Abstract
Background:
At our department, each specimen was assigned a tentative current procedural terminology (CPT) code at accessioning. The codes were subject to subsequent changes by pathologist assistants and pathologists. After the cases had been finalized, their CPT codes went through a final verification step by coding staff, with the aid of a keyword-based CPT code-checking web application. Greater than 97% of the initial assignments were correct. This article describes the construction of a CPT code-predicting neural network model and its incorporation into the CPT code-checking application.
Materials and Methods:
R programming language was used. Pathology report texts and CPT codes for the cases finalized during January 1–November 30, 2018, were retrieved from the database. The order of the specimens was randomized before the data were partitioned into training and validation set. R Keras package was used for both model training and prediction. The chosen neural network had a three-layer architecture consisting of a word-embedding layer, a bidirectional long short-term memory (LSTM) layer, and a densely connected layer. It used concatenated header-diagnosis texts as the input.
Results:
The model predicted CPT codes in both the validation data set and the test data set with an accuracy of 97.5% and 97.6%, respectively. Closer examination of the test data set (cases from December 1 to 27, 2018) revealed two interesting observations. First, among the specimens that had incorrect initial CPT code assignments, the model disagreed with the initial assignments in 73.6% (117/159) and agreed in 26.4% (42/159). Second, the model identified nine additional specimens with incorrect CPT codes that had evaded all steps of checking.
Conclusions:
A neural network model using report texts to predict CPT codes can achieve high accuracy in prediction and moderate sensitivity in error detection. Neural networks may play increasing roles in CPT coding in surgical pathology.
Keywords: Current procedural terminology codes, deep learning, neural network
INTRODUCTION
Surgical pathology practice requires that current procedural terminology (CPT) codes be used for codifying the level of complexity of specimens and pathology procedures performed on specimens. Assignments of CPT codes depend on a combination of factors, mainly tissue source, surgical procedure, final diagnosis, clinical information, and so on.[1] Each specimen is assigned a CPT code depending on the level of complexity, from 88300 to 88309. In some specimens, when additional procedures/tests, such as decalcification, special stains, immunostains, and molecular tests are performed and reported, there are additional CPT codes for these procedures/tests. The model described in this article deals with the initial CPT codes on specimens requiring microscopic examination, including 88302, 88304, 88305, 88307, and 88309.
At our practice, an initial tentative specimen CPT code is assigned for each specimen at the time of accessioning based on the specimen label, surgical procedure, and clinical information. Most of the time (>97%), these presumptive entries are correct. In a minority of cases, the pathologist assistants change the CPT codes based on the gross finding or identifying an error at accessioning. The pathologists change the CPT codes based on the final diagnosis or because of an error at accessioning. After the reports have been finalized, the coding staff reviews each case to verify the CPT codes and correct any errors that have been missed by the pathologist assistants and pathologists. This final step of coding verification is assisted by a CPT code-checking web application capable of detecting coding errors (see materials and methods section). It has been in use by our coding staff for over a year to assist in catching CPT coding errors and in maintaining coding consistency.
In anatomic pathology, deep learning has been utilized to analyze the microscopic images for both disease classification and detection.[2,3,4,5,6,7] It will be interesting to see if applying deep learning to natural language, such as report texts, can produce models that are effective in predicting appropriate CPT code assignments for the specimens.
Programming language R (https://www. r-project.org, accessed January 3, 2019) was selected. R is one of the major languages used by data scientists. With regard to R usage in surgical pathology, there have been reports on using R in statistical analysis[8] and information extraction from the report texts.[9,10]
In early 2018, a deep learning library Keras (https://keras.io, accessed January 3, 2019), originally written in Python (https://www.python.org, accessed January 3, 2019), became available in R (https://keras.rstudio.com, accessed January 3, 2019), making training deep learning models easier for the R programmers. In order to use R Keras package, Python language and Python package Keras need to be installed on the same machine. Anaconda distribution of Python (https://www.anaconda.com/download, accessed January 3, 2019) is a convenient way to obtain Python and many other libraries used in scientific computing and data science, including Keras, with a single download. R Keras package with Google TensorFlow (https://www.tensorflow.org, accessed January 3, 2019) as the back end was used to train the neural network models and to use the selected model for prediction.
In this article, I will describe how a neural network model capable of predicting CPT codes from report texts with high accuracy was constructed as well as the initial observations on the effect of its incorporation into the existing CPT-checking web application.
MATERIALS AND METHODS
The computer workstation used for data retrieval was a desktop personal computer (PC) HP Elitedesk (Hewlett-Packard, Palo Alto, CA, USA) with Intel (R) (Intel, Santa Clara, CA, USA) Core™ i7-670 central processing unit (CPU) at 2.80 GHz and 24.0 GB random-access memory (RAM). The pathology information system was PowerPath 10.0.0.19 (Sunquest Information Systems, Tucson, AZ, USA), with Advanced Material Processing (AMP module). The back end database management system for PowerPath was Microsoft SQL server. A laptop PC (Hewlett-Packard, Palo Alto, CA, USA) with Intel (R) (Intel, Santa Clara, CA, USA) Core™ i7-7500U CPU at 2.70G Hz and 16.0 GB RAM was used for deep learning training.
Open source programming language R version 3.5.1 (https://www.r-project.org, accessed January 3, 2019) was used for interacting with PowerPath database, for data preprocessing and for training neural network model. RStudio Version 1.2.1194 (https://www.rstudio.com, accessed January 3, 2019) was the integrated development environment used for writing and running the R programs.
The process of obtaining data from the pathology database using a database connectivity package (RODBC) is as described previously.[10] Briefly, a connection string containing information on database server address, the name of the database, and user login name and password was constructed. Furnishing connection string and SQL query as two arguments to an RODBC function sqlQuery() retrieved the data of interest from the PowerPath database into R.
The data frame is a two-dimensional R data structure that holds tabular data consisting of rows and columns. The diagnosis header text and diagnosis text for each specimen were extracted from the retrieved report text and concatenated. The header-diagnosis text was then associated with the assigned CPT code for the corresponding specimen. In the resulting data frame, each row represented a specimen and each column represented specific data for the specimens. Relevant columns included clinical information text, header text, diagnosis text, concatenated header-diagnosis text, and CPT code. The data frame thus obtained had 74,477 rows. After the order of the rows was randomized, the first 20,000 rows were used for validation and the remaining 54,477 rows were used for training.
R Keras package (https://keras. rstudio.com, accessed January 3, 2019) was used for model training as well as for using the saved model to predict CPT codes for the specimens in the test data set (data from December 1 to 27, 2018) and for the new specimens encountered daily.
A three-layer neural network model was trained to use the concatenated header-diagnosis texts to predict the correct assignments of CPT codes for both the specimens. The specifics of the process are described below.
Header-diagnosis texts were first tokenized into words, and then each word was converted to a unique nonzero integer by the following R code:
maxlen <- 100
max_words <- 11000
tokenizer <- text_tokenizer (num_words = max_words) %>%
fit_text_tokenizer (mytext$headerNdx)
sequences_headerNdx <- texts_to_sequences (tokenizer, mytext$headerNdx)
headerNdx_matrix <- pad_sequences (sequences_headerNdx, maxlen = maxlen)
For long texts, only the first 100 words were kept (maxlen <- 100). For texts shorter than 100 words, the left side was padded with integer 0 by the function pad_serquences() so that every entry had the same length of 100.
The model architecture for a three-layer neural network model consisting of a word-embedding layer with input length of 100 and with 50-dimensional word vectors as output, a bidirectional LSTM layer with 32 units and a densely connected layer with 5 units were defined by the following R code:
model <- keras_model_sequential() %>%
layer_embedding (input_dim = max_words, input_length = maxlen, output_dim = 50) %>%
bidirectional (layer_lstm (units = 32) %>%
layer_dense (units = 5, activation = ‘softmax’)
The learning process was configured by specifying rmsprop as the gradient decent method (optimizer), sparse categorical cross entropy as the loss function (loss), and classification accuracy as the metrics by the following R code:
model %>% compile(
optimizer = ‘rmsprop’,
loss = ‘sparse_categorical_crossentropy’,
metrics = c(“accuracy”)
)
The batch size for the mini-batch gradient decent was 500.
Functions save_text_tokenizer() and save_model_hdf5() were used to save both the text tokenizer and the trained model; the saved model, with the saved text tokenizer, can be loaded into a different program to predict CPT codes, such as for predicting CPT codes on the test data set and for being incorporated into a web application (see below).
A web application written in R with shiny package (https://shiny.rstudio.com, accessed January 3, 2019) for CPT code checking based on report text parsing has been used at our practice by the coding staff for over a year. A detailed description of that application is beyond the scope of this article. Briefly, the program retrieves information from the PowerPath database and parses report text to identify keywords or the combination of keywords that may suggest incorrect CPT code assignments. For instance, identifying words “fibroepithelial polyp” in the diagnosis text in a skin biopsy with CPT code assignment of 88305 would prompt the user to see if the code needs to be changed to 88304. This web application is hosted on a virtual Windows server (Windows 2012R2, 4 cores and 8 GB of RAM) in the intranet and can be accessed from any PC within the network by using a web browser. The user can query the pathology database by a date range to see if there are any cases flagged for possible errors. The new CPT code-predicting neural network model has been incorporated into the above CPT code-checking web application based on keyword searching. A data table listing the specimens with predicted CPT codes different from the assigned CPT codes is displayed by the browser as an additional output of the application.
RESULTS
The model predicts CPT codes based on the report texts, specifically, diagnosis header, followed by the diagnosis. Figure 1 shows a screenshot of portion of the data frame containing the data for training or validation. Three columns of the data frame are shown as follows: case number concatenated with the ordinal number of the specimen within the case (accNspc), header text followed by diagnosis text (headerNdx), and CPT code (cpt). For instance, S-18-10000_4 denotes the fourth specimen of case S-18-10000. The last three digits of the case numbers were erased for de-identification purposes. The text before the colon (:) is the header, generally consisting of the information about the specimen and the procedure. The text following the colon (:) is the diagnosis. Take the displayed first row as an example, “Antrum bx” is the header, and “Reactive gastropathy” is the diagnosis. They are concatenated into a continuous text. For specimen in row 6, the header is “Subcutaneous mass, left hip, excision,” and the diagnosis is “Unclassified pleomorphic sarcoma. See comment.” Only the beginning portion of the diagnosis is visible in the screenshot.
Figure 1.
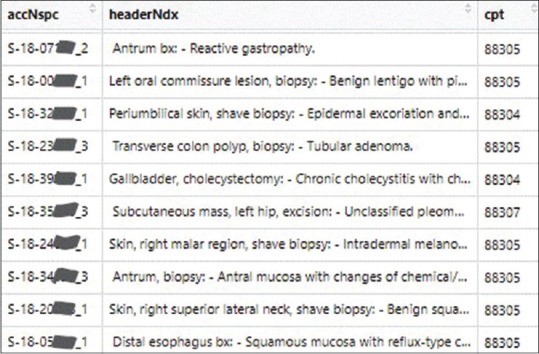
Screenshot of portion of a data frame containing concatenated header-diagnosis texts and CPT codes. Only three columns of the data frame are shown. accNspc: Accession number concatenated with the ordinal number of the specimen within the case. The last three digits of each case number are erased for de-identification, headerNdx: Header text concatenated with the diagnosis text, cpt: The assigned CPT code for the specimen. CPT: Current procedural terminology
As observed from the above two examples, the length of the header-diagnosis text varies. Figure 2 shows the distribution of the length of concatenated header-diagnosis texts. The median length is 12 words. A length of 100 words corresponds to the 98.1th percentile. The first 100 words of the text are used as input for the model. As such, for 98.1% of the cases, the entire header-diagnosis texts are included. For the remaining 1.9% of the specimens with longer texts, only the first 100 words are used.
Figure 2.
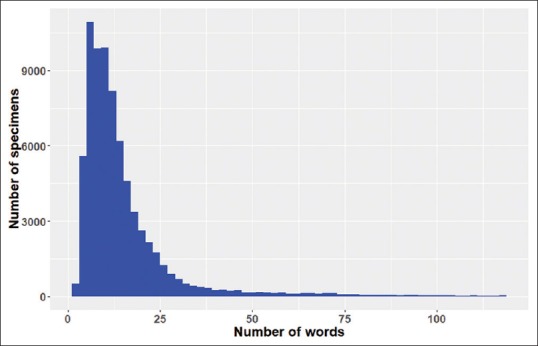
Distribution of the length (number of words) of the concatenated header-diagnosis text. While the horizontal axis denotes the number of words for the concatenated text, the vertical axis is the number of occurrences (number of specimens) for that length
Many model architectures have been explored. Because both LSTM and gated recurrent units (GRU) variants of recurrent neural network are frequently used as layers of neural network for text processing,[11] both were tested. They performed equally well, and LSTM was chosen. Stacking 2 or 3 GRU or LSTM layers in the model to produce models of 4 or 5 layer deep did not increase the accuracy of prediction; therefore, a simpler model of only 3 layers was chosen. Models tested also include models with 2 or 3 branches of inputs, each branch starting with a word-embedding layer, followed by LSTM layer (or GRU layer), and converged at a concatenating layer, and then further followed by a densely connected layer. There was only probably marginal enhancement in the accuracies. In the branched model, each branch processed text from header, diagnosis, and clinical information separately. In regard to the scope of texts to be included as input, the inclusion of the first 10 words of the clinical information did not increase the accuracy; as such, text from clinical information was not included as a component of input. Ultimately, the simplest three-layer model with concatenated header-diagnosis text as input was selected.
Figure 3 shows the training and validation metrics of a representative three-layer model, with 25 epochs of training. After 15 epochs, the model accuracy on the training data set and validation data set was 98.9% and 97.6%, respectively.
Figure 3.
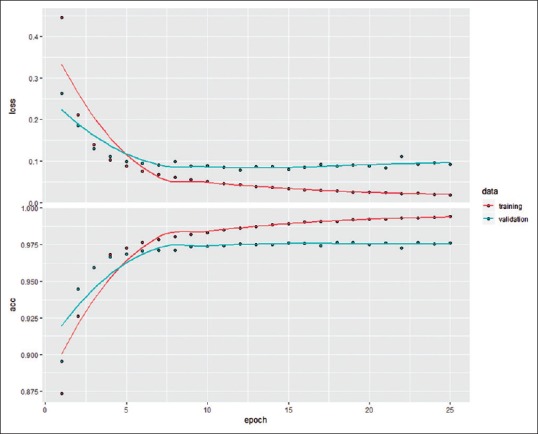
Training and validation metrics for the training of a model. The horizontal axis denotes the number of epochs (iteration over the entire training set once) since the beginning of the training. The y-axis of the top panel denotes the loss (categorical cross entropy); the y-axis of the bottom panel denotes the accuracy of CPT code prediction with reference to the final CPT code assignments, expressed as fraction. The salmon color denotes metrics associated with training data; the teal color denotes the metrics of the validation data. CPT: Current procedural terminology
The text data as shown in Figure 1 needed to be preprocessed into nonzero integer representation. For the final model selected, this preprocessing took 19 s. Iterating over each epoch took between 60 and 104 s. As such, training such a model with 15 epochs took about 20 min.
The chosen model contains 571,573 parameters. Table 1 is a slightly modified printout of the model summary. The modifications were made for the sake of clarity: “Params “ in the first line was changed to “no. of parameters” and “params” in the last three lines were changed to “parameters.#”
Table 1.
Model summary
| Layer (type) | Output shape | Number of parameters |
|---|---|---|
| Embedding_1 (Embedding) | None, 100, 50 | 550,000 |
| Bidirectional_1 (Bidirectional) | None, 64 | 21,248 |
| Dense_1 (Dense) | None, 5 | 325 |
Total parameters: 571,573, Trainable parameters: 571,573, Nontrainable parameters: 0
The file sizes for the model and the text tokenizer are relatively small, 5.5 and 0.5 MB, respectively.
The accuracy on the 20,000-specimen validation set by the selected model was 97.5%. The distribution of predicted CPT codes in the validation set was quite similar to that of the assigned CPT codes [Figure 4]. The model slightly overpredicted 88305; slightly underpredicted 88302, 88304, and 88307; and significantly underpredicted 88309. While there were 131 specimens with assigned CPT code of 88309, there were only 61 specimens with predicted CPT code of 88309.
Figure 4.
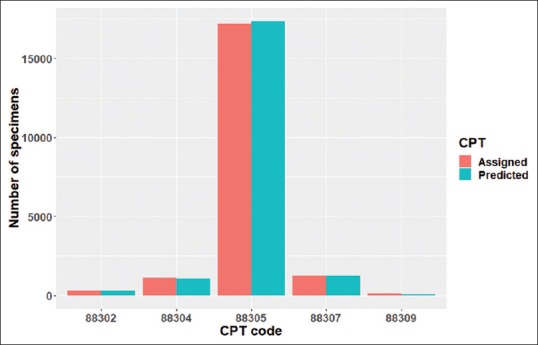
Distributions of assigned versus predicted CPT codes in the validation data set. The height of the bars denotes the number of specimens with the particular CPT codes as shown in the horizontal axis. The salmon color is for the assigned CPT codes; the teal color is for the CPT codes predicted by the model. CPT: Current procedural terminology
The model was then used to predict CPT codes on a test data set. All the cases finalized from December 1 to 27, 2018, were used as the test data set. This end date of December 27 was chosen so as to only include data before the incorporation of the neural network model into the web application. The model prediction differed from the assigned CPT codes in 2.4% of the specimens (152/6240). During this period, excluding the cases with specimen bundling/unbundling and considering only specimens with the five CPT codes (88302–88309), 159 specimens have their initial CPT codes changed (by pathologist assistants, pathologists, or a CPT coding staff). Of these specimens, the model predicted CPT codes different from the initial assignments in only 73.6% (117/159), whereas agreeing with the initial incorrect assignments in the remaining 26.4% (42/159). On the other hand, the model identified nine additional specimens that have incorrect final CPT code assignments not detected by the coding staff. For the purpose of calculation, it is assumed that the number of specimens with CPT codes changed (159), and the nine additional specimens identified by the model constituted 100% of the specimens that had incorrect initial CPT code assignments. Our current practice of verifying 100% of the CPT codes has an error detection rate of 95% (159/[159 + 9]); the error detection rate for the model is 75.0% ([117 + 9]/[159 + 9]), 20 percentage points below that of our current practice.
As mentioned previously, for the final step of CPT code verification at our practice, CPT codes for every specimen were examined by a coding staff and by a CPT code-checking web application. These two approaches are complementary, with coding staff catching more coding errors than the web application (numerically coincidentally 95% vs. 75%, with all the codes modified at the final step of verification as 100% [codes changed by the pathologist assistants and pathologists were not included in this calculation], details not shown).
The neural network model has been newly incorporated into the existing CPT code-checking web application. The prediction by the model and the error detection by rule-based keyword search are both complementary (flagging different specimens) and overlapping (flagging the same specimens). Each approach can identify some specimens that the other approach misses. For instance, the keyword-searching approach caught cases that required specimen unbundling, whereas the model's prediction agreed with the assignments. On the other hand, the model correctly predicted 88305 for a skin with keloid (initially assigned CPT code 88304), 88307 for a neoplastic ovary (initially assigned CPT code 88305), and 88305 for a pleural nodule ultrasound-guided biopsy (initially assigned CPT code 88307), whereas the keyword-searching approach did not flag these specimens. As such, the incorporation of neural network model into the web application has enhanced the application's ability to detect CPT coding errors.
DISCUSSION
Training deep learning model to tackle the real-world problems is frequently a computationally expensive process, requiring multicore GPU.[11] Training on CPU can be excruciatingly slow. Even on multicore GPU, either on local workstation or in the cloud, it can take many hours or even days to train a model.[4] I was pleasantly surprised that the CPT code prediction model took <30 min to train on a conventional laptop without GPU. Perhaps, this was due to the fact that the architecture of the model was relatively simple, consisting of three layers with only roughly half a million parameters. Of course, either a much more powerful workstation with GPU or cloud computing will be needed in order for me to build neural network with higher complexity in future.
A second unexpected finding was that this simple neural network with only three layers and using only up to 100 words of the report text was able to predict the specimen CPT codes with high accuracy, 97.5% and 97.6% for the training data set and test data set, respectively.
In the test data set, although the neural network model predicts CPT codes with a high accuracy of 97.6%, the error detection rate is only 75%, which is not enough for us to change our current practice of verifying CPT codes for every specimen (error detection rate 95%) to only checking the specimens that the model suggests different CPT codes. The reasons for this apparent discrepancy between the high accuracy of CPT code prediction and the moderate capability for error detection became clear after looking at the cases that the model failed to detect the coding errors. The information prompting the changes of CPT codes is frequently quite subtle. While therapeutic appendectomy is 88304, an incidental appendectomy is 88305. The third trimester placentas are 88307, but the first and second trimester placentas are 88305. While most diagnoses on skin specimens are 88305, diagnoses such as cyst or fibroepithelial polyp will necessitate a change to 88304. There are many other similar examples of pairs with subtle differences.
Even with moderate capability to detect coding error, the model was able to identify nine cases with coding errors in the test set; these errors have had evaded vetting by our coding staff. As such, the model is not only able to detect errors that were not detected by our CPT code-checking web application based on keywords, but is also able to identify errors in the specimens after verification by the coding staff. The model has been added to the web application. The preliminary results show that the degree of complementarity is significant. It will be interesting to record the outputs of this application for a period of time so as to enable us to reliably estimate the error detection rate when we combine the deep learning model with the existing keyword-searching approach. With this combined approach and with additional model refining, it is possible that the application may attain high enough sensitivity for error detection so that we only need to check the cases flagged by the program.
CONCLUSIONS
Neural network models using report text to predict CPT codes can achieve high accuracy in prediction and moderate level of sensitivity in error detection. Combining with rule-based approaches, neural networks may play increasing roles in CPT coding in surgical pathology.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
Acknowledgements
I would like to thank Ms. Sheena Braggs, coding staff, and Mr. Michael Tan, supervisor of the gross room, for their daily efforts in insuring correct coding for the specimens and for their assistance in evaluating the model predictions with reference to the actual report text in the test data set. I would like to thank Dr. Limin Yu, dermatopathologist and informaticist at Beaumont Health, Michigan, and Mr. Michael Tan for critically reading the manuscript. I would like to thank Mr. Derek Higgins for furnishing and verifying the specifications of the computing resources.
Footnotes
Available FREE in open access from: http://www.jpathinformatics.org/text.asp?2019/10/1/13/255395
REFERENCES
- 1.American Pathology Foundation. Pathology Service Coding Handbook. Ver. 17.0. American Pathology Foundation. 2017. Jan 01, [Last accessed on 2018 Jan 05]. Available from: http://www.apfconnect.org .
- 2.Janowczyk A, Madabhushi A. Deep learning for digital pathology image analysis: A comprehensive tutorial with selected use cases. J Pathol Inform. 2016;7:29. doi: 10.4103/2153-3539.186902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Djuric U, Zadeh G, Aldape K, Diamandis P. Precision histology: How deep learning is poised to revitalize histomorphology for personalized cancer care. NPJ Precis Oncol. 2017;1:22. doi: 10.1038/s41698-017-0022-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olsen TG, Jackson BH, Feeser TA, Kent MN, Moad JC, Krishnamurthy S, et al. Diagnostic performance of deep learning algorithms applied to three common diagnoses in dermatopathology. J Pathol Inform. 2018;9:32. doi: 10.4103/jpi.jpi_31_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Y, Kohlberger T, Norouzi M, Dahl GE, Smith JL, Mohtashamian A, et al. Artificial intelligence-based breast cancer nodal metastasis detection. Arch Pathol Lab Med. 2018 doi: 10.5858/arpa.2018-0147-OA. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 6.Steiner DF, MacDonald R, Liu Y, Truszkowski P, Hipp JD, Gammage C, et al. Impact of deep learning assistance on the histopathologic review of lymph nodes for metastatic breast cancer. Am J Surg Pathol. 2018;42:1636–46. doi: 10.1097/PAS.0000000000001151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanyal P, Mukherjee T, Barui S, Das A, Gangopadhyay P. Artificial intelligence in cytopathology: A neural network to identify papillary carcinoma on thyroid fine-needle aspiration cytology smears. J Pathol Inform. 2018;9:43. doi: 10.4103/jpi.jpi_43_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cuff J, Higgins JP. Statistical analysis of surgical pathology data using the R program. Adv Anat Pathol. 2012;19:131–9. doi: 10.1097/PAP.0b013e318254d842. [DOI] [PubMed] [Google Scholar]
- 9.Boag A. Extraction and analysis of discrete synoptic pathology report data using R. J Pathol Inform. 2015;6:62. doi: 10.4103/2153-3539.170649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ye JJ. Pathology report data extraction from relational database using R, with extraction from reports on melanoma of skin as an example. J Pathol Inform. 2016;7:44. doi: 10.4103/2153-3539.192822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chollet F, Allaire JJ. Deep Learning with R. Shelter Island: Manning; 2018. pp. 57–9. [Google Scholar]


