Abstract
Background
Wilms’ tumor is also called nephroblastoma and is the most common pediatric renal cancer. Several genetic and epigenetic factors have been found to account for the development of Wilms’ tumor. MiRNAs play important roles in this tumorigenic process. In the present study, we aimed to investigate the role of miR-140-5p in nephroblastoma by identifying its targets, as well as its underlying molecular mechanism of action.
Methods
The miRNA expression profile of nephroblastoma samples was investigated and the targets of miR-140-5p were predicted and validated using the miRNA luciferase reporter method. Moreover, the roles of miR-140-5p in regulating nephroblastoma cell proliferation, migration and cell cycle were analyzed by the CCK8, migration and flow cytometry assays, respectively. The downstream protein of the direct target of miR-140-5p was also identified.
Results
miR-140-5p was downregulated in Wilms’ tumor tissues, whereas in the nephroblastoma cell lines G401 and WT-CLS1 that exhibited high levels of miRNA-140-5p, inhibition of cellular proliferation and metastasis were noted as well as cell cycle arrest at the G1/S phase. TGFBRI and IGF1R were identified as direct target genes for miRNA-140-5p. In addition, SMAD2/3 and p-AKT were regulated by TGFBRI and IGF1R separately and participated in the miRNA-140-5p regulatory network. Ectopic expression of TGFBR1 and IGF-1R could abrogate the inhibitory effect of miR-140-5p.
Conclusion
We demonstrated that miRNA-140-5p participates in the progression of Wilms’ tumor by targeting the TGFBRI/SMAD2/3 and the IGF-1R/AKT signaling pathways.
Keywords: Wilms’ tumor, miRNA-140-5p, IGF1R, TGFBRI, cell signaling
Background
Nephroblastoma is one of the most common solid tumours in children, with an annual incidence rate of 1 case per 100,000 children. This disease comprises 8–10% of all neoplasms in that group [1, 2]. The peak incidence occurs in children, between 1 to 5 years of age [1, 2]. Although the pathogenesis of nephroblastoma remains undiscovered, increasing evidence has suggested that multiple signalling pathways, such as microRNAs, and epigenetic mechanisms play pivotal roles in its progression.
MicroRNAs (miRNAs) are endogenously produced, small (17~25-nucleotides long), non-coding single-stranded RNAs that play important roles in the regulation of crucial biological processes including cell apoptosis, metabolism, inflammation and tumorigenesis, primarily by inhibiting gene expression [3]. MiRNAs regulate the expression of mRNA molecules by binding to the complementary sequence in 3′-untraslated regions (3′UTRs) or the open reading frames of target genes [4, 5]. It has been studied that altered expression of miRNAs contributes to the initiation, invasion and metastasis of various types of cancer [6, 7]. miRNA 140–5p (miR-140-5p) has been shown to participate in various tumor processes. Yang et al. demonstrated that miR-140-5p could suppress tumor progression by targeting TGFBRI and FGF9 in hepatocellular carcinoma [8]. Yunfeng et al. demonstrated that miR-140-5p suppressed tumor growth and metastasis of non-small cell lung cancer by targeting IGF1R [9]. However, the function of miR-140-5p in the progression of nephroblastoma has not been explored.
TGFBRI and IGF1R can participate in the regulation of cell proliferation, differentiation, invasion and migration of cancer cells [10]. IGF1R is a tyrosine kinase receptor, which binds to IGF1 and IGF2. Following binding, the receptor auto-phosphorylates on the corresponding tyrosine residues [11, 12]. TGFBRI is a serine/threonine kinase receptor that is a member of the TGF-β signaling pathway, and exhibits metastatic properties by invading surrounding cells [13]. Previous reports have indicated that the TGF-β and IGF signaling pathways are associated with the development of nephroblastoma [14, 15]. However, the roles of TGFBRI and IGF1R in the progress of nephroblastoma require further investigation.
The purpose of the present study was to detect the potential role of miR-140-5p in the development of nephroblastoma, and the regulatory mechanism of this interaction.
Methods
Cell culture and clinical tissue specimens
The human nephroblastoma cell lines G401 and WT-CLS1 were matained in our labs and both were cultured in McCoy’s 5A medium containing 10% fetal bovine serum, 100 IU/ml penicillin and 100 mg/ml streptomycin. HEK-293 T cell line was maintained in DMEM supplemented with 10% fetal bovine serum, 100 IU/ml penicillin and 100 mg/ml streptomycin. All cells were cultured in a humidified atmosphere containing 5% CO2 at 37 °C.
Nephroblastoma tissue and adjacent non-cancerous tissue (ANT) samples were obtained from nephroblastoma children undergoing tumor resection at the Department of Surgery in the Affiliated Children’s Hospital of the Capital Institute of Pediatrics. The patients were recruited from September 2010 to April 2013. The tissue samples were immediately frozen in liquid nitrogen, and stored at − 80 °C until further analysis [16]. All the patients did not receive chemotherapy before surgical resection. The study was approved by the Ethics Committee for clinical research of the Capital Institute of Pediatrics.
miRNA Array
miRNA expression profile of tumor tissue samples and ANT were detected by miRCURY LNA miRNA chips 16.0 (Exiqon, Vedbaek, Denmark). The assay was conducted following the protocol of the manufacturer. Image analysis was processed by using the image analysis software Genepix Pro 6.0 (Axon Instruments) as described [17].
RNA purification and quantitative Real-Time PCR (qRT-PCR)
Total RNA was isolated from cells and tissue specimens with TRIzol reagent (Invitrogen, USA) as performed by the manufacturer’s instructions [18]. Quantitative real-time RT-PCR analysis was used to determine the level of mature miR-140-5p. One microgram of total RNA sample was reverse-transcribed to first-strand cDNA with the PrimeScript™ RT reagent kit as described by the manufacturer (Takara, Dalian, China). Real-time -PCR was conducted in triplicate, using SYBR Premix Ex Taq™ (Takara). The amplification was carried out on a 7900HT system with the SYBR Premix Ex Taq™ (Takara). The primer sequences used for quantitative real-time -PCR analyses of miRNA-140-5p were as follows: forward, 5’CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCTACCA3’ and reverse, 5′ ACACTCCAGCTGGGCAGTGGTTTTACCCTATG3’. The primer sequences used for small nuclear RNA (U6) were as following: forward, 5′-CAAATTCGTGAAGCGTTCCATAT-3′ and reverse 5′-GTGCAGGGTCCGAGGT-3′. The 2-∆∆Ct method was used to examine the relative expression levels of miRNAs. Specific siRNAs used to silence IGF-1R and TGFBRI gene were obtained from CST (CST, USA).
Lentiviral packaging of miR-140-5p
Oligonucleotides of miRNA-140-5p were synthesized, based on the sequence of human miRNA-140-5p (5′- cagugguuuuacccuaugguag − 3′, MIMAT0000431) from the miRBase database. The oligonucleotides were introduced into a pGCSIL-GFP plasmid (GeneChem Co. Ltd. Shanghai, China). pHelper 1.0 and pHelper 2.0 vector used for lentiviral construction were also obtained from Shanghai GeneChem Co. Ltd.. The generation of lentiviruses and the evaluation of the viral titration were conducted as described previously [19]. Lentiviruses carrying miR-140-5p and/or negative control sequences were packaged following the instructions of the manufacturer (Shanghai Genechem Co., Ltd., Shanghai, China).
Luciferase Reporter Assay
The plasmids used for firefly luciferase reporter assay were packaged by Genechem (Shanghai Genechem, China). And the plasmids designated IGF1R-WT, TGFBRI-WT (wild-type of miR-140-5p, targeting to IGF1R 3′-UTR and TGFBRI 3′-UTR), IGF1R-MU and TGFBRI-MU (mutated miR-140-5p, targeting to IGF1R 3′-UTR and TGFBRI 3′-UTR) were used. The mimic and the mimic negative control of miR-140-5p were purchased from Ribobio (Guangzhou RiboBio, Guangzhou, China). HEK293 cells were cultured in complete medium for 24 h before transfection. 0.05 μg firefly luciferase reporter, 0.05 μg IGF1R/TGFBRI plasmid, and 0.01 μg Renilla luciferase control vector were co-transfected into HEK293 cells by lipofectamine 2000 (Invitrogen, Carlsbad, CA) following the manufacturer’s instructions. At 48 h post-transfection, luciferase activity was detected by using the Dual-Glo luciferase reporter system (Promega Corp., Madison, WI, USA) in accordance with the protocols of manufacturer [20]. The relative luciferase activity value was achieved against the renilla control.
miRNA pull down assay
Biotin-coupled miRNA and mRNA pull-down assays were carried out as described in detail elsewhere [21]. Briefly, Biotinylated miRNA-140-5 probe were generated: (cagugguuuuacccuaugguag-biotin and control biotinylated probe: cuaccauaggguaaaaccacug-biotin, Generay,Shanghai,China). Biotin-coupled RNA probe and G401 cell total RNA were incubated in RNA buffer without RNAase at 4 °C overnight. Then the biotin-coupled RNA complexes were pulled down by using M-280 Streptavidin Dynabeads (Invitrogen, Carlsbad, CA). TGFBR I and IGF1R abundance in the bound fractions was calculated by RT-PCR analysis.
Cell proliferation assay
Cell proliferation viability was measured using the cell counting kit-8 assay kit (Dojindo Laboratories, Japan) as described. Cells were seeded in 96-well plates, and incubated for 48, 72 and 96 h at 37 °C after transfection. The CCK-8 solution was added to each well at the end of incubation, and then cells were further cultured at 37 °C for an additional 1.5 h. Subsequently, the absorbance (450 nm) value of each well was estimated by a microplate reader.
Cell cycle analysis
Flow cytometry was used for cell cycle analysis with a Propidium Iodide (PI) cell cycle detection kit following the manufacturer’s protocols (Transgen Biotech, Beijing, China).
Migration assay
The Ibidi cell migration (gap closure/cell-exclusion zonemigration assays) technology (Ibidi, Martinsried, Germany) was used for the migratory assay. Briefly, G401 or WT-CLS1 cells were cultured, and then miRNA was transfected into the cells. Subsequently, the cells were collected with trypsin and washed with PBS, and then resuspended in DMEM containing 10% FBS (~ 7.0 × 105 cells/ml). Next, 70 μl cell suspension was added to the wells of the culture-insert on a 35-mm dish, cultured in a incubator with 5% CO2 atmosphere at 37 °C. At 24 h after incubation, the culture-insert was removed using sterilized tweezers. Then cell migration was observed over time. At 12 h after culture, the increase of the area was calculated using ImageJ.
Western Blot Analysis
The cells or nephroblastoma tissue were homogenized and lysed in ice-cold RIPA lysis buffer (Kangwei, Beijing, China) including protease and phosphatase inhibitors. The cell debris was removed by centrifuging at 14,000×g for 20 min at 4 °C. The protein samples were prepared and separated by 12% SDS-PAGE, and then electrophoretically transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, Bedford, MA) [16]. Subsequently, the membranes were incubated for 12 h at 4 °C with primary antibodies, including anti-IGF1R, anti-p-AKT, anti-Smad2/3 anti-GAPDH (CST, USA) and anti-TGFBRI (R&D, USA) antibodies following by incubation with secondary antibodies. The binding of all antibodies was visualized using enhanced chemiluminescence (ECL) western blotting detection system (Amersham Life Science, Piscataway, NJ, USA) following the manufacturer’s protocol. GAPDH was utilized as a loading control.
Statistical Analysis
The experiments were performed in triplicate. The data were analyzed by GraphPad Prism 5 (La Jolla, CA, USA). Statistical analysis was performed by using SPSS Statistical Package (v. 13.0, SPSS Inc., Chicago, IL). The Mann-Whitney U test was used for the analysis of the expression data of miR-140-5p. The Student’s t-test was utilized for unpaired sample comparison. Statistical significance levels were set at P < 0.05.
Results
MicroRNA-140-5p is downregulated in nephroblastoma tissues and is associated with clinical outcome
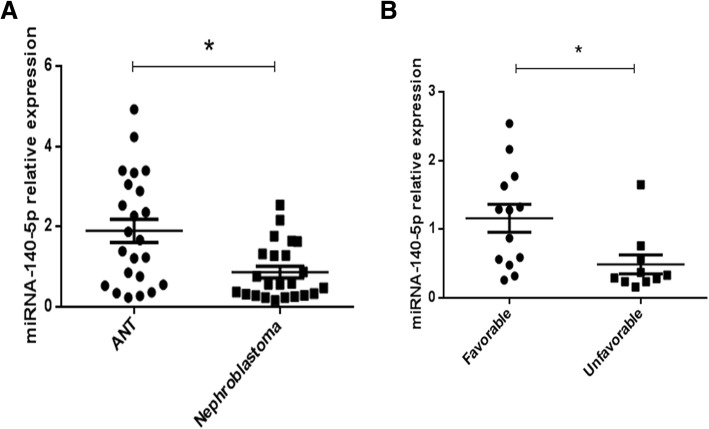
miRCURY LNA microRNA chips from Exiqon was used to analyze three cases of nephroblastoma and adjacent normal tissue (ANT) obtained by surgical removal. miRNA microarray analysis revealed that miR-140-5p was significantly downregulated in nephroblastoma tissues compared with the corresponding levels of expression in ANT. qRT-PCR was performed in 23 cases of nephroblastoma tissues and 23 ANTs. qRT-PCR analysis revealed that the expression of miRNA-140-5p was significantly decreased in nephroblastoma tissues compared with that of the control group (Fig. 1a). To assess the feasibility of miR-140-5p expression in nephroblastoma prognosis, we analyzed the correlation between the clinicopathological characteristics of Wilms’ tumor patients and the miRNA-140-5p levels. The data suggested that patients with higher tumor stage and unfavorable histology exhibited lower levels of miRNA-140-5p (Fig. 1b). The results indicated that miRNA-140-5p may participate in Wilms’ tumor progression.
Fig. 1.
Downregulated miRNA-140-5p expression correlated with poor clinical outcomes of nephroblastoma. Relationship between miRNA-140-5p levels and clinical character of Wilms’ tumor were detected. (a) The expression levels of miRNA-140-5p were analyzed in nephroblastoma and corresponding normal tissues by real-time PCR. U6 served as the control for RNA loading. (b) The clinicopathological characteristics of the nephroblastoma patients were analyzed according to the expression levels of miRNA-140-5p.*P < 0.05
Overexpression of microRNA-140-5p inhibits proliferation and metastasis of nephroblastoma cell lines
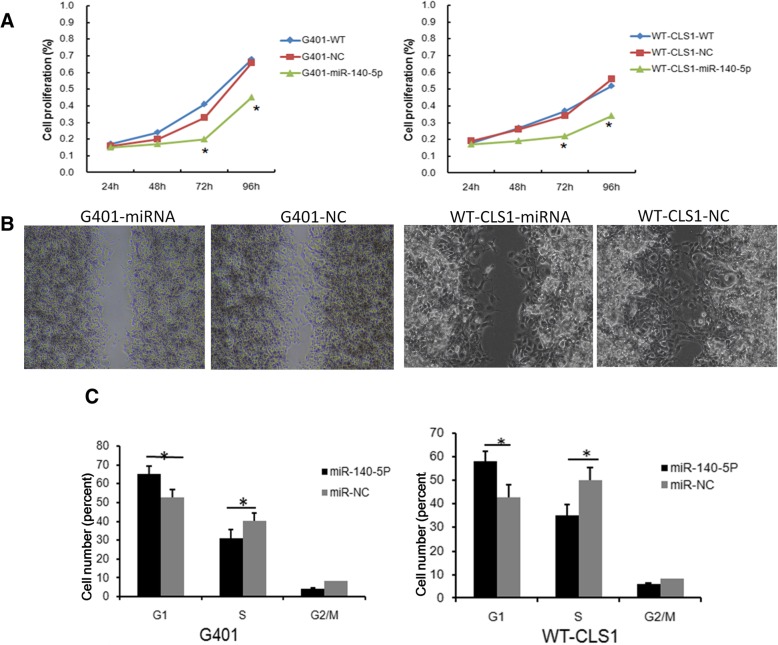
To further assess the impact of miR-140-5p on tumor growth and metastasis, we performed cell proliferation and migration assays. Two nephroblastoma cell lines (G401 and WT-CLS1) were infected with lentivirus containing miR-140-5p sequence, or control lentivirus containing non-specific sequences at an MOI = 10. MiRNA-140-5p expression levels were higher in the miR-140-5p transfection group than those in the control group (data not shown). Cell proliferation and migration assays were performed. The proliferation assay indicated that lentiviral-induced ectopic miR-140-5p resulted in a significant decrease in cell proliferation in both G401 and WT-CLS1 cells (Fig. 2a). The migration assay indicated that overexpression of miR-140-5p significantly suppressed the migratory abilities of the nephroblastoma cell lines (Fig. 2b). We subsequently performed cell cycle analysis and revealed that overexpression of miR-140-5p decreased the percentage of cells in the S phase in G401 and WT-CLS1 cells, while it concomitantly increased the percentage of cells in the G1 phase (P < 0.05) (Fig. 2c).
Fig. 2.
miRNA-140-5p suppresses nephroblastoma cells growth in vitro. Lentivirus containing miRNA-140-5p were transduced into G401 and WT-CLS1 cells, cell proliferation, migratory ability and cell cycle were tested. a The proliferation of G401 and WT-CLS1 cells was downregulated in the miRNA-140-5p group by the CCK8 assay. b The migratory ability of G401 and WT-CLS1 cells were inhibited in the miRNA-140-5p group, representative photographs of cell migration assay were shown. c G401 and WT-CLS1 cells were arrested in the G1/S phase of the cell cycle in the miRNA-140-5p group. *P < 0.05
TGFBR I and IGF-1R are candidate targets for miRNA-140-5p
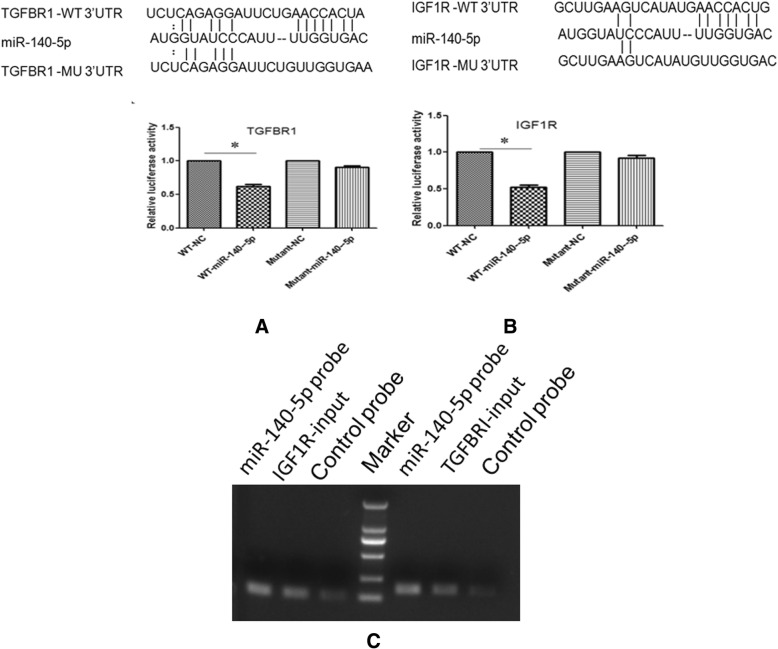
To explore the underlying mechanism of miR-140-5p in nephroblastoma, we performed a bioinformatics search for candidate targets of miR-140-5p in genes that were involved in nephroblastoma pathogenesis using the TargetScan (http://www.targetscan.org), PicTar (http://www.pictar.org/) and miRnada (http://miranda.org.uk/) databases. The analysis indicated that miR-140-5p possibly regulated the IGF1R and TGFBRI genes since their 3′-UTR included the binding sites for the seed region of miR-140-5p (Fig. 3a and b).
Fig. 3.
miRNA-140-5p could target TGFBRI and IGF1R. Validation of TGFBRI and IGF1R as targets of miRNA-140-5p. The sequence of potential binding site of miRNA-140-5p in the 3’UTR of TGFBRI and IGF1R mRNA (a) and (b). Validation of the miRNA-140-5p binding target using luciferase reporter assays. HEK-293 T cells were transfected with an miRNA-140-5p mimic or a control oligonucleotide (NC) and a dual luciferase reporter containing a wild-type promoter or a mutant promoter of TGFBRI and IGF1R. The luciferase activities in the cells with the mutant promoter/NC were significantly higher than those with the miRNA-140-5p mimic-wild-type reporter (a and b). TGFBRI and IGF1R in G401 cell lysis was pulled down and enriched with a miRNA-140-5p-specific probe and then detected using RT-PCR, the amount of IGF1R and TGFBRI were higher in the miR-140-5p probe group than control group(c)
To verify this speculation, HEK293 cells were transfected with miR-140-5p, mimic control, the 3′-UTR wild type of the IGF1R/TGFBRI gene (WT-3′-UTR), and its mutant form (MU-3′-UTR). The results demonstrated that miR-140-5p mimic specifically reduced the activity of a reporter containing the IGF1R/TGFBRI-3′-UTR-WT(P < 0.05), while no effect was apparent in the miR-NC or IGF1R/TGFBRI-3′-UTR-MU groups, implying the inhibitory effect of miR-140-5p on the IGF1R and TGFBRI genes (Fig. 3a and b).
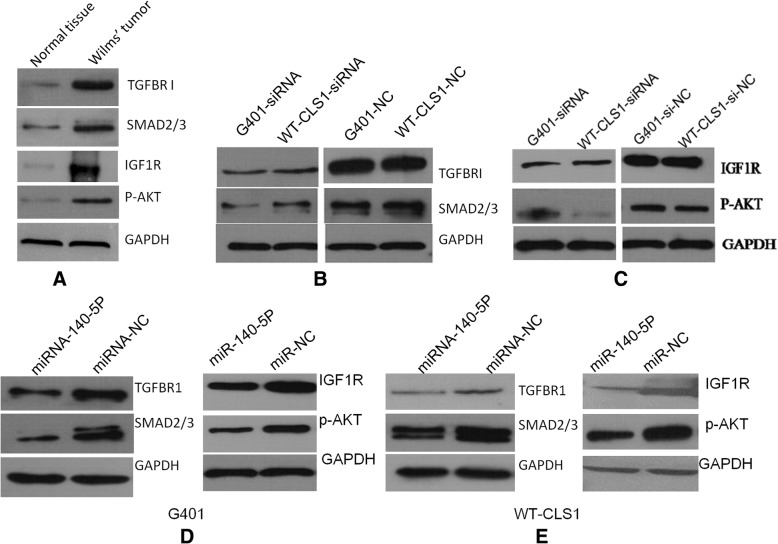
In order to investigate whether miR-140-5p could bind to IGF1R/TGFBRI, we used a miR-140-5p-specific probe to pull down its associated genes in G401 cells. RT-PCR results showed that IGF1R and TGFBRI were greatly enriched in the miR-140-5p precipitation complex (Fig. 3c). Thus, IGF1R and TGFBRI were identified as direct target genes of miR-140-5p. We also found that IGF1R and TGFBRI expression in nephroblastoma tissue were higher than that of adjacent normal tissues (Fig. 4a).
Fig. 4.
MicroRNA-140-5p suppresses IGF-1R and TGFBR I signaling pathways. Expression of the main proteins in IGF-1R and TGFBR I signaling pathways were detected in tumor tissues, siRNA transfected cells and microRNA-140-5p overexpressed cells. TGFBRI/SMAD2/3 and IGF1R/P-AKT expression were detected in the nephroblastoma tissues and adjacent normal tissues, shown were representative results from 3 experiments (a). SMAD2/3 expression was downregulated by inhibition of TGFBRI (b), whereas AKT activation was downregulated by inhibition of IGF1R (c) in G401 and WT-CLS1 cells. MicroRNA-140-5p could suppress TGFBRI/SMAD2/3 and IGF1R/P-AKT expression in G401 (d) and WT-CLS1 cells (e)
MiRNA-140-5p represses IGF-1R and TGFBR I signaling to suppress nephroblastoma growth and metastasis
Based on the finding that miRNA-140-5p could suppress cell proliferative and metastatic activities, we further evaluated the function of the target genes IGF-1R and TGFBR1 in G401 and WT-CLS1 cells by specific siRNA-silencing. IGF-1R-siRNA and TGFBR1-siRNA could decrease cell proliferation and metastasis (data not shown). The expression levels of SMAD2/3 and p-AKT were further evaluated in siRNA-transfected cells. SMAD2/3 is a downstream mediator of TGFBR1 and the activation of AKT is considered the main downstream signal of IGF-1R, both SMAD2/3 and p-AKT participate in the process of cancer progression by promoting cell growth, anti-apoptotic effects, and cell invasion [22, 23]. To verify importance of both proteins in Wilms’ tumor, we detected expression of SMAD2/3 and p-AKT and found that both were highly expressed in the tumor tissues compared with adjacent normal tissue (Fig. 4a). We also found that SMAD2/3 and p-AKT were downregulated in TGFBR1- knockdown cells and IGF-1R-knockdown cells, respectively (Fig. 4b and c). Furthermore, SMAD2/3 and p-AKT levels were both downregulated in miR-140-5p-overexpressing G401 and WT-CLS1 cells (Fig. 4d and e).
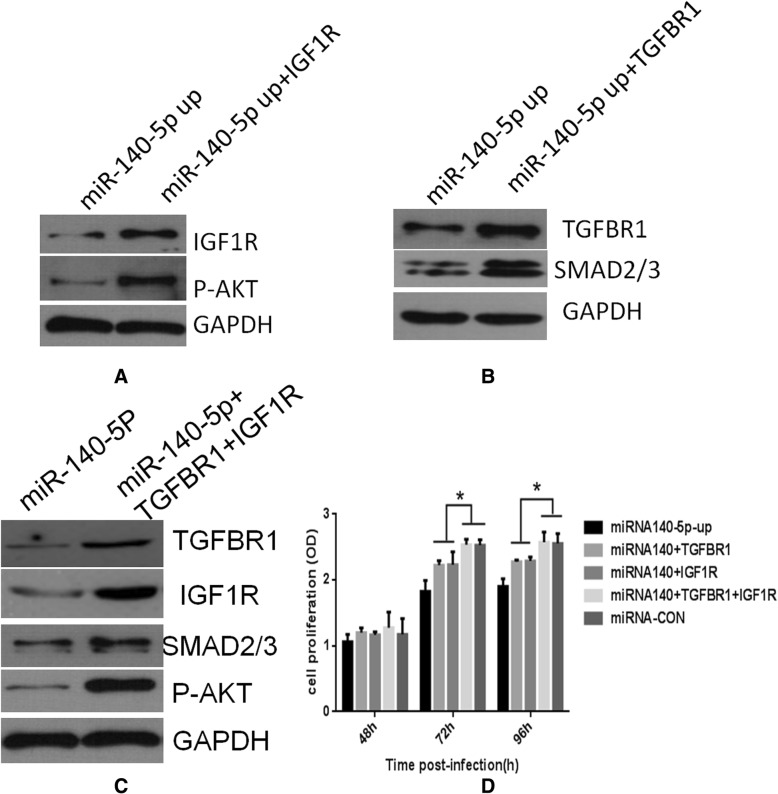
Having shown that TGFBR1 and IGF-1R are direct targets of miR-140-5p and that they participate in cell proliferation, we further tested whether ectopic expression of TGFBR1 and IGF-1R could rescue the effect caused by miR-140-5p. Ectopic expression of TGFBR1 or IGF-1R in miR-140-5p-transduced G401 cells could abrogate inhibition of SMAD2/3 or p-AKT by miR-140-5p (Fig. 5 a, b), and partially attenuate the inhibitory effects of miR-140-5p on cell proliferation (Fig. 5 d). In contrast to these observations, overexpression of both TGFBR1 and IGF-1R could rescue SMAD2/3 and p-AKT expression and cell proliferation completely (Fig. 5 c and d). In summary, we propose that miR-140-5p can regulate oncogenic receptor-related cell invasiveness and proliferation in Wilms’ tumor.
Fig. 5.
Overexpression of TGFBRI and IGF1R could abolish the inhibitory effects of microRNA-140-5p. Exogenous TGFBRI and IGF1R were introduced into microRNA-140-5p overexpressed G401 cells. Overexpression of IGF1R could rescue activation of AKT and overexpression of TGFBRI could rescue SMAD2/3 expression (a, b and c). Overexpression of TGFBRI and IGF1R could rescue cell proliferation rate inhibited by miR-140-5p in G401 cells. *P < 0.05 (d)
Discussion
Nephroblastoma also called Wilms’ tumor, is one of the most common solid tumours in children. Increasing evidence has suggested that multiple signaling, epigenetic and miRNA pathways are involved in its progression [24]. Aberrant miRNA expression is closely associated with various types of cancer [25, 26], and numerous miRNAs have significant functions in cancer cell proliferation, apoptosis, migration and neoplastic transformation [8, 27, 28]. In the present study, we investigated the miRNA expression profile of nephroblastoma specimens and concluded that miR-140-5p was downregulated in the tumor site of these samples.
MiR-140-5p levels have been shown to be decreased in multiple cancer types, such as hepatocellular carcinoma [8], colorectal cancer [29] and gastric cancer [30]. MiR- 140-5p upregulation can inhibit cell proliferation. Yang et al. demonstrated that miR-140-5p levels were decreased in HCC tissues and that miR-140-5p could regulate the activity of ERK/MAPK signaling by directly targeting TGFBRI during the metastatic process of hepatocellular carcinoma [8]. Yunfeng et al. demonstrated that miR-140-5p could suppress tumor growth and metastasis of non-small cell lung cancer by targeting IGF1R [9]. However, the function of miR-140-5p in the proliferation of Wilms’ tumor has not been previously studied. In the present study, we proved that TGFBR I and IGF1R are direct target of mi-140-5p, whereas the data further indicated that overexpression of miR-140-5p could modulate their expression and the proliferation and metastasis of nephroblastoma cells.
TGF-β signaling regulates the growth, differentiation, and metastasis of various types of cells. It functions both as a tumor suppressor and tumor promoter [31, 32]. In the early stages of cancer, TGF-β serves as a tumor suppressor [31]. However, in the advanced stages of cancer, TGF-β facilitates the progression and metastasis of tumors. In the present study, we found that inhibition of TGFBRI by siRNA could suppress cell proliferation and invasion that was in accordance with previous studies that examined the function of miR-140-5p during the advanced stages of cancer.
Zhai et al. identified SMAD2 as a direct target of miR-140-5p in CRC cells [33]. The data of the present study indicated that SMAD2/3 levels were reduced in WT-CLS1 and G401 cells when high levels of miR-140-5p were present. We further demonstrated that exogenous TGFBRI expression could recover the activity of SMAD2/3 in miR-140-5p-overexpressing WT-CLS1 and G401 cells. Considering that SMAD2/3 belongs to the TGF-β signaling pathway and that it is a downstream mediator of TGFBRI [34], we reasoned that inhibition of SMAD2/3 could be attributed notably to TGFBRI reduction. This finding indicated that miR-140-5p was not a direct target in Wilms’ tumor and that its function in carcinogenesis may be disease-specific. This finding indicated that SMAD2/3 was not a direct target of miR-140-5p in Wilms’ tumor and that its function in carcinogenesis may be disease-specific.
Several studies have shown that overexpression of the IGF-1 receptor (IGF-1R) constitutes a typical hallmark of the majority of cancer types [9]. Overexpression of IGF1R was associated with poor outcome in Wilms’ tumors, whereas the inhibition of IGF1R activity could decrease cancer malignancy [35]. One of the main downstream signals of IGF1R is AKT, which acts as a key regulator in cancer progression by promoting cell growth, anti-apoptotic effects, and cell invasion [36]. We demonstrated in the present study that miR-140-5p could inhibit IGF1R in G401 and WT-CLS1 cells, and that activation of p-AKT was also significantly decreased.
In the present study, we demonstrated that IGF-1R and TGFBRI were highly expressed in Wilms’ tumor tissues and that they participated in Wilms’ tumor progression. Overexpression of miR-140-5p inhibited cell progression via the IGF-1R/AKT and TGFBRI/SMAD2/3 pathways. We further demonstrated that both exogenous IGF-1R and TGFBRI in miR-140-5p overexpression cells could recover cell proliferation to normal levels, while ectopic either could not. Our data suggested that miR-140-5p regulated Wilms’ tumor progression and TGFBRI/SMAD2/3 and IGF-1R/AKT signaling pathways participate in this process.
Conclusion
miRNA-140-5p expression was downregulated in Wilms’ tumor tissues, and miR-140-5p could regulate Wilms’ tumor progression via the IGF-1R/ AKT and TGFBRI/SMAD2/3 pathways and that it might have tumor suppressive functions with regard to Wilms’ tumor progression and metastasis.
Acknowledgements
We would like to thank all of our colleagues for their contribution to this study.
Funding
This work was supported by the CAMS Central Public Welfare Scientific Research Institute Basal Research Expenses (2016ZX310182–3) and CAMS Initiative for Innovative Medicine (CAMS-I2M). The funding body had no role in the design of the study and collection, analysis and interpretation of data and in writing the manuscript.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request for privacy reasons.
Abbreviations
- 3′UTRs
3′-untraslated regions
- ANT
Adjacent non-cancerous tissue
- IGF1R
Insulin-like growth factor 1 receptor
- TGFBRI
Transforming growth factor-beta type I receptor
Authors’ contributions
ZL and FH contributed the cell culture and treatment for the study. SOY and YL performed the data collection and analysis. FM and HC carried out the luciferase report assay and western-blot assay. JW designed the work. DC and JW wrote the manuscript and contributed the critical revision of the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The study was approved by the Ethics Committee for Clinical Research of Capital Institute of Pediatrics, Beijing, China.
Written informed consent for the use and obtaining of tissue samples from all patients was provided by the patients’ parents and/or legal guardians.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Zhuo Liu, Phone: +86 01085695578, Email: liuzhuozhuo2005@163.com.
Feng He, Email: hefengfeng2001@163.com.
Shengrong OuYang, Email: cipbiochem@163.com.
Yuanyuan Li, Email: liyuanyuan075@sina.com.
Feifei Ma, Email: mafei0817@163.com.
Huibo Chang, Email: huibochang2012@163.com.
Dingding Cao, Email: cddzlm@163.com.
Jianxin Wu, Phone: +86 01085695593, Email: cipbiolab@163.com.
References
- 1.Hohenstein P, Pritchard-Jones K, Charlton J. The yin and yang of kidney development and Wilms' tumors. Genes Dev. 2015;29(5):467–482. doi: 10.1101/gad.256396.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Apoznanski W, Sawicz-Birkowska K, Palczewski M, Szydelko T. Extrarenal nephroblastoma. Central Eur J Urol. 2015;68(2):153–156. doi: 10.5173/ceju.2015.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pekarsky Y, Balatti V, Palamarchuk A, Rizzotto L, Veneziano D, Nigita G, Rassenti LZ, Pass HI, Kipps TJ, Liu CG, et al. Dysregulation of a family of short noncoding RNAs, tsRNAs, in human cancer. Proc Natl Acad Sci U S A. 2016;113(18):5071–5076. doi: 10.1073/pnas.1604266113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu C, Liu R, Zhang D, Deng Q, Liu B, Chao HP, Rycaj K, Takata Y, Lin K, Lu Y, et al. MicroRNA-141 suppresses prostate cancer stem cells and metastasis by targeting a cohort of pro-metastasis genes. Nat Commun. 2017;8:14270. doi: 10.1038/ncomms14270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leichter AL, Sullivan MJ, Eccles MR, Chatterjee A. MicroRNA expression patterns and signalling pathways in the development and progression of childhood solid tumours. Mol Cancer. 2017;16(1):15. doi: 10.1186/s12943-017-0584-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhayadia R, Krowiorz K, Haetscher N, Jammal R, Emmrich S, Obulkasim A, Fiedler J, Schwarzer A, Rouhi A, Heuser M, et al. Endogenous Tumor Suppressor microRNA-193b: Therapeutic and Prognostic Value in Acute Myeloid Leukemia. J Clin Oncol. 2018;36(10):1007–1016. doi: 10.1200/JCO.2017.75.2204. [DOI] [PubMed] [Google Scholar]
- 7.Lei W, Yan C, Ya J, Yong D, Yujun B, Kai L. MiR-199a-3p affects the multi-chemoresistance of osteosarcoma through targeting AK4. BMC Cancer. 2018;18(1):631. doi: 10.1186/s12885-018-4460-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang H, Fang F, Chang R, Yang L. MicroRNA-140-5p suppresses tumor growth and metastasis by targeting transforming growth factor beta receptor 1 and fibroblast growth factor 9 in hepatocellular carcinoma. Hepatology. 2013;58(1):205–217. doi: 10.1002/hep.26315. [DOI] [PubMed] [Google Scholar]
- 9.Yuan Y, Shen Y, Xue L, Fan H. miR-140 suppresses tumor growth and metastasis of non-small cell lung cancer by targeting insulin-like growth factor 1 receptor. PLoS One. 2013;8(9):e73604. doi: 10.1371/journal.pone.0073604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gong Y, Ren J, Liu K, Tang LM. Tumor suppressor role of miR-133a in gastric cancer by repressing IGF1R. World J Gastroenterol. 2015;21(10):2949–2958. doi: 10.3748/wjg.v21.i10.2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen G, Fang T, Huang Z, Qi Y, Du S, Di T, Lei Z, Zhang X, Yan W. MicroRNA-133a Inhibits Osteosarcoma Cells Proliferation and Invasion via Targeting IGF-1R. Cell Physiol Biochem. 2016;38(2):598–608. doi: 10.1159/000438653. [DOI] [PubMed] [Google Scholar]
- 12.Sun X, Li J, Sun Y, Zhang Y, Dong L, Shen C, Yang L, Yang M, Li Y, Shen G, et al. miR-7 reverses the resistance to BRAFi in melanoma by targeting EGFR/IGF-1R/CRAF and inhibiting the MAPK and PI3K/AKT signaling pathways. Oncotarget. 2016;7(33):53558–53570. doi: 10.18632/oncotarget.10669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang LK, Hsiao TH, Hong TM, Chen HY, Kao SH, Wang WL, Yu SL, Lin CW, Yang PC. MicroRNA-133a suppresses multiple oncogenic membrane receptors and cell invasion in non-small cell lung carcinoma. PLoS One. 2014;9(5):e96765. doi: 10.1371/journal.pone.0096765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maschietto M, Charlton J, Perotti D, Radice P, Geller JI, Pritchard-Jones K, Weeks M. The IGF signalling pathway in Wilms tumours--a report from the ENCCA Renal Tumours Biology-driven drug development workshop. Oncotarget. 2014;5(18):8014–8026. doi: 10.18632/oncotarget.2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amarante MK, de Oliveira CEC, Ariza CB, Sakaguchi AY, Ishibashi CM, Watanabe MAE. The predictive value of transforming growth factor-beta in Wilms tumor immunopathogenesis. Int Rev Immunol. 2017;36(4):233–239. doi: 10.1080/08830185.2017.1291639. [DOI] [PubMed] [Google Scholar]
- 16.Dang Z, Xu WH, Lu P, Wu N, Liu J, Ruan B, Zhou L, Song WJ, Dou KF. MicroRNA-135a inhibits cell proliferation by targeting Bmi1 in pancreatic ductal adenocarcinoma. Int J Biol Sci. 2014;10(7):733–745. doi: 10.7150/ijbs.8097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murn J, Alibert O, Wu N, Tendil S, Gidrol X. Prostaglandin E2 regulates B cell proliferation through a candidate tumor suppressor, Ptger4. J Exp Med. 2008;205(13):3091–3103. doi: 10.1084/jem.20081163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ning ZQ, Lu HL, Chen C, Wang L, Cai W, Li Y, Cao TH, Zhu J, Shu YQ, Shen H. MicroRNA-30e reduces cell growth and enhances drug sensitivity to gefitinib in lung carcinoma. Oncotarget. 2017;8(3):4572–4581. doi: 10.18632/oncotarget.13944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He F, Yao H, Wang J, Xiao Z, Xin L, Liu Z, Ma X, Sun J, Jin Q, Liu Z. Coxsackievirus B3 engineered to contain microRNA targets for muscle-specific microRNAs displays attenuated cardiotropic virulence in mice. J Virol. 2015;89(2):908–916. doi: 10.1128/JVI.02933-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cha Y, He Y, Ouyang K, Xiong H, Li J, Yuan X. MicroRNA-140-5p suppresses cell proliferation and invasion in gastric cancer by targeting WNT1 in the WNT/beta-catenin signaling pathway. Oncol Lett. 2018;16(5):6369–6376. doi: 10.3892/ol.2018.9480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li X, Liu CX, Xue W, Zhang Y, Jiang S, Yin QF, Wei J, Yao RW, Yang L, Chen LL. Coordinated circRNA Biogenesis and Function with NF90/NF110 in Viral Infection. Mol Cell. 2017;67(2):214–227 e217. doi: 10.1016/j.molcel.2017.05.023. [DOI] [PubMed] [Google Scholar]
- 22.Alsina-Sanchis E, Figueras A, Lahiguera A, Vidal A, Casanovas O, Graupera M, Villanueva A, Vinals F. The TGFbeta pathway stimulates ovarian cancer cell proliferation by increasing IGF1R levels. Int J Cancer. 2016;139(8):1894–1903. doi: 10.1002/ijc.30233. [DOI] [PubMed] [Google Scholar]
- 23.Subbiah V, Brown RE, Jiang Y, Buryanek J, Hayes-Jordan A, Kurzrock R, Anderson PM. Morphoproteomic profiling of the mammalian target of rapamycin (mTOR) signaling pathway in desmoplastic small round cell tumor (EWS/WT1), Ewing's sarcoma (EWS/FLI1) and Wilms' tumor (WT1) PLoS One. 2013;8(7):e68985. doi: 10.1371/journal.pone.0068985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gadd S, Huff V, Walz AL, Ooms A, Armstrong AE, Gerhard DS, Smith MA, Auvil JMG, Meerzaman D, Chen QR, et al. A Children's Oncology Group and TARGET initiative exploring the genetic landscape of Wilms tumor. Nat Genet. 2017;49(10):1487–1494. doi: 10.1038/ng.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dvinge H, Git A, Graf S, Salmon-Divon M, Curtis C, Sottoriva A, Zhao Y, Hirst M, Armisen J, Miska EA, et al. The shaping and functional consequences of the microRNA landscape in breast cancer. Nature. 2013;497(7449):378–382. doi: 10.1038/nature12108. [DOI] [PubMed] [Google Scholar]
- 26.Mao B, Wang G. MicroRNAs involved with hepatocellular carcinoma (Review) Oncol Rep. 2015;34(6):2811–2820. doi: 10.3892/or.2015.4275. [DOI] [PubMed] [Google Scholar]
- 27.Chen M, Liu Y, Varley P, Chang Y, He XX, Huang H, Tang D, Lotze MT, Lin J, Tsung A. High-Mobility Group Box 1 Promotes Hepatocellular Carcinoma Progression through miR-21-Mediated Matrix Metalloproteinase Activity. Cancer Res. 2015;75(8):1645–1656. doi: 10.1158/0008-5472.CAN-14-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin CW, Li XR, Zhang Y, Hu G, Guo YH, Zhou JY, Du J, Lv L, Gao K, Zhang Y, et al. TAp63 suppress metastasis via miR-133b in colon cancer cells. Br J Cancer. 2014;110(9):2310–2320. doi: 10.1038/bjc.2014.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li J, Zou K, Yu L, Zhao W, Lu Y, Mao J, Wang B, Wang L, Fan S, Song B, et al. MicroRNA-140 Inhibits the Epithelial-Mesenchymal Transition and Metastasis in Colorectal Cancer. Mol Ther Nucleic Acids. 2018;10:426–437. doi: 10.1016/j.omtn.2017.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Fang Z, Yin S, Sun R, Zhang S, Fu M, Wu Y, Zhang T, Khaliq J, Li Y. miR-140-5p suppresses the proliferation, migration and invasion of gastric cancer by regulating YES1. Mol Cancer. 2017;16(1):139. doi: 10.1186/s12943-017-0708-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith AL, Robin TP, Ford HL. Molecular pathways: targeting the TGF-beta pathway for cancer therapy. Clin Cancer Res. 2012;18(17):4514–4521. doi: 10.1158/1078-0432.CCR-11-3224. [DOI] [PubMed] [Google Scholar]
- 32.Li L, Shi JY, Zhu GQ, Shi B. MiR-17-92 cluster regulates cell proliferation and collagen synthesis by targeting TGFB pathway in mouse palatal mesenchymal cells. J Cell Biochem. 2012;113(4):1235–1244. doi: 10.1002/jcb.23457. [DOI] [PubMed] [Google Scholar]
- 33.Zhai H, Fesler A, Ba Y, Wu S, Ju J. Inhibition of colorectal cancer stem cell survival and invasive potential by hsa-miR-140-5p mediated suppression of Smad2 and autophagy. Oncotarget. 2015;6(23):19735–19746. doi: 10.18632/oncotarget.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bierie B, Moses HL. TGF-beta and cancer. Cytokine Growth Factor Rev. 2006;17(1–2):29–40. doi: 10.1016/j.cytogfr.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 35.Bielen A, Box G, Perryman L, Bjerke L, Popov S, Jamin Y, Jury A, Valenti M, Brandon Ade H, Martins V, et al. Dependence of Wilms tumor cells on signaling through insulin-like growth factor 1 in an orthotopic xenograft model targetable by specific receptor inhibition. Proc Natl Acad Sci U S A. 2012;109(20):E1267–E1276. doi: 10.1073/pnas.1105034109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Y, Zhang J, Liu Y, Zhang B, Zhong F, Wang S, Fang Z. MiR-30a-5p confers cisplatin resistance by regulating IGF1R expression in melanoma cells. BMC Cancer. 2018;18(1):404. doi: 10.1186/s12885-018-4233-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request for privacy reasons.