natl. Acad. Sci. USA 112 (51) (2015) E7093–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Abdelmohsen K, Gorospe M, Posttranscriptional regulation of cancer traits by HuR, Wiley Interdiscip. Rev. RNA 1 (2) (2010) 214–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lukong KE et al. , RNA-binding proteins in human genetic disease, Trends Genet. 24 (8) (2008) 416–425. [DOI] [PubMed] [Google Scholar]
- [16].Wurth L, Versatility of RNA-Binding Proteins in Cancer, Comp. Funct. Genomics 2012 (2012) 178525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Galante PA et al. , A comprehensive in silico expression analysis of RNA binding proteins in normal and tumor tissue: Identification of potential players in tumor formation, RNA Biol. 6 (4) (2009) 426–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Mazan-Mamczarz K et al. , Post-transcriptional gene regulation by HuR promotes a more tumorigenic phenotype, Oncogene 27 (47) (2008) 6151–6163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Mazan-Mamczarz K et al. , Identification of transformation-related pathways in a breast epithelial cell model using a ribonomics approach, Cancer Res. 68 (19) (2008) 7730–7735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Mukherjee N et al. , Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability, Mol. Cell 43 (3) (2011) 327–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Papadaki O et al. , Control of thymic T cell maturation, deletion and egress by the RNA-binding protein HuR, J. Immunol 182 (11) (2009) 6779–6788. [DOI] [PubMed] [Google Scholar]
- [22].Mukherjee N et al. , Coordinated posttranscriptional mRNA population dynamics during T-cell activation, Mol. Syst. Biol 5 (2009) 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Chaudhury A et al. , CELF1 is a central node in post-transcriptional regulatory programmes underlying EMT, Nat. Commun 7 (2016) 13362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Lee AV, Oesterreich S, and Davidson NE, MCF-7 cells–changing the course of breast cancer research and care for 45 years. J Natl Cancer Inst, 2015. 107(7). [DOI] [PubMed] [Google Scholar]
- [25].Hooper ML, Tumour suppressor gene mutations in humans and mice: parallels and contrasts, EMBO J. 17 (23) (1998) 6783–6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Rangarajan A et al. , Species- and cell type-specific requirements for cellular transformation, Cancer Cell 6 (2) (2004) 171–183. [DOI] [PubMed] [Google Scholar]
- [27].Hahn WC et al. , Creation of human tumour cells with defined genetic elements, Nature 400 (6743) (1999) 464–468. [DOI] [PubMed] [Google Scholar]
- [28].Kendall SD, Adam SJ, Counter CM, Genetically engineered human cancer models utilizing mammalian transgene expression, Cell Cycle 5 (10) (2006) 1074–1079. [DOI] [PubMed] [Google Scholar]
- [29].Ouellette MM et al. , The establishment of telomerase-immortalized cell lines representing human chromosome instability syndromes, Hum. Mol. Genet 9 (3) (2000) 403–411. [DOI] [PubMed] [Google Scholar]
- [30].Counter CM et al. , Dissociation among in vitro telomerase activity, telomere maintenance, and cellular immortalization, Proc. Natl. Acad. Sci. USA 95 (25) (1998) 14723–14728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Hahn WC et al. , Enumeration of the simian virus 40 early region elements necessary for human cell transformation, Mol. Cell. Biol 22 (7) (2002) 2111–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Yeh E et al. , A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells, Nat. Cell Biol 6 (4) (2004) 308–318. [DOI] [PubMed] [Google Scholar]
- [33].Anderson P, Kedersha N, Visibly stressed: the role of eIF2, TIA-1, and stress granules in protein translation, Cell Stress Chaperones 7 (2) (2002) 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Wigington CP et al. , Post-transcriptional regulation of programmed cell death 4 (PDCD4) mRNA by the RNA-binding proteins human antigen R (HuR) and T-cell intracellular antigen 1 (TIA1), J. Biol. Chem 290 (6) (2015) 3468–3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Mayr C, Bartel DP, Widespread shortening of 3’UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells, Cell 138 (4) (2009) 673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Fumagalli D et al. , Principles Governing A-to-I RNA Editing in the Breast Cancer Transcriptome, Cell Rep. 13 (2) (2015) 277–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Warzecha CC et al. , An ESRP-regulated splicing programme is abrogated during the epithelial-mesenchymal transition, EMBO J. 29 (19) (2010) 3286–3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Hopkins TG et al. , The RNA-binding protein LARP1 is a post-transcriptional regulator of survival and tumorigenesis in ovarian cancer, Nucleic Acids Res. 44 (3) (2016) 1227–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Xu Y et al. , Cell type-restricted activity of hnRNPM promotes breast cancer metastasis via regulating alternative splicing, Genes Dev. 28 (11) (2014) 1191–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Huschtscha LI et al. , Loss of p16INK4 expression by methylation is associated with lifespan extension of human mammary epithelial cells, Cancer Res. 58 (16) (1998) 3508–3512. [PubMed] [Google Scholar]
- [41].Gazdar AF, Butel JS, Carbone M, SV40 and human tumours: myth, association or causality?, Nat Rev. Cancer 2 (12) (2002) 957–964. [DOI] [PubMed] [Google Scholar]
- [42].Shah KV, SV40 and human cancer: a review of recent data, Int. J. Cancer 120 (2) (2007) 215–223. [DOI] [PubMed] [Google Scholar]
- [43].Boichuk S et al. , Multiple DNA damage signaling and repair pathways deregulated by simian virus 40 large T antigen, J. Virol 84 (16) (2010) 8007–8020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Hein J et al. , Simian virus 40 large T antigen disrupts genome integrity and activates a DNA damage response via Bub1 binding, J. Virol 83 (1) (2009) 117–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Ali SH, DeCaprio JA, Cellular transformation by SV40 large T antigen: interaction with host proteins, Semin. Cancer Biol 11 (1) (2001) 15–23. [DOI] [PubMed] [Google Scholar]
- [46].Skoczylas C, Fahrbach KM, Rundell K, Cellular targets of the SV40 small-t antigen in human cell transformation, Cell Cycle 3 (5) (2004) 606–610. [PubMed] [Google Scholar]
- [47].Castello A et al. , Insights into RNA biology from an atlas of mammalian mRNA-binding proteins, Cell 149 (6) (2012) 1393–1406. [DOI] [PubMed] [Google Scholar]
- [48].Castello A et al. , Comprehensive Identification of RNA-Binding Proteins by RNA Interactome Capture, Methods Mol. Biol 1358 (2016) 131–139. [DOI] [PubMed] [Google Scholar]
- [49].Strein C et al. , A versatile assay for RNA-binding proteins in living cells, RNA 20 (5) (2014) 721–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].He C et al. , High-resolution mapping of RNA-binding regions in the nuclear proteome of embryonic stem cells, Mol. Cell 64 (2) (2016) 416–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Munschauer M et al. , High-resolution profiling of protein occupancy on polyadenylated RNA transcripts, Methods 65 (3) (2014) 302–309. [DOI] [PubMed] [Google Scholar]
- [52].Nicholson CO, Friedersdorf M, Keene JD, Quantifying RNA binding sites transcriptome-wide using DO-RIP-seq, RNA 23 (1) (2017) 32–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Nicholson CO et al. , DO-RIP-seq to quantify RNA binding sites transcriptome-wide, Methods (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Dolken L et al. , High-resolution gene expression profiling for simultaneous kinetic parameter analysis of RNA synthesis and decay, RNA 14 (9) (2008) 1959–1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Ingolia NT et al. , The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments, Nat. Protoc 7 (8) (2012) 1534–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]