Abstract
Facioscapulohumeral dystrophy type 1 (FSHD-1) is the most common autosomal dominant form of muscular dystrophy with a prevalence of ∼1 in 8000 individuals. It is considered a late-onset form of muscular dystrophy and leads to asymmetric muscle weakness in the facial, scapular, trunk and lower extremities. The prevalent hypothesis on disease pathogenesis is explained by misexpression of a germ line, primate-specific transcription factor DUX4-fl (double homeobox 4, full-length isoform) linked to the chromosome 4q35. In vitro and in vivo studies have demonstrated that very low levels of DUX4-fl expression are sufficient to induce an apoptotic and/or lethal phenotype, and therefore modeling of the disease has proved challenging. In this study, we expand upon our previously established injection model of DUX4 misexpression in zebrafish and describe a DUX4-inducible transgenic zebrafish model that better recapitulates the expression pattern and late onset phenotype characteristic of FSHD patients. We show that an induced burst of DUX4 expression during early development results in the onset of FSHD-like phenotypes in adulthood, even when DUX4 is no longer detectable. We also utilize our injection model to study long-term consequences of DUX4 expression in those that fail to show a developmental phenotype. Herein, we introduce a hypothesis that DUX4 expression during developmental stages is sufficient to induce FSHD-like phenotypes in later adulthood. Our findings point to a developmental role of DUX4 misexpression in the pathogenesis of FSHD and should be factored into the design of future therapies.
Introduction
Facioscapulohumeral muscular dystrophy type 1 (FSHD-1) is one of the most common form of myopathy, affecting ∼1 in 8000 individuals (1). FSHD is characterized by an asymmetrical skeletal muscle wasting and weakness in the facial, scapular and humeral region, with progression to the lower limbs. FSHD-1 is inherited in an autosomal dominant pattern and results from deletion of the D4Z4 tandem repeats on chromosome 4q35, leading to chromatin hypomethylation (2). The resulting chromatin contraction leads to epigenetic de-repression of transcription factor double homeobox 4 (DUX4), a germ line-specific protein found in the thymus and testis that becomes uniquely activated in skeletal muscle cells of patients with FSHD (3,4). High expression of DUX4 in muscle cells inevitably leads to cell death while low-expression levels inhibit muscle cell differentiation and fusion by repressing myogenic regulators MyoD and PAX7 and their targets (5–7).
FSHD poses many unique clinical and molecular features that together make the disease difficult to model in animals. These challenges include the spectrum of clinical variability, stochastic-nature of DUX4 expression and the toxicity of DUX4 misexpression resulting in widespread lethality (8). An additional complexity is posed by the fact that the DUX4 retrotransposon is specific to primates and thus raises the possibility that non-primate models may not harbor the same molecular players needed to recapitulate the disease. Nevertheless, several attempts to model FSHD using DUX4 and non-DUX4 means have been published that recapitulate various aspects of disease pathology.
Invertebrate FSHD models in fruit flies (Drosophila melanogaster) include DUX4 and FRG1 transgenic models, while vertebrate models of FSHD have been attempted in frogs (Xenopus laevis; FRG1 injection model), zebrafish (Danio rerio; DUX4 injection model) and mice (DUX4, FRG1, FAT1, PITX1, patient xenograft model) (9). DUX4 expression models in mice include AAV6-DUX4 overexpression (10), transgenic insertion of 2.5 copies of permissive D4Z4 units (D4Z4-2.5) (11) and two variations of doxycycline-inducible DUX4 on the X-chromosome, iDUX4(2.7) (12) and iDUX4pA (7). Another model, iDUX4pA, has come closest to recapitulating both the clinical and molecular aspects of FSHD, that is, the low and infrequent expression of DUX4 leading to a progressive myopathy with signs of inflammation, fibrosis, loss of muscle regeneration and hearing (7). The latter, FLExDUX4 mouse (conditional floxed DUX4-fl), is becoming the most widely used inducible transgenic mouse model. This model is easily manipulated to produce controllable levels of DUX4 and produces phenotypic changes representative of disease in muscle (13).
Each of the models mentioned above have their associated pros and cons, and no single model has entirely recapitulated the human disease. Existing models to date are too mild or severe, fail to mimic the late-onset associated with human FSHD and/or do not result in the same muscle and non-muscle phenotypes observed in patients. However, what each of these models has taught us so far is the importance of considering dosage, timing and localization of DUX4 expression to best achieve a progressive muscle-wasting phenotype while minimizing lethality and/or toxicity.
In our previous studies, we presented an injected DUX4 messenger RNA (mRNA) zebrafish model that successfully recapitulated several key FSHD-1 phenotypes including asymmetric abnormalities such as reduced pigmentation of the eyes, altered morphology of hearing structures, developmental abnormality of fin muscle, disorganization of facial musculature and degeneration of trunk muscle later in development (9). In this manuscript, we investigated the late-onset disease pathology in a zebrafish model that is more representative of the human disease. We show that within a cohort of DUX4-injected larvae, a subgroup exhibited a severe muscle phenotype as early as 4 days post-fertilization (dpf) (DUX4i_L) but that the remaining subgroup that was initially considered unaffected larvae began to show signs of skeletal muscle abnormalities between 8 and 12 weeks (DUX4i_A). To complement these injection model studies, we generated a skeletal muscle-specific DUX4-inducible transgenic model larvae (DUX4t_L) and adult (DUX4t_A) (see nomenclature in Table 1). This model allowed us to investigate the consequences of DUX4 expression during development and the unexpected adult onset and muscle pathology that developed after DUX4 expression was no longer detected.
Table 1.
Nomenclatures for injection and transgenic zebrafish models
| Injection zebrafish model (DUX4i) | Nomenclature |
|---|---|
| Larvae | DUX4i_L |
| Adult with no phenotype at 5 dpf | DUX4i_A |
| Transgenic zebrafish model (DUX4t) | |
| Larvae | DUX4t_L |
| Adult | DUX4t_A |
The utility of both models has led us to formulate a novel hypothesis that FSHD is a developmental disease and that bursts of DUX4 expression at early stages of embryonic development are sufficient to induce FSHD-like phenotypes in adult stages. Our findings highlight the importance of monitoring long-term effects evoked by low levels of embryonic DUX4 expression on muscle development and provide a more accurate model of FSHD for future drug screens and therapeutic discovery.
Results
Generation and characterization of transgenic DUX4 zebrafish model (DUX4t)
To enable control of DUX4-fl expression timing and dosage, we generated a DUX4-fl-inducible transgenic line driven by a myosin light chain 2 promoter and crossed this with a transgenic line expressing ubiquitous 4-hydroxytamoxifen-controlled Cre recombinase from the ubi:CreERt2 transgene (Fig. 1A and B) (14,15). Addition of (Z)-4-hydroxytamoxifen induces CreERt2-mediated loxP excision and subsequent activation of the DUX4-mCherry fusion transgene. In the absence of tamoxifen and CreERt2-driven recombination, myofibers are EGFP-positive but become EGFP-negative upon loxP excision. CreERt2-mediated loxP system turns on DUX4 but it is not designed to turn off DUX4 expression. This dose-dependent CreERt2-mediated activity enables us to titer levels of DUX4-fl expression for viability in the context of developmental and aging studies as well as planning for future drug screens.
Figure 1.
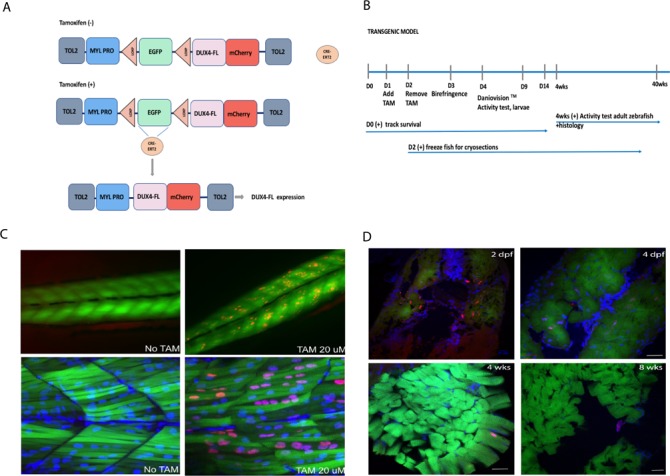
(A) Scheme of tamoxifen-inducible Cre–LoxP construct of transgenic zebrafish. The loxP-EGFP-loxP stop cassette and DUX4-fl coding sequence are placed downstream of fast muscle-specific mylz2 promoter. The transgene was inserted into zebrafish genome by Tol2 transposon-mediated transgenesis. Expression of DUX4-fl-mCherry fusion protein (mCherry is fused to the C-terminus of DUX4-fl) occurs when tamoxifen-inducible CreERt2 recombinase cuts off LoxP sites. (B) Timeline of the experiments from 0 dpf to 40 weeks. (C) Top panel shows a low-magnification image of a control (left) and tamoxifen-treated fish (right) at 3 dpf. DUX4 distribution is scattered and expressed asymmetrically along the length of the fish. Bottom panel shows a high-magnification image of a control (left) and tamoxifen-treated fish (right). DUX4 positive nuclei are shown in red while all nuclei are denoted by DAPI staining in blue. Tamoxifen treatment results in mosaic expression of DUX4 across myofibers and also varying levels of DUX4 expression as observed by degrees of `redness’. (D) EGFP (green; myosin light chain 2 promoter), Hoechst (blue; nuclear stain), mCherry (Red; DUX4 stain) at 2 and 4 dpf in DUX4t_L and DUX4t_A at 4 and 8 weeks; 20 μm scale.
DUX4-fl expression declined over time becoming undetectable in the majority of the fish at 8 weeks old and undetectable in all fish at 16 weeks old (Fig. 1C and D). Our results suggest that developmental activation of DUX4-fl results in consequences that lead to manifestation of FSHD-like phenotypes in later adulthood.
Abnormalities in the skeletal muscle structure in DUX4t_L
To activate DUX4-fl expression, embryos were treated with 20 μM tamoxifen at 1 dpf, for 24 h. Control embryos were treated with the same dosage of tamoxifen. Tamoxifen dosage was optimized for both the survival of embryos as well as the expression of the DUX4-fl mutation. Upon activation of DUX4-fl, expression pattern (appearance of mCherry) was visualized and evaluated by the use of confocal microscopy. After 24 h, embryos were washed with water to remove tamoxifen and allowed to incubate in normal conditions prior to observation of the effect of DUX4 expression. Interestingly, DUX4 activation was not homogenous across all myofibers and demonstrated a mosaic expression. In these embryos, DUX4 activation was observed in multiple nuclei in ∼70% of fibers. This muscle-specific mosaic expression pattern mimics the expression pattern of DUX4 in patient muscle cells. To further characterize the effect of DUX4 expression in DUX4t_L muscle, embryos were evaluated for muscle structure (birefringence, hatching time), locomotor function (automated movement tracking, DanioVision™, ThermoFisher Scientific, Walthman, MA, USA) and DUX4-fl-mCherry intensity (Arraycan™, ThermoFisher Scientific, Walthman, MA, USA). The exact timeline of the consecutive experiments is presented in Figure 1. A marked drop in embryos survival was observed when tamoxifen dosage exceeded 20 μm; therefore, we selected 20 μm as the most optimal dosage. All fish subjected for either short or long-term experiments were treated only once with 20 μm tamoxifen.
DUX4t_L zebrafish survival was tracked for 2 weeks following the treatment with tamoxifen and activation of DUX4-fl transgene. No significant differences in lifespan were observed in the DUX4t_L zebrafish in comparison to control zebrafish (Fig. 2A). To investigate the effect of DUX4 expression on muscle structure, birefringence (3–4 dpf) assay was used. About 30% of DUX4t_L displayed abnormal, patchy birefringence as early as at 3 dpf whereas control fish showed normal birefringence (Fig. 2B). Fish exhibiting abnormalities by birefringence assay were subjected for functional mobility test (DanioVision™) to assess their swimming behavior (Fig. 2C). Moreover, to explore the relationship between the amount of DUX4 expression and swimming behavior of larval fish, DUX4-fl-mCherry intensity assessment (Arrayscan™) was performed. We observed that the group of fish exhibiting patchy birefringence at 4 dpf displayed higher level of DUX4-fl (measured on Arrayscan™) and swam a significantly shorter distance when compared to the control group (Fig. 2D and E). To examine the skeletal muscle histology, we performed hematoxylin and eosin (H&E) staining on cryosectioned DUX4t_L zebrafish. Interestingly, we noticed an accumulation of centrally located nuclei as early as 7–9 dpf in DUX4t_L indicating that expression of DUX4 during embryonic development leads to skeletal muscle abnormalities during early developmental stages in zebrafish (Supplementary Material, S1).
Figure 2.
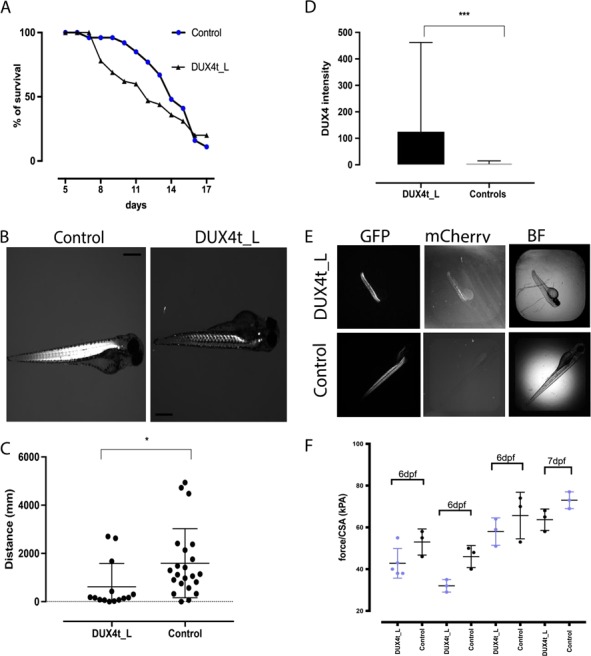
(A) Survival curve of the DUX4t_L and control zebrafish from 5 to 18 dpf. (B) Birefringence images of DUX4t_L and control zebrafish at 3 dpf. (C) Daniovision™ activity test of DUX4t_L with abnormal birefringence and control zebrafish at 4 dpf. (D) Muscle fiber tetanic force measurement per CSA in DUX4t_L and control zebrafish at day 6 and 7; data collected in triplicates for day 6 and 7; P < 0.05. (E) DUX4t_L intensity measurements (2.5× objective) by Arrayscan™ at 4 dpf. (F) Images (GFP, mCherry, BF) of DUX4t_L and control zebrafish, Arrayscan™; P < 0.001.
Muscle contractility in DUX4t_L
Twitch and tetanic force were measured in axial muscle of control and DUX4t_L fish. Four different clutches of fish, each consisting of three controls and three to five DUX4t_L, were studied. Fish were 6 dpf except for one clutch where they were 7 dpf. Data were analyzed using a two-way analysis of variance with main effects of clutch number and genotype. For both twitch and tetanic force, there were significant clutch number (P < 0.0001 for twitch and tetanic force) and genotype effects (P = 0.0168 and 0.0018 for twitch and tetanic force, respectively) but no interaction. The differences in muscle fiber tetanic force measurement per cross-section area (CSA) in DUX4t_L and control zebrafish at day 6 and 7 were significant; however, the mean was consistently lower for DUX4t_L fish. Thus, while force pooled across genotype varied across clutches, preparations from DUX4-expressing fish consistently showed lower twitch and tetanic force (Fig. 2F).
Abnormalities in the skeletal muscle structure in adult transgenic (DUX4t_A) zebrafish
To further characterize the effect of DUX4 misexpression in adult skeletal muscle, DUX4t_A zebrafish were analyzed at 12 months post tamoxifen induction of DUX4-fl expression. Zebrafish were analyzed by electron microscopy and by H&E, trichrome (collagen), neutral lipid and fluorescent staining to evaluate muscle structure disorganization and DUX4-fl expression in adult muscles. To evaluate the ultrastructure of DUX4t_A zebrafish, electron microscopy was performed on 12-month-old fish. Control and DUX4t_A zebrafish skeletal muscle showed no significant differences in sarcomeric organization at the larval stage (4 dpf). Interestingly, aged fish (12 months old) exhibited swollen sarcoplasmic reticulum and occasional accumulation of lipid bodies as observed in human FSHD patients (Fig. 3). DUX4t_A zebrafish (14 dpf to 48 weeks of age) exhibited a varying level of mild inflammation as well as muscle fiber replacement by fat and/or collagen (Fig. 4A and B). This observed difference in the severity of the phenotype closely correlates to the observed heterogeneity of FSHD-1. Collagen accumulation was also significantly higher in DUX4t_A (8–12 weeks old) compared to the controls with a tendency for increased accumulation over time. Moreover, we observed a trend of left to right asymmetry in collagen deposition in 8 weeks old DUX4t_A versus age-matched control zebrafish (Fig. 4C and D). More collagen accumulation was observed in the left myotome in these fish in comparison to the right myotomal area. Additionally, the disease phenotype is more pronounced in skeletal muscles located in proximity to the caudal fins. Interestingly, asymmetry of the caudal fin and muscles abnormalities close to that region were only observed in fish ranging from 8 weeks to 10 months old, but not at earlier stages (Fig. 4E).
Figure 3.
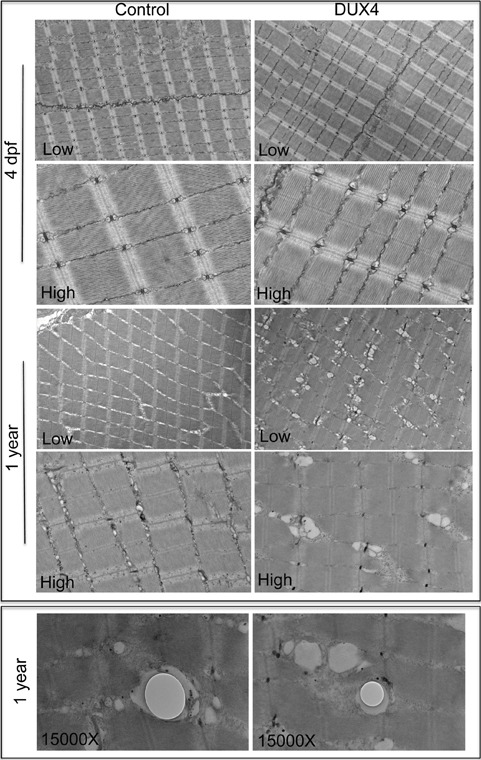
Electron micrographs of longitudinal sections at 4 dpf (DUX4t_L) and 1 year (DUX4t_A) transgenic zebrafish (n = 2 for each group). Control samples are tamoxifen-treated DUX4-inducible fish without Cre transgene, which effectively do not express DUX4. DUX4 samples are tamoxifen-treated DUX4-inducible fish with a Cre transgene to enable DUX4 transgene expression. Low- and high-magnification images of control and DUX4t_L fish at 4 dpf show normal sarcomeric striations and with discernable differences. However, low- and high-magnification images of 1-year aged samples show DUX4t_A have swollen endoplasmic reticulum (ER) compared to control. Presence of lipid droplets was also observed in DUX4i_A zebrafish.
Figure 4.
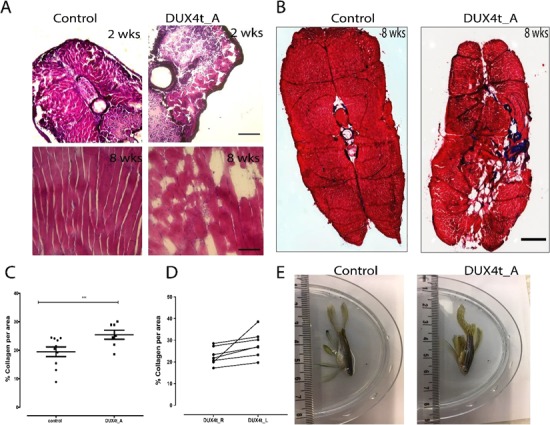
(A) H&E staining of control and adult DUX4 transgenic zebrafish (DUX4t_A) cryosections of muscles close to caudal fin, at 2 and 8 weeks; 20× objective; 500 μm scale. (B) Trichrome staining (collagen in blue, cytoplasm in red) of control and DUX4t_A cryosections of adult zebrafish muscles close to the caudal fin at 8 weeks; abnormal collagen accumulation seen for DUX4t_A; 20× objective; 500 um scale. (C) Collagen quantifications for DUX4t_A (n = 10) and tamoxifen treated controls (n = 7); P < 0.05. (D) Trend in right (DUX4t_R) to left (DUX4t_L) side asymmetry in collagen accumulation in DUX4t_A zebrafish; P > 0.05. (E) Asymmetry of the muscle structure and caudal fin in 1 year, DUX4t_A versus control zebrafish.
Asymmetrical fat accumulation was detected in 4-month-old DUX4t_A (Fig. 5).
Figure 5.
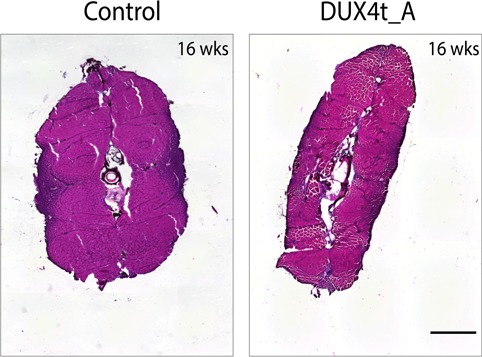
H&E staining of control and adult DUX4 transgenic (DUX4t_A) zebrafish muscles located close to the caudal fin, at 16 weeks; abnormal, asymmetrical fat accumulation is seen between myofibers in DUX4t_A only; 500 μm scale.
Abnormalities in the skeletal muscle function in DUX4t_A zebrafish
In order to assess the impact of DUX4 overexpression on adult fish, we measured maximal swimming speed (Umax) of DUX4t_A fish at 32 weeks of age. Transgenic expression of DUX4 had no effect on body length and mass as these variables were similar for DUX4t_A and control fish. There was a tendency (P = 0.089) for Umax speed to be reduced in the DUX4t_A fish as Umax of the adult control fish averaged 15.1 ± 0.3 body lengths (BL)/s (n = 9) compared to 13.3 ± 0.9 BL/s (n = 9) for transgenic fish (Fig. 6, left). However, there was considerably more variability in Umax of transgenic fish compared to control fish. The range of Umax for the transgenic fish was twice that of control fish with over half of the transgenic fish showing a slower Umax than the slowest control fish (16).
Figure 6.
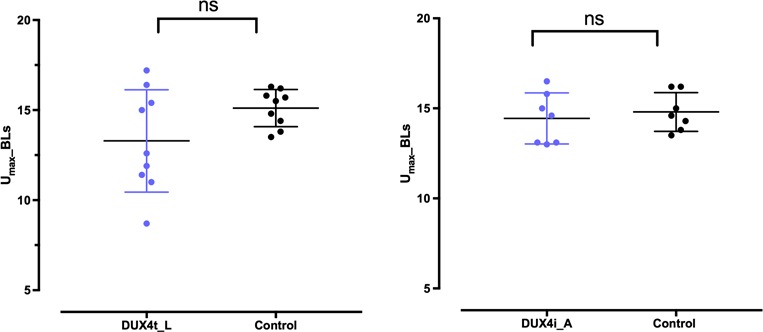
Umax DUX4 transgenic adult zebrafish (DUX4t_A) at 32 weeks (P < 0.05; left) and on DUX4-injected adult zebrafish (DUX4i_A) at 16 weeks (P > 0.05; right).
Abnormalities in the skeletal muscle function in DUX4i_A zebrafish
To evaluate the effect of DUX4 misexpression during development on swimming performance as adults, Umax was evaluated in DUX4i_A fish. There was no difference in the Umax of control fish receiving no injection as embryos (14.5 ± 0.7 BL/s; n = 4) and those receiving an injection of vehicle (14.3 ± 0.4 BL/s; n = 3) so these two groups were combined for comparison to the DUX4i adults. DUX4-injected fish reached exhaustion at an average flow velocity of 14.4 ± 0.5 BL/s (n = 7) that was similar to that of the control fish (Fig. 6, right). However, while most of the DUX4-injected fish had Umax values within the range of the control fish, a cluster of three injected fish had velocities that placed them at the very low-end of normal Umax values (16). This variability, we think, presumably depends on the stage of the ongoing muscle degeneration and regeneration process that is visually distinguishable for DUX4i_A and control animals. Here, we emphasize that for that activity test we selected DUX4i_A that did not display a strongly affected muscle phenotype close to the caudal fins, which we assessed by eye at 8 weeks old. We assumed that zebrafish with such abnormalities were too severely affected to undergo the Umax evaluation.
Abnormalities in the skeletal muscle structure in DUX4i_L and DUX4i_A zebrafish
To support our hypothesis that DUX4-fl expressed at early stages is sufficient to induce FSHD-like phenotypes later in life, we used our previously published DUX4-fl-injected zebrafish model (9) and utilized a subset for aging and downstream experiments as described in Figure 7A. Briefly, we injected 0.2 pg DUX4-fl mRNA (∼1–2 × 105 copies) into one-cell stage embryos and monitored the onset of abnormal phenotypes (9). We noticed a shorter lifespan as well as delayed hatching in DUX4i_L compared to controls (Fig. 7B and C). Both lifespan and hatching rates appear to be more affected in the DUX4i_L than in DUX4t_L zebrafish. The DUX4i_L resulted in ∼30% of fish that displayed a spectrum of abnormal phenotypes (body shape abnormalities, abnormal fin development and altered eye pigmentation) at 3–5 dpf whereas the remaining 70% of DUX4i_L had either died or showed no observable abnormalities upon hatching. The subset of DUX4i_L showing no gross developmental abnormalities were aged for 12 weeks in order to monitor their long-term fate and a potential delay in phenotypic onset. Our aging experiments indeed revealed muscle disorganization within proximity to the caudal fin, as well as muscle regeneration in proximity to the dorsal fin via H&E staining (Fig. 8A). Additionally, abnormal collagen and fat deposition in these 12-week-old DUX4i_A-injected fish were also detected (Fig. 8B, C and D).
Figure 7.
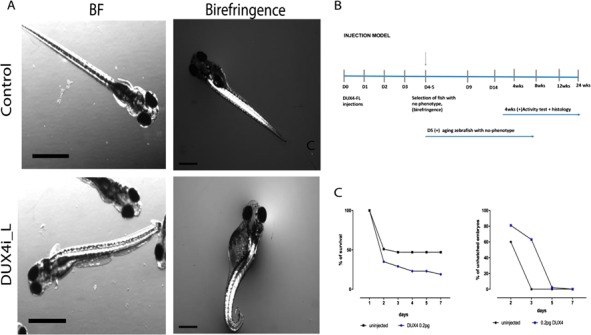
(A) Phenotype characterization of DUX4-injected larvae (DUX4i_L); BF and birefringence at 4 dpf; 120 μm scale. (B) Timeline of the experiments from 0 dpf to 3–4 weeks. (C) Survival curve (%) for DUX4(FL) 0.2 pg injected and uninjected controls (left); % of unhatched embryos in DUX4i_L and uninjected embryos (right).
Figure 8.
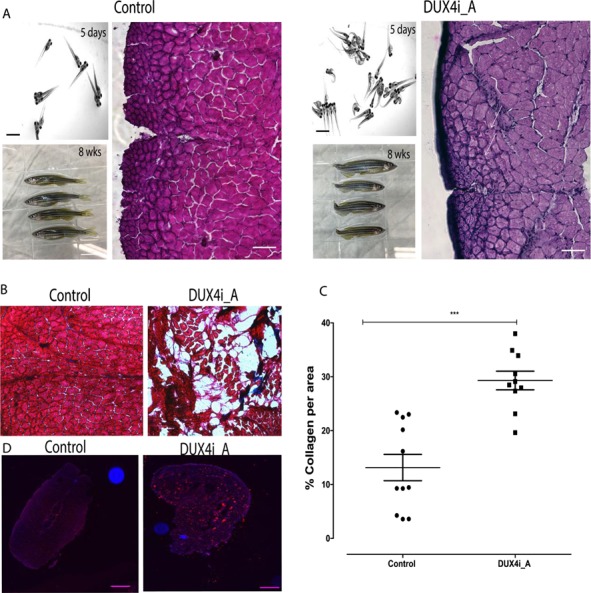
(A) Zebrafish were injected with DUX4 at 0 dpf, phenotype was assessed at 5 dpf for both control and DUX4 injected fish; 120 μm scale. At 8 weeks, abnormal tail fins were observed in fish that did not display phenotype at day 5; 1 cm scale. H&E staining for 8 week control and adult DUX4-injected zebrafish (DUX4i_A) presents regenerated nuclei only in DUX4i_A 8 weeks versus control zebrafish; 500 μm scale. (B) Trichrome staining of control and DUX4i_A, red for cytoplasm and blue for collagen; 20× objective. (C) Quantifications of collagen accumulation in control, DUX4 transgenic and injection model at 12 weeks. (D) Neutral lipid stain of control and DUX4i_A, red for lipid stain and blue for DAPI; 500 μm scale.
Discussion
FSHD has proven to be a difficult disease to model in vertebrate animals due to the primate-specific origins of DUX4 and the complex epigenetic components of the disease. Although several animal models of the disease exist, none of those fully recapitulate the phenotypic spectrum of human FSHD. The heterogeneity associated with FSHD including variable age of onset, disease severity and progression as well as the unique non-muscle phenotypes (hearing and vision loss) adds to the difficulties in modeling the disease in the laboratory (17). Another puzzling feature of the disease is the lack of correlation between the temporal and spatial consequences of DUX4 expression, with disease onset and affected muscle groups, respectively (18). In this manuscript, we have explored several methods of inducing DUX4 expression in zebrafish and as a result report a more accurate model of FSHD, particularly in relation to DUX4 expression causing a delayed onset of asymmetric muscle phenotypes. Zebrafish are a powerful laboratory tool to study muscle development and modeling of muscle diseases.
They are advantageous over other species during their developmental stage (embryo and larval) where they are transparent and allow for muscle phenotypes to be easily observed. During this developmental window, zebrafish also permit uptake of small molecule compounds from surrounding water and thus prove optimal for administration of pharmacological agents for drug testing or activation of gene expression. The limitation, however, might be the fact that some embryos absorb drugs irregularly which might lead to variations in the DUX4 expression levels in our transgenic model as shown in Figure 2D.
In our laboratory, we have exploited this property to observe DUX4-inducible expression through administration of tamoxifen in fish water during the developmental window. Addition of tamoxifen induces irreversible excision and recombination events that activate muscle-specific DUX4 expression in a mosaic manner. This mosaic nature of DUX4-positive nuclei in our model is more representative of DUX4 expression in human FSHD muscle, where DUX4 expression occurs in random `bursts’ (19,20). We believe this mosaic expression of DUX4 in our model allows the survival and phenotypic characterization into adulthood, instead of premature embryonic lethality typically associated with DUX4 overexpression models. Characterization of this model has led us to conclude that it represents a mild form of FSHD, where collagen and fat deposition exist but are not as prominent as in more severe forms of FSHD, such as that represented in the advanced stages of our adult DUX4 injection model (Supplementary Material, S2). Characterization of our transgenic model revealed variable histological phenotypes associated with fat and collagen accumulation as well as rather rarely seen muscle inflammation. Interestingly, functional testing performed using this model delivered uniform results and, regardless, may serve as a valuable outcome measurement for prospective drug screenings.
In this manuscript, we have also reported further characterization of our previously published DUX4 injection model (9), where injected larvae that appear phenotypically indistinguishable from controls are selected for further aging (DUX4i_A). The subset of injected larvae exhibiting phenotype early at development represents a `severe form of FSHD’ and resembles rare infantile form of FSHD (21). The adult DUX4 injection model, however, is more representative of the typical adult onset FSHD and is a more severe model relative to our transgenic inducible model. The FSHD-like phenotype we observe in adult DUX4-injected zebrafish is more uniform and severe histologically, where we observed that collagen and fat accumulate at a higher percentage compared to the transgenic model (Supplementary Material, S2). However, in contrast to our transgenic inducible model, functional evaluation of adult DUX4-injected zebrafish varies and presumably depends on the stage of muscle regeneration and degeneration close to the caudal fin that occurs usually between 8 and 12 weeks. We noticed that some of DUX4i_A (Fig. 4D) form abnormal caudal fins that consist of caudal fin musculature, skeleton, musculature (12 muscles that are arranged in a superficial and a deep muscle layer), vasculature, fat, nerves and rays (principal and percurrent). We need to perform more studies to understand which of these components are primarily linked to the phenotype we observe. Caudal fin formation is divided into five stages related to the number of fin rays forming over time. Stage 5 is the last one (when 18–24 fin rays are developed) and has been characterized for adult zebrafish (90 days). It is assumed that muscle formation as well as caudal fin formation is completed when fish became 3 months old. Since our fish were collected at 8 weeks old (which was the earliest timepoint we found abnormal tail fin formation), we think that caudal fin might still have been undergoing the natural developmental process which could have been disturbed by DUX4 expression.
Moreover, our unpublished yet gene ontology analysis from chromatin Immunoprecipitation (CHIP) sequencing of DUX4-injected embryos at 12 h (6 somite stage) revealed a group of genes being involved in the fin regeneration process that indicates that DUX4 expression might disturb fin formation early in development. To fully understand the role of DUX4 in abnormal fin formation in zebrafish, more studies have to be conducted (22).
In concordance with our observations, activation of DUX4 misexpression during embryonic development results in a mild form of FSHD in adulthood using our transgenic model and a more severe form of FSHD using our adult DUX4-injected model. This difference can be attributed to several factors—firstly, the global expression of DUX4 in the injected model compared to the muscle-specific expression in the transgenic model; secondly, differences in DUX4 dosage per cell (tamoxifen-inducible Cre versus human DUX4 mRNA); thirdly, difference in time window of DUX4 expression (1-cell stage for injected model and 26-somite to prim-6 stage for transgenic inducible model). Nevertheless, both models successfully recapitulate late-onset muscle degenerative phenotype and asymmetry associated with FSHD and are suitable for future candidate drug screens to ameliorate disease-associated phenotypes.
Interestingly, embryonic DUX4 activation resulted in differences in phenotypic onset between the transgenic inducible (4 weeks to 10 months, although in some fish even earlier–7 to 14 days) and adult DUX4-injected models (5 days). The transgenic model demonstrates a consistent decline in DUX4 expression over time and the subsequent development of abnormal muscle phenotypes in the form of unilateral collagen deposition, muscle fiber replacement with fat and mild inflammation. Together, our findings highlight that DUX4 expression during early muscle development is important to induce FSHD pathology in later life. Both models characterized in our study point to a potential developmental role in FSHD pathogenesis, where early DUX4 expression may not necessarily manifest in detectable symptoms until adulthood. The comparison of two zebrafish models is summarized in Table 2. These findings, when translated to human FSHD, suggest that although FSHD clinical symptoms are detected in early adulthood, patient muscles were `primed’ for degeneration as a result of DUX4 misexpression during their early life. This is the first time that a developmental role for DUX4 in FSHD such has been proposed and was only achieved through our continued attempt to model this disease in zebrafish. Our results also raise the potential concern for use of DUX4-target therapies in adulthood that may not be as efficacious as expected if the molecular signals underlying FSHD pathogenesis has commenced in early life.
Table 2.
A comparison of DUX4t_L, DUX4t_A, DUX4i_L and DUX4i_A zebrafish models
| DUX4 injection model (DUX4i) | Dux4 transgenic model (DUX4t) | |
|---|---|---|
| Timing of DUX4 expression | One-cell stage embryo | Twenty-six somite to prime six |
| Dosage and location of DUX4 expression | 0.2 pg DUX4-fl mRNA (∼1 – 2 × 105 copies); whole body | Muscle-specific DUX4 activation, ∼70% fibers positive for DUX4 |
| Early phenotype (3–5 dpf) | DUX4i_L: abnormal eye and ear formation, fin asymmetry, abnormal birefringence, late hatching | DUX4t_L: occasionally curved bodies, abnormal birefringence in ∼30% fish, late hatching |
| Muscle function and mechanistic (3–5 dpf) | DUX4i_L: muscle activity and mechanistic not tested | DUX4t_L: swim with less distance, lower twitch and tetanic force |
| Late phenotype (adult) | DUX4i_A: neutral lipid and collagen accumulation, inflammation, fin and muscle regeneration | DUX4t_L: asymmetrical fat deposition, collagen accumulation, occasionally inflammation |
| Muscle function (adult) | DUX4i_A: not measured in severely affected, normal in mildly affected | DUX4t_L: fully abnormal |
| Disease severity | DUX4i_L: mild–severe DUX4i_A: mild–severe | DUX4t_L: mildDUX4t_A: mild |
Materials and Methods
Fish strains and maintenance
Zebrafish wild-type AB strain used for controls and DUX4-fl injections as well as our DUX4-fl transgenic lines were maintained in the Boston Children’s Hospital Zebrafish facility under approved protocols. Zebrafish embryos were raised at 28.5°C and cared for as described previously (23).
Generation of DUX4 transgenic line
Zebrafish myosin light chain 2 (mylz2) promoter sequence was PCR (Polymerase Chain Reaction) amplified from zebrafish AB line genomic deoxyribonucleic acid (DNA) with primers 5′-ACCGCTCGAGATTCGCCACAGAGGAATGAGCC-3′ and 5′-TCAAGCTTGTCGAGACGGTATGTGTGAAGTC-3′ and cloned into pENTR5′-TOPO vector (Invitrogen). The mylz2 promoter sequence was digested with XhoI and HindIII and subcloned into pENTR5′_ubi:loxP-EGFP_loxP (14) to obtain P5′E: Mylz:loxP_EGFP_loxP. The DUX4 transgenic line was generated using Tol2-mediated transgenesis (Tol2-transposable element of Oryzias latipes, number 2) (24,25). Transgenesis vectors (P5′E: Mylz:loxP_EGFP_loxP, P3′E: mCherry-pA, pME: DUX4-FL, pDestTol2A2) were assembled by MultiSite Gateway system using LR Clonase II Plus (Invitrogen) (26). The resulting DNA construct, pDestTol2A2_mylz2:loxP-EGFP-loxP-DUX4-fl-mCherry, was injected with Tol2 transposase mRNA into one-cell-stage zebrafish eggs obtained from AB wild-type crosses. Injected fish were grown to adulthood and F0 parent generation (filial 0) founders were outcrossed. Positive first filial generation (filial 1) individuals were screened for their transgenesis marker (mylz2:EGFP) using UV (ultraviolet) filter glasses. The resulting transgenic line was crossed with the ubi:CreERt2 line (gift from Zon laboratory) to enable DUX4 induction via tamoxifen treatment.
Tamoxifen treatment for CreERt2 induction
4-Hydroxytamoxifen (tamoxifen, Sigma) was dissolved in 100% ethanol to achieve a final concentration of 10 mM and stored in −20 C aliquots. To induce Cre-recombinase activity in DUX4-Cre lines, 1 dpf embryos (20–30) were placed in 6 well culture plates and tamoxifen was added to fish water at a final concentration of 20 μm.
Muscle birefringence and motor function of DUXt_L zebrafish larvae
Muscle structure was analyzed by birefringence at 3–4 dpf as previously described (23). Briefly, we placed anesthetized zebrafish on a glass-polarizing filter then subsequently covered them with a second polarizing filter that enabled detection of normal or abnormal birefringence. Images were taken on a Nikon SMZ1500 stereomicroscope with Openlab Software version 3.1.5 (Improvision, Nikon, Melville, NY, USA). Automated activity test was performed by an automated infrared movement detector DanioVision™ (Noldus, Sturbrige, MA, USA), designed for the high-throughput testing of zebrafish larvae in multi-well plates.
Fluorescence imaging of DUX4t_L
DUX4t_L zebrafish were snap frozen, stored at −80 C and sectioned in 10 μm increments on a Leica cryostat (from tail to head). Immunostaining was performed on zebrafish cryosections (10 μm) for nuclear visualization with Hoechst (1:5000). EGFP (green) and mCherry (red) labeling DUX4 integrated into the construct and did not require any additional antibodies for visualization. To visualize DUX4 expression, images of fluorescence slides were captured on a Zeiss LSM 700 laser scanning confocal microscope [located in Cellular Imaging Core (BCH)].
DUX4-fl-mCherry intensity quantifications in DUX4t_L
For in vitro quantifications of DUX4-fl-mCherry intensities in DUX4t_L, an image-based high-content analysis (HCA) platform—Thermo Fisher Arrayscan XTi—was used to monitor expression in zebrafish larvae. Both DUX4-fl and control zebrafish were anesthetized at 4 dpf and submerged in 2% gelatin in a well of a 384 well plate. Plates were scanned by use of the 2.5× objective for EGFP, mCherry, as well as bright field (BF) signals. Intensities were calculated by use of the adjusted algorithms determined by HCA software.
Injection model
DUX4-injected zebrafish model has been recently established in our laboratory by Mitsuhashi, H. et al. (9). Briefly, human DUX4 mRNA (0.2 pg; ∼1–2 × 105 copies) was introduced into one-cell stage fertilized zebrafish eggs. As shown previously, ∼30% of injected fish displayed abnormal spectrum of phenotypes whereas the rest did not manifest any clear abnormalities. The first group of fish with clear phenotype was directly subjected for structural and functional tests. The second group of fish with no distinct phenotype was selected for further studies and monitored for their long-term effects of DUX4 exposure.
Histology: H&E, collagen and neutral lipid staining
Zebrafish cryosections of 10 μm were obtained and stored at −80 C as described above. Sections were taken for nuclear (hematoxylin) and cytoplasmic (eosin) staining. Sections were briefly washed with 1× PBS (phosphate-buffered saline), incubated for 3 min in hematoxylin solution (VWR, Radnor, PA, USA; no. 95057-844), washed 3× in water, incubated for 3 min in eosin (VWR, Radnor, PA, USA; no. 95057-848) and dehydrated in 70, 80, 90 and 100% ethanol solutions for 30 s. Next, cryosections were incubated in xylenes (Sigma-Aldrich, USA, no. 534056) for 10 min to remove any traces of ethanol and mounted in a xylene-based mounting medium (Cytoseal™ 60, Thermo-scientific, Runcorn, UK; Ref 8310-16).
Fat staining was performed on 10 μm cryosections with the HCS LipidTOX™ Red Neutral Lipid Stain (Invitrogen, H34476) with minor modifications to standard protocols. Cryosections were air dried for 1 h, fixed in 4% PFA (paraformaldehyde) for 1 h and incubated for 2 h in neutral lipid solution (1:200). Sections were then mounted using mounting medium containing DAPI (Vector Laboratories, Vectashield Mounting Medium with DAPI H-1200).
For collagen visualization, cryosections (10 μm) were stained with Sigma Masson’s Trichrome (no. HT15-1KT, Sigma-Aldrich) with slight modifications. Briefly, sections were fixed overnight in Bouin’s solution, immediately stained with Biebrich acid staining and destained with phosphotungstic–phosphomolybdic acid solution. Next, sections were submerged in aniline blue and were not differentiated in 1% acetic acid at the completion of the staining. Nuclei were not stained so that images could be quantified by densitometry of red:blue pixels. Slides were cover-slipped with cytoseal™60 Thermo-scientific, Runcorn, UK; Ref 8310-16 and allowed to dry prior to imaging. The quantification method comprises deconvolution of image channels by grouping pixels with a similar color with FIJI (ImageJ). The area of each image is calculated by normalizing the number of pixels of each channel to 100% and subtracting the relative background and non-stained areas as described by Ruifrok et al. (27). Calculations were then performed to determine the percentage of red pixels (myofibers) to the percentage of blue pixels (collagen). All images (H&E, fat and collagen staining) were taken with Nikon Ti Eclipse Inverted Microscope (BCH).
Muscle mechanics of DUX4t_L zebrafish
Larvae (6–7 dpf) were anesthetized with tricaine. The head was removed and body was submerged in a fish bicarbonate buffer that was maintained at 25 C and equilibrated with 95% O2 and 5% CO2 as Widrick et al. (28). One end of the preparation, aligned with the gastrointestinal opening, was attached to a titanium wire extending from an isometric force transducer using 10-0 monofilament suture. The other end, aligned several myotomes proximal from the tip of the tail, was attached in a similar manner to the arm of a position motor. Supramaximal pulses (200 μs in duration) were generated by a constant current muscle stimulator and delivered to platinum electrodes flanking the preparation. Brief tetni (30 ms at 300 Hz), separated by ≥60 s to minimize fatigue, was used to establish the optimal length for tension (Lo). Once Lo had been established, twitch and tetanic forces were recorded for analysis. The CSA of the preparation was calculated as an ellipse by measuring maximal preparation width at its attachment to the force transducer, carefully rotating the preparation 90° and measuring preparation depth. Active twitch and tetanic force were normalized to CSA as described previously (28).
Measurement of Umax in DUX4t_A zebrafish
Adult zebrafish were studied in a flume designed specifically for zebrafish. Individual fish were evaluated in a 15 cm long cylindrical polycarbonate swimming section with an interior CSA of 5.07 cm2. Water passed through an array of plastic straws to ensure that flow was laminar in the swimming section. All water exiting the swimming section passed through flow meters. Data from the meters were processed by an open-source microcontroller (Arduino model 101) to yield an observed flow rate (averaged every 5 s). The protocol flow rate was controlled by the microcontroller which was pre-programmed to modulate the output of two spherical impeller pumps that drew water through the flume. Umax was determined at a water temperature of 25°C using a modification of the protocol described by Gilbert et al. (29). Fish were given a 20–30 min acclimatization period at a flow of ≈2.5 cm/s. Flow was then increased to 10 cm/s followed by 2.5 cm/s increments every 30 s. Exhaustion occurred when the fish could no longer maintain its position in the swimming section, was swept back against the downstream screen and would not resume swimming despite several sharp taps to the side of the swimming section. Umax was calculated as the average of the three flow measurements immediately preceding exhaustion. After 5 min of recovery (at ≈2.5 cm/s), the fish was removed from the flume and lightly anesthetized with tricaine. Standard length (the distance from the tip of the snout to the caudal peduncle) and body mass were recorded. The fish was then returned to its aquarium for recovery (30).
Electron microscopy
Zebrafish embryos (DUX4t_L at 4 dpf) were fixed in formaldehyde–glutaraldehyde–picric acid in cacodylate buffer overnight at 48°C followed by osmication and uranyl acetate staining. Subsequently, embryos were dehydrated in a series of ethanol washes and finally embedded in Taab epon (Marivac Ltd, Nova Scotia, Canada). Ninety-five nanometer sections were cut with a Leica ultracut microtome, picked up on 100 m formvar-coated Cu grids and stained with 0.2% lead citrate. Sections were viewed and imaged under the Philips Tecnai BioTwin Spirit Electron Microscope (Electron Microscopy Core, Harvard Medical School).
Statistical analysis
All statistical analysis and graphs were performed on Graphpad Prism Version 7. All figures were prepared in Adobe Illustrator CC 2017.
Supplementary Material
Acknowledgements
We thank the Harvard Medical School Electron Microscopy Facility for their service in preparation of our samples. We thank Koichi Kawakami (National Institute of Genetics, Japan) for the Tol2 transposon system. We also thank Lee Barret from Boston Intellectual and Developmental Disabilities Research Center, Human Neuron Core (U54HD090255) and Anthony Hill from Boston Children’s Cellular Imaging core for facilitating authors a high-quality zebrafish imaging. We would like to thank Emanuela Gussoni for her intellectual input and valuable comments. We would like to thank Christian Lawrence for his superb management of the aquatics facility.
Conflict of Interest statement. None declared.
Funding
Senator Paul D. Wellstone Muscular Dystrophy Cooperative Research Center (U54HD060848).
References
- 1. Deenen J.C., Arnts H., Maarel S.M., Padberg G.W., Verschuuren J.J., Bakker E., Weinreich S.S., Verbeek A.L. and Engelen B.G. (2014) Population-based incidence and prevalence of facioscapulohumeral dystrophy. Neurology, 83, 1056–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Overveld P.G., Enthoven L., Ricci E., Rossi M., Felicetti L., Jeanpierre M., Winokur S.T., Frants R.R., Padberg G.W. and Maarel S.M. (2005) Variable hypomethylation of D4Z4 in facioscapulohumeral muscular dystrophy. Ann. Neurol., 58, 569–576. [DOI] [PubMed] [Google Scholar]
- 3. Geng L.N., Yao Z., Snider L., Fong A.P., Cech J.N., Young J.M., Maarel S.M., Ruzzo W.L., Gentleman R.C., Tawil R. and Tapscott S.J. (2012) DUX4 activates germline genes, retroelements, and immune mediators: implications for facioscapulohumeral dystrophy. Dev. Cell, 22, 38–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jones T.I., Chen J.C., Rahimov F., Homma S., Arashiro P., Beermann M.L., King O.D., Miller J.B., Kunkel L.M., Emerson C.P. Jr., Wagner K.R. and Jones P.L. (2012) Facioscapulohumeral muscular dystrophy family studies of DUX4 expression: evidence for disease modifiers and a quantitative model of pathogenesis. Hum. Mol. Genet., 21, 4419–4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Knopp P., Krom Y.D., Banerji C.R., Panamarova M., Moyle L.A., Hamer B., Maarel S.M. and Zammit P.S. (2016) DUX4 induces a transcriptome more characteristic of a less-differentiated cell state and inhibits myogenesis. J. Cell Sci., 129, 3816–3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Banerji C.R.S., Panamarova M., Hebaishi H., White R.B., Relaix F., Severini S. and Zammit P.S. (2017) PAX7 target genes are globally repressed in facioscapulohumeral muscular dystrophy skeletal muscle. Nat. Commun., 8, 2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bosnakovski D., Chan S.S.K., Recht O.O., Hartweck L.M., Gustafson C.J., Athman L.L., Lowe D.A. and Kyba M. (2017) Muscle pathology from stochastic low level DUX4 expression in an FSHD mouse model. Nat. Commun., 8, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lek A., Rahimov F., Jones P.L. and Kunkel L.M. (2015) Emerging preclinical animal models for FSHD. Trends Mol. Med., 21, 295–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mitsuhashi H., Mitsuhashi S., Lynn-Jones T., Kawahara G. and Kunkel L.M. (2013) Expression of DUX4 in zebrafish development recapitulates facioscapulohumeral muscular dystrophy. Hum. Mol. Genet., 22, 568–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wallace L.M., Garwick S.E., Mei W., Belayew A., Coppee F., Ladner K.J., Guttridge D., Yang J. and Harper S.Q. (2011) DUX4, a candidate gene for facioscapulohumeral muscular dystrophy, causes p53-dependent myopathy in vivo. Ann. Neurol., 69, 540–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Krom Y.D., Thijssen P.E., Young J.M., Hamer B., Balog J., Yao Z., Maves L., Snider L., Knopp P., Zammit P.S. et al. (2013) Intrinsic epigenetic regulation of the D4Z4 macrosatellite repeat in a transgenic mouse model for FSHD. PLoS Genet., 9, e1003415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dandapat A., Bosnakovski D., Hartweck L.M., Arpke R.W., Baltgalvis K.A., Vang D., Baik J., Darabi R., Perlingeiro R.C., Hamra F.K. et al. (2014) Dominant lethal pathologies in male mice engineered to contain an X-linked DUX4 transgene. Cell Rep., 8, 1484–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jones T. and Jones P.L. (2018) A cre-inducible DUX4 transgenic mouse model for investigating facioscapulohumeral muscular dystrophy. PLoS One, 13, e0192657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mosimann C., Kaufman C.K., Li P., Pugach E.K., Tamplin O.J. and Zon L.I. (2011) Ubiquitous transgene expression and Cre-based recombination driven by the ubiquitin promoter in zebrafish. Development, 138, 169–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hans S., Kaslin J., Freudenreich D. and Brand M. (2009) Temporally-controlled site-specific recombination in zebrafish. PLoS One, 4, e4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Seebacher F. and Walter I. (2012) Differences in locomotor performance between individuals: importance of parvalbumin, calcium handling and metabolism. J. Exp. Biol., 215, 663–670. [DOI] [PubMed] [Google Scholar]
- 17. Tonini M.M., Passos-Bueno M.R., Cerqueira A., Matioli S.R., Pavanello R. and Zatz M. (2004) Asymptomatic carriers and gender differences in facioscapulohumeral muscular dystrophy (FSHD). Neuromuscul. Disord., 14, 33–38. [DOI] [PubMed] [Google Scholar]
- 18. Rahimov F., King O.D., Leung D.G., Bibat G.M., Emerson C.P. Jr., Kunkel L.M. and Wagner K.R. (2012) Transcriptional profiling in facioscapulohumeral muscular dystrophy to identify candidate biomarkers. Proc. Natl. Acad. Sci. U. S. A., 109, 16234–16239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rickard A.M., Petek L.M. and Miller D.G. (2015) Endogenous DUX4 expression in FSHD myotubes is sufficient to cause cell death and disrupts RNA splicing and cell migration pathways. Hum. Mol. Genet., 24, 5901–5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Block G.J., Narayanan D., Amell A.M., Petek L.M., Davidson K.C., Bird T.D., Tawil R., Moon R.T. and Miller D.G. (2013) Wnt/β-catenin signaling suppresses DUX4 expression and prevents apoptosis of FSHD muscle cells. Hum. Mol. Genet., 22, 4661–4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Klinge L., Eagle M., Haggerty I.D., Roberts C.E., Straub V. and Bushby K.M. (2006) Severe phenotype in infantile facioscapulohumeral muscular dystrophy. Neuromuscul. Disord., 16, 553–558. [DOI] [PubMed] [Google Scholar]
- 22. Singleman C. and Holtzman N.G. (2014) Growth and maturation in the zebrafish, Danio rerio: a staging tool for teaching and research. Zebrafish, 11, 396–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kawahara G., Guyon J.R., Nakamura Y. and Kunkel L.M. (2010) Zebrafish models for human FKRP muscular dystrophies. Hum. Mol. Genet., 19, 623–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Urasaki A., Morvan G. and Kawakami K. (2006) Functional dissection of the Tol2 transposable element identified the minimal cis-sequence and a highly repetitive sequence in the subterminal region essential for transposition. Genetics, 174, 639–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kawakami K., Takeda H., Kawakami N., Kobayashi M., Matsuda N. and Mishina M. (2004) A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev. Cell, 7, 133–144. [DOI] [PubMed] [Google Scholar]
- 26. Kwan K.M., Fujimoto E., Grabher C., Mangum B.D., Hardy M.E., Campbell D.S., Parant J.M., Yost H.J., Kanki J.P. and Chien C.B. (2007) The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn., 236, 3088–3099. [DOI] [PubMed] [Google Scholar]
- 27. Ruifrok A.C. and Johnston D.A. (2001) Quantification of histochemical staining by color deconvolution. Anal. Quant. Cytol. Histol., 23, 291–299. [PubMed] [Google Scholar]
- 28. Widrick J.J., Alexander M.S., Sanchez B., Gibbs D.E., Kawahara G., Beggs A.H. and Kunkel L.M. (2016) Muscle dysfunction in a zebrafish model of Duchenne muscular dystrophy. Physiol. Genomics, 48, 850–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gilbert M.J., Zerulla T.C. and Tierney K.B. (2014) Zebrafish (Danio rerio) as a model for the study of aging and exercise: physical ability and trainability decrease with age. Exp. Gerontol., 50, 106–113. [DOI] [PubMed] [Google Scholar]
- 30. Widrick J.J., Gibbs D.E., Sanchez B., Gupta V.A., Pakula A., Lawrence C., Beggs A.H. and Kunkel L.M. (2018) An open source microcontroller based flume for evaluating swimming performance of larval, juvenile, and adult zebrafish. PLoS One, 13, e0199712. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


